Abstract
We used a rapid scanning stop-flow technique to study the kinetics of reduction of cytochrome P450 3A4 (CYP3A4) by the flavin domain of cytochrome P450-BM3 (BMR), which was shown to form a stoichiometric complex (KD = 0.48 µM) with CYP3A4. In the absence of substrates only about 50% of CYP3A4 was able to accept electrons from BMR. Whereas the high-spin fraction was completely reducible, the reducibility of the low-spin fraction did not exceed 42%. Among four substrates tested (testosterone, 1-pyrenebutanol, bromocriptine, or α-naphthoflavone (ANF)) only ANF is capable of increasing the reducibility of the low-spin fraction to 75%. Our results demonstrate that the pool of CYP3A4 is heterogeneous, and not all P450 is competent for electron transfer in the complex with reductase. The increase in the reducibility of the enzyme in the presence of ANF may represent an important element of the mechanism of action of this activator.
Keywords: cytochrome P450 3A4, cytochrome P450BM-3 flavin domain (BMR), electron transfer kinetics, heterogeneity, spin equilibrium, allostery, testosterone, 1-pyrenebutanol, bromocriptine, α-naphthoflavone
Introduction
Homo- and heterotropic cooperativity is now well recognized as an inherent feature of numerous mammalian drug metabolizing cytochromes P450 [1–9]. The most prominent examples of cooperativity are observed with human CYP3A4 [3, 10–18]. The initial concept of the mechanisms of P450 cooperativity was based on the presence of multiple binding loci in a large substrate-binding pocket. It was assumed that microsomal drug metabolizing cytochromes P450 sometimes require more than one substrate molecule to assure a productive binding orientation of at least one of them [10–12]. There is convincing evidence for the presence of up to three substrate binding sites in the same molecule of cytochromes P450 exhibiting cooperativity [17–23]. Recent efforts from several laboratories have provided important details on the location of the binding sites, as well as on the mechanisms of effector-induced transitions in the enzyme involved in cooperativity [17, 24–29].
The multiple substrate-binding sites paradigm taken alone fails to explain the whole body of complex and mutually incompatible data on of CYP3A4 cooperativity [15, 30–35]. At present there are numerous strong lines of evidence linking the mechanism of cooperativity to persistent conformational heterogeneity of the enzyme [28, 36–38]. In particular, our recent studies on the kinetics of dithionite-dependent reduction showed that CYP3A4 is distributed among several populations with different rate constants of reduction [28]. These populations, which do not appear to interconvert in the time frame of the experiment, have different spin states (or, more precisely, a different position of spin equilibrium). The fraction reduced in the fast phase is represented by the low-spin hemeprotein only, while the enzyme reduced in the third (slowest) phase has the spin equilibrium completely shifted to the high-spin state. Our interpretation of these results is based on the hypothesis of heterogeneity of CYP3A4 caused by aggregation of the enzyme into oligomers, where the subunits differ in spin equilibrium and accessibility to the reducing agent (anion-monomer of dithionite) [28].
This finding appears to be closely related to the observations of the biphasicity of NADPH-dependent reduction of microsomal cytochromes P450, CYP2B4 in particular [39–43]. Importantly, the fast phase of NADPH-dependent reduction in the presence of substrate (benzphetamine) was shown to correspond solely to the reduction of the high-spin heme protein both in P450 2C11 reconstituted with NADPH-cytochrome P450 reductase (CPR) in phospholipid vesicles [44] and in cytochrome P450 (predominately CYP2B4) in liver microsomes of phenobarbital-treated rabbits [45]. This observation is quite unexpected in view of the fact that the extremely rapid rate of spin transitions in cytochromes P450 [46–49] suggests that the spin equilibrium should stay unchanged during the reduction. This apparent inconsistency has also been interpreted as evidence of distribution of CYP2B4 between two populations with different positions of spin equilibrium that do not interconvert in the time frame of the experiment [45]. It is logical to assume therefore that conformational heterogeneity of CYP3A4 revealed in our studies on the kinetics of dithionite-dependent reduction may also be reflected in the kinetics of NADPH-dependent reduction of the heme protein.
The present study addresses possible differences among multiple conformers of CYP3A4 in their affinity for reductase and/or rate constants of first electron transfer. However, studies using NADPH-cytochrome P450 reductase (CPR) are complicated by the necessity to reconstitute a membranous, multienzyme system [41, 50–54] or to use detergents [55–57] to achieve efficient interactions and electron transfer between CPR and P450. Reconstitution of CYP3A4 in the presence of phospholipids and detergents is especially challenging [58–60]. Therefore we took advantage of the use of the soluble flavin domain of P450BM-3 from Bacillus megaterium (BMR) as an electron donor for CYP3A4. Purified recombinant BMR has been shown to be capable of supporting the activity of BMP, the heme domain of P450BM-3 [61], and of efficient reduction of CYP2B4 through similar mechanisms as CPR [57]. Recently BMR has been successfully utilized to construct functionally active chimeric proteins with several human cytochromes P450 including CYP3A4 [62].
In the present study we demonstrate with the use of a rapid scanning stop-flow technique that BMR can be used to study the kinetics of electron transfer to CYP3A4 in a simple, soluble model system. We investigate the effect of allosteric and non-allosteric substrates on the kinetics of reduction of the low- and high-spin states of the heme protein in order to assess possible differences among multiple conformers of the enzyme in their interactions with the reductase and in the rates of first electron transfer. Our results demonstrate that BMR is capable of complete reduction of the high-spin fraction of heme protein, while the reduction of the low-spin fraction of CYP3A4 was considerably incomplete. Among the substrates probed only ANF was found to increase the reducibility of the low-spin fraction of CYP3A4 to over 75%. This unique effect of ANF is thought to be related to its action as a heterotropic activator of CYP3A4.
Materials and Methods
Materials
Bromocriptine (BCT) mesylate, glucose oxidase, catalase, glucose-6-phosphate dehydrogenase, glucose-6-phosphate, NADPH, protocatechuic acid and protocatechuate-3,4-dioxygenase were from Sigma Chemicals (St. Louis, MO). 1-Pyrenebutanol was a product of Invitrogen (Carlsbad, CA). α-Naphthoflavone (ANF), testosterone, and glucose were obtained from Aldrich. All other chemicals used were of the highest grade available from commercial sources and were used without further purification.
Expression and purification of CYP3A4 and BMR
CYP3A4 was expressed as the His-tagged protein in Escherichia coli TOPP3 [12, 63, 64], purified as described previously [24, 27], and stored at −80 °C in 100 mM HEPES buffer (pH 7.4), containing 10% glycerol (v/v), 1 mM DTT, and 1 mM EDTA (ethylenediaminetetraacetic acid). The following protocol for the expression and the purification of BMR reflects minor modifications of two previously published methods [65, 66]. The plasmid pT7-7-BMR was obtained from Dr. T. M. Poulos (University of California-Irvine). E. coli BL21(DE3) cells were transformed and grown in Terrific Broth supplemented with 1.0 mM FMN and 200 µg/ml AMP at 37 °C, with 250 rpm stirring. At OD600 between 0.6 and 1.0, BMR production was induced by addition of IPTG to a final concentration of 1 mM. After 18–24 hours of incubation at 28 °C and at 190 rpm stirring the cells were harvested by centrifugation (6000 rpm 15 min at 4 ° C) and resuspended at 4 °C in 50 mM Tris-acetate (pH 8.0), 0.25 mM DTT, 0.25 mM EDTA, 250 mM sucrose and 20% (v/v) glycerol supplemented with fresh lysozyme at a final concentration of 0.1 mg/ml. After 30 minutes of incubation at 4 °C with continuous stirring the cells were precipitated by centrifugation and resuspended in the same buffer supplemented with a cocktail of protease inhibitors (0.4 KIU/ml aprotinin, 0.2 mM AEBSF, 0.01 mg/ml leupeptin, and 0.05 mg/ml benzamidine). After 4 × 45 s (1 minute rest interval) sonication at 4 °C the suspension was centrifuged at 35,000 rpm at 4 °C for 1 hour, and the pellet was discarded. After addition of KCl to a final concentration of 500 mM the supernatant was applied to a 5-ml column of 2′–5′ ADP Sepharose 4B (GE Healthcare Bio-Sciences AB, Uppsala, Sweden) equilibrated with 20 mM K-phosphate buffer (pH 8.0) containing 1.0 µM FMN, 0.1 mM DTT, 0.1 mM EDTA, and 10% glycerol (v/v) (buffer A). The column was washed with 5 volumes of buffer A containing 5 mM adenosine and with 5 volumes of 500 mM K-phosphate buffer (pH 7.8) containing 1.0 µM FMN, 0.1 mM DTT, 0.1 mM EDTA, and 10% glycerol (v/v). BMR was eluted with buffer A containing 15 mM AMP. The colored fractions (OD 456 nm > 0.01) were collected and dialyzed against 50 mM KP (pH 7.6), 0.1 mM EDTA ,and 20% glycerol (v/v) (storage buffer). Determination of the enzyme concentration was performed on the basis of flavin absorbance at 456 nm using the extinction coefficient of 21.4 mM−1 cm−1 [67]. The turnover rate of purified BMR in the reduction of cytochrome c was found to be around 3800 min−1, which is consistent with the value of 3600 min−1 reported earlier [68].
Tandem cell titration and reduction kinetics experiments
To determine the dissociation constant of the CYP3A4-BMR complex we used the counter-flow continuous variation (Job’s) titration technique [19], which makes it possible to probe the stoichiometry of the interaction and increases the accuracy of measurement of BMR-induced changes in the spin state of CYP3A4 by eliminating the contribution of the spectral overlap of absorbance of BMR with the bands of CYP3A4. At the beginning of the titration a solution of BMR and an equal volume of buffer were added to two separate compartments of a tandem cell placed into the spectrophotometer in such a way that the light passes through both compartments consecutively. The baseline was recorded, and the content of the second compartment was replaced by the solution of CYP3A4 at a concentration equal to the concentration of BMR in the first compartment. Titration was performed by reciprocal mutual displacements of 10 – 40 µl aliquots between the compartments and recording of the absorbance spectra at each step. This procedure results in a gradual equalization of the concentrations of interacting proteins in the two compartments, so that the molar fraction of CYP3A4 increases in the first and decreases in the second compartment, while the total concentrations of the proteins remains unchanged. In these and other titration experiments experiments we used a S2000 CCD rapid scanning spectrometer from Ocean Optics, Inc. (Dunedin, FL) equipped with an L7893 UV-VIS fiber-optics light source from Hamamatsu Photonics K. K. (Hamamatsu City, Shizuoka, Japan).
