Abstract
Tumor cells respond to the harsh hypoxic microenvironment, in part, by transcriptional regulation of specific target genes. We found that hypoxia-mediated activation of selected genes occurs amidst widespread repression of transcription that is neither cell type-specific nor HIF-1-dependent. Despite overall repression, hypoxia induces a pool of histone modifications typically associated with transcriptional activation or repression. Chromatin immunoprecipitation analyses showed that this global mixture of hypoxia-modified histones is sorted in a gene-specific manner to correlate with transcriptional response to hypoxia. Exceptions to this were unexpected increases in H3K4me3 levels, typically associated with transcriptional activation, and decreased H3K27me3 levels, generally a marker of transcriptional silencing, at core promoters of both hypoxia-activated and -repressed genes. These data suggest that a novel signature of chromatin modifications is induced under hypoxic stress, which may play a role in gene regulatory switches active in proliferating tumor cells undergoing cycles of hypoxia and reoxygenation.
1. Introduction
Tissue hypoxia, or low oxygen tension, occurs as a result of inadequate blood flow to tissues or reduced oxygen transport capacity. Within normal cells, hypoxic stress generally leads to cell cycle arrest, apoptosis, and/or necrosis [1]. However, cells within tumors, which often have transient and chronic areas of hypoxia or anoxia, can survive and proliferate in this adverse environment by inducing genes that increase angiogenesis, glycolysis and/or oppose cell death. These hypoxia-induced changes in gene expression result in a more clinically aggressive phenotype with increased resistance to typical cancer treatments [2].
Stress-induced changes in gene expression are accompanied by or a result of changes in chromatin structure; however, the relationship between hypoxia-regulated alterations of chromatin and downstream effects on transcription is addressed in relatively few studies [3]. It is known that hypoxia-inducible factor 1 (HIF-1) drives much of hypoxic gene activation and interacts with histone acetyltransferases p300, CBP and SRC-1 [4]. Although the interaction between HIF-1 and CBP/p300 is critical for the activation of a large percentage of hypoxia-inducible genes [5], there are few determinations of histone acetylation levels at the promoters of hypoxia-responsive genes [6]. Additionally, the variety of histone modifications investigated globally and at the promoters of hypoxia-responsive genes is even more limited.
In this study, we found that hypoxia causes widespread repression of total RNA and mRNA synthesis, independently of HIF-1, and globally induces a mixture of histone modifications, typically associated with either transcriptional activation or repression. In general, our chromatin immunoprecipitation (ChIP) analyses of core promoters of hypoxia-repressed and -activated genes showed that gene-specific profiles of repressed or activated chromatin structure correlate with hypoxia-regulated, decreased or increased gene expression. However, at each hypoxic-responsive promoters tested, whether activated or repressed, we found increased H3K4me3 and decreased H3K27me3 induced by hypoxia. These specific exceptions to the generalities of the “histone code” [7] reveal a hypoxia-induced signature of histone modifications that may be indicative of the often transient state of tumor hypoxia.
2. Materials and Methods
2.1. Cells and treatments
Hepa 1–6 (ATCC) were cultured as in [8]. HIF-1α. null MEFs and HIF-1β deficient (Hepa-1C4) cell lines were obtained as in [9]. To achieve <0.01% oxygen, cells were treated as in [10]. Other oxygen concentrations were generated in a Ruskinn Invivo2 400 Hypoxia Workstation. 150µM CoCl (in H2O, Sigma, CAS# 7791-13-1) or 10µg/mL actinomycin D (Sigma, CAS# 50-76-0)was added to fresh media.
2.2. Cell extracts and immunoblot analysis
Preparation of histone-enriched fractions is described in supplemental information. Proteins were separated on a 15% SDS-PAGE gel, transferred onto a PVDF membrane, and probed with antibodies indicated in supplemental information.
2.3. Pulse-labeling and RNA synthesis analysis
Cells were labeled with 2–4µCi/mL of tritiated uridine (5-3H uridine, Amersham) for 1 hour. RNA was isolated with TRIzol (Invitrogen) (Fig. 1A) or TCA precipitated and collected on GF/C filters (Milipore) (Fig. 1B and C). Poly-adenylated RNA was purified via an Oligotex mRNA miniprep kit (Qiagen).
