Abstract
Base excision repair (BER) provides relief from many DNA lesions. While BER enzymes have been characterized biochemically, BER functions within cells are much less understood, in part because replication bypass and double-strand break (DSB) repair can also impact resistance to base damage. To investigate BER in vivo, we examined the repair of methyl methanesulfonate (MMS) induced DNA damage in haploid G1 yeast cells, so that replication bypass and recombinational DSB repair cannot occur. Based on the heat-lability of MMS-induced base damage, an assay was developed that monitors secondary breaks in full-length yeast chromosomes where closely spaced breaks yield DSBs that are observed by pulsed-field gel electrophoresis. The assay detects damaged bases and abasic (AP) sites as heat-dependent breaks as well as intermediate heat-independent breaks that arise during BER. Using a circular chromosome, lesion frequency and repair kinetics could be easily determined. Monitoring BER in single and multiple glycosylase and AP-endonuclease mutants confirmed that Mag1 is the major enzyme that removes MMS-damaged bases. This approach provided direct physical evidence that Apn1 and Apn2 not only repair cellular base damage but also prevent break accumulation that can result from AP sites being channeled into other BER pathway(s).
INTRODUCTION
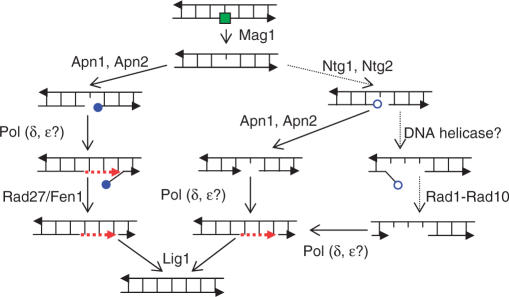
Base excision repair (BER) is the major repair pathway for non-bulky lesions in DNA. These include alkylated and oxidized bases arising from endogenous or exogenous sources as well as spontaneously formed apurinic/apyrimidinic (AP) sites (1,2). BER is a step-wise process involving several enzymes. It is initiated by specific DNA N-glycosylases that remove the damaged bases yielding AP sites (3,4). The budding yeast Saccharomyces cerevisiae has proven a model eukaryote for genetic dissection of several steps in BER of alkylation damages (Figure 1). The major pathway for removing AP sites in yeast is carried out by AP endonuclease Apn1 or Apn2 cutting 5′- of an AP site and yielding single-strand breaks (SSBs) with a 5′-deoxyribose phosphate (5′-dRP) [reviewed in (1)]. Although there are many BER functions in common with mammalian cells, yeast appears to lack an enzyme that, similar to the DNA polymerase (Pol) β, can function as a Pol as well as an AP lyase in BER. However, in yeast and in mammalian cells the removal of 5′-dRP can be accomplished through the successive action of a Pol (presumably δ or ɛ) and a 5′-flap endonuclease (Rad27/Fen1) followed by sealing a DNA strand with a DNA ligase (presumably Lig 1). Alternatively, an AP site can be incised on the 3′-side by a glycosylase-associated AP lyase to form a 3′-α,β-unsaturated aldehyde (3′-dRP). These ends represent a major problem for DNA integrity because they cannot be extended by a DNA polymerase or ligated. Based on genetic studies, endonucleases Apn1 or Apn2 are the major contributors to removal of 3′-dRP (5–7). In the absence of these endonucleases, the nucleotide excision repair (NER) genes RAD1 and RAD10 become essential suggesting that NER components can provide a backup pathway for 3′-dRP removal (5,8).
Figure 1.
BER of alkylation DNA damage in S. cerevisiae [modified from (1)]. An alkylated base is shown as a filled green square. The 5′- and 3′-deoxyribose phosphates (5′-dRP and 3′-dRP) are shown as filled and open blue circles, respectively. Arrowheads on DNA strands correspond to 3'-ends. Red dashed lines represent repair-associated DNA synthesis.
A major category of DNA lesions subject to BER is alkylation damage generated by endogenous and environmental agents as well as by many anti-cancer drugs (9,10). Most alkylation lesions are cytotoxic and mutagenic, posing a threat to genome integrity (9). Methyl methanesulfonate (MMS) is a classic alkylating agent that has been widely used to investigate pathways of repair and tolerance of alkylation damage in DNA. The predominant lesions are N-methylation adducts N7-methylguanine (N7-MeG) and N3-methyladenine (N3-MeA). While N7-MeG is thought to be relatively innocuous, N3-MeA is cytotoxic due to blockage of replication fork progression (11,12).
The enzymatic steps of BER, including the BER of alkylation base damage have been studied in detail in vitro utilizing purified proteins as well as cell extracts (13,14). However, the redundancy of BER enzymes and versatility of BER pathways make it difficult to understand the actual cellular mechanisms. It is also clear that the efficiency of in vivo BER could depend on chromatin organization which may influence lesion accessibility as well as repair activity (15). Other factors, such as coordination with the cell cycle, regulation of BER gene expression and delivery of BER proteins into the nucleus may also impact repair.
Information about BER in vivo is typically pursued through genetic studies. Hundreds of genes have been found that affect MMS-resistance in eukaryotic cells (16–18). Some genes may encode BER enzymes and cofactors or be involved in BER regulation and cellular compartmentalization. However, MMS tolerance is determined not only by BER efficiency, but also by downstream DNA transactions, such as replication bypass and double-strand break (DSB) repair (19).
In order to identify and characterize genetic defects that specifically affect BER efficiency among alkylation damage sensitive mutants an in vivo assay is needed to monitor and quantify BER. We present here the development and characterization of such an assay in the budding yeast S. cerevisiae. MMS induction of AP sites and BER are monitored in G1haploid cells, thereby avoiding competing processes associated with replication bypass and recombinational DSB repair. Methylation base damage was detected by secondary breakage (presumably at the sites of closely spaced lesions) of full-size yeast chromosomes that arose in the course of preparation and/or pulsed-field gel electrophoresis (PFGE). The assay measures the formation and repair of damaged bases and/or AP sites as well as the accumulation of nicks during BER. With the use of this assay we obtained physical evidence that Apn1 and Apn2 not only provide for BER of AP sites but also prevent the formation of unrepairable nicks in chromosomal DNA treated with MMS.
MATERIALS AND METHODS
Yeast strain construction
Standard methods of yeast genetics were as described (20). All strains used in this study are derivatives of the two isogenic strains ALE1000 and ALE1001 (MATα leu2-3112 ade5-1 his7-2 ura3Δ trp1-289 [(chr II) lys2::Alu-DIR-LEU2-lys2Δ5′]) (21). In addition to the mutant copy of the complete LEU2 gene in chromosome III the strain contains a copy of the wild-type LEU2 integrated in the vicinity of the LYS2 in chromosome II. MWJ49 (derived from ALE1000) and MWJ50 (derived from ALE1001) strains with circular chromosome III were generated by DSB-induced homologous recombination between the HML and HMR silent cassettes of the MAT locus using GAL1-I-SceI CORE cassette (22,23). Briefly, the unique (Ya) part of the HMRa locus was first replaced with the GAL1-I-SceI CORE cassette in sir2-null derivatives of ALE1000 and ALE1001 to allow expression of GAL1-I-SceI CORE. A site-specific DSB was induced by incubating cells in synthetic complete media with 2% galactose with vigorous shaking for 4 h. After plating on YPDA (10 g/l yeast extract, 20 g/l Bacto peptone, 20 g/l glucose, 10 mg/l adenine) and growing for 1 day, colonies that lost the CORE marker URA3 were selected on medium with 5-fluoroorotic acid. CORE loss was verified by loss of the second marker, G418-R. Selected strains with circularized chromosome III were verified by PCR amplification of the junction sequence with primers located upstream of HMR and downstream of HML as well as by PFGE, as described in ‘Results’. SIR2+ strains with circular chromosome III were obtained from the spore progeny after dissecting the meiotic products from the cross with the isogenic MATa strain.