Scanning stop-flow kinetic experiments were performed using a rapid scanning MC2000-2 CCD-spectrometer (Ocean Optics, Inc., Dunedin, FL) combined with a SF-MiniMixer Stopped-Flow apparatus (KinTek Corporation Austin, TX) as described earlier [28]. The anaerobiosis was achieved using an oxygen scavenging system consisting of glucose oxidase (30 units/ml), 60 mM glucose, and 2000 units/ml catalase. These components were added as a concentrated stock solution to CO-saturated buffer. In some of the experiments this system was replaced by protecatechuate-3,4-dioxygenase (0.5 units/ml) and 5 mM protocatechuic acid. The quality of anaerobiosis was assessed using a FOXY-18G optical oxygen probe connected to an S2000 spectrometer (Ocean Optics, Dunedin, FL). Both oxygen scavengers decreased the concentration of oxygen below the level detectable by the sensor. The reaction was initiated by mixing equal volumes of the mixture of CYP3A4 with BMR and a 0.2 mM solution of NADPH containing glucose-6-phosphate dehydrogenase (2 units/ml) and 20 mM glucose-6-phosphate. If not otherwise specified, the concentrations of CYP3A4 and BMR in the reaction cell were equal to 2.5 and 5 µM respectively. When the reduction was studied in the presence of substrates, BCT, 1-PB, testosterone, or ANF were added to both syringes of the stop-flow apparatus to reach concentrations of 5, 30, 200 and 25 µM respectively. All experiments were performed at 25 °C in 100 mM HEPES buffer (pH 7.4) containing 1 mM DTT and 1 mM EDTA. BCT mesylate was prepared as a 300 µM stock solution in 20 mM Na-acetate buffer (pH 7.4). Stock solutions (10 – 20 mM) of 1-PB, ANF, and TST were prepared in acetone. The concentration of acetone did not exceed 0.2% in our experiments with 1-PB and ANF or 1% in the experiments with TST. Control experiments showed that there was no effect of these concentrations of acetone on the spin state of CYP3A4 under the conditions of our experiments.
Data processing
The series of absorbance spectra obtained in titration experiments were analyzed using principal component analysis (PCA) combined with the fitting of the principal component spectra to a set of spectral standards, as described previously [28, 69, 70]. Briefly, the PCA procedure was applied to a set of difference spectra obtained by subtraction of the first spectrum in the series (zero time point) from all subsequent spectra. To interpret the resulting series in terms of the changes in concentrations of the ferric low-spin, ferric high-spin, and the ferrous carbonyl complexes of P450 and P420 states of the heme protein the basis spectra were approximated with a linear combination of the respective spectral standards combined with a low-order (≤4) polynomial function. The latter was introduced to compensate for the baseline fluctuations during the experiment. The high-ranking components characterized by a square correlation coefficient (ρ2) above the threshold, which was set to 0.5 in most cases, were considered significant and used to calculate the changes in the concentrations of the P450 species. This technique was described in detail earlier [69, 70]. The set of spectral standards consisted of the spectra of ferric low- and high-spin P450 species of CYP3A4 [17] and the spectra of the carbonyl complexes of ferrous P450 and P420 states of CYP3A4. The latter were refined by studying the pressure-induced P450-to-P420 conversion of the CYP3A4 ferrous carbonyl complex, similarly to the technique used for CYP2B4 and CYP1A2 [57, 71]. The complete set of the standards used in this study is available in Supporting Information. These kinetic curves were fitted to a multiexponential equation using a combination of Marquardt and Nailder-Mead algorithms. All data fitting and spectral analysis procedures were performed using our SPECTRALB software [69].
Results
Interaction of BMR with CYP3A4
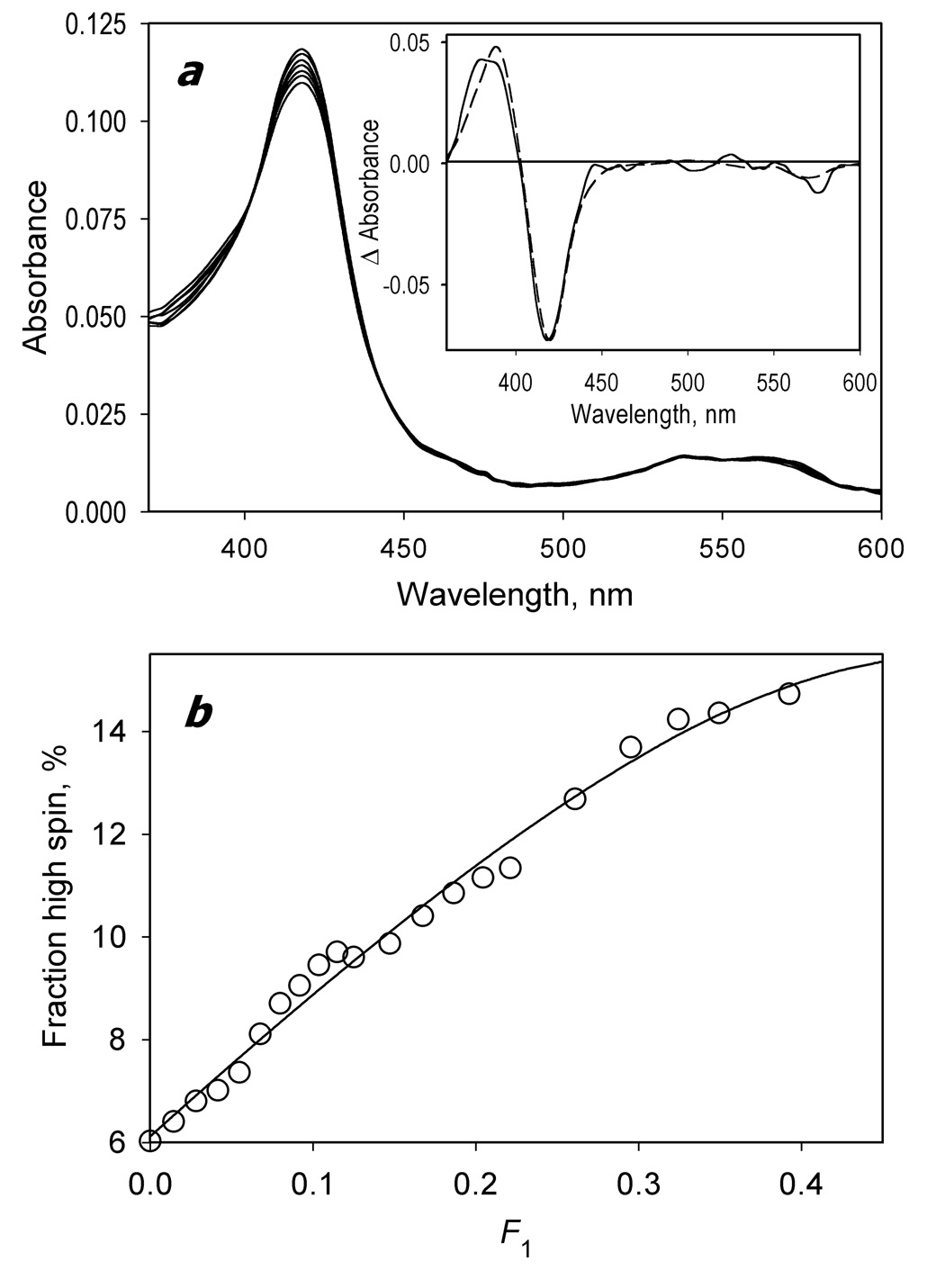
Figure 1a shows a series of absorbance spectra obtained in a counter-flow continuous variation experiment. As seen from the inset, the first principal spectrum obtained from the application of PCA to this dataset reveals a distinct low-to-high-spin shift of CYP3A4, which reflects the formation of the complexes of the heme protein with BMR, as observed earlier for the interactions of cytochromes P450 with CPR [72, 73]. The titration curve is represented by a one-half of a symmetric bell-shaped dependency expected for 1:1 stoichiometry of the complex formation (Fig. 1b). Fitting of the titration curve to the equation for the equilibrium of bimolecular association (p 73, eq. II-53 in [74]) adapated for the case of the counter-flow continuous variation titration (eq. 3 in [19]) gives a maximal amplitude of the BMR-induced spin shift of 11.9 ± 0.3% and the KD value of 48 ± 0.06 µM. This value is in good agreement with the value of 0.37 µM reported earlier for the interactions of BMR with CYP2B4 under similar conditions (0.1M Na-Hepes buffer, ionic strength of 0.058 M – see fig. 6b in [57]).
Fig. 1.
Formation of the CYP3A4 complex with BMR studied by a counter-flow continuous variation technique. At the beginning of the experiment the first and the second compartments of a 2 × 5 mm tandem cell contained equal volumes of 2.8 µM solutions of BMR and CYP3A4, respectively. The interactions were monitored by the changes in the concentration of P450 low-spin and high-spin states. Mixing of the interacting proteins was achieved by reciprocal gradual displacement of small aliquots of the solutions between compartments, so that the molar fraction of CYP3A4 in the first compartment (F1) changed from 0 to 0.5. The experiment was performed at 25 °C in 0.1 mM Na-HEPES buffer (pH 7.4) containing 1 mM EDTA and 1 mM DTT. A series of absorbance spectra recorded at F1 values of 0, 0.08, 0.22, 0.32, 0.42, 0.48, and 0.49 are shown in panel a. The inset shows the spectrum of the first principal component of the observed changes found by PCA (solid line). The dashed line in the inset represents the standard spectrum of the CYP3A4 high-to-low-spin shift. Panel b shows the dependence of the high-spin fraction of CYP3A4 on F1 obtained from the analysis of the spectra. The solid line represents the results of the fitting of the titration curve to the equation for the equilibrium of bimolecular association suited to the case of the counter-flow titration (eq. (3) in [19]).