Fig. 1. Hypoxia induces global repression of total RNA and mRNA synthesis.
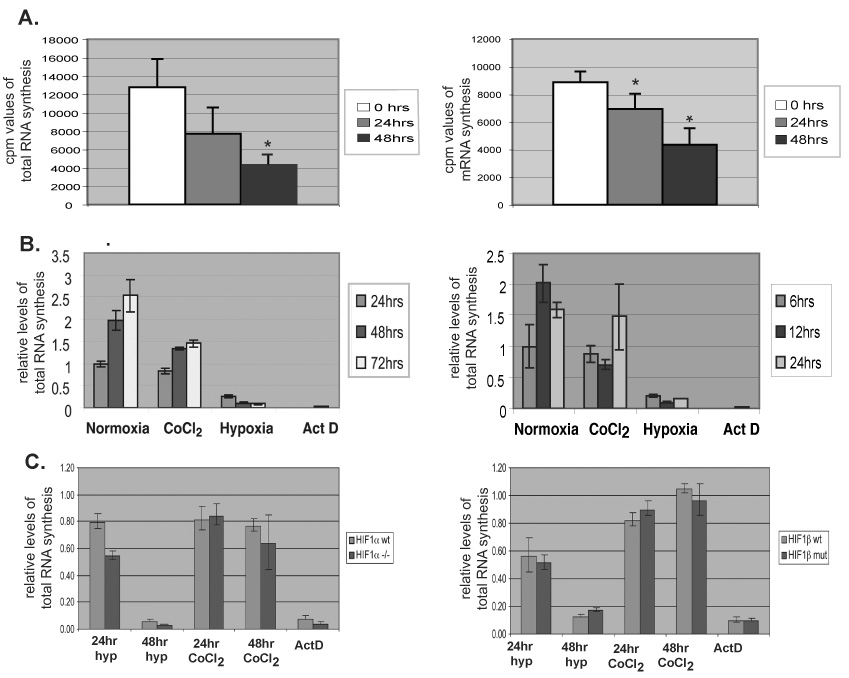
A) Total RNA (left) and mRNA (right) labeled with tritiated uridine (cpm) during 1 hr synthesis in Hepa 1–6 cells subjected to normoxia or a time-course of 0.2% oxygen. Values, normalized to cell number, represent an average of 5 replicates. Error bars denote SD. *P<0.005. B) Relative total RNA synthesis in three replicates, Hepa 1–6 cells (left) or normal human fibroblasts (right) subjected to indicated time of normoxia, CoCl2 treatment, <0.01% oxygen (hypoxia), or actinomycin D. C) Relative total RNA synthesis in immortalized HIF-1α-null MEFs (left) or HIF-1β mutant hepatoma cells (right) exposed for indicated time as in (B). Values normalized to normoxic controls.
2.4. RT-PCR
Quantitative PCR was performed and analyzed on an ABI 7500 Fast instrument, from cDNA prepared as described [8]. Primers and PCR conditions are listed in supplemental information.
2.5. Chromatin immunoprecipitation
ChIPs were performed as in [8] with antibodies listed in supplemental information. Fragmentation of chromatin to less than 300bp was verified by electrophoresis and ethidium bromide staining. DNA was analyzed by quantitative PCR; Taqman probes are listed in supplemental information.
3. RESULTS AND DISCUSSION
3.1. Hypoxia induces global repression of transcription independently of HIF1
Gene-specific response to hypoxia is often mediated by the HIF transcription factor and includes both repression and activation of expression [2]. Here, we assessed general transcription levels to define a global, cellular response to hypoxic stress. We pulse-labeled Hepa 1–6 cells subjected to normoxia or a time-course of hypoxia with tritiated uridine in order to measure ongoing total RNA and mRNA synthesis. After 24h or 48h at 0.2% oxygen, total RNA synthesis was repressed 42% and 65%, respectively (Fig. 1A), and mRNA synthesis was repressed 22% and 51%, respectively (Fig. 1B). Repression of RNA synthesis is dependent upon the severity and duration of hypoxic exposure, as 75% of transcription is repressed in cells exposed to 24h of <0.01% oxygen (Fig 1B, left). Hypoxia-induced, widespread transcriptional repression proved independent of cell type-specificity or tumor origin, as repression of total RNA synthesis occurs in normal primary fibroblasts (Fig. 1B, right). We assessed potential, indirect effects of cellular growth and density by plating cells to achieve cultures of equal density under normoxic and hypoxic incubation conditions (Fig. 1A) or by establishing equal numbers of cells at t=0 and growth to sub-confluent levels (Fig. 1B and C). In all cases, effects of hypoxia in mediating repression of global transcription were apparent over time.