MMS treatment
Yeast were grown in rich YPDA medium at 30°C for 48 h with vigorous shaking. At this stage >95% of cells were in the G1 stage of cell cycle based on the absence of buds. Cells were washed with distilled water and re-suspended in phosphate-buffered saline (PBS; 10 mM phosphate, NaCl, 0.138 M; KCl, 0.0027 M, pH 7.4) at a density of 1.5 × 108 cells/ml. After treatment with 11.8 mM (0.1%) MMS at 30°C for 15 or 30 min with vigorous shaking, the MMS was neutralized in 5% sodium thiosulfate, by mixing 1:1 (v/v) ratio with 10% Na2S2O3. Cells were washed twice with dH2O, and re-suspended in PBS. A part of each sample was immediately processed for DNA preparation in agarose for PFGE (DNA plugs) as described subsequently. To monitor the repair, another volume of the MMS-treated or control yeast cells was kept in PBS buffer for 24 h at 30°C with constant shaking referred to as liquid holding (LH). Thereafter, cells were processed for DNA plug preparation and PFGE analysis.
DNA preparation and PFGE
Preparation of DNA plugs containing chromosome-sized genomic DNA embedded in agarose was performed as suggested by the manufacturer (CHEF Yeast Genomic DNA Plug Kit, Bio-Rad, Richmond, CA) with minor modifications. Briefly, control and MMS-treated yeast cells were immobilized in 0.6% agarose and treated with 1 mg/ml Zymolyase (100 U/mg, MP Biochemicals, Solon, OH) for 2 h at 30°C in a ‘spheroplasting’ solution (1 M Sobitol, 20 mM EDTA, 10 mM Tris pH7.5) to remove the cell wall. This was followed by digestion with proteinase K (10 mM Tris, pH 8.0, 100 mM EDTA, 1.0% N-lauroylsarcosine, 0.2% sodium deoxycholate, 1 mg/ml Proteinase K) for 24 h at 30°C or 55°C. The electrophoresis was performed using a Bio-Rad CHEF-Mapper XA PFGE system. Samples were resolved in a 1% agarose gel at 6 V/cm for 20 h with a 60–120 s switch time ramp (14°C). Gels were stained with ethidium bromide (0.5 μg/ml for 2 h) and destained with 0.5×TBE for 1 h.
Southern blot and hybridization
Southern transfer from CHEF gels to Hybond N+ (Amersham, Piscataway, NJ) was done with the PosiBlot transfer apparatus (Stratagene, La Jolla, CA) in neutral conditions (10×SSC). Hybridization was carried out with the 288 nt fragment of the LEU2 gene, copies of which were present in the chromosome III as well as in the chromosome II, as described before. The fragment was amplified from yeast genomic DNA with the following primers
LEU2-pr1-5′ – 5′-TGTCAGAGAATTAGTGGGAGG-3′ and LEU2-pr1-3′ – 5′-ATCATGGCGGCAGAATCAAT-3′.
The probe was labeled with 32P using the PrimeIt random primer labeling kit (Stratagene, La Jolla, CA) and incubated with the filter overnight at 70°C. Autoradiographs were digitized and densitometric analysis was performed using Kodak 1D software (version 4.0). The band mass was calculated from the plotted band profile under a Gaussian fit to its intensity using the internal function of the software. The ratio between the band masses of chromosome II and III was used to calculate chromosome breakage as described in the Results.
Formula for the dose dependence of chromosomal breakage resulting from closely spaced single-strand breaks in opposite DNA strands
Calculations were based on the assumption that SSBs are distributed evenly between the two DNA strands of a chromosome. We also assume that in order for a given SSB to form a DSB, a second SSB must appear on the opposite strand and it should be separated from the first SSB by a distance ≤S. The probability of at least one SSB falling into a region 2S within one strand of a chromosome, where there are N SSBs distributed randomly across each chromosome (approximated by a Poisson distribution) of length L, can be expressed by the following equation:
| (1) |
The total expected number (D) of DSBs in one chromosome would be
| (2) |
Taking the first two terms of the Taylor expansion
then:
| (3) |
Thus, with an SSB-inducing agent, where N is proportional to dose, the expected dose-dependence for DSB induction would follow second order kinetics.
RESULTS
Quantifying breakage of full size yeast chromosomes by PFGE
The induction of single-strand lesions and repair events can potentially be measured by detecting modifications in DNA. Our approach to detecting SSBs is based on the appearance of DSBs that arise from SSBs in close proximity. The SSBs could be actual nicks in DNA or lesions such as heat-labile sites that under certain conditions can lead to nicks (24).
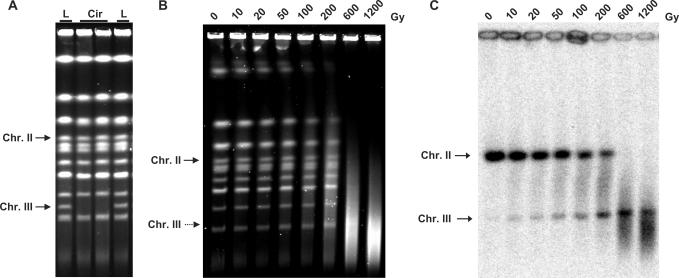
The system for quantifying DSBs was developed based on PFGE measurements of full length, chromosome-size DNA in non-dividing G1 (or G2) yeast cells. (S-phase cells are not good material for PFGE analysis since chromosomes with replicating structures do not enter the gel.) The mobility of linear molecules in PFGE is strictly size-dependent, so that broken linear molecules exhibit faster mobility. An important advantage of PFGE for studies of DNA damage and repair is that there is minimal disruption of chromosomal DNA, which is important for the detection of damage-associated SSBs as discussed subsequently. The strains also contained a circularized chromosome III. While full-size linear yeast chromosomes can be separated by PFGE, a circular chromosome does not migrate from the plug preparation into the gel (Figure 2A). However, a single DSB allows the chromosome to enter the gel as a unique band which can be detected by Southern hybridization [Figure 2B and C and (25)].
Figure 2.
Measuring chromosome breaks using PFGE in strains containing circular chromosome III. Positions of linearized chromosome III (Chr III) and linear chromosome II (Chr II) are indicated on each gel. (A) Circular chromosome cannot migrate into the gel. ‘L’ – yeast strains with linear chromosome III; ‘Cir’ – yeast strains with circular chromosome III. (B) In vitro treatment of DNA plugs with γ-radiation. Agarose plugs with genomic DNA from G1 haploid cells were placed in buffer (20 mM Tris, pH 8.0 and 50 mM EDTA) and γ -irradiated in the Shepherd Irradiator Model 431 with a 137Cs source at a dose rate of 23 Gy/min. Dose (Gy) is indicated above each lane. Chromosomal DNA in (A) and (B) was visualized by ethidium bromide staining after PFGE. Linearized chromosome III is not visible as a separate band on panel (B); its position is indicated by Southern hybridization. (C) Southern hybridization of the gel shown on panel (B) with a LEU2-probe, which identifies both chromosomes II (Chr II) and III (Chr III).