Kinetics of reduction of CYP3A4 by BMR
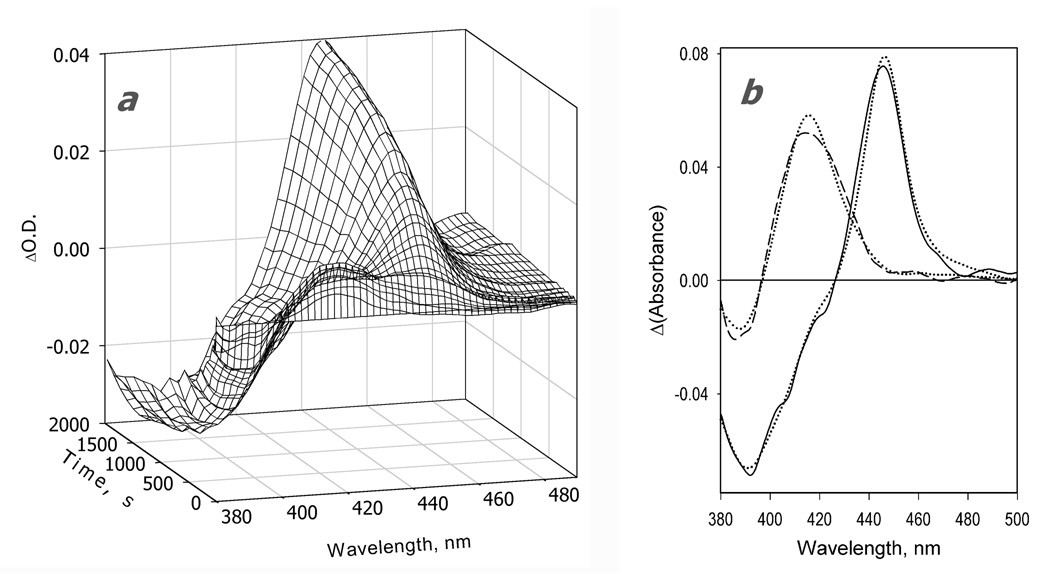
A series of difference spectra showing the changes in absorbance after addition of NADPH to the mixture of CYP3A4 with BMR is exemplified in Fig. 2a. Appearance of the absorbance maximum of the ferrous carbonyl complex of CYP3A4 at 448 nm indicates heme protein reduction, which takes about 2000 seconds for completion. In addition to this major change, a decrease in the absorbance band around 390 nm accompanied by an increase in the band at 418 nm is clearly seen in the initial stages of the process. These changes reveal a high-to-low-spin shift in CYP3A4, which takes place immediately after addition of NADPH and prior to reduction of the heme protein. Such displacement of the P450 spin state in response to the reduction of the interacting reductase by NADPH has been never been reported previously. This high-to-low-spin shift may reflect an inherent difference in the complexes of reduced vs. oxidized BMR with CYP3A4 or a lower affinity of reduced than oxidized BMR for CYP3A4, leading to partial dissociation of the BMR-CYP3A4 complex. Changes in the mode of interaction of CPR with P450 upon the reduction of the flavoprotein have been proposed to explain an anomalous dependence of the kinetics of reduction on the P450-to-reductase ratio in reconstituted systems [75].
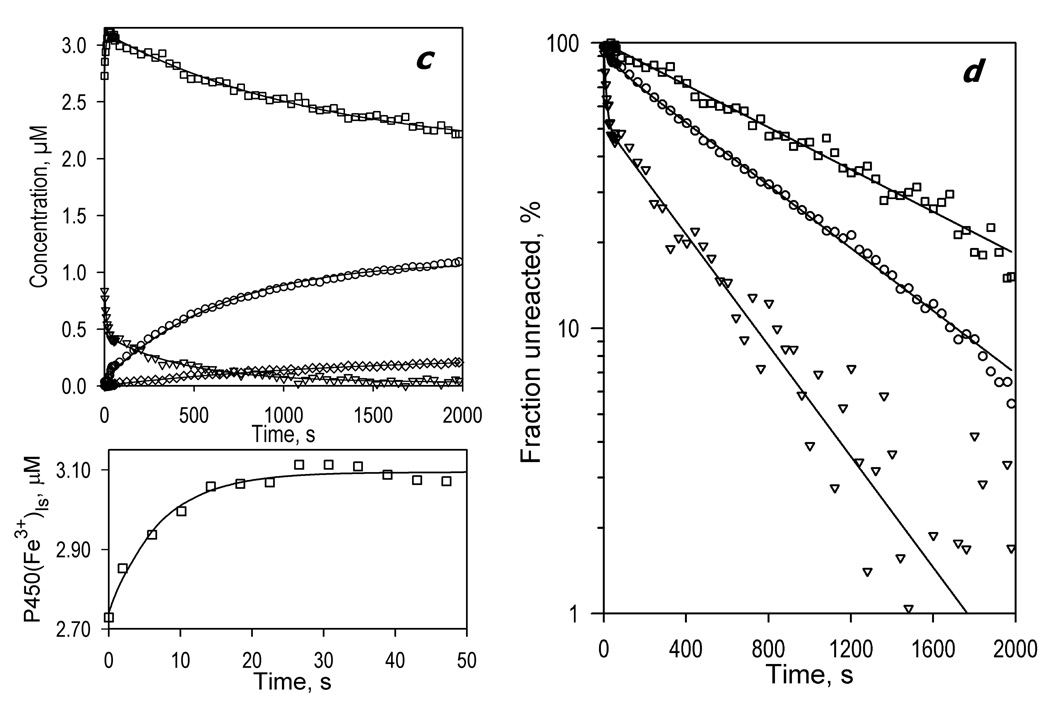
Figure 2.
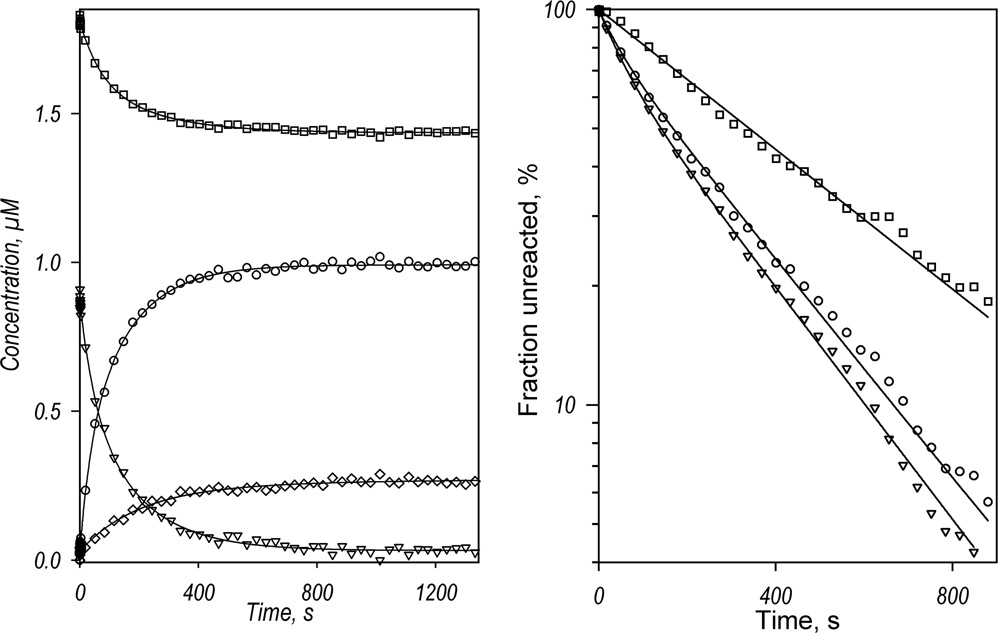
Kinetics of CYP3A4 reduction by BMR in the absence of substrate recorded with a rapid scanning stop-flow technique. Conditions: 3.5 µM 3A4, 7 µM BMR, 1mM NADPH, CO-saturated 0.1 M Na-Hepes buffer (pH 7.4), 1 mM DTT, 1 mM EDTA, and a glucose oxidase/catalase oxygen-scavenging system (see Materials and Methods), 25 °C. Spectra were recorded in a stop-flow cell with 5 mm optical path length. a: Changes in absorbance in the Soret region during the reduction. The spectrum measured at time of origin is subtracted. b: The spectra of the first (solid line) and the second (dashed line) principal components of the observed changes found by PCA. The spectra are scaled to correspond to a transition in 1 µM heme protein. Dotted lines represent the approximation of these spectra by a combination of the CYP3A4 spectral standards (see Materials and Methods). The time course of the changes in the concentrations of the ferrous carbonyl complexes of CYP3A4 P450 (circles) and P420 (diamonds) states and the low-spin (squares) and the high-spin (triangles) states of ferric CYP3A4 are shown in panel c (top). The initial part of the kinetic curve for the low-spin CYP3A4(Fe3+) is shown in the bottom plot of this panel, where the solid line represents the results of the fitting of the data set to the first order kinetic equation with the rate constant of 0.14 s−1. Panel d shows the curves for the ferric enzyme species and the ferrous P450 carbonyl complex in semi-logarithmic coordinates. The limits (maximal amplitudes) of the changes used to scale the data in this plot were taken from the results of fitting of the kinetic curves.
Application of PCA to the series of spectra obtained in these experiments gives two principal components that cover over 99% of the overall spectral changes. As seen from Fig. 2b, while the first principal component reflects the reduction of the (primarily high-spin) heme protein, the second component reveals a high-to-low-spin transition of the ferric enzyme. This component results from both the initial high-to-low-spin shift and a difference in the kinetics of reduction of the high- and low-spin heme protein discussed below.
The changes in the concentration of the high- and low-spin states of the ferric CYP3A4 and the ferrous carbonyl complexes of the P450 and P420 states deduced from the results of PCA are shown in Fig. 2c. Some conversion to P420 was observed concomitant with the reduction process. Fig 2d shows the changes in the concentrations of the high- and low-spin P450(Fe3+) and the P450(Fe2+)-CO states of the enzyme in semi-logarithmic coordinates. In contrast to mono-exponential kinetics of the reduction of CYP2B4 by BMR [57], the kinetics of CYP3A4 reduction reveals at least two exponential phases, the rate constants of which differ by over an order in magnitude. The amplitude of the fast phase comprises around 12% of the maximal amplitude of the reduction (see Table 1).
Table 1.