Treatment with CoCl2 mimics hypoxia by inducing HIF-1α protein stability; however, it did not induce substantial transcriptional repression (Fig. 1B), suggesting that HIF-1α is not required for this effect. To test this hypothesis, we measured RNA synthesis in mouse embryonic fibroblast (MEF) cells derived from HIF-1α-null mouse embryos (Fig. 1C left) and in hepatoma cells mutant for HIF-1β (Fig. 1C right). Analyses over a time course of severe hypoxia (<0.01% oxygen) or CoCl2 treatment show that neither loss of functional HIF-1α nor HIF-1β inhibited hypoxia-induced repression of total RNA synthesis. A HIF-1-independent mechanism of widespread transcription repression may facilitate energy conservation during hypoxia, an ATP-limiting stress [11], and allow hypoxic tumor cells to channel their energy toward activation of genes that promote survival.
3.2. Hypoxia-induced global histone modifications
To determine a potential mechanism for the observed, widespread repression of transcription, we conducted an analysis of histone modifications present in extracts of normoxic (21% oxygen) or hypoxic (0.2% oxygen) hepatoma-derived Hepa 1–6 cells (Fig. 2 and Supplemental Fig. 1). During hypoxia, there was 32% loss of H3K9ac, an event generally associated with transcriptional repression; however, we observed an increase in H3K14ac (1.9-fold) and induced histone arginine methylation (H4R3me2, 2.5-fold), which can facilitate acetylation of histones normally associated with activation of transcription [12]. Methylation of lysine residues of histones is associated with either repression or activation of transcription, depending on the specific residue modified by methylation and the extent of methylation. We observed an increase in H3K4me2 (2-fold), H3K4me3 (3.6-fold), and H3K79me2 (1.4-fold) in response to hypoxia, all of which generally correlate with transcriptional activation [13]. Analysis with an antibody that recognizes multiple acetylated lysines on histone H4 showed little quantifiable change between normoxic and hypoxic conditions.
Fig. 2. Hypoxia induces altered post-translational modifications of total histone populations.
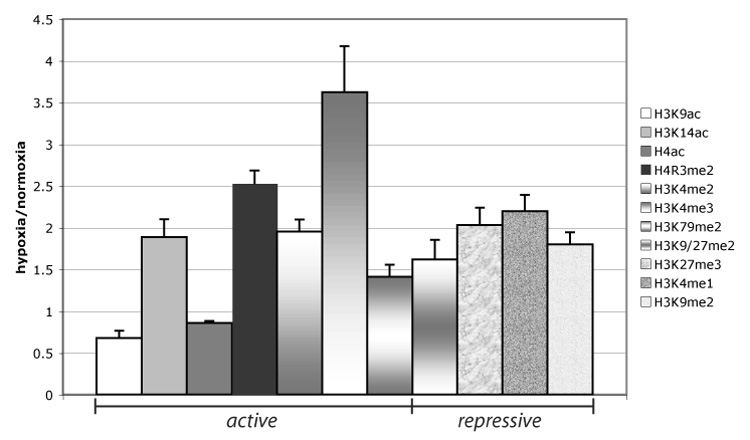
Modifications of histones, associated with transcriptional activation and repression, were determined by immunoblotting of protein extracts of Hepa 1–6 cells grown under normoxia or 0.2% oxygen for 48 hrs (representative blots, Supplemental Fig. 1). Quantified values, generated by NIH image, were normalized to H3-C-terminus, and fold change (H/N) is graphed. The average was generated from quantified values, obtained by NIH image, from three independent immunoblots of three independently obtained normoxic and hypoxic extracts. Error bars denote the SEM.
