Abstract
Genetic events often require proteins to be activated by interacting with two DNA sites, trapping the intervening DNA in a loop. While much is known about looping equilibria, only a few studies have examined DNA-looping dynamics experimentally. The restriction enzymes that cut DNA after interacting with two recognition sites, such as FokI, can be used to exemplify looping reactions. The reaction pathway for FokI on a supercoiled DNA with two sites was dissected by fast kinetics to reveal, in turn: the initial binding of a protein monomer to each site; the protein–protein association to form the dimer, trapping the loop; the subsequent phosphodiester hydrolysis step. The DNA motion that juxtaposes the sites ought on the basis of Brownian dynamics to take ∼2 ms, but loop capture by FokI took 230 ms. Hence, DNA looping by FokI is rate limited by protein association rather than DNA dynamics. The FokI endonuclease also illustrated activation by looping: it cut looped DNA 400 times faster than unlooped DNA.
INTRODUCTION
Many events on DNA are mediated by proteins that bind two sites at separate genetic loci, and then interact with each other to trap the intervening DNA in a loop (1,2). Other processes are governed by multimeric proteins that trap loops directly by binding two DNA sites at the same time. Processes that involve DNA looping include the replication, repair, recombination and restriction of DNA (2–5) and the regulation of gene expression (1,6).
The factors governing looping equilibria have been analysed extensively from both theoretical and experimental standpoints (1,7–9). In contrast, very little experimental data are currently available about looping dynamics on unconstrained DNA in free solution. Loop capture can occur only when thermal fluctuations in DNA configuration bring the sites close together in 3D space, so that the distance between the sites can be spanned by the protein(s). Numerous theoretical studies have estimated the time needed for two sites to become juxtaposed in this manner. While the most rigorous approaches refer to linear rather than supercoiled (SC) DNA (10), fluctuations in SC DNA have been modelled by Brownian dynamics to calculate average times to the first juxtaposition event, typically around 2 ms (11–13). Experiments measuring the relative rates of loop capture at varied site separations concurred with these simulations (14) but actual capture rates have seldom been measured.
At present, looping rates have been measured most often by tethering the DNA between a bead and a glass surface and then assessing the frequency with which the length of the tether shortens upon loop formation (15–18). Loop closure times from these studies usually fall between 5 and 75 s, very much longer than that predicted for site juxtaposition by Brownian dynamics (10–13). The slow rates may be due to the limited conformational freedom of the tethered DNA relative to unconstrained DNA: repulsion of the bead from the glass surface results in the tethered DNA spending most of its time in an extended configuration that precludes loop capture between sites that are distant from each other along the DNA.
Rates of loop formation on untethered DNA are currently available from only two experimental systems. In one, streptavidin was used to capture loops on a linear DNA that had been tagged with two biotin moieties 500 bp apart (19): this yielded capture times of about 10 ms, approaching theoretical expectations but on an unnatural system for DNA looping. The other examined the rates for forming synaptic complexes prior to site-specific recombination by resolvase: some DNA molecules yielded productive synapses in <10 ms but others took >100 s (20,21). The slow rates observed on some of the DNA are probably due to dissociation of abortive complexes before re-attempting to establish the productive complex. Resolvase possesses considerable potential for non-productive complexes as its reaction demands a unique set of interactions between six molecules of the protein bound to two separate DNA sites, to the exclusion of all other sets (4). There is thus a pressing need to develop a natural system suitable for a kinetic analysis of looping on unconstrained DNA.
Perhaps the most amenable test systems for DNA looping are the Type II restriction endonucleases that bridge two recognition sites (2,5). Type II enzymes recognize specific DNA sequences and cut both strands at or near their targets (22). However, many of them need to interact with two cognate sites before they can cut the DNA (2). Such enzymes trap loops on DNA molecules with two copies of the target sequence (14,17). Proteins that bind two sites generally have higher affinities for sites in cis, on the same DNA molecule, over sites in trans, on separate molecules, due to the concentration of one site in the vicinity of another being higher for sites in cis (1,7,10). The Type II enzymes that need two sites thus usually cleave two-site substrates by acting in cis, with two sites on the same molecule of DNA, but they normally have to act in trans on one-site substrates, synapsing two separate molecules of the DNA. Consequently, these enzymes generally cut DNA with two or more sites more rapidly than DNA with a single site (23–27). Most restriction enzymes that recognize asymmetric—as opposed to palindromic—sequences act in this way (24,26). One example is FokI (24,27,28).
The FokI restriction enzyme recognizes an asymmetric sequence, 5′-GGATG-3′. In the presence of Mg2+, it cuts ‘top’ and ‘bottom’ strands downstream of this site, 9 and 13 bases away, respectively (22). The restriction enzymes that act at palindromic sequences usually employ two subunits per site in a symmetrical arrangement, with each subunit recognizing one half of the site and cleaving one strand of the DNA (29). The rate constants for cutting the two strands are normally equal to each other though in heterodimeric enzymes the subunits can operate at different rates (30–32). In contrast, FokI exists as a monomer in solution and when bound to its asymmetric DNA site (33). A single monomer engulfs the entire recognition sequence (34). The protein contains two domains, one for DNA recognition and one for catalysis, but its catalytic domain has the functions for cleaving only one phosphodiester bond at a time (34,35). Even so, the monomer bound to the specific site fails to cut either strand until it forms a dimer (36). The dimer is formed by association of the catalytic domains to give a structure in which the two active sites are each positioned to cleave one strand (35).
On DNA with one FokI site, the second monomer must come from free solution but, due to the small area of the dimer interface (35,36), DNA-bound and free monomers associate weakly (27). Only a small fraction of the enzyme is then active dimer, so one-site DNA is cleaved slowly. However, the dimeric form of FokI can bind two copies of its target DNA simultaneously (28). On DNA with two FokI sites, it cleaves one site rapidly, more rapidly than one-site substrates and the second at the same slow rate as one-site DNA (24). The fast reaction on two-site DNA involves a complex whose stability varies cyclically as the length of DNA between the sites is varied (27). The changes in stability displayed a periodicity of about 10 bp, similar to the helical repeat of DNA, a classical signature for looping (1,7,8). It had initially been proposed that a monomer bound to one recognition site on a DNA with two sites associates with a second monomer from free solution to form a dimer at that site: the dimer then uses its free DNA-recognition domain to capture the second site (24). However, subsequent studies raised the possibility of an alternative scheme in which two monomers bound to separate sites on the same DNA interact with each other via their catalytic domains to form a dimer spanning both sites, looping out the intervening DNA (27). In either case, the dimer formed across sites in cis will be more stable than at a single site, as its subunits are tethered together by the intervening DNA.