Kinetic parameters of CYP3A4 reduction by BMR.
| P450(Fe2+)-CO | P450(Fe3+) high-spina | P450(Fe3+) low-spin | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Substrate | k1×103, s−1 | k2×103, s−1 | F1, % | Reducibility, % | k1×103, s−1 | k2×103, s−1 | F1, % | k1×103, s−1 | k2×103, s−1 | F1, % | Reducibility, % |
| No substrate | 22±1 | 0.9±0.2 | 12±7 | 50±9 | 15±9 | 1.2±0.3 | 41±6 | - | 1.0±0.3 | 0 | 42±7 |
| BCT | 13±4 | 3.7±1.0 | 41±20 | 64±7 | 19±8 | 2.8±0.9 | 51±17 | 18±9 | 3.7±0.7 | 66±30 | 48±12 |
| 1-PB | 27±13 | 3.2±0.4 | 25±6 | 39±4 | 19±10 | 2.9±0.4 | 26±3 | 10±6 | 2.6±1.2 | 37±12 | 26±3 |
| TST | 20±7 | 4.0±2.5 | 65±27 | 34±5 | 12±2 | 3.5±1.7 | 74±22 | 12±4 | 2.2±0.8 | 71±22 | 16±4 |
| ANF | 6.9±3.4 | 1.6±0.4 | 36±9 | 78±3 | 9.1±2.8 | 1.5±0.5 | 35±9 | 18±8 | 2.3±0.7 | 11±7 | 75±10 |
Represents the relative amplitude of formation of the CO complex of ferrous CYP3A4 (either in P450 or P420 states). The maximal amplitude observed for the P420-CO complex formation was never higher than 13%.
The values given in the table were obtained by averaging the results of 3 – 10 experiments.
As seen from Fig. 2b and 2c, the kinetics of reduction of the high-spin heme protein obeys a bi-exponential equation with kinetic constants similar to those derived from the kinetics of the appearance of P450(Fe2+)-CO, although the fraction of the fast phase here was as high as 41±6%. In contrast, the kinetics of reduction of the low-spin heme protein obeys a simple first-order kinetic equation with a rate constant similar to that of the slow phase of the total reduction process. (It should be noted that in order to prevent the fitting results from being affected by the initial high-to-low-spin shift, the data points corresponding to the initial ten seconds of the rreactin were omitted in fitting of the kinetic curves for high and low-spin P450.) Therefore, the heme protein reduced in the fast phase is represented by the high-spin state only. These results are in strong agreement with our earlier conclusion on functional and conformational heterogeneity of CYP3A4 in solution based on the barotropic behavior of the enzyme [38] and on the kinetics of its reduction by dithionite [28].
Analysis of the amplitudes of the reduction process provides further evidence of the heterogeneity of the CYP3A4. As seen from Fig 2c, the process of CYP3A4 reduction monitored by the appearance of ferrous carbonyl complex was essentially incomplete. Only about 50% of the total heme protein may be reduced by BMR with the formation of either P450(Fe2+) or P420(Fe2+) carbonyl complexes. At the same time, the high-spin heme protein state disappears completely during the reduction process, while the maximal amplitude of the low-spin CYP3A4 reduction was only 42% (Table 1). Therefore, there is an important (~50%) fraction of CYP3A4 that is incapable of being reduced by BMR and appears to be represented by the low-spin state of the enzyme only In this context it should be noted that both the kinetics and the relative amplitude of CYP3A4 reduction by BMR were found to be unaffected by molar ratios of BMR to CYP3A4 ranging from 1:1 to 4:1: (P450 concentration = 5 µM), as well as on CYP3A4 concentrations in the range from 2 to 5 µM (2:1 BMR to CYP3A4 molar ratio) (data not shown). This observation suggests that in the conditions of our experiments the rate of formation of the CYP3A4-BMR complexes was not rate limiting for either the fast or the slow phases of reduction.
Effect of substrates on the kinetics of reduction
In order to probe possible modulation of CYP3A4 heterogeneity by ligands we studied the kinetics of reduction of CYP3A4 by BMR in the presence the non-allosteric substrate BCT and the allosteric substrates 1-PB, testosterone, and ANF. ANF is also a prominent heterotropic activator of the enzyme [37, 76–78]. In absorbance titration experiments addition of all four substrates to CYP3A4 causes a Type-I spin shift. The maximal amplitudes of substrate-induced spin shift observed with BCT, 1-PB, TST, and ANF observed in the presence of BMR under conditions similar to those in our reduction experiments were found to be 39 ± 2%, 43 ± 2%, 40 ± 1%, and 36 ± 2% respectively.
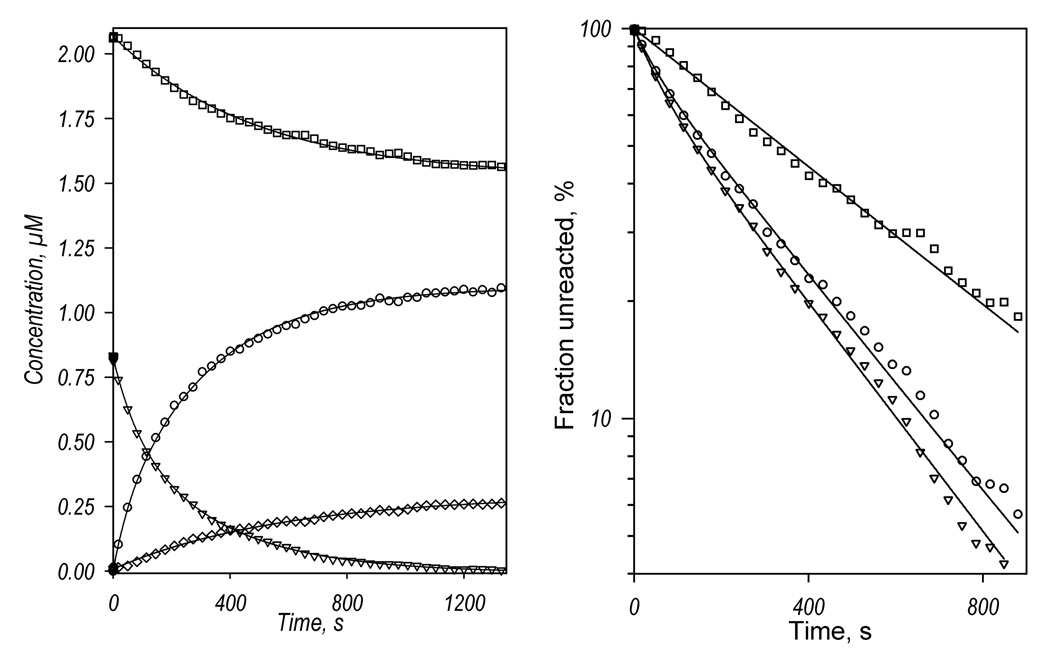
Qualitatively, the spectral changes observed in the presence of BCT are similar to those observed in the absence of substrate. Two principal components deduced by PCA cover over 99.8% of the total spectral changes. However, with BCT the first principal component, which reflects the main process of the reduction of the heme protein, represents over 99.5 ± 0.2% of the observed spectral changes compared with 97 ± 1% in the absence of substrate. Therefore, the weight of the second component, which describes the difference in the kinetic behavior between the high- and low-spin heme protein, is considerably diminished. In agreement with this observation, the kinetic curves reflecting the changes in the ferric high- and low-spin heme protein and its ferrous carbonyl complex are nearly similar in shape (Fig. 3a). All three kinetic curves obey a bi-exponential equation with the fraction of the fast phase around 20–26% (Fig. 3a, Table 1). Therefore, binding of BCT to CYP3A4 largely eliminates the difference in the kinetics of reduction of the high- and low-spin CYP3A4 states. Similar to observations in the absence of substrate the reducibility of the low-spin state remains incomplete, while the high-spin heme protein is completely reducible. Accordingly, as the addition of BCT shifts CYP3A4 towards the high-spin state, the overall reducibility of the enzyme increases from 50% in the absence of substrate to 64% in the presence of BCT.
Fig. 3.
Kinetics of CYP3A4 reduction by BMR in the presence of bromocryptine (a), testosterone (b) and 1-pyrenebutanol (c). Left panels show the changes in the concentrations of the ferrous carbonyl complexes of CYP3A4 P450 (circles) and P420 (diamonds) states and the low-spin (squares) and the high-spin (triangles) states of ferric CYP3A4. Right panels show the curves for the ferric enzyme species and the ferrous P450 carbonyl complex in semi-logarithmic coordinates.
Surprisingly, addition of 1-PB and testosterone (both possessing heterotropic cooperativity with CYP3A4) decreases considerably the overall reducibility of the enzyme to 39 and 34% respectively (Fig. 3b,c, Table 1). This decrease is due to decreased reducibility of the low-spin state fraction of the enzyme to only 16% in the presence of testosterone and 26% in the presence of 1-PB (Table 1). As with BCT, the kinetics of reduction of the high- and low-spin enzyme in the presence of 1-PB or TST is nearly the same. However, addition of testosterone increases considerably the fraction of the fast phase in the kinetic curves registered by either disappearance of the high- or low-spin ferric enzyme, or by formation of the ferrous carbonyl complex of CYP3A4.