Additional complexity emerged when we investigated histone modifications associated with transcriptional repression. H3K9/27me2 and H3K9me2 levels, which are concentrated at repressed euchromatin and heterochromatin [14], increased 1.6-fold and 1.8-fold, respectively, after hypoxic exposure. Other histone modifications correlated with repression of transcription, H3K27me3 (2-fold) and monomethylated H3K4 (H3K4me1, 2.2-fold), were increased during hypoxia. Taken together, we found that the cellular pool of bulk histones, readily extracted from nuclei and chromatin, is altered in response to hypoxia and comprises a mixture of histone modifications typically associated with transcriptional activation or repression.
3.3. H3 occupancy inversely correlates with gene activity
To assess a direct relationship between hypoxia-induced alterations of chromatin and regulation of transcription at specific genes, we analyzed chromatin structure, at the level of nucleosomal occupancy and histone modifications, at the core promoters of hypoxia-repressed and -activated genes. We analyzed both albumin (ALB), a constitutively expressed hepatic gene, and alpha-fetoprotein (AFP) gene, a marker of hepatocellular carcinoma, as models of hypoxia-repressed genes [10]. To investigate hypoxia-activated genes, we used early growth response 1 (EGR1), which has both oncogenic and tumor-suppressor properties, and vascular endothelial growth factor (VEGF), an angiogenesis-promoting gene, as models of hypoxia-activated genes [15,16]. We found that endogenous ALB and AFP are repressed and EGR1 and VEGF are induced in Hepa 1–6 cells during 24 hours of the indicated hypoxic treatments (Fig. 3A).
Fig. 3. Nucleosome-occupancy at specific core promoters inversely correlates with transcription activation in response to hypoxia.
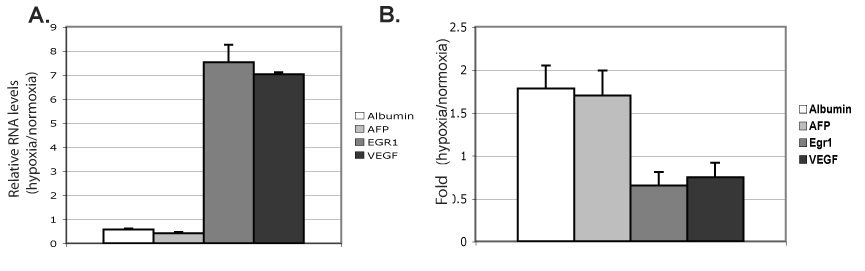
A) Relative levels of Albumin, AFP, EGR1, VEGF, and Albumin expression (hypoxia/normoxia), generated by quantitative RT-PCR, in Hepa 1–6 cells exposed for 48h to normoxia or 0.2% oxygen; each sample normalized to 18S. An average of at least four experiments is shown, and error bars denote SD. B) ChIP of histone H3 in Hepa 1–6 cells after 48h at normoxia or 0.2% oxygen, followed by quantitative PCR of core promoter regions of Albumin, AFP, EGR1, and VEGF. The %-input bound by histone H3, minus background (IgG), in hypoxia relative to normoxia shown. An average of at least three independent experiments shown, each PCR performed in duplicate, with error bars representing SD.
Whether active core promoters of mammalian genes lie within nucleosome-free regions and/or density of nucleosomes changes with transcriptional activity has been somewhat controversial [17]. To determine if hypoxia influences nucleosomal occupancy at the core promoters of these genes, we performed ChIP analyses with an antibody that recognizes the C-terminal domain of histone H3 (total histone H3). We observed an increase in total histone H3 occupancy at core promoters of repressed AFP and ALB, and a decrease at hypoxia-activated VEGF and EGR1 (Fig. 3B). Thus, we saw an inverse relationship between nucleosome occupancy and transcriptional activation that shifts proportionally as a regulatory response to hypoxia. These findings suggest a model of hypoxia-responsive remodeling of chromatin structure with direct links to downstream regulation of transcription.