In this study, the reaction pathway for FokI on DNA with two target sites was dissected into individual steps by rapid-reaction single-turnover kinetics. The steps included: the initial binding of first one monomer of FokI to one recognition site and then another to the second site, in two independent stages; the subsequent association of the two monomers bound to their separate sites to form the dimer and trap the loop; finally, phosphodiester hydrolysis by the looped complex. The kinetics reveal whether the rate of loop closure is diffusion-limited by the internal motion of the DNA or is reaction-limited by protein assembly. In addition, the rates of phosphodiester hydrolysis by FokI in its looped and unlooped complexes reveal the extent to which a DNA-binding protein can be activated by looping.
MATERIALS AND METHODS
Materials
FokI endonuclease was purified from an over-producing strain of Escherichia coli (from W. Jack, New England Biolabs) to >95% homogeneity as before (27). Concentrations were determined by UV absorbance and are given in terms of monomeric protein.
The plasmids pIF185 and pIF190 (27) were purified from transformants of E. coli HB101 that had been cultured in minimal media containing [methyl-3H]thymidine by CsCl density gradient centrifugations (23–26). Typically, 85–95% of each preparation was the SC monomeric plasmid and the rest nicked or dimeric forms.
Buffer R is 20 mM Tris–acetate (pH 7.9), 50 mM potassium acetate, 1 mM DTT, 0.2 mM EDTA and 100 μg/ml BSA. Magnesium acetate (MgOAc) was added as required.
Methods
Reactions were carried out at 37°C in a Hi-Tech RQF-63 quench flow apparatus (http://www.tgkscientific.com/). For post-mix reactions starting with enzyme and DNA in separate solutions, 80 µl of FokI in Buffer R with 10 mM MgOAc was mixed with an equal volume of either pIF185 or pIF190, also in Buffer R with 10 mM MgOAc. For pre-mix reactions, which were started by adding Mg2+ to a solution containing both enzyme and DNA, FokI was added to the plasmid in Buffer R and the solution incubated at 37°C before mixing with an equal volume of 20 mM MgOAc in Buffer R. With both procedures, the final reactions contained varied concentrations of FokI enzyme (6.25–250 nM) and fixed concentrations of both plasmid (2.5 nM) and MgOAc (10 mM). After the requisite time, the reactions were quenched by mixing with 80 µl of 0.1 M EDTA. Aliquots from the quenched samples were analysed by electrophoresis through agarose to separate the SC substrate from the cleaved DNA products (23–27). The concentration of the SC DNA in each sample was assessed by scintillation counting. The rate constants that gave the best fits of the data to single or multiple exponential decays were obtained by non-linear regression in GRAFIT (http://www.erithacus.com/grafit/). For the reaction scheme in Figure 3b, the differential rate equations for the change in substrate concentration with time were solved by numerical integration in version 8.0.1 of BERKELEY MADONNA (http://www.berkeleymadonna.com/).
Figure 3.
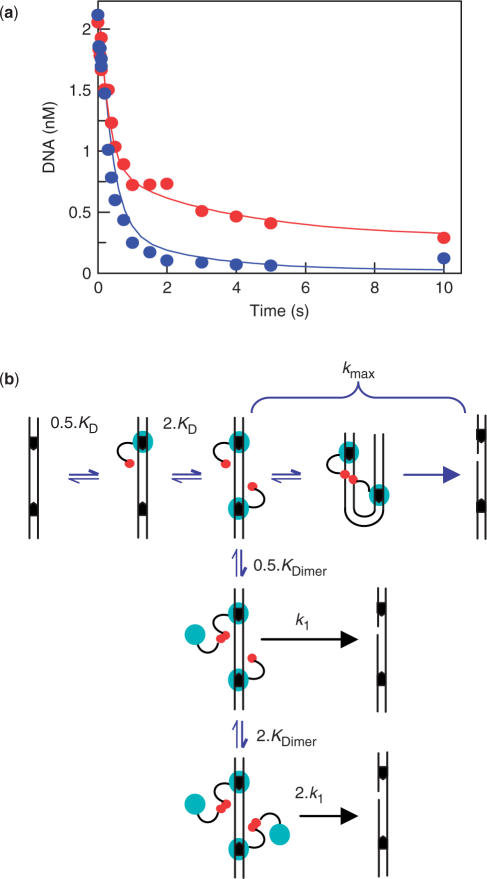
Dimers at separate sites. (a) Reactions were as in Figure 2a, except with FokI at 125 nM: red circles denote the residual concentrations of SC DNA at the times indicated. For comparison, the reaction from Figure 2a at 50 nM FokI is reproduced here (blue circles). The red and the blue lines are theoretical lines calculated as below. (b) The pathway in Figure 2c was extended for reactions at [E0] > KDimer. The DNA carrying a monomer at each site either forms the looped complex and cleaves the DNA at the kmax rate (as in Figure 2c), or it binds monomer(s) from free solution to form dimer(s) at individual site(s), which then cleave the DNA at the k1 rate (as in Figure 1). (The multipliers of 0.5 or 2 applied to KD, KDimer and k1 are the statistical factors arising from, respectively, the first and second binding events.) The rate equations for this scheme were solved by numerical integration and the residual concentration of SC substrate calculated using the following parameters: 1 × 109 M−1s−1 and 4 s−1 for the forward and reverse rate constants for the DNA-binding step (to give KD = 4 nM); 5 × 106 M−1s−1 and 0.5 s−1 for the forward and reverse rate constants for the dimerization step (to give KDimer = 100 nM), 3 s−1 for kmax; and 0.05 s−1 for k1. The calculations were for 2.5 nM pIF185 with either 125 nM FokI [red line in (a)] or 50 nM FokI [blue line in (a)].