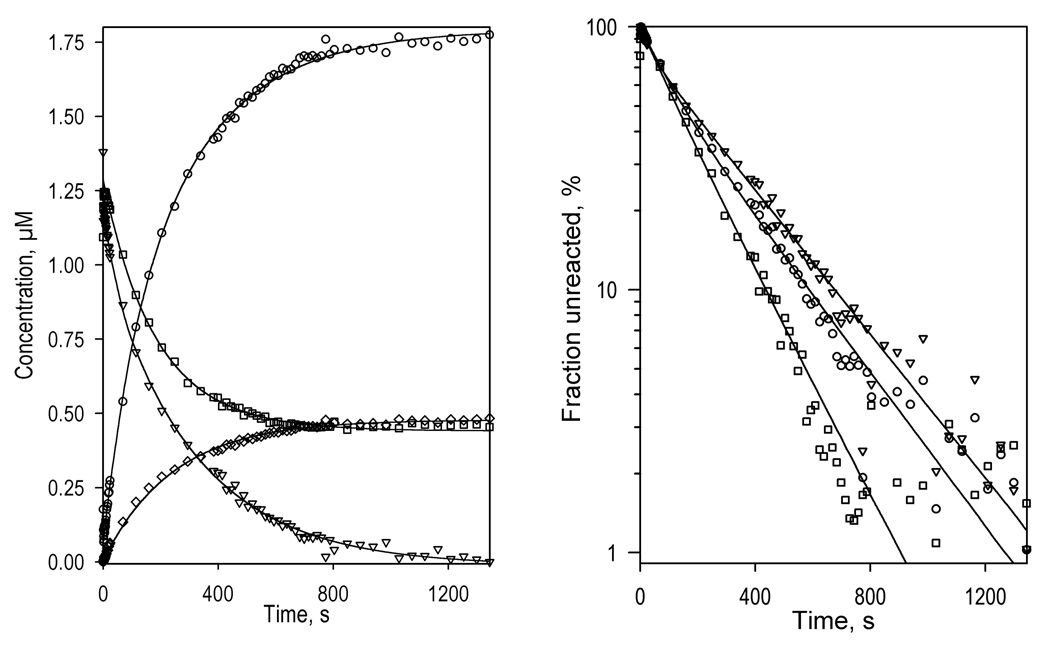
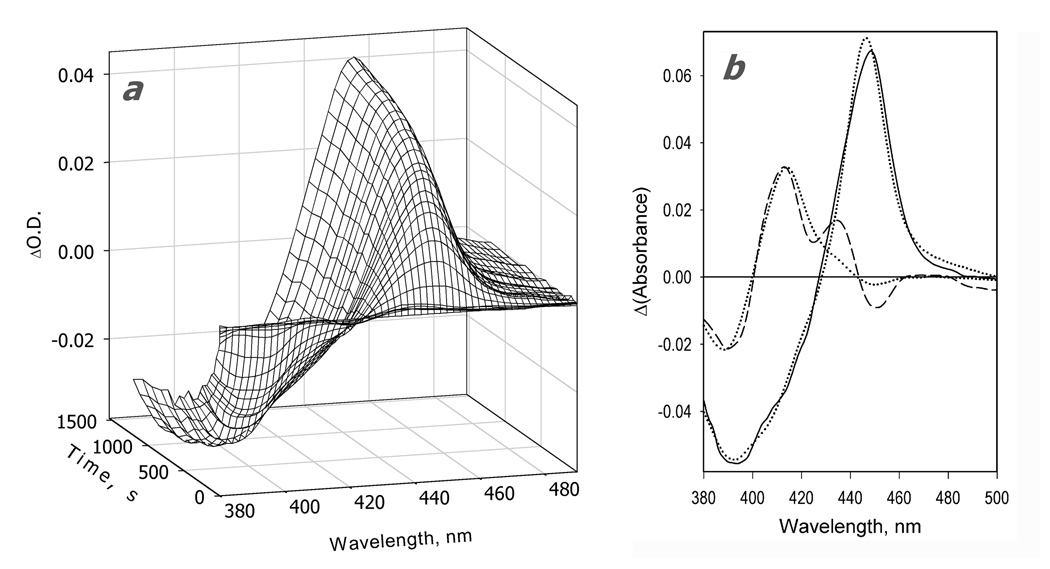
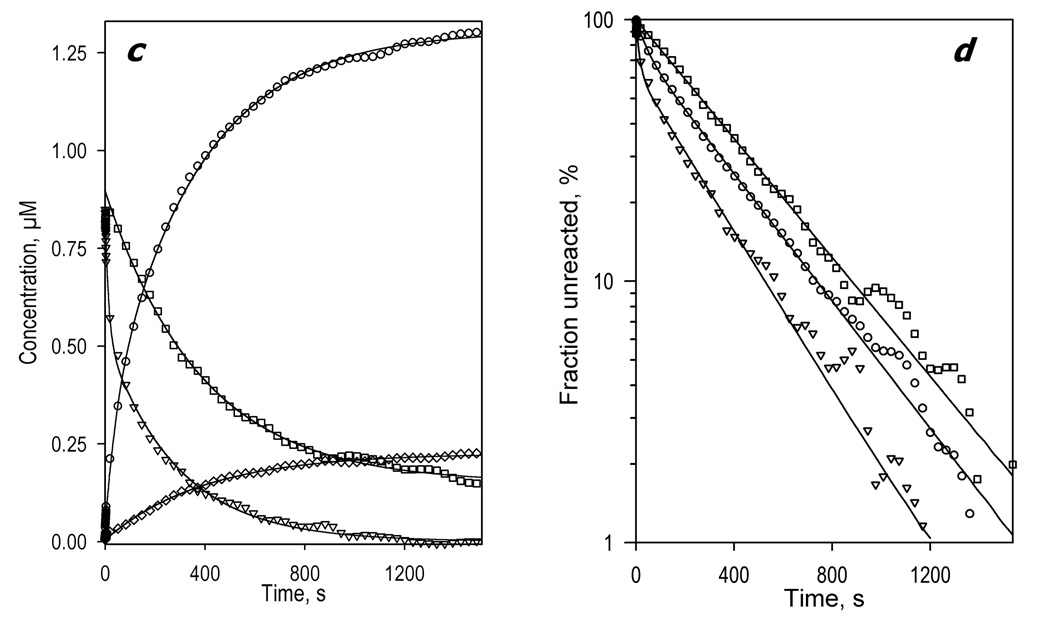
The changes in the kinetics of reduction caused by the addition of ANF are very different from those resulting from the addition of 1-PB or testosterone. Among the four substrates studied here, ANF is the only one that increases the reducibility of the low-spin fraction of the enzyme. In contrast to the substrate-free CYP3A4 or the enzyme complexed with BCT, 1-PB, or testosterone, where BMR was able to reduce no more than 48% of the low-spin enzyme, the ANF-bound enzyme exhibits 75% reducibility of the low-spin heme protein (Fig. 4, Table 1). Therefore, consistent with the hypothesis of Koley and co-authors [37, 79], our results suggest that the interactions of CYP3A4 with ANF cause a redistribution of the P450 pool among several functionally different conformers and activate an otherwise inactive subpopulation of predominantly low-spin heme protein by making it capable ofo accepting electrons from BMR. None of the other three substrates probed here exhibited such an effect.
Fig. 4.
Kinetics of CYP3A4 reduction by BMR in the presence of ANF. The reaction mixture contained 1.75 µM 3A4, 3.5 µM BMR, and 25µM ANF. Other conditions as indicated in Fig. 2. a: Changes in absorbance in the Soret region during the reduction. The spectrum measured at time of origin is subtracted. b: The spectra of the first (solid line) and the second (dashed line) principal components of the observed changes found by PCA. The spectra are scaled to correspond to a transition in 1 µM heme protein. Dotted lines represent the approximation of the spectra by a combination of the CYP3A4 spectral standards. The time course of the changes in the concentrations of the ferrous carbonyl complexes of CYP3A4 P450 (circles) and P420 (diamonds) states and the low-spin (squares) and the high-spin (triangles) states of ferric CYP3A4 are shown in panel c. Panel d shows the curves for the ferric enzyme species and the ferrous P450 carbonyl complex in semi-logarithmic coordinates.
Discussion
Our absorbance spectroscopy experiments with a counter flow Job’s titration setup confirmed that BMR forms a stoichiometric 1:1 complex with CYP3A4 with a KD of of 0.48 µM, where the spin equilibrium of the heme protein is shifted towards the high-spin state, similar to observations with other microsomal cytochromes P450 and CPR [72, 73]. Consistent with this observation, BMR was able to reduce ferric CYP3A4 with the formation of the ferrous carbonyl complex of the enzyme. The lack of dependence of the kinetics of reduction on the molar ratio of BMR to CYP3A4 or CYP3A4 concentration under the conditions of our experiments suggests that the formation of the electron transfer complexes is not rate limiting and the saturation of the heme protein by BMR is nearly complete. The latter inference is consistent with a calculation based on the KD value that over 86% of the heme protein is present in the complex with the reductase at the concentrations of 2.5µM CYP3A4 and 5µM BMR used in most of our experiments.
In view of apparent saturation with BMR, it was striking that the maximal level of reduction of CYP3A4 was considerably below 100%. However, review of the literature revealed numerous examples of incomplete reduction of microsomal cytochromes P450 by CPR In particular, studies of purified CYP2B4 reduction by CPR in the presence of 0.25 g/l Emulgen-913 [56, 57] or in DLPC vesicles [53] demonstrated that about half of the P450 was reducible in the absence of substrates and 70–80 % in the presence of benzphetamine. Reduction of CYP2B4 by BMR in the presence of 0.25 g/l Emulgen-913 was also found to be essentially imcomplete [57]. Likewise, careful re-examination of kinetic curves of reduction of CYP3A4 by CPR published in prior studies suggests reducibility of the heme protein in the range of 35 – 50% [80, 81]. It should be noted, however, that the literature contains no discussion of a possibility that such incomplete reduction may represent an inherent feature of the P450 enzymes, at least in some reconstituted systems. Rather incomplete reduction has been interpreted as a drawback of the experimental technique used in the study. In particular, Kanaeva and co-authors [56] hypothesized that such incomplete reducibility may result from possible inactivation of the heme protein and formation of cytochrome P420 in conditions of their experiments. However, the use of a rapid scanning stop-flow technique in the present study allowed us to discount such a possibility and demonstrate that the maximal overall amplitude of the reduction of CYP3A4 with the formation of either P450 or P420 ferrous carbonyl complexes in the absence of substrate was 50%. More important, the ability to monitor independently the disappearance of low- and high-spin ferric enzyme allowed us to show for the first time that the reduction of high-spin enzyme was always complete, whereas reducibility of the low-spin form ranged from 16 to 75 % depending on the substrate present.
The above observations raise the important question of whether it is valid to apply a simple concept of spin equilibrium to the pool of CYP3A4 in solution as if it were a single entity. Rather, consistent with our earlier results on CYP3A4 [27, 28, 38] and CYP2B4 [45, 69, 82], the data on reduction by BMR suggest that the pool of CYP3A4 diverges into several persistent populations with different position of spin equilibrium and functional properties. Our results indicate that ~50% of the heme protein is represented by a conformer in which the transition from the low- to high-spin state is completely prohibited, so that this fraction is represented by the low-spin state only. This hypothetical state of the heme protein, which we designate here as HS(−) conformer, appears to be refractory to enzymatic reduction.
The simplest explanation of the inability of the HS(−) conformer to accept electrons from the reductase is an unfavorable redox potential, as suggested by early studies on P450cam [83, 84]. However, subsequent studies of microsomal P450 enzymes have called into question any direct connection between the spin state and redox potential [43, 53, 80, 85]. Another possibility is that the BMR complex of the HS(−) conformer suffers from improper orientation and/or conformation of the two protein partners. Such unproductive complexes between P450 and CPR were hypothesized by Backes and Eyer to account for biphasicity in the kinetics of CYP2B4 reduction [53]. It was suggested that the heme protein is represented by two conformations existing in slow equilibrium, where both bind to CPR but only one allows electron transfer. Although this model accounts for a slow phase of reduction, it does not explain incomplete reducibility of the heme protein. To explain the results presented here we have to suggest that some part of CYP3A4 (the HS(−) conformer) is not in equilibrium with a functionally competent conformer.