3.4. Hypoxia-induced, gene-specific histone modifications
We performed ChIP analyses with antibodies that recognize H3K9ac and H3K4me3, markers of transcriptional activation, and H3K9/27me2 and H3K27me3, associated with transcriptional repression, at hypoxia-repressed AFP and ALB, hypoxia-activated EGR1 and VEGF, and the silent, hypoxia-nonresponsive Brn3b gene. These data were quantified as %-input precipitated by each modified histone antibody, histone H3 antibody, and nonspecific IgG, at each region (Supplemental Fig. 2). Hypoxia induced little change in overall H3K9ac levels at the AFP and ALB core promoters, relative to normoxia (Suppl. Figs. 2 and 3). However, per bound nucleosome, hypoxia induced a 1.8-fold decrease in H3K9ac and a 3.4-fold increase in H3K9/27me2 at AFP (Fig. 4A) and similar trends at repressed ALB (Fig. 4B), indicating active changes in these modifications that correlate with transcriptional repression. Interestingly, Brn3-b, which does not alter in its expression in response to hypoxia, showed little alteration in nucleosomal occupancy or in histone modifications with the exception of a 3.5-fold increase in repression marks H3K9/27me2 (Supplemental Fig. 2E). In contrast, at hypoxia-activated EGR1 and VEGF, induction of transcription was marked by increased H3K9ac levels per bound histone H3 and a decrease in total, quantified H3K9/27me2 at both VEGF and EGR1 promoters during hypoxia (Fig. 4C and D; Supplemental Figs. 2 and 3). When H3K9/27me2 was normalized per bound histone H3, EGR1 exhibited a 41% loss at its core promoter (Fig. 4B), and VEGF was relatively unchanged (Fig. 4C) during hypoxia.
Fig. 4. Specific histone modifications induced at hypoxia-responsive genes.
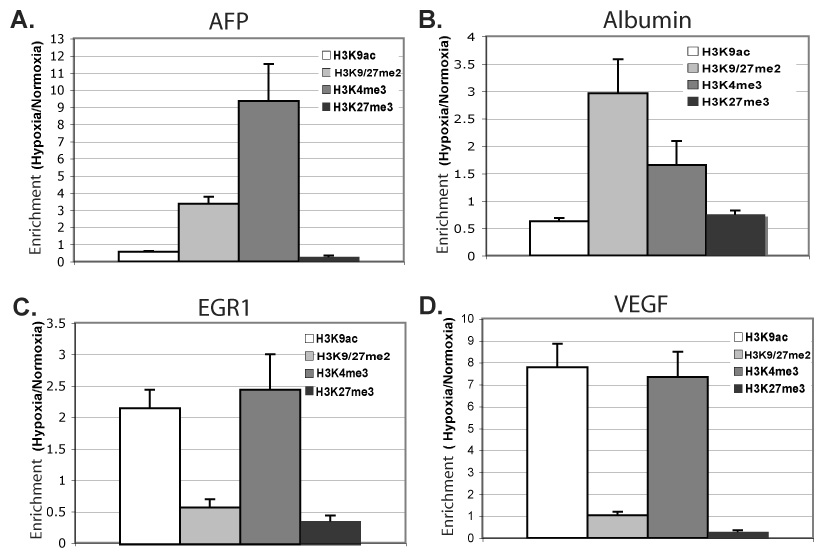
ChIP of indicated modified histones in Hepa 1–6 cells after 48h at normoxia or 0.2% oxygen, followed by quantitative PCR for core promoter regions of AFP (A), Albumin (B), EGR1 (C), and VEGF (D). The %-input bound by each modified histone H3, minus background (IgG), was normalized by total bound histone H3; fold-change hypoxia versus normoxia is graphed. Averages represent of two to three independent experiments, each experiment quantified in duplicate, with error bars representing SEM.
Unexpectedly, we saw a marked increase in H3K4me3 levels at all core promoters, whether activated or repressed (Fig. 4A–D), but little was detected at the hypoxia-inert Brn3b gene (Suppl. Fig. 2E). The relative change in H3K4me3 induced by hypoxia varied in a gene-specific manner (AFP>VEGF>EGR1>ALB), rather than by regulatory response of activation versus repression, likely reflecting gene-specific paradigms of chromatin regulation interwoven with global, hypoxic response. A measured decrease in bound H3K27me3 at all four hypoxia-responsive promoters was equally unanticipated, as H3K27me3 is normally associated with gene repression (Fig. 4A–D). The combination of histone modifications induced by hypoxia, increased H3K4me3 and decreased H3K27me3, is especially surprising at hypoxia-repressed AFP and ALB promoters.