RESULTS
Experimental strategy
The reaction pathway for FokI on a DNA with one target site was elucidated previously by single-turnover kinetics (27). The aim of this study was to identify the pathway for FokI on plasmids with two sites, in order to determine which steps differed between the one- and two-site substrates and so account for its enhanced reactivity on the two-site DNA. A particular goal was to characterize the loop capture event during its reactions on two-site plasmids.
For single turnovers on the one-site plasmid, the nuclease had been added to a solution of SC plasmid and Mg2+, to give reactions with excess enzyme ([E0]) over plasmid. Samples were removed at various times (10–300 s) and mixed immediately with EDTA to stop the reaction, prior to determining the concentrations of the intact DNA, the nicked DNA cut in one strand and the linear form cut in both strands. Some reactions employed an alternative procedure, ‘pre-mix’ instead of ‘post-mix’: post-mixes were initiated by adding enzyme to DNA in the presence of Mg2+; for pre-mixes, the enzyme was first incubated with the DNA in the absence of Mg2+, to permit binding, before adding MgOAc. Post- and pre-mix reactions on the one-site plasmid were identical, so product formation is rate limited by DNA cleavage, not by DNA binding. In single turnovers, product formation denotes enzyme-bound rather than free products.
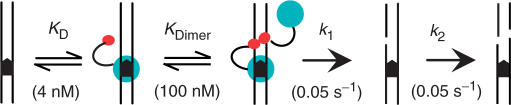
FokI acts on one-site DNA as follows (Figure 1). First, the monomeric protein binds to the recognition site with a relatively high (but at that time unknown) affinity, to give an inactive complex. Next, a second monomer associates with the DNA-bound monomer to give the active dimer with two catalytic sites. The association of the monomer in free solution with that on the DNA is relatively weak. It had a much weaker equilibrium dissociation constant (KDimer = 100 nM) than that for the initial association to the DNA (KD). Consequently, FokI concentrations >>100 nM are required to drive dimerization towards completion (for typical restriction reactions, [E0] ≤1 nM, when hardly any of the FokI protein would be active dimer). The dimer then cuts both strands while remaining bound to the DNA, presumably using one active site on each strand (35). Despite only one of the two subunits of the dimer being bound to the recognition sequence, the two strands are cut at equal rates, 0.05 s−1 in both cases (27).
Figure 1.
FokI on DNA with one recognition site. The catalytic and DNA-binding domains of the FokI endonuclease are shown as red and blue circles, respectively, the DNA duplex as parallel lines, and the FokI recognition site as a black arrowhead. One monomer of FokI binds to the recognition site through its DNA-binding domain, with an equilibrium dissociation constant KD = 4 nM. A second monomer then associates with the DNA-bound protein through its catalytic domain, with KDimer = 100 nM. The dimer proceeds to cut first one and then the second strand, with rate constants k1 and k2, respectively (both 0.05 s−1). For clarity, the enzyme bound to cleaved DNA is not shown, nor the subsequent dissociation of the enzyme. Published values are cited for KDimer, k1 and k2 (27); that for KD is from this study (Table 1).
FokI dimers bridging two sites
FokI reactions at two sites were examined on two 3.0-kb plasmids. In one case, pIF185, the sites were separated by 185 bp; in the other, pIF190, by 190 bp. The 185-bp separation corresponds to a minimum in the cyclical variation of loop stability with inter-site distance while the 190-bp spacing is at a maximum (27). The restriction enzymes that act at two asymmetric sites can show markedly different levels of activity depending on whether the sites are in inverted or directly repeated orientation (37). To eliminate this factor, both pIF185 and pIF190 were constructed with FokI sites in inverted (head-to-head) orientation. In addition, to nullify any variation that could be introduced by different flanking sequences around each site, the two FokI sites on both plasmids were embedded within the same 35-bp sequence as that around the target on the one-site plasmid: the region of identity encompasses not only the immediate vicinity of the recognition sequence but also the downstream sites of DNA cleavage.
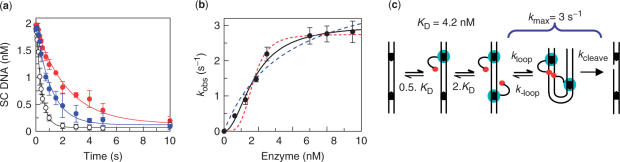
Single turnovers of FokI on two-site substrates were initially carried out in post-mix mode, using pIF185 as the substrate. FokI had typically taken >100 s to cleave all of the one-site plasmid (27) but the two-site plasmids were mostly cleaved in <5 s (Figure 2a). Single turnovers on two-site DNA therefore required a rapid quench-flow device (38). This was used to mix together FokI and DNA, with MgOAc in both solutions. After the requisite time delay, the reaction was stopped and the samples analysed by electrophoresis to separate the following forms of the DNA (23–26): the intact (SC) substrate; open-circle DNA cut in one strand at one or both sites; the linear species a double-strand break at one site, with or without a single-strand break at the other site; the two final products with double-strand breaks at both sites. Rather than estimating the rates for cleaving each bond from the transient formation and decay of all of these species (23,25), the kinetics were evaluated from the decline in the concentration of the SC substrate, which denotes the rate at the first of the four bonds cut by the enzyme.
Figure 2.