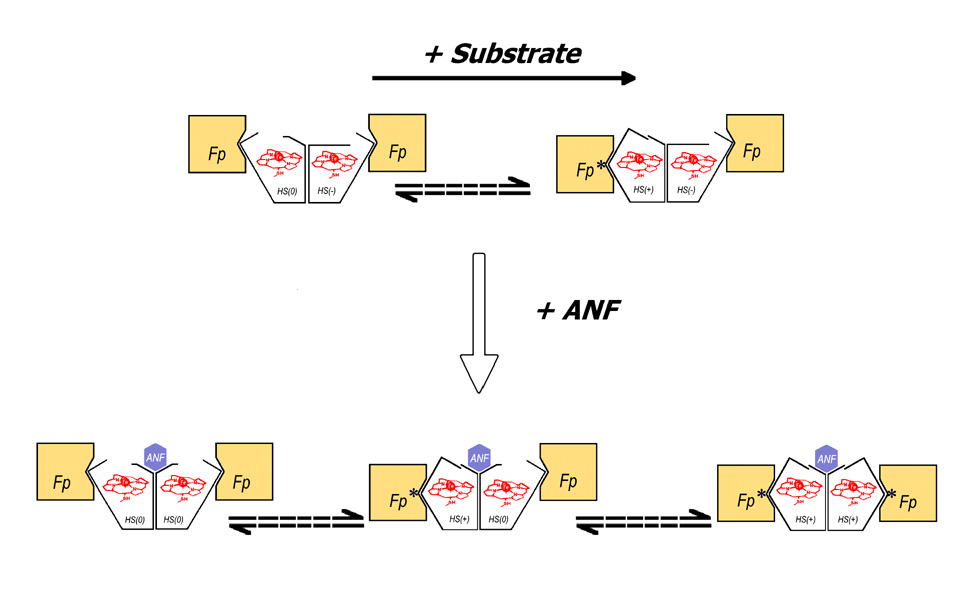
Since the reduction of the total and high-spin CYP3A4 in the absence of substrate exhibits a fast phase, which is missing in the kinetics of reduction of the low-spin state, the functionally competent conformer appears to be enriched in the high-spin state. We designate this conformer as HS(+). The third, slowly reducible population of CYP3A4 is predominately represented by the low-spin state. This fraction we designate as HS(0). The rate of reduction of this conformer appears to be limited by a slow conformational transition to the HS(+) state, in accordance with the model of Backes and Eyer. Binding of substrates appears to displace the equilibrium between HS(+) and HS(0) states towards the former. A substrate-induced conversion of the reducible HS(0) ↔ HS(+) fraction of the heme protein would leave the remaining low-spin fraction enriched in the functionally incompetent HS(−) conformer. Thus, the reducibility of the low-spin enzyme would be decreased, as clearly seen in the presence of 1-PB and testosterone. In contrast to these substrates, ANF increased the reducibility of the low-spin form to 75 ± 11 %, while the overall reducibility of the heme protein increased to 78 ± 3 %. Therefore, the interactions of CYP3A4 with this heterotropic activator appear to induce a redistribution of P450 pool between HS(−) and HS(+)↔HS(0) conformers towards the latter two. The effect of BCT is somewhat intermediate between those observed with 1-PB and TST on the one hand and ANF on the other hand.
According to our earlier hypothesis [28] partitioning of the hemoprotein between several persistent conformers with different functional properties may be due to oligomerization. The orientation and/or conformation of the subunits in such an oligomer (“aggregate”) may be uneven, and the conformational flexibility of some subunits may be restricted, thus preventing a transition between conformers without dissociation of the oligomer. Binding of allosteric effectors such as ANF is therefore hypothesized to cause a reorganization of the oligomer and change the partitioning between the conformers.
Our overall hypothesis, which is illustrated in Fig. 5, is completely compatible with the model proposed in our publication on the kinetics of dithionite-dependent reduction of CYP3A4 [28]. There we demonstrated that kinetics of dithionite-dependent reduction of CYP3A4 in solution and in liposomes with high density of the hemoprotein in the membrane obeys a three-exponential equation, where the high-spin fraction is reduced in the slow phase (in contrary to the BMR-supported reduction). While the binding of BCT decreases the amplitude of the fast phase of dithionite-dependent reduction concomitant with an increase in the amplitude of the slow phase, the middle phase remains unaffected. According to our current interpretation (Fig. 5), the fraction that is slowly reducible by dithionite corresponds to the HS(+) conformer, which is rapidly reducible by BMR, while the middle phase of the dithionite-dependent reduction represents the HS(−) conformer, and the fraction reduced in the fast phase corresponds to HS(0) conformer.
Fig. 5.
A suggested scheme of conformational transitions in CYP3A4, which explains the heterogeneity of the enzyme observed in the kinetics of NADPH-dependent reduction in the complex with BMR. Oligomers of the heme protein are shown as dimers for simplicity only; their actual size is likely to be considerably larger. CYP3A4 (open polygons) is represented by three conformational states: HS(+), HS(0), and HS(−). The states HS(+) and HS(0) exist in slow equilibrium (dashed arrows), which is displaced towards HS(+) in response to substrate binding. The state HS(−) is stabilized by subunit interactions, being therefore excluded from the conformational equilibrium. All three conformers are able to bind BMR (flavoprotein, Fp), but only the HS(+) conformer forms productive electron-transfer complexes, which are designated with an asterisk. The rate of reduction of the HS(0) conformer is therefore defined by the rate of the HS(0) → HS(+)transition. The binding of an allosteric effector (ANF) causes a reorganization of an oligomer, which eliminates the HS(−) conformer so that the whole enzyme pool becomes involved in the HS(+)↔HS(0) equilibrium.
Slow reduction of the substrate-bound high-spin enzyme by dithionite is in remarkable contrast to rapid reduction of the high-spin heme protein in the BMR-supported reaction. This difference between NADPH- and dithionite-dependent processes is attributable to the fact that the rate of dithionite-dependent reduction appears to be limited by the accessibility of the heme to the reducing agent, anion monomer [28, 86], while the rate of the NADPH-dependent electron transfer is not directly linked to the accessibility of the heme. Therefore, the substrate-promoted and high-spin-enriched HS(+) conformer, which is rapidly reducible by BMR, appears to represent a “closed” conformation where the accessibility of the heme for small molecules, such as , is hindered. Along with the dithionite reduction studies [28], this inference is supported by our recent observation that the interactions with substrates results in stabilization of the high-spin state of CYP3A4 at elevated hydrostatic pressure. This finding indicates a drastic decrease in water accessibility of the heme pocket induced by substrate binding [27]. This conclusion is also supported by the observation that CYP3A4 interactions with benzp[a]pyrene and, to a much larger extent the binding of ANF, result in a substantial decrease in the rate of recombination of ferrous CYP3A4 carbonyl complex after flash photolysis [37, 79].
Therefore, according to our model, oligomerization of cytochrome P450 results in incomplete reducibility of the enzyme by a flavoprotein partner. An important fraction of the enzyme appears to be unable to accept electrons from the electron donor without dissociation of the enzyme oligomers or their reorganization caused by allosteric effectors, such as ANF. A similar mechanism may also take place in the membrane, where the presence of cytochrome P450 oligomers is well established [87–92]. However, the reversibility of oligomerization in the membrane would blunt the manifestations of such heterogeneity, so that the whole population of the enzyme is expected to be (slowly) reducible via dissociation and reassembly of the oligomers.
At first glance, the biological advantage of hindering electron flow to a fraction of the enzyme and rendering it non-functional is unclear. However, physiological relevance of this mechanism becomes understandable when one considers that the endoplasmic reticulum contains multiple species of cytochrome P450 with different substrate specificity. There are several reports showing clear reciprocal influence of different isoforms of P450 in reconstituted systems and microsomes on activity and other properties (see [93] for review), which prompted some authors to consider direct association between different P450 species (i.e., formation of their mixed oligomers or “aggregates”) [90, 92, 94, 95] as an important determinant of the properties of microsomal monooxygenases [57, 93, 95–99]. If the distribution of these species between active (HS(0) ↔ HS(+)) and hindered (HS(−)) conformers in mixed oligomers is modulated by specific substrates [57, 95, 99], incomplete reducibility of the cytochrome P450 pool in the membrane would allow a redirection of the electron flow to a particular cytochrome P450 species in response to its specific substrate in the cell. This regulatory mechanism would block "idle" isoforms of P450 by interrupting unproductive electron flow and enable rapid adaptation to exposure to the changing spectrum of xenobiotics. The potential importance of this mechanism becomes evident in view of the poor coupling of electron flow to monooxygenation in drug-metabolizing cytochromes P450, leading to significant production of reactive oxygen species (ROS) (see [100] for review). Therefore, the suggested regulatory mechanism based on incomplete reducibility of P450 in the oligomers may have evolved to maintain the balance between the monooxygenase activity of cytochromes P450 and the production of ROS.
In summary, our results provide important evidence that the mechanism of action of heterotropic activators, such as ANF, involves redistribution of the CYP3A4 among several stable conformers and activation of an otherwise inactive subpopulation of the enzyme, as hypothesized by Koley and co-authors [36]. Admittedly, the use of oligomers of CYP3A4 in solution in a pair with BMR as a model electron donor does not allow direct extrapolation of our results to the membranous microsomal system. Nonetheless, strict parallels found between the present results and those obtained in our studies of dithionite-dependent reduction in CYP3A4-enriched liposomes [28] suggest that the general regularities revealed here are also maintained in the membranous system. In conjunction with our results on dithionite-dependent reduction kinetics, our data suggest that the heterogeneity of CYP3A4 may originate from the formation of cytochrome P450 oligomers, where the subunits may differ in their conformation and/or orientation. Hampering the electron flow to a fraction of cytochrome P450 pool is hypothesized to play a role in redirecting electron flow in the microsomal monooxygenase system containing several cytochrome P450 species. Further compelling evidence for the applicability of the above model to membranous systems is expected to be provided in our ongoing studies of the effect of ANF on the kinetics of electron transfer from BMR to CYP3A4 incorporated into model liposomal and microsomal membranes.