It is speculated that nucleosomes bearing H3K4me3 reside at genes poised for activation by specific stimuli [18,19]. Genes that are inducible, though minimally transcribed, in resting T-cells contain relatively high levels of H3K4me3 [19]. Notably, bivalent domains consisting of stretches of H3K27me3 and pockets of H3K4me3 are found within mouse embryonic stem cells at silenced, developmentally regulated genes, which are poised for regulatory response during differentiation [18]. In hypoxic cells, which quickly respond to changes in oxygen tension by altering gene expression levels [1], repressed chromatin that is detectably enriched in H3K4me3 and low in H3K27me3 may be rapid activated once hypoxic conditions are reversed.
Our findings of hypoxia-induced, widespread repression of transcription suggest that limited energy resources are directed, independently of HIF-1 activation, toward survival-promoting gene responses under hypoxic stress. Interestingly, the recently discovered H3K27 demethylases can associate as protein complexes with enzymes that methylate H3K4, offering a two-pronged response to stimuli [20]. Hypoxia may additionally mediate rapid control of these histone and protein modifiers by directly altering the reaction components of their enzymatic processes. Both amine oxidases and JmjC-domain proteins demethylate by oxidation or hydroxylation, processes directly affected under hypoxia [21]. One potential mechanism driving increased H3K4me3 may be hypoxia-induced inactivation of H3K4me3-specfic histone demethylases, which utilize molecular oxygen during enzymatic removal of methyl groups [22–24]. The observed modifications of histones provide a hypoxia-regulated platform for binding and interactions of additional chromatin modifiers, remodelers or regulators of transcription, further amplifying gene-specific response to hypoxic stress.
A bivalent, chromatin signature of increased H3K4me3 and decreased H3K27me3 under hypoxia may poise hypoxia-responsive chromatin and facilitate downstream flexibility in response to induced or inhibited trans-acting regulators of gene expression. Identification of “hypoxia-signature” alterations in chromatin structure may further guide use or development of therapeutic, “epigenetic” agents that target specific histone modifications and shift hypoxia-regulated gene expression patterns. Further studies are needed to understand how chromatin responds to the demands of fluctuating signals, such as cycles of hypoxia and reoxygenation, and its role in mechanisms that mediate rapid changes in gene regulation.
Supplementary Material
Acknowledgements
We apologize to our colleagues whose work we failed to cite, due to space limitations. We are especially grateful to P. Corn and M. Czyzyk-Krzeska for use of their hypoxic incubators in times of need. This work was supported in part by grant GM053686 to M.C.B. and grant CA100132 to N.D. from the National Institutes of Health. A.B.J. is a trainee supported by grant T32-CA009299 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hockel M, Vaupel P. Tumor hypoxia: definitions and current clinical, biologic, and molecular aspects. J Natl Cancer Inst. 2001;93:266–276. doi: 10.1093/jnci/93.4.266. [DOI] [PubMed] [Google Scholar]
- 2.Vaupel P, Harrison L. Tumor hypoxia: causative factors, compensatory mechanisms, and cellular response. Oncologist. 2004;9 Suppl 5:4–9. doi: 10.1634/theoncologist.9-90005-4. [DOI] [PubMed] [Google Scholar]
- 3.Johnson AB, Barton MC. Hypoxia-induced and stress-specific changes in chromatin structure and function. Mutat Res. 2007 doi: 10.1016/j.mrfmmm.2006.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrero P, Okamoto K, Coumailleau P, O'Brien S, Tanaka H, Poellinger L. Redox-regulated recruitment of the transcriptional coactivators CREB-binding protein and SRC-1 to hypoxia-inducible factor 1alpha. Mol Cell Biol. 2000;20:402–415. doi: 10.1128/mcb.20.1.402-415.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kasper LH, Boussouar F, Boyd K, Xu W, Biesen M, Rehg J, Baudino TA, Cleveland JL, Brindle PK. Two transactivation mechanisms cooperate for the bulk of HIF-1-responsive gene expression. Embo J. 2005;24:3846–3858. doi: 10.1038/sj.emboj.7600846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jung JE, Lee HG, Cho IH, Chung DH, Yoon SH, Yang YM, Lee JW, Choi S, Park JW, Ye SK, Chung MH. STAT3 is a potential modulator of HIF-1-mediated VEGF expression in human renal carcinoma cells. Faseb J. 2005;19:1296–1298. doi: 10.1096/fj.04-3099fje. [DOI] [PubMed] [Google Scholar]