Dimers bridging two sites. (a) FokI endonuclease and pIF185 (85% SC), both in Buffer R with 10 mM MgOAc, were mixed to give solutions containing 2.5 nM pIF185 and FokI at: 6.25 nM, red circles; 12.5 nM, blue circles; 50 nM, open circles. After the times indicated, reactions were quenched with 0.1 M EDTA, and the residual concentrations of SC DNA measured. Error bars indicate standard errors of the mean from ≥3 independent repeats. Reactions were fitted to first-order rate constants (kobs): the red, blue and black lines indicate the best fits to the corresponding data. (b) Values of kobs (bars denote standard errors from ≥3 repeats) were plotted against [E0] and the data fitted to Equation (2) with one of the following values for n: 1, dashed blue line; 2, black line; 4, dashed red line. (c) Pathway for FokI on DNA with two sites, at [E0] < KDimer. The catalytic and recognition domains of FokI, the DNA and the recognition sites are all shown as in Figure 1. One monomer of FokI binds to each site with the same KD (4 nM) though the actual values for first and second monomers are 0.5 × KD and 2 × KD, respectively (see text). The monomers associate with each other to trap the loop and cleave the DNA, initially at one phosphodiester bond: the subsequent products cleaved at 2, 3 and 4 bonds are not shown. The apparent rate constant for DNA cleavage (kmax = 3 s−1) incorporates both loop capture (kloop) and phosphodiester hydrolysis (kcleave).
The single turnovers of FokI on pIF185 employed fixed concentrations of DNA and Mg2+ but varied [E0], though with protein monomers always in excess of DNA sites. However, in reactions with monomers in excess of recognition sites, there exists the possibility that the enzyme will dimerize at individual sites rather than forming the dimer across two sites. To minimize this situation, the range for [E0] was initially limited to concentrations below the dimerization constant at individual sites (KDimer = 100 nM). Throughout this range, the amount of SC DNA substrate declined exponentially during the reactions (Figure 2a). For each [E0], an apparent first-order rate constant (kobs) was evaluated from the decline. The values for kobs increased with increasing [E0] up to a maximum at saturating levels, but in a sigmoidal rather than a hyperbolic fashion (Figure 2b).
If a substrate (S) is cleaved to product (P) after binding n molecules of enzyme to independent but equal sites,
| 1 |
then, provided that [E0] > [S] and that the binding steps are faster than the subsequent cleavage event,
| 2 |
where kmax is the rate constant for cleavage at saturation with enzyme and KD the equilibrium dissociation constant for the enzyme at a single site.
To test whether FokI reactions on two-site DNA follow the scheme in Equation (1), a series of values were selected for n and, with each value, the sigmoidal variation in kobs with [E0] was fitted to Equation (2) to find the optimal values for kmax and KD at that particular n (Figure 2b). The data fitted better with n = 2 (i.e. one monomer per recognition site: χ2 = 0.07) than with n at either 1 (one monomer between two sites: χ2 = 0.30) or 4 (two monomers per site: χ2 = 0.26).
The best-fit values (Table 1) show firstly that the initial binding of the monomer to a single recognition site, KD = 4 nM, is much tighter than the association of a second monomer from free solution, KDimer = 100 nM. Hence, on DNA with two sites, FokI monomers will bind to both sites more readily than forming a dimer at one site. Previously, two alternative routes had been suggested for the formation of a FokI dimer across two distant DNA sites: either a dimer at one site captures the second site through its free DNA-binding domain, or two monomers bound to the separate sites associate with each other to form the dimer (24,27). The data presented here (Figure 2c) show that the dimer spanning two sites is actually formed by the association of two DNA-bound monomers. However, the effective KD for the first binding event—to either site—will be half the intrinsic KD and that for the second event; to the unoccupied site—twice the intrinsic KD. These factors arise because the first monomer can bind either site but the second can only bind the remaining free site, while the converse applies to dissociation; two pathways for the E2S → E1S step, one for E1S → E + S.
Table 1.
Kinetic parameters
| Reaction type | Parameter | Plasmid | |
|---|---|---|---|
| pIF185 | pIF190 | ||
| Post-mix | KD (nM) | 4.2 ± 0.2 | 4.0 ± 0.1 |
| kmax (s−1) | 3.0 ± 0.1 | 2.5 ± 0.1 | |
| Pre-mix | k for fast phase (s−1) | 23 ± 5 | 17 ± 7 |
Second, kmax refers to the rate of conversion of the intact DNA, with one monomer at each site, to DNA cut at one or more of its scissile phosphodiester bonds. However, this stage includes at least two steps (Figure 2c): the protein-protein association to trap the DNA loop (kloop), followed by phosphodiester hydrolysis (kcleave). The value for kmax may reflect either of these processes.
Post-mix reactions also utilized pIF190 as the substrate (data not shown). This plasmid is identical to pIF185 except its two FokI sites are 190 bp apart, a local maximum for loop stability, as opposed to 185 bp, a minimum (27). The single-turnover reactions on pIF190 kept [E0] < KDimer so as to minimize dimerization at individual sites. In these reactions, the concentration of the SC form of pIF190 declined exponentially with time (as in Figure 2a), and the values of kobs varied sigmoidally with [E0] (as in Figure 2b).
Like pIF185, the sigmoidal curve fitted better to Equation (2) with n = 2 (χ2 = 0.03) than with n at either 1 (χ2 = 0.08) or 4 (χ2 = 0.30), which in turn yielded best-fit values for kmax and KD (Table 1). As expected, given that the FokI sites on pIF190 and on pIF185 have identical flanking sequences, both plasmids gave similar values for KD. The two plasmids also gave similar values for kmax but as kmax incorporates both loop closure and phosphodiester hydrolysis, this only shows that the slower of these two steps is not significantly affected by the change in site separation.
FokI dimers at solitary sites
To see if dimer formation at individual site(s) can affect reactions on a DNA with two recognition sites, single turnovers were also carried out with FokI concentrations above the dimerization constant. The reactions used the post-mix method with pIF185 as the substrate, as above. Figure 3a shows two reaction records at different enzyme concentrations: in one (in blue), with [E0] < KDimer; in the other (in red), with [E0] > KDimer.
At elevated [E0], the decline in the concentration of the SC substrate no longer followed a single exponential as had been observed at lower [E0], but was instead markedly biphasic (Figure 3a). Some of the DNA was cleaved rapidly, at a similar rate to that for the dimer spanning two sites (kmax in Figure 2). The remainder was cleaved more slowly, at a rate approaching those for a dimer at a single site (k1 in Figure 1). Similar results were obtained with further increases in [E0] though the increases caused a decline in the proportion of rapidly cleaved DNA and a corresponding rise in the proportion cleaved slowly (data not shown).