Supplementary Material
Acknowledgements
The authors thank Dr. T. M. Poulos at University of California-Irvine for the kind donation of plasmid DNA of BMR and Ms. N. Davydova for her assistance in the purification of CYP3A4. This research was supported by NIH grant GM54995 (JRH) and Center grant ES06676 (JRH).
Appendix A. Supplementary data
Supplementary data associated with this article include the set of spectral standards of ferric and ferrous CYP3A4 states, which can be found in the online version.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ingelman-Sundberg M, Johansson I, Hansson A. Acta Biol. Med. Ger. 1979;38:379–388. [PubMed] [Google Scholar]
- 2.Kiselev PA, Garda G, Finch SA, Stir A, Khatyleva MS, Akhrem AA. Biochemistry (Moscow) 1990;55:1535–1544. [PubMed] [Google Scholar]
- 3.Guengerich FP. Annu. Rev. Pharmacol. Toxicol. 1999;39:1–17. doi: 10.1146/annurev.pharmtox.39.1.1. [DOI] [PubMed] [Google Scholar]
- 4.Hutzler JM, Hauer MJ, Tracy TS. Drug Metab. Dispos. 2001;29:1029–1034. [PubMed] [Google Scholar]
- 5.Ariyoshi N, Miyazaki M, Toide K, Sawamura Y, Kamataki T. Biochem. Biophys. Res. Commun. 2001;281:1256–1260. doi: 10.1006/bbrc.2001.4524. [DOI] [PubMed] [Google Scholar]
- 6.Ekins S, Stresser DM, Williams JA. Trends Pharm. Sci. 2003;24:161–166. doi: 10.1016/s0165-6147(03)00049-x. [DOI] [PubMed] [Google Scholar]
- 7.Yoon MY, Campbell AP, Atkins WM. Drug Metab. Rev. 2004;36:219–230. doi: 10.1081/dmr-120033998. [DOI] [PubMed] [Google Scholar]
- 8.Locuson CW, Gannett PM, Tracy TS. Arch. Biochem. Biophys. 2006;449:115–129. doi: 10.1016/j.abb.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 9.Atkins WM. Expert Opinion Drug Metab. Toxicol. 2006;2:573–579. doi: 10.1517/17425255.2.4.573. [DOI] [PubMed] [Google Scholar]
- 10.Ueng YF, Kuwabara T, Chun YJ, Guengerich FP. Biochemistry. 1997;36:370–381. doi: 10.1021/bi962359z. [DOI] [PubMed] [Google Scholar]
- 11.Korzekwa KR, Krishnamachary N, Shou M, Ogai A, Parise RA, Rettie AE, Gonzalez FJ, Tracy TS. Biochemistry. 1998;37:4137–4147. doi: 10.1021/bi9715627. [DOI] [PubMed] [Google Scholar]
- 12.Harlow GR, Halpert JR. Proc. Natl. Acad. Sci. USA. 1998;95:6636–6641. doi: 10.1073/pnas.95.12.6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hosea NA, Miller GP, Guengerich FP. Biochemistry. 2000;39:5929–5939. doi: 10.1021/bi992765t. [DOI] [PubMed] [Google Scholar]
- 14.Atkins WM, Wang RW, Lu AYH. Chem. Res. Toxicol. 2001;14:338–347. doi: 10.1021/tx0002132. [DOI] [PubMed] [Google Scholar]
- 15.Houston JB, Galetin A. Arch. Biochem. Biophys. 2005;433:351–360. doi: 10.1016/j.abb.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 16.Denisov IG, Grinkova YV, Baas BJ, Sligar SG. J. Biol. Chem. 2006;281:23313–23318. doi: 10.1074/jbc.M605511200. [DOI] [PubMed] [Google Scholar]
- 17.Fernando H, Halpert JR, Davydov DR. Biochemistry. 2006;45:4199–4209. doi: 10.1021/bi052491b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Denisov IG, Baas BJ, Grinkova YV, Sligar SG. J. Biol. Chem. 2007;282:7066–7076. doi: 10.1074/jbc.M609589200. [DOI] [PubMed] [Google Scholar]
- 19.Davydov DR, Femando H, Halpert JR. Biophys. Chem. 2006;123:95–101. doi: 10.1016/j.bpc.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 20.Cameron MD, Wen B, Allen KE, Roberts AG, Schuman JT, Campbell AP, Kunze KL, Nelson SD. Biochemistry. 2005;44:14143–14151. doi: 10.1021/bi051689t. [DOI] [PubMed] [Google Scholar]
- 21.Roberts AG, Campbell AP, Atkins WM. Biochemistry. 2005;44:1353–1366. doi: 10.1021/bi0481390. [DOI] [PubMed] [Google Scholar]
- 22.Baas BJ, Denisov IG, Sligar SG. Arch. Biochem. Biophys. 2004;430:218–228. doi: 10.1016/j.abb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 23.He YA, Roussel F, Halpert JR. Arch. Biochem. Biophys. 2003;409:92–101. doi: 10.1016/s0003-9861(02)00484-8. [DOI] [PubMed] [Google Scholar]
- 24.Tsalkova TN, Davydova NY, Halpert JR, Davydov DR. Biochemistry. 2007;46:106–119. doi: 10.1021/bi061944p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lampe JN, Atkins WM. Biochemistry. 2006;45:12204–12215. doi: 10.1021/bi060083h. [DOI] [PubMed] [Google Scholar]
- 26.Isin EM, Guengerich FP. J. Biol. Chem. 2007;282:6863–6874. doi: 10.1074/jbc.M610346200. [DOI] [PubMed] [Google Scholar]
- 27.Davydov DR, Baas BJ, Sligar SG, Halpert JR. Biochemistry. 2007;46:7852–7864. doi: 10.1021/bi602400y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davydov DR, Fernando H, Baas BJ, Sligar SG, Halpert JR. Biochemistry. 2005;44:13902–13913. doi: 10.1021/bi0509346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davydov DR, Botchkareva AE, Kumar S, He YQ, Halpert JR. Biochemistry. 2004;43:6475–6485. doi: 10.1021/bi036260l. [DOI] [PubMed] [Google Scholar]
- 30.Houston B, Galetin A. Drug Metab. Rev. 2003;35:393–415. doi: 10.1081/dmr-120026870. [DOI] [PubMed] [Google Scholar]
- 31.Schrag ML, Wienkers LC. Arch. Biochem. Biophys. 2001;391:49–55. doi: 10.1006/abbi.2001.2401. [DOI] [PubMed] [Google Scholar]
- 32.Galetin A, Clarke SE, Houston JB. Drug Metab. Dispos. 2002;30:1512–1522. doi: 10.1124/dmd.30.12.1512. [DOI] [PubMed] [Google Scholar]
- 33.Egnell AC, Houston JB, Boyer CS. J. Pharmacol. Exp. Ther. 2005;312:926–937. doi: 10.1124/jpet.104.078519. [DOI] [PubMed] [Google Scholar]
- 34.Kenworthy KE, Clarke SE, Andrews J, Houston JB. Drug. Metab. Dispos. 2001;29:1644–1651. [PubMed] [Google Scholar]
- 35.Kenworthy KE, Bloomer JC, Clarke SE, Houston JB. Brit. J. Clin. Pharmacol. 1999;48:716–727. doi: 10.1046/j.1365-2125.1999.00073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koley AP, Robinson RC, Markowitz A, Friedman FK. Biochem. Pharm. 1997;53:455–460. doi: 10.1016/s0006-2952(96)00836-2. [DOI] [PubMed] [Google Scholar]
- 37.Koley AP, Buters JTM, Robinson RC, Markowitz A, Friedman FK. J. Biol. Chem. 1997;272:3149–3152. doi: 10.1074/jbc.272.6.3149. [DOI] [PubMed] [Google Scholar]
- 38.Davydov DR, Halpert JR, Renaud JP, Hui Bon Hoa G. Biochem. Biophys. Res. Commun. 2003;312:121–130. doi: 10.1016/j.bbrc.2003.09.247. [DOI] [PubMed] [Google Scholar]
- 39.Peterson JA, Ebel RE, O'keeffe DH, Matsubara T, Estabrook RW. J. Biol. Chem. 1976;251:4010–4016. [PubMed] [Google Scholar]
- 40.Matsubara T, Baron J, Peterson LL, Peterson JA. Arch. Biochem. Biophys. 1976;172:463–469. doi: 10.1016/0003-9861(76)90099-0. [DOI] [PubMed] [Google Scholar]
- 41.Blanck J, Smettan G, Ristau O, Ingelman-Sundberg M, Ruckpaul K. Eur. J. Biochem. 1984;144:509–513. doi: 10.1111/j.1432-1033.1984.tb08495.x. [DOI] [PubMed] [Google Scholar]
- 42.Davydov DR, Kurganov BI. Biokhimiia. 1982;47:1476–1482. [PubMed] [Google Scholar]
- 43.Guengerich FP, Johnson WW. Biochemistry. 1997;36:14741–14750. doi: 10.1021/bi9719399. [DOI] [PubMed] [Google Scholar]
- 44.Backes WL, Tamburini PP, Jansson I, Gibson GG, Sligar SG, Schenkman JB. Biochemistry. 1985;24:5130–5136. doi: 10.1021/bi00340a026. [DOI] [PubMed] [Google Scholar]
- 45.Karyakin AV, Davydov DR. Vestn. Akad. Med. Nauk SSSR. 1988;1988:53–62. [PubMed] [Google Scholar]
- 46.Cole PE, Sligar SG. FEBS Lett. 1981;133:252–254. doi: 10.1016/0014-5793(81)80517-0. [DOI] [PubMed] [Google Scholar]
- 47.Ziegler M, Blanck J, Ruckpaul K. FEBS Lett. 1982;150:219–222. [Google Scholar]
- 48.Fisher MT, Sligar SG. Biochemistry. 1987;26:4797–4803. doi: 10.1021/bi00389a029. [DOI] [PubMed] [Google Scholar]
- 49.Narasimhulu S. Endocr. Res. 1993;19:223–258. doi: 10.1080/07435809309026680. [DOI] [PubMed] [Google Scholar]