In order to evaluate the utility of our system for DSB quantification, we utilized ionizing radiation to induce DSBs. As shown in Figure 2B and C, gamma irradiation of chromosomal DNA contained within plugs leads to loss of the unique linear chromosomal bands. The radiation also transforms circular chromosome III to unique linear molecules that can be detected in the gel with a LEU2 probe (Figure 2C). As expected, the intensity of the singly broken chromosome III increased with increasing dose over the range examined.
While the full-sized linear broken chromosome III could be detected in the gel, it is not possible to determine the efficiency of DSB induction because the initial amount of circular chromosome molecules contained in the plug prior to PFGE could not be accurately evaluated. In order to quantify DSBs, we introduced an additional copy of LEU2 sequence into the linear chromosome II which is 2.9 times the size of circular chromosome III, so that the bands are well separated in our PFGE conditions. The increasing intensity of the linear chromosome III band combined with reduction of the chromosome II band provides a convenient means for estimating the frequency of DSBs in chromosomal DNA (Figure 2B).
The frequency of DSBs was determined based on the following assumptions: (i) linear chromosomal DNAs migrate in PFGE strictly on the basis of size; (ii) DSBs are distributed randomly throughout the genome; (iii) the DSBs follow a Poisson distribution. Thus, for chromosome III the distribution of numbers of breaks can be approximated as:
where (PChr III(A)) is the probability of (A) number of breaks in this chromosome and (X) is the average number of DSBs in chromosome III. In order to create a full size linearized chromosome III, exactly one DSB is required (A = 1). The full size molecules of chromosome II would be found in the gel if no breaks have occurred (A = 0). Considering the ratio of the lengths of chromosome II and circular chromosome III as 2.9, the probabilities of full size molecules of a chromosome migrating in the gel would be
The absolute values for PChrIII (1) and PChr.II (0) could be estimated by comparing band intensities of the corresponding chromosomes in treated versus untreated cells assuming that the same amount of DNA enters the gel. However, for the case of the circular chromosomes, little or no unbroken DNA enters the gel. Also, there might be differences in amounts of DNA that enter the gels due to differences in sample preparation. These issues can be avoided by calculating the ratio of radioactivity contained in the band representing the circular chromosome III with one break and the band representing unbroken chromosome II. Both bands are identified by the LEU2 probe within the same lane. The average number of DSBs in chromosome III (X) subjected to PFGE was calculated from the transcendental equation below which can be solved by iteration:
Using this approach, the amount of DSBs per genome equivalent induced by gamma-irradiation of plugs of chromosomal DNA was determined (Table 1). Based on the experiments with ionizing radiation as well as on MMS studies (described later) we limited the range of DSB measurements to those variants where the intensities of chromosome bands can be reliably detected by computer analysis of digitized autoradiographs and where the band peak is at least three times greater than a corresponding adjacent region (i.e. 10–200 Gy). With the typical amount of background in our experiments, the chromosome III band was not detectable when the number of DSBs per haploid genome was less than two, while the chromosome II band was not detectable when DSB number was greater than 40 per genome (Figure 2C and Table 1). Thus the range of reliable measurements was 2–40 breaks per haploid genome.
Table 1.
DSB quantification based on Southern analysisa
| Parameter | γ dose (Kr)b | MMSc | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 5 | 10 | 20 | 60 | 120 | exp.1 | exp. 2 | |
| Chr II to Chr III ratio | >35 | 15.9 | 7.3 | 4.6 | 3.3 | 1.1 | <0.1 | <0.1 | 0.4 | 0.2 |
| DSB per genome | <2 | 2.4 | 4.7 | 6.9 | 8.7 | 17.3 | >40 | >40 | 29.8 | 39.0 |
aThe number of DSBs in yeast chromosomes separated by PFGE was calculated as described in the text. The Chr II: Chr III ratio is the ratio of the amount of unbroken chromosome II to the amount of chromosome III with a single break (i.e. a unique band since chromosome III was originally a circle), where both chromosomes are identified with the LEU2 probe. The sizes of chromosome II, linearized circular chromosome III missing two telomere-proximal regions (HMRa-Tel and HMLα-Tel) and the haploid G1 size the yeast genome were assumed to be 813 178 bp, 280 500 bp and 12 156 678 bp, respectively (calculated based on the data from http://www.yeastgenome.org).
bResults are derived from the experiment presented in Figure 2.
cThe two intermediate levels of chromosome breakage that were not observed in the γ-irradiation experiments were obtained from experiments (exp. 1 and exp. 2), where wild-type yeast were treated with MMS for 30 min (Table 2, described later in the ‘Results’).
Monitoring MMS-induced base damage and repair in full-size yeast chromosomes
It has been established that AP-sites in DNA are heat-labile (26). These heat-labile AP-sites can be converted into strand breaks at the elevated temperatures often used in the processing of chromosomal DNA for PFGE. The heat-labile sites can result in DSBs that are detectable with PFGE (24,27) due to secondary DSBs that are considered to result from closely spaced nicks in DNA. Initial MMS base damage (i.e. N3-MeA and N7-MeG) are prone to non-enzymatic depurination at high temperature (19) and thus also can also result in heat-labile sites.
Based on these properties, our assay was refined to quantify MMS-induced DNA damage and repair by BER in G1 haploid yeast which has only one copy of the genome thereby preventing opportunities for recombinational repair [or even the HR process associated with this repair (28)], as well as the bypass repair associated with replication.
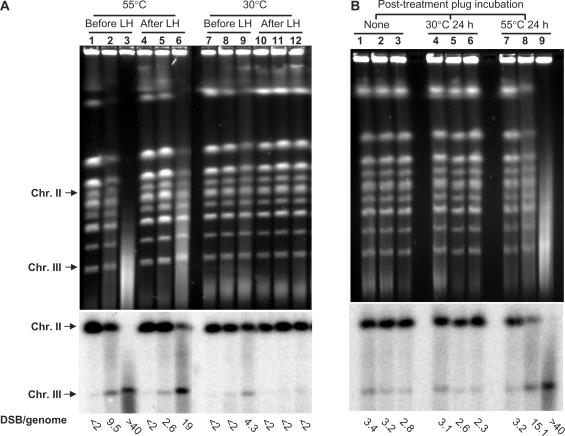
Cells were treated for 15–30 min with 11.8 mM (0.1%) MMS. To allow repair, they were washed and kept in buffer for 24 h (liquid holding, abbreviated as LH). Chromosome size DNA was prepared by embedding cells in agarose (DNA plug) before the DNA isolation process to prevent artificial shearing. MMS-induced damage was assessed by the appearance of secondary DSBs that can be detected with PFGE. As shown in Figure 3A, massive breakage was observed based on the disappearance of full size yeast chromosome bands (lanes 2 and 3). Repair is evident after LH (lanes 5 and 6) based on reappearance or increased intensity of chromosomal bands in the ethidium-strained gel, the decreased intensity of linearized chromosome III band and the increased intensity of full-size chromosome II band revealed by Southern hybridization. More than 70% of the DSBs disappeared after LH of cells exposed to MMS for 15 min (compare lanes 2 and 5). Based on our results described subsequently, as well as on previous reports (24,27), broken DNA molecules were the consequence of secondary DSBs that resulted from closely spaced single-strand nicks. The secondary DSBs arose when the DNA plugs were heated at 55°C during proteinase K digestion. When the plugs were processed at lower temperature (30°C), DSB formation was reduced over 90% and approached the level of untreated controls (Figure 3A, lanes 7–12). Although fewer DSBs were detected when plugs were processed at the lower temperature, lesions were still present (see Southern hybridization, lanes 8 and 9).
Figure 3.