To accommodate the reactions on two-site DNA at high [E0], the previous scheme for the reactions at relatively low FokI concentrations (Figure 2c) was extended to include dimer formation at solitary sites (Figure 3b). In the extended model, the DNA with a monomer at each site has two possible fates: either the monomers associate with each other to give the looped complex which then proceeds to cut the DNA at the kmax rate, as in Figure 2c; or the monomers at the separate sites associate with free monomers to give a dimer at either one or both sites, and that each dimer then proceeds to cut the DNA at the characteristic rate for a solitary dimer, k1 (Figure 1) (or 2 × k1 for two dimers, one at each site).
To test whether the model in Figure 3b can accommodate the biphasic reactions at high [E0], the decline in concentration of SC substrate with time was calculated by assigning values for the rate constants for each individual step and then solving numerically the differential rate equations for substrate utilization. The values selected for the rate constants were either known values (kmax = 3 s−1, k1 = 0.05 s−1) or were chosen so that the ratio of forward and reverse rate constants for a particular step matched the known equilibrium constant for that step (KD = 4 nM, KDimer = 100 nM). The calculations were carried out for both the high and the low enzyme concentrations shown in Figure 3a. In both cases, the theoretical curves fell close to the experimental data. Dimerization at individual sites can thus account for the biphasicity.
DNA looped by FokI
The rate constant (kmax) for the conversion of the intact DNA with a monomer at each site to the cleaved product incorporates at least two steps: the protein–protein association to trap the DNA loop (kloop), followed by phosphodiester hydrolysis (kcleave). Which of these limits kmax to 3 s−1 has yet to be established.
The above reactions were all conducted by the post-mix procedure, by mixing separate solutions of enzyme and DNA in the presence of Mg2+ and then monitoring the subsequent cleavage of the DNA. An alternative is the pre-mix method in which enzyme and DNA are equilibrated together in the absence of Mg2+ before initiating the reaction by adding the Mg2+. The latter has the potential to bypass the binding of the enzyme to the DNA and the loop capture. If in the absence of Mg2+ the enzyme binds both sites and loops out the DNA, the pre-mix reaction will then start from the looped DNA–protein complex, which would cut the DNA directly upon adding Mg2+, at a rate that corresponds explicitly to kcleave.
As FokI has a low affinity for DNA in the absence of metal ions (28), high enzyme concentrations were used in the pre-mixes, to ensure that it bound to the DNA during the pre-equilibrium, even though these concentrations lead to dimer formation at individual sites. Both pIF185 and pIF190 were tested, with similar results (Table 1).
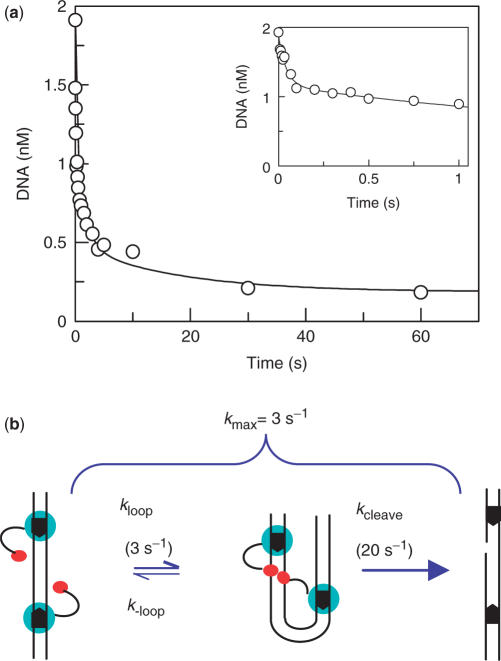
Following the addition of Mg2+ to the enzyme–DNA mixture, about 40% of the DNA was cleaved rapidly, within ∼0.2 s, while the overall reaction took >60 s to reach completion (Figure 4a). With both substrates, at least three exponentials were required to accommodate the complete reaction: an initial rapid phase with a rate constant of about 20 s−1 (Table 1), largely complete within 0.2 s (Figure 4a, insert); an intermediate phase at about 0.6 s−1, the predominant process from 0.2 to 5 s (Figure 4a, both panels); a slow phase from 5 to >60 s, with a rate constant of about 0.06 s−1 (Figure 4a, main panel). Further increases in [E0] had no significant effect on these rate constants, which demonstrates that these reflect saturation rates.
Figure 4.
Pre-looped DNA. (a) FokI and 3H-labelled pIF185 in Buffer R was mixed with an equal volume of MgOAc in Buffer R to give a mixture that contained 2.5 nM DNA (80% SC), 125 nM FokI and 10 mM MgOAc. After various times, the reactions were quenched with EDTA and the residual concentrations of SC DNA measured. The insert shows data from reactions over 1 s, the main panel reactions over 60 s. The line through the data points corresponds to the equation for a triple exponential decay, with values of 23 ± 5 s−1, 0.6 ± 0.2 s−1 and 0.06 ± 0.03 s−1 for the rate constants. The first and second were the best fits to the data in the insert, the third to the data in the main panel. (b) The data in (a) show that kcleave must be about 20 s−1 and that the process which limits kmax to 3 s−1 is kloop.
The rate constant for the fast phase of DNA cleavage in the pre-mix reaction, ∼20 s−1, is larger than the maximal rate constant from the post-mix reactions, kmax = 3 s−1. This fast phase must therefore be due to a complex formed during the pre-equilibration of enzyme and DNA. If the protein had not bound DNA during the pre-equilibrium, there would be no difference between pre- and post-mix reactions. Moreover, the DNA–protein complex that cuts the DNA at 20 s−1 upon binding Mg2+ must be located in the pathway after the step that limits kmax to 3 s−1. Otherwise, there would again have been no difference between pre- and post-mix reactions. The rate constant of 20 s−1 can thus be assigned to kcleave, the constant for phosphodiester hydrolysis by the looped complex and the 3 s−1 constant to kloop, for loop closure (Figure 4b). Given the relationship k = ln2/τ½, a rate constant of 3 s−1 corresponds to a τ½ (reaction half-time) of 230 ms.