- 7.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 8.Wilkinson DS, Ogden SK, Stratton SA, Piechan JL, Nguyen TT, Smulian GA, Barton MC. A direct intersection between p53 and transforming growth factor beta pathways targets chromatin modification and transcription repression of the alpha-fetoprotein gene. Mol Cell Biol. 2005;25:1200–1212. doi: 10.1128/MCB.25.3.1200-1212.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ameri K, Hammond EM, Culmsee C, Raida M, Katschinski DM, Wenger RH, Wagner E, Davis RJ, Hai T, Denko N, Harris AL. Induction of activating transcription factor 3 by anoxia is independent of p53 and the hypoxic HIF signalling pathway. Oncogene. 2007;26:284–289. doi: 10.1038/sj.onc.1209781. [DOI] [PubMed] [Google Scholar]
- 10.Denko N, Wernke-Dollries K, Johnson AB, Hammond E, Chiang CM, Barton MC. Hypoxia actively represses transcription by inducing negative cofactor 2 (Dr1/DrAP1) and blocking preinitiation complex assembly. J Biol Chem. 2003;278:5744–5749. doi: 10.1074/jbc.M212534200. [DOI] [PubMed] [Google Scholar]
- 11.Liu L, Cash TP, Jones RG, Keith B, Thompson CB, Simon MC. Hypoxia-induced energy stress regulates mRNA translation and cell growth. Mol Cell. 2006;21:521–531. doi: 10.1016/j.molcel.2006.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang S, Litt M, Felsenfeld G. Methylation of histone H4 by arginine methyltransferase PRMT1 is essential in vivo for many subsequent histone modifications. Genes Dev. 2005;19:1885–1893. doi: 10.1101/gad.1333905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 14.Sims RJ, 3rd, Nishioka K, Reinberg D. Histone lysine methylation: a signature for chromatin function. Trends Genet. 2003;19:629–639. doi: 10.1016/j.tig.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 15.Finkenzeller G, Technau A, Marme D. Hypoxia-induced transcription of the vascular endothelial growth factor gene is independent of functional AP-1 transcription factor. Biochem Biophys Res Commun. 1995;208:432–439. doi: 10.1006/bbrc.1995.1356. [DOI] [PubMed] [Google Scholar]
- 16.Yan SF, Lu J, Zou YS, Soh-Won J, Cohen DM, Buttrick PM, Cooper DR, Steinberg SF, Mackman N, Pinsky DJ, Stern DM. Hypoxia-associated induction of early growth response-1 gene expression. J Biol Chem. 1999;274:15030–15040. doi: 10.1074/jbc.274.21.15030. [DOI] [PubMed] [Google Scholar]
- 17.Workman JL. Nucleosome displacement in transcription. Genes Dev. 2006;20:2009–2017. doi: 10.1101/gad.1435706. [DOI] [PubMed] [Google Scholar]
- 18.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 19.Roh TY, Cuddapah S, Cui K, Zhao K. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci U S A. 2006;103:15782–15787. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swigut T, Wysocka J. H3K27 demethylases, at long last. Cell. 2007;131:29–32. doi: 10.1016/j.cell.2007.09.026. [DOI] [PubMed] [Google Scholar]
- 21.Cheng X, Zhang X. Structural dynamics of protein lysine methylation and demethylation. Mutat Res. 2007;618:102–115. doi: 10.1016/j.mrfmmm.2006.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wissmann M, Yin N, Muller JM, Greschik H, Fodor BD, Jenuwein T, Vogler C, Schneider R, Gunther T, Buettner R, Metzger E, Schule R. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptordependent gene expression. Nat Cell Biol. 2007;9:347–353. doi: 10.1038/ncb1546. [DOI] [PubMed] [Google Scholar]
- 23.Klose RJ, Yan Q, Tothova Z, Yamane K, Erdjument-Bromage H, Tempst P, Gilliland DG, Zhang Y, Kaelin WG., Jr The Retinoblastoma Binding Protein RBP2 Is an H3K4 Demethylase. Cell. 2007 doi: 10.1016/j.cell.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 24.Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Heng Qi H, Whetstine JR, Bonni A, Roberts TM, Shi Y. The X-Linked Mental Retardation Gene SMCX/JARID1C Defines a Family of Histone H3 Lysine 4 Demethylases. Cell. 2007 doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