PFGE of yeast chromosomes after MMS treatment. The calculated number of chromosome breaks is shown under each lane. (A) MMS-treatment of yeast cells induces heat-labile base damages that can be detected by formation of secondary DSBs. G1 haploid yeast were treated with 0.1% MMS for 15 min (lanes 2, 5, 8, 11) or 30 min (lanes 3, 6, 9, 12) followed by immediate DNA purification and PFGE (lanes 1–3 and 7–9) or PFGE after LH or 24 h in a buffer (LH) (lanes 4–6 and 10–12); lanes 1, 4, 7 and 10 are mock-treated controls. DNA purification with proteinase K digestion was performed either at 55°C (lanes 1–6) to measure heat-dependent breaks (HDBs) or at 30°C (lanes 7–12) to measure heat-independent breaks (HIBs). Chromosomes were visualized by ethidium bromide staining and breaks were quantified based on Southern hybridization. The numbers of DSBs per haploid yeast genome, calculated as described in the text are shown under each lane. (B) MMS treatment of chromosomal DNA within the plug produces heat labile sites. Plugs containing genomic DNA from G1 were treated with 11.8 mM MMS for 15 min (lanes 2, 5, 8) or 30 min (lanes 3, 6, 9); lanes 1, 4 and 7 are mock-treated controls. Lanes 1–3 contain DNA-plugs with no post-treatment incubation; lanes 4–6 contain DNA-plugs that were post-incubated at 30°C for 24 h; lanes 7–9 contain DNA-plugs that were post-incubated at 55°C for 24 h.
To confirm that MMS does not directly produce DSBs but instead leads to heat-labile sites that give rise to secondary DSBs, DNA plugs prepared from untreated cells were treated with MMS. The DNA plugs were then processed in three ways: (i) no further treatment (Figure 3B, lanes 1–3); (ii) incubation at 30°C for 24 h (Figure 3B, lanes 4–6); (iii) incubation at 55°C for 24 h (Figure 3B, lanes 7–9). As revealed by PFGE and Southern hybridization, a significant increase in DNA breakage in MMS-treated plugs over untreated controls was observed only if they were processed at 55°C.
In summary, the heat-labile property of MMS-induced base damage can be observed as heat-dependent breaks (HDBs) in full size yeast chromosomes separated by PFGE. Monitoring and quantifying HDBs provides a useful way to study alkylation damage and its repair in vivo. A large dose of MMS also caused small, but reproducible amounts of heat-independent breaks (HIBs; Figure 3A, lane 9). As shown subsequently, HIBs provide a measure of nicks that are directly associated with cellular lesions or their repair.
Mag1 glycosylase is required for the repair of MMS-induced heat-labile sites in haploid G1 yeast cells
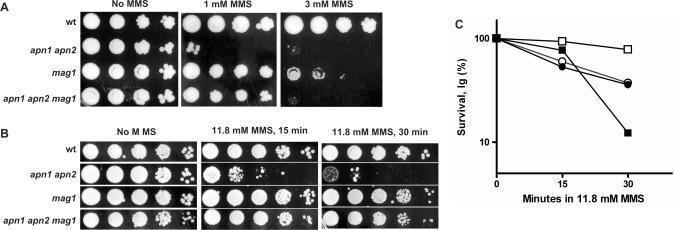
The most abundant lesions caused by MMS are N7-MeG (80–85%) and N3-MeA (9–12%) (29). Based on the strong MMS-sensitivity of the S. cerevisiae Mag1-glycosylase null mutants, this activity was proposed to be important in the removal of both types of damaged bases (30). In our strain background the mag1-null mutants were also sensitive when plated to media containing 3 mM MMS (Figure 4A). However, survival of G1 haploid mag1-null cells treated with 11.8 mM MMS under conditions used for our damage-detection experiments was indistinguishable from the wild-type (Figure 4B and C). (Note: increased growth delay compared to wild-type was retained in cells derived from (Figure 4B) mag1-null colonies that arose from cells plated onto YPDA after 30 min 11.8 mM MMS.) However, logarithmically growing mag1-null cells were sensitive to MMS (Figure 4C), similar to a previous report where cells with a different background were treated under the same condition and MMS concentration for the mag1 and wild-type (7). The similarity in MMS-killing of the mag1-null and wild-type G1 haploid cells might be due to either high capacity for downstream damage bypass and/or DSB repair or by the action of other glycosylases that are secondary to Mag1 in dealing with MMS damaged bases (Discussion).
Figure 4.
MMS-induced killing of different yeast genotypes used in this study. (A) MMS sensitivity of yeast strains as determined by plating from serial 10-fold dilutions onto rich (YPDA) medium containing 1 mM or 3 mM MMS. (B) Sensitivity of yeast strains treated with 11.8 mM (0.1%) MMS for 15 or 30 min in PBS and then plated to rich (YPDA) medium from serial 10-fold dilutions. (C) MMS sensitivity of G1 yeast or logarithmically growing cells: G1 cells wt (open circle) and mag1 (filled circle); log phase wt (open square) and mag1 (filled square). All experiments were done with two isogenic derivatives of wild-type yeast strains MWJ49 and MWJ50 or two mutant strains derived from these wild types. Plating in (A) and (B) was done 3–4 times for each strain. All numbers in (C) represent the average for two isogenic strains, which differed from each other by no more than 30%.
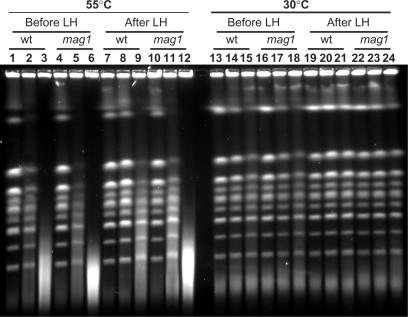
In order to clarify the role of Mag1 in removal of MMS-damaged bases we directly assessed its contribution to induction of HDBs and their repair in G1 haploid cells (Figure 5 and Table 2). MMS induced high levels of HDBs in wild-type and mag1-null yeast (Figure 5, lanes 1–12), whereas only small amounts of HIBs were observed (Figure 5, lanes 13–24). As expected there was considerable repair in the wild-type cells as manifested by the large reduction in HDBs following LH after both doses of MMS (compare lanes 2–3 with lanes 8–9). The repair was much less in the mag1-null cells (compare lanes 5–6 with lanes 11–12). Quantification of breakage based on Southern blot analysis in repeat experiments (Table 2) showed that the number of HDBs after repair was always less in the wild-type than in mag1-null (PH0 < 0.05 by Mann–Whitney test).
Figure 5.
A mag1 deletion causes severe blockage in the repair of MMS-induced heat-labile sites. Formation and repair of HDBs (lanes 1–12) and HIBs (lanes 13–24) were assessed as described in the text. Genotypes are indicated. Each group of three lanes (from left to right) contains mock-treated control, 15 min and 30 min MMS-treatment.
Table 2.