The slow phases of the pre-mix reactions may be due to FokI binding first as a monomer to one or both sites but that, during the pre-equilibrium of the DNA with high [E0] in the absence of Mg2+, a fraction of the DNA ends up with FokI dimers at individual sites rather than the dimer across two sites (as in Figure 3b). Upon adding Mg2+, the dimers at solitary sites could cleave DNA at their characteristic rate (k1 = 0.05 s−1; Figure 1), thus accounting for the 0.06 s−1 process, the slowest stage of the pre-mix reactions. Alternatively, the dimers at the separate sites may release one subunit to form DNA-bound monomers, which can then cleave DNA via the looped complex: if so, the 0.6 s−1 process—the intermediate stage in the pre-mix reactions—reflects the rate of subunit dissociation from dimers at individual sites. The numerical model for the scheme in Figure 3b employed a rate constant of 0.5 s−1 for subunit dissociation.
DISCUSSION
A pathway has been established for the reaction of the FokI restriction endonuclease on SC plasmids with two target sites. The DNA first binds two monomers of FokI, one at each site, in two separate but equal reactions. The two monomers bound to the same molecule of DNA then associate in cis, looping out the intervening DNA. Finally, the dimeric enzyme in the looped complex hydrolyses the scissile phosphodiester bonds much more rapidly than in the unlooped complex (20 s−1 c.f. 0.05 s−1). The looped complex that cleaves the DNA rapidly is formed with a τ½ of 230 ms. The protein–protein association that traps the loop must take place within this time span, together with whatever protein conformational changes (if any) occur upon looping. This analysis sheds light on two general aspects of DNA looping that are relevant to essentially all looping processes: the dynamics of loop capture and the activation of protein function relative to the same protein at a solitary site.
Loop capture
A DNA loop between two sites can be trapped only when the sites are sufficiently close in 3D space to allow the protein(s) to bridge the sites. Brownian dynamics have been used to estimate the times taken for the distance between two specific sites in a SC DNA to fall to an appropriate level (11–13). The simulations modelled DNA molecules of ∼3.0 kb and computed an average time of ∼2 ms for the first occasion that two sites—separated by ∼500 bp along the DNA—become juxtaposed to within 10 nm. The DNA used in these calculations is the same size as the plasmids used here, and the reaction diameter is similar to the distance between the two DNA-binding domains in the FokI dimer (28,35). However, the simulations were for longer inter-site distances than the 185- or 190-bp distances examined here, but this should have no more than a 2-fold effect on juxtaposition time (39).
In both site-specific recombination by resolvase and in the looping of biotinylated DNA by streptavidin (19–21), some looped complexes were established within 10 ms. In these cases, the loop capture rates approach that for site juxtaposition. In contrast, with FokI, the τ½ for loop capture is about 10 times longer than the juxtaposition time. The rate of loop capture by FokI therefore cannot be diffusion-limited by the internal motion of the DNA. Instead, it must be reaction-limited, maybe by the protein–protein association after the juxtaposition of the two DNA-bound proteins (or alternatively, as noted above, by protein conformational changes). However, the protein conformational changes that accompany the binding of FokI to its recognition site affect primarily the relative positions of the two domains rather than the conformation within each domain (34,35). The conformational changes do not perturb significantly the dimerization surface in the catalytic domain, so are unlikely to limit the rate of the protein–protein association. Recently, loop formation by a transcription factor was also shown to be reaction rather than diffusion-limited, albeit indirectly without measuring the capture rate (40).
Supercoiled plasmids with 185 or 190 bp between the sites gave similar rates for loop closure (Table 1), despite these spacings being at, respectively, minima and maxima in the variation of loop stability with site separation (27). The DNA in the 185-bp loop will thus be twisted 180° out of register relative to the 190-bp loop. However, the torsional relaxation time of DNA, about 20 ns (41), is much shorter than any time constant for the global re-configuration of the DNA molecule (11). Consequently, during the time while the two DNA-bound proteins remain in close proximity of each other, which is particularly long on SC DNA in its native—tightly interwound—configuration (12), the two complexes will sample all possible rotational orientations. They can thus encounter each other in an appropriate orientation for the protein–protein interaction regardless of inter-site spacing. Since the 185- and the 190-bp separations give rise to loops of unequal stability but equal capture rates, the different stabilities must be due to different rates for loop release. The deformation of twist in the 185-bp loop must therefore drive release rather than limiting capture.
Enzyme activation
Many proteins that interact with two DNA sites have virtually no activity when bound to a single site and are only active after binding two sites (2,4–6). However, enzymes that can loop DNA by binding two sites in cis are often capable of binding—albeit less readily—two sites in trans (7). Consequently, it can be difficult to determine whether the activity of a looping protein on a DNA with one target site is due to a synaptic complex with two sites in trans or to the enzyme bound to that solitary site (42,43). However, FokI definitely cleaves one-site DNA by acting at that solitary site, and not by spanning sites in trans (27,28,36). For example, the dimer formed between a wild-type subunit and a mutant incapable of binding DNA (36) cannot bind two sites at the same time yet it cleaves one-site DNA at the same rate as the wild-type dimer (27). FokI thus constitutes a test system that allows for an unambiguous measurement of the extent of enzyme activation by DNA looping.
On a DNA with a single recognition site, the active assembly for FokI consists of a dimer of catalytic domains, each of which is attached to a monomeric recognition domain (35), but only one of the recognition domains contacts the specific DNA (Figure 1). Thus, unlike homodimeric enzymes at palindromic sites (29–31), the two subunits in the FokI dimer are not equivalent to each other. Nevertheless, the catalytic domains attached to the free and to the bound recognition domains cleave their target phosphodiester bonds at equal rates, 0.05 s−1, so whatever conformational change is induced by DNA binding to one subunit must be relayed to the other subunit (27). This rate is, however, slow compared to other restriction enzymes (29–32), which typically hydrolyse phosphodiester bonds at about 1 s−1. Even so, the dimer on a one-site DNA cannot activate itself by forming a ‘trans’ dimer spanning sites on separate DNA molecules, on account of the weakness of the dimerization equilibrium KDimer.