- 50.Oprian DD, Vatsis KP, Coon MJ. J. Biol. Chem. 1979;254:8895–8902. [PubMed] [Google Scholar]
- 51.Taniguchi H, Imai Y, Iyanagi T, Sato R. Biochim. Biophys. Acta. 1979;550:341–356. doi: 10.1016/0005-2736(79)90220-7. [DOI] [PubMed] [Google Scholar]
- 52.Blanck J, Janig GR, Schwarz D, Ruckpaul K. Xenobiotica. 1989;19:1231–1246. doi: 10.3109/00498258909043175. [DOI] [PubMed] [Google Scholar]
- 53.Backes WL, Eyer CS. J. Biol. Chem. 1989;264:6252–6259. [PubMed] [Google Scholar]
- 54.Ingelman-Sundberg M, Blanck J, Smettan G, Ruckpaul K. Eur. J. Biochem. 1983;134:157–162. doi: 10.1111/j.1432-1033.1983.tb07546.x. [DOI] [PubMed] [Google Scholar]
- 55.Blanck J, Behlke J, Jänig GR, Pfeil D, Ruckpaul K. Acta Biol. Med. Ger. 1979;38:11–21. [PubMed] [Google Scholar]
- 56.Kanaeva IP, Nikityuk OV, Davydov DR, Dedinskii IR, Koen YM, Kuznetsova GP, Skotselyas ED, Bachmanova GI, Archakov AI. Arch. Biochem. Biophys. 1992;298:403–412. doi: 10.1016/0003-9861(92)90428-y. [DOI] [PubMed] [Google Scholar]
- 57.Davydov DR, Petushkova NA, Archakov AI, Hui Bon Hoa G. Biochem. Biophys. Res. Commun. 2000;276:1005–1012. doi: 10.1006/bbrc.2000.3596. [DOI] [PubMed] [Google Scholar]
- 58.Gillam EM, Baba T, Kim BR, Ohmori S, Guengerich FP. Arch. Biochem. Biophys. 1993;305:123–131. doi: 10.1006/abbi.1993.1401. [DOI] [PubMed] [Google Scholar]
- 59.Ingelman-Sundberg M, Hagbjork AL, Ueng YF, Yamazaki H, Guengerich FP. Biochem. Biophys. Res. Commun. 1996;221:318–322. doi: 10.1006/bbrc.1996.0593. [DOI] [PubMed] [Google Scholar]
- 60.Shaw PM, Hosea NA, Thompson DV, Lenius JM, Guengerich FP. Arch. Biochem. Biophys. 1997;348:107–115. doi: 10.1006/abbi.1997.0378. [DOI] [PubMed] [Google Scholar]
- 61.Boddupalli SS, Hasemann CA, Ravichandran KG, Lu JY, Goldsmith EJ, Deisenhofer J, Peterson JA. Proc. Natl. Acad. Sci. USA. 1992;89:5567–5571. doi: 10.1073/pnas.89.12.5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dodhia VR, Fantuzzi A, Gilardi G. J Biol. Inorg. Chem. 2006;11:903–916. doi: 10.1007/s00775-006-0144-3. [DOI] [PubMed] [Google Scholar]
- 63.Domanski TL, He YA, Harlow GR, Halpert JR. J. Pharmacol. Exp. Ther. 2000;293:585–591. [PubMed] [Google Scholar]
- 64.Domanski TL, He YA, Khan KK, Roussel F, Wang Q, Halpert JR. Biochemistry. 2001;40:10150–10160. doi: 10.1021/bi010758a. [DOI] [PubMed] [Google Scholar]
- 65.Govindaraj S, Poulos TL. J. Biol. Chem. 1997;272:7915–7921. doi: 10.1074/jbc.272.12.7915. [DOI] [PubMed] [Google Scholar]
- 66.Jamakhandi AP, Jeffus BC, Dass VR, Miller GP. Arch. Biochem. Biophys. 2005;439:165–174. doi: 10.1016/j.abb.2005.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.French JS, Coon MJ. Arch. Biochem. Biophys. 1979;195:565–577. doi: 10.1016/0003-9861(79)90383-7. [DOI] [PubMed] [Google Scholar]
- 68.Sevrioukova I, Peterson JA. Biochimie. 1996;78:744–751. doi: 10.1016/s0300-9084(97)82532-1. [DOI] [PubMed] [Google Scholar]
- 69.Davydov DR, Deprez E, Hui Bon Hoa G, Knyushko TV, Kuznetsova GP, Koen YM, Archakov AI. Arch. Biochem. Biophys. 1995;320:330–344. doi: 10.1016/0003-9861(95)90017-9. [DOI] [PubMed] [Google Scholar]
- 70.Renaud JP, Davydov DR, Heirwegh KPM, Mansuy D, Hui Bon Hoa G. Biochem. J. 1996;319:675–681. doi: 10.1042/bj3190675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Davydov DR, Knyushko TV, Hui Bon Hoa G. Biochem .Biophys.Res.Commun. 1992;188:216–221. doi: 10.1016/0006-291x(92)92372-5. [DOI] [PubMed] [Google Scholar]
- 72.French JS, Guengerich FP, Coon MJ. J. Biol. Chem. 1980;255:4112–4119. [PubMed] [Google Scholar]
- 73.Bösterling B, Trudell JR. J. Biol. Chem. 1982;257:4783–4787. [PubMed] [Google Scholar]
- 74.Segel IH. Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems. New York: Wiley-Interscience; 1975. [Google Scholar]
- 75.Reed JR, Hollenberg PF. J. Inorg. Biochem. 2003;97:276–286. doi: 10.1016/s0162-0134(03)00310-6. [DOI] [PubMed] [Google Scholar]
- 76.Tang W, Stearns RA. Curr. Drug Metab. 2001;2:185–198. doi: 10.2174/1389200013338658. [DOI] [PubMed] [Google Scholar]
- 77.Harlow GR, Halpert JR. J. Biol. Chem. 1997;272:5396–5402. doi: 10.1074/jbc.272.9.5396. [DOI] [PubMed] [Google Scholar]
- 78.Shou M, Grogan J, Mancewicz JA, Krausz KW, Gonzalez FJ, Gelboin HV, Korzekwa KR. Biochemistry. 1994;33:6450–6455. doi: 10.1021/bi00187a009. [DOI] [PubMed] [Google Scholar]
- 79.Koley AP, Buters JTM, Robinson RC, Markowitz A, Friedman FK. Biochimie. 1996;78:706–713. doi: 10.1016/s0300-9084(97)82528-x. [DOI] [PubMed] [Google Scholar]
- 80.Yamazaki H, Johnson WW, Ueng YF, Shimada T, Guengerich FP. J. Biol. Chem. 1996;271:27438–27444. doi: 10.1074/jbc.271.44.27438. [DOI] [PubMed] [Google Scholar]
- 81.Isin EM, Guengerich FP. J. Biol. Chem. 2006;281:9127–9136. doi: 10.1074/jbc.M511375200. [DOI] [PubMed] [Google Scholar]
- 82.Davydov DR, Darovsky BV, Dedinsky IR, Kanaeva IP, Bachmanova GI, Blinov VM, Archakov AI. Arch. Biochem. Biophys. 1992;292:304–313. doi: 10.1016/0003-9861(92)90677-o. [DOI] [PubMed] [Google Scholar]
- 83.Sligar SG, Gunsalus IC. Biochemistry. 1979;18:2290–2295. doi: 10.1021/bi00578a024. [DOI] [PubMed] [Google Scholar]
- 84.Fisher MT, Sligar SG. J. Am. Chem. Soc. 1985;107:5018–5019. [Google Scholar]
- 85.Guengerich FP. Biochemistry. 1983;22:2811–2820. doi: 10.1021/bi00281a007. [DOI] [PubMed] [Google Scholar]
- 86.Davydov DR, Karyakin AV, Binas B, Kurganov BI, Archakov AI. Eur. J. Biochem. 1985;150:155–159. doi: 10.1111/j.1432-1033.1985.tb09001.x. [DOI] [PubMed] [Google Scholar]
- 87.Greinert R, Finch SA, Stier A. Biosci. Rep. 1982;2:991–994. doi: 10.1007/BF01122166. [DOI] [PubMed] [Google Scholar]
- 88.Kawato S, Gut J, Cherry RJ, Winterhalter KH, Richter C. J. Biol. Chem. 1982;257:7023–7029. [PubMed] [Google Scholar]
- 89.Schwarz D, Pirrwitz J, Meyer HW, Coon MJ, Ruckpaul K. Biochem. Biophys. Res. Commun. 1990;171:175–181. doi: 10.1016/0006-291x(90)91373-z. [DOI] [PubMed] [Google Scholar]
- 90.Alston K, Robinson RC, Park SS, Gelboin HV, Friedman FK. J. Biol. Chem. 1991;266:735–739. [PubMed] [Google Scholar]
- 91.Szczesna-Skorupa E, Mallah B, Kemper B. J. Biol. Chem. 2003;278:31269–31276. doi: 10.1074/jbc.M301489200. [DOI] [PubMed] [Google Scholar]
- 92.Schwarz D, Chernogolov L, Kisselev P. Biochemistry. 1999;38:9456–9464. doi: 10.1021/bi990584l. [DOI] [PubMed] [Google Scholar]
- 93.Backes WL, Kelley RW. Pharm. Ther. 2003;98:221–233. doi: 10.1016/s0163-7258(03)00031-7. [DOI] [PubMed] [Google Scholar]
- 94.Cawley GF, Zhang SX, Kelley RW, Backes WL. Drug Metab. Dispos. 2001;29:1529–1534. [PubMed] [Google Scholar]
- 95.Kelley WK, Reed JR, Backes WL. Biochemistry. 2005;44:2632–2641. doi: 10.1021/bi0477900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kaminsky LS, Guengerich FP. Eur. J. Biochem. 1985;149:479–489. doi: 10.1111/j.1432-1033.1985.tb08950.x. [DOI] [PubMed] [Google Scholar]
- 97.Yamazaki H, Inoue K, Mimura M, Oda Y, Guengerich FP, Shimada T. Biochem. Pharmacol. 1996;51:313–319. doi: 10.1016/0006-2952(95)02178-7. [DOI] [PubMed] [Google Scholar]
- 98.Backes WL, Batie CJ, Cawley GF. Biochemistry. 1998;37:12852–12859. doi: 10.1021/bi980674a. [DOI] [PubMed] [Google Scholar]
- 99.Davydov DR, Petushkova NA, Bobrovnikova EV, Knyushko TV, Dansette P. Adv. Exp. Med. Biol. 2001;500:335–338. doi: 10.1007/978-1-4615-0667-6_53. [DOI] [PubMed] [Google Scholar]
- 100.Zangar RC, Davydov DR, Verma S. Toxicol. Appl. Pharm. 2004;199:316–331. doi: 10.1016/j.taap.2004.01.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.