Median number of DSBs in chromosomes caused by MMSa
| Type of breaks | Genotype | Before LH | After LH | ||||
|---|---|---|---|---|---|---|---|
| 0 min | 15 min | 30 min | 0 min | 15 min | 30 min | ||
| Heat-dependent breaks (HDBs) | Wild type (15b) | <2 | 12.3 | >40 | <2 | 5.2 | 13.6 |
| (55°C)c | (<2) | (9.8–19.1) | (>40) | (<2) | (3.6–7.1) | (11.8–19.3) | |
| [<2–3.9] | [6.1–30.6] | [10.3–>40] | [<2–2.4] | [2.1–11.2] | [5.9–29.4] | ||
| mag1 (4) | <2 | 23.9 | >40 | <2 | 17.9 | >40 | |
| (<2) | (18.6–31.6) | (>40) | (<2–4.1) | (11.5–29.1) | (>40) | ||
| apn1 apn2 (4) | <2 | 29.3 | >40 | <2 | 15.7 | >40 | |
| (<2) | (16.4–>40) | (>40) | (<2–2.6) | (9.2–16.8) | (18.9–>40) | ||
| apn1 apn2 mag1 (3) | 2.4 | 18.1 | >40 | 2.2 | 13.3 | >40 | |
| (<2–2.5) | (12.6–33.4) | (>40) | (<2–2.9) | (12.8–14.8) | (>40) | ||
| Heat-independent | Wild type (13) | <2 | 2.4 | 4.7 | <2 | 2.1 | 3.0 |
| breaks (HIBs) (30°C)d | (<2) | (<2–3.6) | (3.4–7.6) | (<2–2.6) | (<2–3.7) | (<2–4.2) | |
| [<2–3.0] | [<2–5.7] | [<2–9.5] | [<2–3.7] | [<2–4.6] | [2–5.9] | ||
| mag 1 (3) | 2.2 | 3.2 | 7.6 | 3.3 | 5.2 | 6.6 | |
| (<2–3.8) | (<2–3.7) | (4.5–14.7) | (<2–5.1) | (<2–6.8) | (3.5–6.8) | ||
| apn1 apn2 (4) | <2 | 8.5 | >40 | <2 | 15.9 | >40 | |
| (<2–2.19) | (2.9–11.3) | (16–>40) | (<2–5.11) | (5.7–26.5) | (30.7–>40) | ||
| apn1 apn2 mag1 (3) | <2 | <2 | 4.3 | 2.6 | 4.3 | 6.5 | |
| (<2) | (<2–3.3) | (3.9–9.1) | (<2–2.9) | (4.1–4.8) | (6.4–7.3) | ||
aNumber of DSBs per haploid genome in chromosomes separated by PFGE was calculated in each experiment as described in the text. Presented in parentheses below median values are either 95% CI of the median (44, 45) for the wild type or ranges for all mutant strains. Presented in brackets under the 95% CI for the wild type are the ranges. In the cases where both borders of the interval were either >40 or <2, only one border of the range or CI is shown.
bNumber of independent experiments is presented in parentheses below each genotype.
cPlug preparation included treatment for 24 h at 55°C before PFGE.
dAll stages of plug preparation were performed at 30°C.
As shown in Table 2, the number of HDBs in the mag1-null mutant following 15 min MMS treatment (before LH) was higher than in the wild-type cells (PH0 < 0.05 by Mann–Whitney test). The number of DSBs observed after 30 min MMS treatment was out of the range of reliable measurements. Since the induction of lesions is likely to be the same, the difference is most likely due to fast repair of base damage in wild-type cells before lysis. The initiation of BER, including the suggested fast repair, would be blocked or slowed down in mag1-null yeast. In this mutant the 15 min MMS-treatment resulted in around 24 closely spaced lesions in the genome, revealed as HDBs (Table 2). Since only approximately 5 HDBs/genome were left in wild-type cells after LH repair, around 80% of lesions that could contribute to HDBs were repaired. Thus, Mag1 glycosylase defines a major pathway for repair of MMS-induced base damage in yeast. There also appeared to be limited repair of HDBs in three out of four experiments (data not shown) (approximately 25% based on comparing median values); however, more experiments are required to establish statistical significance of this observation. There was no apparent difference in HIBs between wild-type and mag1-null yeast (Figure 5, lanes 13–24) suggesting that most lesions in this mutant are preserved as methylated bases without conversion into AP sites or SSBs within the cell. This underscores the major role that Mag1 plays in the initial stage of repair of N3 and N7 alkylation base damages.
Mag1-dependent accumulation of HIBs after MMS treatment in cells lacking AP-endonucleases Apn1 and Apn2
Following removal of methylated bases by Mag1, repair of AP sites in yeast usually proceeds via cleavage by the redundant AP endonucleases Apn1 and Apn2 in yeast (6,31–33). Deletion of both APN1 and APN2 renders yeast extremely MMS-sensitive [(5,7,34) and Figure 4A and B]. The studies of MMS-sensitivity in various yeast mutants suggested that abasic sites accumulated in the apn1 apn2 double mutant can be processed by a secondary pathway involving glycosylase/lyase activities encoded by the NTG1 and NTG2 genes [Figure 1; (5,7,34) and results subsequently].
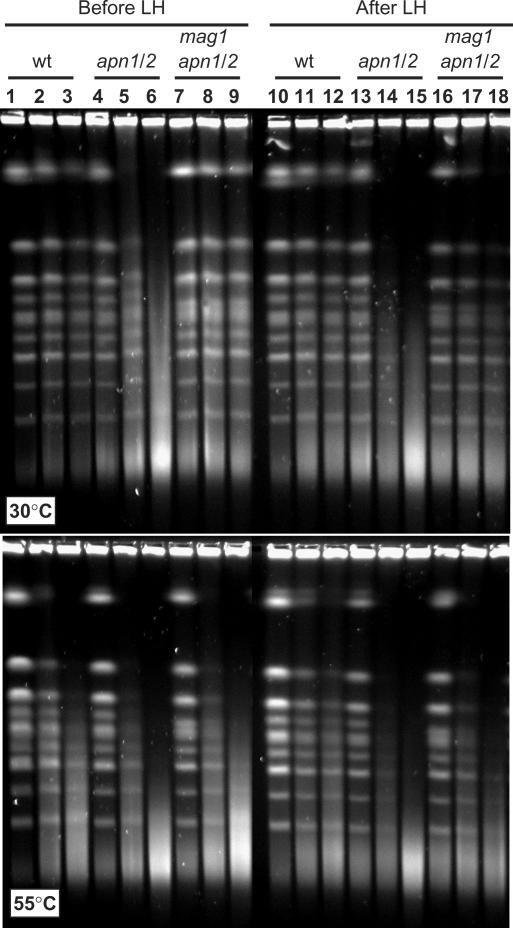
While deletion of either APN1 or APN2 did not produce any noticeable effect on repair of MMS damage (data not shown), the double mutant accumulated HIBs that could not be repaired in the G1 cells (Figure 6 and Table 2). Breaks were observed in MMS-treated apn1 apn2 double mutants when DNA plugs were processed at 30°C [Figure 6 upper panel (lanes 5 and 6), and Table 2] while MMS-treated wild-type yeast showed very few HIBs (Figure 6 upper panel, lanes 2 and 3 and Table 2). Importantly, HIBs even increased during LH of the double mutant, while they remained at the same low level in the wild-type cells (Figure 6, compare lanes 5 and 14). Based on quantification of HIBs (Table 2) in each of four independent experiments breaks were added during LH. The amount of breaks added in the course of LH was comparable with the number of initially induced breaks (15 min MMS treatment, Table 2).
Figure 6.
Break accumulation in apn1 apn2 double mutants is modulated by a mag1 deletion. Formation and repair of HIBs (upper panel) and HDBs (lower panel) were assessed as described in the text. Genotypes are indicated above lanes, apn1 apn2 double mutant is abbreviated as apn1/2. Each group of three lanes (from left to right) contains mock-treated control, 15 min and 30 min MMS-treatment.