On a DNA with two FokI sites, enzyme monomers at individual sites associate more readily with each other than with protein from free solution, as the DNA-bound monomers are held close together by the DNA itself. In this case, the resultant assembly again consists of a dimer of catalytic domains but now both recognition domains are bound to specific DNA (27,28). The dimer with both recognition domains bound to DNA cuts the first of its four target phosphodiester bonds at 20 s−1, very much faster than the dimer with only one recognition domain bound to DNA, which cuts scissile bonds at 0.05 s−1. When both recognition domains are bound to the cognate sequence, the dimer of catalytic domains must be in a much more active conformation than when only one recognition domain is bound to DNA. As the dimer of FokI can bind two DNA sites at the same time only when the sites are in cis, on which it must trap a DNA loop, looping by FokI is concomitant with a 400-fold enhancement of catalytic activity.
ACKNOWLEDGEMENTS
We thank Bill Jack, Jurate Bitinaite and Abigail Welsh for materials, Ted King (TgK Scientific) for help with the quench-flow, and Marks Dillingham and Szczelkun for comments and advice. This work was funded by The Wellcome Trust (grant 078794/Z/06). Funding to pay the Open Access publication charges for this article was also provided by the Wellcome Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.Schleif R. DNA looping. Annu. Rev. Biochem. 1992;61:199–223. doi: 10.1146/annurev.bi.61.070192.001215. [DOI] [PubMed] [Google Scholar]
- 2.Halford SE, Welsh AJ, Szczelkun MD. Enzyme-mediated DNA-looping. Annu. Rev. Biophys. Biomol. Struct. 2004;33:1–24. doi: 10.1146/annurev.biophys.33.110502.132711. [DOI] [PubMed] [Google Scholar]
- 3.Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem. Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 4.Grindley NDF, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 5.Gemmen GJ, Millin R, Smith DE. Tension-dependent DNA cleavage by restriction endonucleases: two-site enzymes are “switched off” at low force. Proc. Natl Acad. Sci. USA. 2006;103:11555–11560. doi: 10.1073/pnas.0604463103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ptashne M. Regulation of transcription: from lambda to eukaryotes. Trends Biochem. Sci. 2005;30:275–279. doi: 10.1016/j.tibs.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 7.Bellomy GR, Record M.T., Jr. Stable DNA loops in vivo and in vitro: roles in gene regulation at a distance and in biophysical characterization of DNA. Prog. Nucleic Acid Res. Mol. Biol. 1990;39:81–128. doi: 10.1016/s0079-6603(08)60624-8. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, McEwen AE, Crothers DM, Levene SD. Statistical-mechanical theory of DNA looping. Biophys. J. 2006;90:1903–1912. doi: 10.1529/biophysj.105.070490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allemand JF, Cocco S, Douarche N, Lia G. Loops in DNA: an overview of experimental and theoretical approaches. Eur. Phys. J. E, Soft Matter. 2006;19:293–302. doi: 10.1140/epje/i2005-10073-y. [DOI] [PubMed] [Google Scholar]
- 10.Jun S, Bechhoefer J, Ha B-Y. Diffusion-limited loop formation of semiflexible polymers: Kramer's theory and the intertwined time scales of chain relaxation and closing. Europhys. Lett. 2003;64:420–426. [Google Scholar]
- 11.Jian H, Schlick T, Vologodskii A. Internal motion of supercoiled DNA: Brownian dynamic simulations of site juxtaposition. J. Mol. Biol. 1998;284:287–296. doi: 10.1006/jmbi.1998.2170. [DOI] [PubMed] [Google Scholar]
- 12.Huang J, Schlick T, Vologodskii A. Dynamics of site juxtaposition in supercoiled DNA. Proc. Natl Acad. Sci. USA. 2001;98:968–973. doi: 10.1073/pnas.98.3.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klenin KV, Langowski J. Intrachain reactions of supercoiled DNA simulated by Brownian dynamics. Biophys. J. 2001;81:1924–1929. doi: 10.1016/S0006-3495(01)75843-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Embleton ML, Vologodskii AV, Halford SE. Dynamics of DNA loop capture by the Sfil restriction endonuclease on supercoiled and relaxed DNA. J. Mol. Biol. 2004;339:53–66. doi: 10.1016/j.jmb.2004.03.046. [DOI] [PubMed] [Google Scholar]
- 15.Finzi L, Gelles J. Measurement of lactose repressor-mediated loop formation and breakdown in single DNA molecules. Science. 1995;267:378–380. doi: 10.1126/science.7824935. [DOI] [PubMed] [Google Scholar]
- 16.Lia G, Bensimon D, Croquette V, Allemand JF, Dunlap D, Lewis DE, Adhya S, Finzi L. Supercoiling and denaturation in Gal repressor/heat unstable nucleoid protein (HU)-mediated DNA looping. Proc. Natl Acad. Sci. USA. 2003;100:11373–11377. doi: 10.1073/pnas.2034851100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van den Broek B, Vanzi F, Normanno D, Pavone FS, Wuite GJ. Real-time observation of DNA looping dynamics of Type IIE restriction enzymes NaeI and NarI. Nucleic Acids Res. 2006;34:167–174. doi: 10.1093/nar/gkj432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gemmen GJ, Millin R, Smith DE. Dynamics of single DNA looping and cleavage by Sau3AI and effect of tension applied to the DNA. Biophys. J. 2006;91:4154–4165. doi: 10.1529/biophysj.106.088518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bussiek M, Klenin K, Langowski J. Kinetics of site-site interactions in supercoiled DNA with bent sequences. J. Mol. Biol. 2002;322:707–718. doi: 10.1016/s0022-2836(02)00817-3. [DOI] [PubMed] [Google Scholar]
- 20.Parker CN, Halford SE. Dynamics of long-range interactions on DNA: the speed of synapsis during site-specific recombination by resolvase. Cell. 1991;66:781–791. doi: 10.1016/0092-8674(91)90121-e. [DOI] [PubMed] [Google Scholar]
- 21.Oram M, Marko JF, Halford SE. Communications between distant sites on supercoiled DNA from the non-exponential kinetics of DNA synapsis by resolvase. J. Mol. Biol. 1997;270:396–412. doi: 10.1006/jmbi.1997.1109. [DOI] [PubMed] [Google Scholar]
- 22.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE - enzymes and genes for DNA restriction and modification. Nucleic Acids Res. 2007;35:D269–D270. doi: 10.1093/nar/gkl891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nobbs TJ, Szczelkun MD, Wentzell LM, Halford SE. DNA excision by the SfiI restriction endonuclease. J. Mol. Biol. 1998;281:419–432. doi: 10.1006/jmbi.1998.1966. [DOI] [PubMed] [Google Scholar]
- 24.Bath AJ, Milsom SE, Gormley NA, Halford SE. Many type IIs restriction endonucleases interact with two recognition sites before cleaving DNA. J. Biol. Chem. 2002;277:4024–4033. doi: 10.1074/jbc.M108441200. [DOI] [PubMed] [Google Scholar]
- 25.Gowers DM, Bellamy SRW, Halford SE. One recognition sequence, seven restriction enzymes, five reaction mechanisms. Nucleic Acids Res. 2004;32:3469–3479. doi: 10.1093/nar/gkh685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marshall JM, Gowers DM, Halford SE. Restriction endonucleases that bridge and excise two recognition sites from DNA. J. Mol. Biol. 2007;387:419–431. doi: 10.1016/j.jmb.2006.12.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Catto LE, Ganguly S, Milsom SE, Welsh AJ, Halford SE. Protein assembly and DNA looping by the FokI restriction endonuclease. Nucleic Acids Res. 2006;34:1711–1720. doi: 10.1093/nar/gkl076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vanamee ES, Santagata S, Aggarwal AK. FokI requires two specific DNA sites for cleavage. J. Mol. Biol. 2001;309:69–78. doi: 10.1006/jmbi.2001.4635. [DOI] [PubMed] [Google Scholar]
- 29.Perona JJ. Type II restriction endonucleases. Methods. 2002;28:353–364. doi: 10.1016/s1046-2023(02)00242-6. [DOI] [PubMed] [Google Scholar]
- 30.Erskine SG, Baldwin GS, Halford SE. Rapid-reaction analysis of plasmid DNA cleavage by the EcoRV restriction endonuclease. Biochemistry. 1997;36:7567–7576. doi: 10.1021/bi970155s. [DOI] [PubMed] [Google Scholar]
- 31.Wright DJ, Jack WE, Modrich P. The kinetic mechanism of EcoRI endonuclease. J. Biol. Chem. 1999;274:31896–31902. doi: 10.1074/jbc.274.45.31896. [DOI] [PubMed] [Google Scholar]
- 32.Bellamy SRW, Milsom SE, Scott DJ, Daniels LE, Wilson GG, Halford SE. Cleavage of individual DNA strands by the different subunits of the heterodimeric restriction endonuclease BbvCI. J. Mol. Biol. 2005;348:641–653. doi: 10.1016/j.jmb.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 33.Skowron P, Kaczorowski T, Tucholski J, Podhajska AJ. Atypical DNA-binding properties of class-IIS restriction endonucleases: evidence for recognition of the cognate sequence by a FokI monomer. Gene. 1993;125:1–10. doi: 10.1016/0378-1119(93)90738-o. [DOI] [PubMed] [Google Scholar]
- 34.Wah DA, Hirsch JA, Dorner LF, Schildkraut I, Aggarwal AK. Structure of the multimodular endonuclease FokI bound to DNA. Nature. 1997;388:97–100. doi: 10.1038/40446. [DOI] [PubMed] [Google Scholar]
- 35.Wah DA, Bitinaite J, Schildkraut I, Aggarwal AK. Structure of FokI has implications for DNA cleavage. Proc. Natl Acad. Sci. USA. 1998;95:10564–10569. doi: 10.1073/pnas.95.18.10564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bitinaite J, Wah DA, Aggarwal AK, Schildkraut I. FokI dimerization is required for DNA cleavage. Proc. Natl Acad. Sci. USA. 1998;95:10570–10575. doi: 10.1073/pnas.95.18.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kingston IJ, Gormley NA, Halford SE. DNA supercoiling enables the Type IIS restriction enzyme BspMI to recognise the relative orientation of two DNA sequences. Nucleic Acids Res. 2003;31:5221–5228. doi: 10.1093/nar/gkg743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barman TE, Bellamy SRW, Gutfreund H, Halford SE, Lionne C. The identification of chemical intermediates in enzyme catalysis by the rapid quench-flow technique. Cell Mol. Life Sci. 2006;63:2571–2583. doi: 10.1007/s00018-006-6243-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Podtelezhnikov AA, Vologodskii AV. Dynamics of small loops in DNA molecules. Macromolecules. 2000;33:2767–2771. [Google Scholar]
- 40.Polikanov YS, Bondarenko VA, Tchernaenko V, Jiang YI, Lutter LC, Vologodskii A, Studitsky VM. Probability of the site juxtaposition determines the rate of protein-mediated DNA looping. Biophys. J. 2007;93:2726–3271. doi: 10.1529/biophysj.107.111245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Millar DP, Robbins RJ, Zewail AH. Direct observation of the torsional dynamics of DNA and RNA by picosecond spectroscopy. Proc. Natl Acad. Sci. USA. 1980;77:5593–5597. doi: 10.1073/pnas.77.10.5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zaremba M, Sasnauskas G, Urbanke C, Siksnys V. Conversion of the tetrameric restriction endonuclease Bse634I into a dimer: oligomeric structure-stability-function correlations. J. Mol. Biol. 2005;348:459–478. doi: 10.1016/j.jmb.2005.02.037. [DOI] [PubMed] [Google Scholar]
- 43.Bellamy SRW, Milsom SE, Kovacheva YS, Halford SE. A switch in the mechanism of communication between the two DNA-binding sites in the SfiI restriction endonuclease. J. Mol. Biol. 2007;373:1169–1183. doi: 10.1016/j.jmb.2007.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]