HIBs in the apn1 apn2 double mutant could accumulate if abasic sites generated by Mag1 were converted into single-strands breaks that were not further repaired. Indeed, MMS-induced HIB formation in apn1 apn2 mag1 triple mutant was almost completely blocked when Mag1 was eliminated (Figure 6 and Table 2). The slightly elevated amount of HIBs in apn1 apn2 mag1 triple mutants after LH (when compared to that of before LH) was most likely due to spontaneous depurination or formation of AP sites via backup pathways. In contrast to HIB induction, the level of MMS-induced breakage after processing DNA plugs of the triple mutant at 55°C was high (comparable to the high level of HDBs in the mag1 single mutant (Table 2). Consistent with prior studies (7) and our result that deletion of mag1 prevents formation of unrepairable breaks in apn1 apn2 yeast, deletion of MAG1 reduced the level of MMS-sensitivity of the double mutant (Figure 4A, 1 mM MMS and Figure 4B).
DISCUSSION
To fully understand cellular components and dynamics, DNA repair process should be examined in vivo. We have developed a method to monitor BER of MMS damage to the yeast chromosomes. Discussed subsequently are the rationale and capabilities of this approach along with important features of yeast BER revealed by our study.
In vivo monitoring of alkylation DNA damage and repair
Our study confirmed that MMS does not break DNA directly, either in vivo or in vitro at 30°C. In addition to AP sites arising during repair, heat-induced formation of breaks at 55°C can be due to the accelerated spontaneous depurination of methylated N3 and N7 purines, leading to the formation of AP sites in vitro which are also heat unstable and can be further converted into SSBs (19,26). The fact that MMS-induced damage is heat-labile and can be converted into breaks in vitro has been employed in this study to investigate in vivo repair processing of methylated base damage by analyzing HDBs (converted in vitro from methylated bases and AP sites) and HIBs (resulting from in vivo enzyme activity). DSB analysis was based on chromosome breaks detected and quantified by PFGE with the use of a circular chromosome. Estimates of DSB induction were obtained by comparing the amount of linearized circular chromosome formed after exposure with the amount of linear chromosome that remained unbroken. Since comparisons were made within the same lane, the estimate does not depend on the amount of DNA that enters the gel, a problem that may arise with other methods when comparing bands between different lanes where the amounts of DNA entering the gel may vary. Our method allows for comparison of chromosome breakage between different lanes even if unequal amounts of cells are loaded and/or lysed during plug processing. The limitations on the accuracy of DSB measurements result from background associated with Southern blot hybridization as well as spontaneous chromosome breakage within the cell or during plug preparation and PFGE. Regardless, we established a dynamic range that covers from 2–40 DSBs per haploid yeast genome in our experimental conditions.
It was proposed that MMS-induced heat-labile sites that can be detected as DSBs with PFGE result from closely spaced SSBs located on opposite DNA strands (24). If so, the appearance of HDBs should be related to MMS dose by second-order kinetics [Equation (3) in ‘Material and Methods’]. The median values of MMS-induced HDBs after 15 min and 30 min treatment of wild-type yeast are in good agreement with this prediction (Table 2). Since there is a lack of information on the relationship between the distance separating SSBs and likelihood of appearance of a DSB during PFGE, we can only estimate heat-labile SS damage. If SSBs separated by 10 or less nucleotides result in broken chromosomal DNA, the number of HDBs observed after 15 min of treatment with 11.8 mM MMS would correspond to one damaged base per 2 kb as determined from Equation (2) in ‘Material and Methods’. This corresponds to approximately 6000 damaged bases per yeast genome or 1.7 × 10−4 lesions/mM·h, which is close to the value of 10−4 lesions/mM·h determined in MMS-treated mammalian cells (35,36). Direct measurement of MMS damage in yeast by independent techniques is required for more precise calculations.
Our assay is set to identify genetic controls that affect BER among the vast collection of damage-sensitive mutants identified in genome-wide screens (16,17). This relates not only to BER enzymes that directly participate in repair, but also to cell elements that enable and regulate the in vivo damage recognition and repair in the context of highly organized chromatin. Only mutants that affect BER would be detected in our assay. Other mutants that impact downstream events, such as damage bypass or DSB-repair would be seen as wild-type in our system. For example while the rad52 mutant deficient in recombinational repair of DSBs is extremely sensitive to MMS, it was comparable to wild-type in our in vivo BER assay (Supplementary Figure 1). As described subsequently, our assay can also dissect cellular metabolism of alkylation DNA damage and downstream AP sites.
Crucial role of Mag1 glycosylase in the repair of MMS damage
We found that MMS killing of mag1-null G1 cells is comparable to that of wild-type yeast. The MMS-resistance of the mutant might be due to alternative pathways for removal of N3-MeA or N7-MeG. Based on the increased MMS-sensitivity of the triple mag1 ntg1 ntg2 mutants it has been suggested that the bi-functional glycosylases Ntg1 and Ntg2 have a broad range of substrate specificity and their glycosylase function provides a backup activity for Mag1 to protect yeast from alkylation damage (34). Alternatively, the synthetic effect could be due to the role of Ntg1 and Ntg2 in repair of AP sites occurring by spontaneous depurination of N3-MeA or N7-MeG nucleotides (see subsequently). Other studies suggested that NER might be a backup to BER in removing methylated bases. This was surmised from the observation that deletion of NER genes rendered mag1-null mutants far more sensitive to the toxic effects of MMS in budding (37,38) and fission yeast (39). However, the hypersensitivity does not necessarily mean that any of the suggested pathways is directly involved in repairing alkylated bases or AP sites (discussed subsequently) when BER is defective. For example, alkylated bases might be converted into other types of lesions during DNA replication or damage bypass that requires NER for further repair.
Our results (Figure 5 and Table 2) indicate that Mag1 is the major and may be the only DNA glycosylase responsible for the removal of MMS-induced base damage in vivo in G1 haploid yeast cells. In the absence of Mag1, most methylated bases were not removed based on the retention of HDBs (at least 75%) following LH. The small amount of repair (no more than 25%) after LH is most likely due to spontaneous depurination to form AP sites, which can be quickly repaired by the downstream enzymes. The half-lives for spontaneous depurination of N3-MeA and N7-MeG at 39°C are approximately 30 and 70 h, respectively (40). Considering that 9–12% of MMS lesions are N3-MeA and 80–85% are N7-MeG and (29), the possible amount of repair (up to 25%) after 24 h of LH (30°C) is well correlated with the in vitro depurination rate.
The observation that deletion of MAG1 almost completely abolishes the removal of methylated bases in G1 haploid cells excludes the possibility that alternative players such as NER or Ntg1 and Ntg2 are involved to a significant extent in the initial step of BER. The lack of significant MMS-killing of mag1-null G1 haploid yeast could be explained by high capacity of downstream damage bypass (41) and/or DSB repair (39).
Break accumulation can be caused by interrupted BER of MMS damage
Severe MMS sensitivity of BER mutants lead to propose that these defects may cause production of SSBs as a result of disrupted repair. For example SSB that remains unrepaired until DNA replication could lead to a DSB. Unrepaired SSBs would accumulate at a higher rate if one end of the break contains an AP-site, thereby precluding ligation. Break accumulation was suggested as an explanation for the severe MMS-sensitivity of mammalian cells lacking the 5′-dRP lyase activity associated with DNA polymerase β (42). This generally-accepted explanation accounts for the strong MMS sensitivity caused by defects in the yeast 5'-flap endonuclease Rad27/Fen1 (Figure 1 and ref. (1)). In support of this, the double mutant apn1 rad27 was reported to be less MMS-sensitive than the rad27 single mutant (43). Apparently, abasic sites are not as toxic as 5'-flaps that can accumulate after cleavage of an abasic site with AP-endonuclease. Single apn1 mutants have only marginal MMS-sensitivity, presumably because of the remaining Apn2 activity.
Deficiency in both AP-endonucleases results in extreme sensitivity to MMS and other DNA base-damaging agents (7,34). As indicated in Figure 1, the Ntg1 or Ntg2 lyases could have access to the abasic sites that arise in the absence of AP-endonucleases and cause accumulation of SSBs with 3'-dRP ends. The 3'-dRPs are highly toxic due to the inability of DNA polymerases to extend them. Importantly, the apn1 apn2 double mutants were less MMS sensitive when formation of abasic sites was blocked by deletion of MAG1 [Figure 4A and B and (7)]. (Note: mag1 apn1 apn2 triple mutants were still significantly more sensitive than single mag1 mutants when incubated on plates with higher concentration of MMS (3 mM), possibly because AP sites could be formed at low rate in the absence of Mag1.) Through monitoring base damage and repair in the chromosomal DNA of G1 haploid yeast we have provided direct evidence for accumulation of breaks that are refractory to repair in the apn1 apn2 double mutant (Figure 6 and Table 2).
Breaks accumulated in the double mutant could be detected even in chromosomes that were processed at low temperature (HIBs). Therefore the breaks are clearly distinguishable from base damage and/or abasic sites. The breaks are likely to have arisen by enzymatic incision of AP sites since the formation of breaks was prevented by a mag1 deletion mutant that abolishes AP formation. Based on the synthetic lethality between the double mutation apn1 apn2 and either rad1 or rad10, it has been suggested that the 3′-endonuclease activity of Rad1-Rad10 can release 3′-blocked termini thereby providing an alternative branch of BER (1,5,8). The apn1 apn2 double mutant carries wild-type RAD1 and RAD10 and still showed accumulation of breaks, rather than repair. Therefore, the RAD1/10 pathway does not appear to play a significant role in repair of MMS base damage, at least in G1 haploid yeast. However, BER of spontaneously occurring DNA lesions might still involve RAD1/10 pathway as an important backup, which would explain synthetic lethality of apn1 apn2 rad1 (or rad10).
The approaches presented here open the way to quantify alkylation base damage and to assess repair in full-size chromosomes. While the assay has been developed for methylation damage, the information obtained here should also apply to the repair of oxidative or other types of base damage where an AP site is the intermediate in repair. Since DNA alkylation is a common form of environmental damage and many anti-cancer drugs act by alkylating DNA, it is important to understand how the cell deals with such challenges. Importantly, the assay discriminates initial base damages from SSBs formed as downstream intermediates in repair. Prior to this report there has been a considerable amount of indirect evidence for in vivo SSB-accumulation resulting from faulty or misrouted DNA repair. Our study provided direct physical evidence of accumulation of DNA strand breaks resulting from a block in an intermediate stage of BER.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors are grateful to Joan Sterling for help and advice in the experiments; to Matt Rushing and Eric Steele (NIEHS Science Computing lab) for creating Excel macros for DSB calculation and for advice on the software; to Jim Westmoreland, Dr Julie Horton, Dr Chris Halweg and Dr Vladimir Poltoratsky for critical reading of the manuscript and suggestions. This work was supported by the Intramural Research Program of the NIEHS (NIH, DHHS). Funding to pay the Open Access publication charges for this article was provided by the Intramural Research Program of the NIEHS (NIH, DHHS).
Conflict of interest statement. None declared.
REFERENCES
- 1.Boiteux S, Guillet M. Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair (Amst) 2004;3:1–12. doi: 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Lindahl T. Suppression of spontaneous mutagenesis in human cells by DNA base excision-repair. Mutat. Res. 2000;462:129–135. doi: 10.1016/s1383-5742(00)00024-7. [DOI] [PubMed] [Google Scholar]
- 3.Krokan HE, Standal R, Slupphaug G. DNA glycosylases in the base excision repair of DNA. Biochem. J. 1997;325(Pt 1):1–16. doi: 10.1042/bj3250001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scharer OD, Jiricny J. Recent progress in the biology, chemistry and structural biology of DNA glycosylases. Bioessays. 2001;23:270–281. doi: 10.1002/1521-1878(200103)23:3<270::AID-BIES1037>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 5.Guillet M, Boiteux S. Endogenous DNA abasic sites cause cell death in the absence of Apn1, Apn2 and Rad1/Rad10 in Saccharomyces cerevisiae. Embo J. 2002;21:2833–2841. doi: 10.1093/emboj/21.11.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson RE, Torres-Ramos CA, Izumi T, Mitra S, Prakash S, Prakash L. Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes Dev. 1998;12:3137–3143. doi: 10.1101/gad.12.19.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao W, Chow BL, Hanna M, Doetsch PW. Deletion of the MAG1 DNA glycosylase gene suppresses alkylation-induced killing and mutagenesis in yeast cells lacking AP endonucleases. Mutat. Res. 2001;487:137–147. doi: 10.1016/s0921-8777(01)00113-6. [DOI] [PubMed] [Google Scholar]
- 8.Karumbati AS, Deshpande RA, Jilani A, Vance JR, Ramotar D, Wilson TE. The role of yeast DNA 3′-phosphatase Tpp1 and Rad1/Rad10 endonuclease in processing spontaneous and induced base lesions. J. Biol. Chem. 2003;278:31434–31443. doi: 10.1074/jbc.M304586200. [DOI] [PubMed] [Google Scholar]
- 9.Sedgwick B, Bates PA, Paik J, Jacobs SC, Lindahl T. Repair of alkylated DNA: recent advances. DNA Repair (Amst) 2007;6:429–442. doi: 10.1016/j.dnarep.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 10.Drablos F, Feyzi E, Aas PA, Vaagbo CB, Kavli B, Bratlie MS, Pena-Diaz J, Otterlei M, Slupphaug G, et al. Alkylation damage in DNA and RNA–repair mechanisms and medical significance. DNA Repair (Amst) 2004;3:1389–1407. doi: 10.1016/j.dnarep.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 11.Boiteux S, Laval J. Mutagenesis by alkylating agents: coding properties for DNA polymerase of poly (dC) template containing 3-methylcytosine. Biochimie. 1982;64:637–641. doi: 10.1016/s0300-9084(82)80103-x. [DOI] [PubMed] [Google Scholar]
- 12.Larson K, Sahm J, Shenkar R, Strauss B. Methylation-induced blocks to in vitro DNA replication. Mutat. Res. 1985;150:77–84. doi: 10.1016/0027-5107(85)90103-4. [DOI] [PubMed] [Google Scholar]
- 13.Fortini P, Pascucci B, Parlanti E, D'rrico M, Simonelli V, Dogliotti E. The base excision repair: mechanisms and its relevance for cancer susceptibility. Biochimie. 2003;85:1053–1071. doi: 10.1016/j.biochi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 14.Lindahl T. Keynote: past, present, and future aspects of base excision repair. Prog. Nucleic Acid Res. Mol. Biol. 2001;68:xvii–xxx. doi: 10.1016/s0079-6603(01)68084-x. [DOI] [PubMed] [Google Scholar]
- 15.Jagannathan I, Cole HA, Hayes JJ. Base excision repair in nucleosome substrates. Chromosome Res. 2006;14:27–37. doi: 10.1007/s10577-005-1020-7. [DOI] [PubMed] [Google Scholar]
- 16.Begley TJ, Rosenbach AS, Ideker T, Samson LD. Damage recovery pathways in Saccharomyces cerevisiae revealed by genomic phenotyping and interactome mapping. Mol. Cancer Res. 2002;1:103–112. [PubMed] [Google Scholar]
- 17.Begley TJ, Rosenbach AS, Ideker T, Samson LD. Hot spots for modulating toxicity identified by genomic phenotyping and localization mapping. Mol. Cell. 2004;16:117–125. doi: 10.1016/j.molcel.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 18.Horton JK, Wilson SH. Hypersensitivity phenotypes associated with genetic and synthetic inhibitor-induced base excision repair deficiency. DNA Repair (Amst) 2007;6:530–543. doi: 10.1016/j.dnarep.2006.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wyatt MD, Pittman DL. Methylating agents and DNA repair responses: methylated bases and sources of strand breaks. Chem. Res. Toxicol. 2006;19:1580–1594. doi: 10.1021/tx060164e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rose MD, Winston F, Hieter P. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 21.Jin YH, Obert R, Burgers PM, Kunkel TA, Resnick MA, Gordenin DA. The 3′–>5′ exonuclease of DNA polymerase delta can substitute for the 5′ flap endonuclease Rad27/Fen1 in processing Okazaki fragments and preventing genome instability. Proc. Natl Acad. Sci. USA. 2001;98:5122–5127. doi: 10.1073/pnas.091095198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Storici F, Resnick MA. The delitto perfetto approach to in vivo site-directed mutagenesis and chromosome rearrangements with synthetic oligonucleotides in yeast. Methods Enzymol. 2006;409:329–345. doi: 10.1016/S0076-6879(05)09019-1. [DOI] [PubMed] [Google Scholar]
- 23.Storici F, Durham CL, Gordenin DA, Resnick MA. Chromosomal site-specific double-strand breaks are efficiently targeted for repair by oligonucleotides in yeast. Proc. Natl Acad. Sci. USA. 2003;100:14994–14999. doi: 10.1073/pnas.2036296100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lundin C, North M, Erixon K, Walters K, Jenssen D, Goldman AS, Helleday T. Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res. 2005;33:3799–3811. doi: 10.1093/nar/gki681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Game JC, Sitney KC, Cook VE, Mortimer RK. Use of a ring chromosome and pulsed-field gels to study interhomolog recombination, double-strand DNA breaks and sister-chromatid exchange in yeast. Genetics. 1989;123:695–713. doi: 10.1093/genetics/123.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindahl T, Andersson A. Rate of chain breakage at apurinic sites in double-stranded deoxyribonucleic acid. Biochemistry. 1972;11:3618–3623. doi: 10.1021/bi00769a019. [DOI] [PubMed] [Google Scholar]
- 27.Stenerlow B, Karlsson KH, Cooper B, Rydberg B. Measurement of prompt DNA double-strand breaks in mammalian cells without including heat-labile sites: results for cells deficient in nonhomologous end joining. Radiat. Res. 2003;159:502–510. doi: 10.1667/0033-7587(2003)159[0502:mopdds]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 28.Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beranek DT. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat. Res. 1990;231:11–30. doi: 10.1016/0027-5107(90)90173-2. [DOI] [PubMed] [Google Scholar]
- 30.Chen J, Derfler B, Samson L. Saccharomyces cerevisiae 3-methyladenine DNA glycosylase has homology to the AlkA glycosylase of E. coli and is induced in response to DNA alkylation damage. Embo J. 1990;9:4569–4575. doi: 10.1002/j.1460-2075.1990.tb07910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sander M, Ramotar D. Partial purification of Pde1 from Saccharomyces cerevisiae: enzymatic redundancy for the repair of 3'-terminal DNA lesions and abasic sites in yeast. Biochemistry. 1997;36:6100–6106. doi: 10.1021/bi970048y. [DOI] [PubMed] [Google Scholar]
- 32.Bennett RA. The Saccharomyces cerevisiae ETH1 gene, an inducible homolog of exonuclease III that provides resistance to DNA-damaging agents and limits spontaneous mutagenesis. Mol. Cell Biol. 1999;19:1800–1809. doi: 10.1128/mcb.19.3.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Popoff SC, Spira AI, Johnson AW, Demple B. Yeast structural gene (APN1) for the major apurinic endonuclease: homology to Escherichia coli endonuclease IV. Proc. Natl Acad. Sci. USA. 1990;87:4193–4197. doi: 10.1073/pnas.87.11.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanna M, Chow BL, Morey NJ, Jinks-Robertson S, Doetsch PW, Xiao W. Involvement of two endonuclease III homologs in the base excision repair pathway for the processing of DNA alkylation damage in Saccharomyces cerevisiae. DNA Repair (Amst) 2004;3:51–59. doi: 10.1016/j.dnarep.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 35.Horton JK, Joyce-Gray DF, Pachkowski BF, Swenberg JA, Wilson SH. Hypersensitivity of DNA polymerase beta null mouse fibroblasts reflects accumulation of cytotoxic repair intermediates from site-specific alkyl DNA lesions. DNA Repair (Amst) 2003;2:27–48. doi: 10.1016/s1568-7864(02)00184-2. [DOI] [PubMed] [Google Scholar]
- 36.Sobol RW, Watson DE, Nakamura J, Yakes FM, Hou E, Horton JK, Ladapo J, Van Houten B, Swenberg JA, et al. Mutations associated with base excision repair deficiency and methylation-induced genotoxic stress. Proc. Natl Acad. Sci. USA. 2002;99:6860–6865. doi: 10.1073/pnas.092662499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao W, Chow BL. Synergism between yeast nucleotide and base excision repair pathways in the protection against DNA methylation damage. Curr. Genet. 1998;33:92–99. doi: 10.1007/s002940050313. [DOI] [PubMed] [Google Scholar]
- 38.Torres-Ramos CA, Johnson RE, Prakash L, Prakash S. Evidence for the involvement of nucleotide excision repair in the removal of abasic sites in yeast. Mol. Cell Biol. 2000;20:3522–3528. doi: 10.1128/mcb.20.10.3522-3528.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Memisoglu A, Samson L. Contribution of base excision repair, nucleotide excision repair, and DNA recombination to alkylation resistance of the fission yeast Schizosaccharomyces pombe. J. Bacteriol. 2000;182:2104–2112. doi: 10.1128/jb.182.8.2104-2112.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osborne MR, Phillips DH. Preparation of a methylated DNA standard, and its stability on storage. Chem. Res. Toxicol. 2000;13:257–261. doi: 10.1021/tx990182e. [DOI] [PubMed] [Google Scholar]
- 41.Johnson RE, Yu SL, Prakash S, Prakash L. A role for yeast and human translesion synthesis DNA polymerases in promoting replication through 3-methyl adenine. Mol. Cell. Biol. 2007;27:7198–7205. doi: 10.1128/MCB.01079-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sobol RW, Prasad R, Evenski A, Baker A, Yang XP, Horton JK, Wilson SH. The lyase activity of the DNA repair protein beta-polymerase protects from DNA-damage-induced cytotoxicity. Nature. 2000;405:807–810. doi: 10.1038/35015598. [DOI] [PubMed] [Google Scholar]
- 43.Wu X, Wang Z. Relationships between yeast Rad27 and Apn1 in response to apurinic/apyrimidinic (AP) sites in DNA. Nucleic Acids Res. 1999;27:956–962. doi: 10.1093/nar/27.4.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dixon WJ, Massey F.J., Jr. Introduction to Statistical Analysis. Inc., New York: McGraw-Hill; 1969. p. 349. [Google Scholar]
- 45.Foster PL. Methods for determining spontaneous mutation rates. Methods Enzymol. 2006;409:195–213. doi: 10.1016/S0076-6879(05)09012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]