Abstract
Heme oxygenase 1 (HO-1) is the first and rate-controlling enzyme in heme degradation. Bach1 is a mammalian transcriptional repressor of HO-1. To understand how zinc mesoporphyrin (ZnMP) induces the expression of HO-1, we investigated the effects of ZnMP on Bach1 mRNA and protein levels in human hepatoma Huh-7 cells by quantitative RT-PCR and Western blots. We found that ZnMP markedly up-regulated HO-1 mRNA and protein levels, and rapidly and significantly decreased Bach1 protein levels by increasing degradation of Bach1 protein [half life (t1/2) from 19 h to 45 min], whereas ZnMP did not influence Bach1 mRNA levels. The proteasome inhibitors, epoxomicin and MG132, significantly inhibited degradation of Bach1 by ZnMP in a dose-dependent fashion, indicating the degradation of Bach1 by ZnMP is proteasome dependent. Purified Bach1 C-terminal fragment bound heme, but there was no evidence for binding of ZnMP to the heme-binding region of Bach1. In conclusion, ZnMP produces profound post-transcriptional down-regulation of Bach1 protein levels and transcriptional up-regulation of HO-1. Our results indicate that ZnMP up-regulates HO-1 gene expression by markedly increasing Bach1 protein degradation in a proteasome-dependent manner.
Keywords: Zinc mesoporphyrin, Post-transcriptional regulation, Proteasome, Bach1, HO-1, Heme
1. Introduction
Heme oxygenase (HO) is the rate-controlling enzyme in heme degradation, generating ferrous iron, carbon monoxide and biliverdin, which have anti-oxidant and anti-inflammatory activities in vivo [1–4]. HO has two isoforms: HO-1 and HO-2. The HO-1 gene is highly inducible by chemical and physical stress [e.g., reactive oxygen species (ROS), arsenicals, transition metals, heat shock]; indeed, another name for HO-1 is heat shock protein 32 (Mr = 32 kDa) [4]. HO-1 is also highly inducible by heme, its physiologic substrate, and by other selected metalloporphyrins (MePns), including cobalt protoporphyrin (CoPP) [5, 6].
Bach1, a basic leucine zipper (bZip) mammalian transcriptional repressor, is a sensor and effector of heme [7, 8]. Bach1 forms antagonizing heterodimers with the Maf-related oncogene family. These heterodimers bind to Maf recognition elements (MAREs) and suppress expression of genes (e.g., HO-1 and NQO1) that respond to Maf-containing heterodimers and other positive transcriptional factors [8–10]. After binding heme, Bach1 loses its DNA binding activity, after which it is exported out of nuclei [11]. Recent work has established that heme and CoPP lead to the up-regulation of the HO-1 gene by influencing intranuclear levels and binding of Bach1 and small Maf proteins to key regulatory sites of the HO-1 promoter [5, 12]. These studies show that heme enters the nuclei, binds to CP-containing motifs of the C-terminal region of Bach1 (amino acids 417–739), and leads to the dissociation of Bach1 from a heterodimeric repressor complex with small Maf proteins [8]. A separate synergistic effect of MePns is to stabilize Nrf2, which then can bind to the small Maf proteins and enhance gene transcription [5, 13].
It is well documented that zinc mesoporphyrin (ZnMP), an analogue of heme, is a competitive inhibitor of HO activity [14, 15]. In previous tudies, we have characterized the regulation of the HO-1 promoter activities and mRNA levels by MePns. Recently we found that ZnMP up-regulates the HO-1 promoter activity in human hepatoma Huh-7 cells, but does not significantly influence HO-1 promoter activity or mRNA levels in chick hepatoma LMH and primary cultures of chick embryo liver cells [6]. Other recent studies from our laboratory have shown that ZnMP up-regulates HO-1 mRNA and protein levels in NIH 3T3 cells (unpublished). However, little is known about the mechanism by which this occurs. To understand how ZnMP induces the expression of HO-1, in the work reported here we investigated the effects of ZnMP on Bach1 mRNA and protein levels. We found that ZnMP markedly decreases Bach1 protein levels but does not influence its mRNA levels. In addition, we expressed and purified the recombinant C-terminal heme binding region of Bach1, containing amino acids 417–739, named Bach1C, to investigate whether ZnMP is able to bind to Bach1. Binding assays showed that ZnMP does not bind to the heme-binding region of Bach1. Proteasome inhibitors, epoxomicin and MG132, significantly block the degradation of Bach1 by ZnMP. Our results indicate that ZnMP up-regulates human HO-1 gene expression by inducing Bach1 protein degradation in a proteasome-dependent manner.
2. Materials and Methods
2.1. Reagents and materials
Plasmid pCMV-Bach1 harboring the mouse Bach1 cDNA sequence was kindly provided by K. Igarashi (Tohoku University School of Medicine, Sendai, Japan). Prokaryotic expression vectors pQE-30 and pQE-40 were purchased from Qiagen (Valencia, CA). Ferric (Fe3+)-protoporphyrin IX•Cl (hemin), ZnMP and tin mesoporphyrin (SnMP) were purchased from Frontier Scientific (Logan, UT). TRIzol was from Invitrogen (Carlsbad, CA). Goat anti-human Bach1, goat anti-human GAPDH polyclonal antibodies, mouse anti-rabbit IgG, and rabbit anti-goat IgG were purchased from Santa Cruz Biotechnology, Inc (Santa Cruz, CA). Rabbit anti-human HO-1 polyclonal antibody was purchased from StressGen (Victoria, BC Canada). ECL-Plus was purchased from Amersham Biosciences Corp (Piscataway, NJ). Epoxomicin and MG132 were from Sigma-Aldrich (St. Louis, MO). Dimethyl sulfoxide (DMSO) was purchased from Fisher Biotech (Fair Lawn, NJ). BCA protein assay reagent was from Pierce (Rockford, IL). Dulbecco's modified Eagle's medium (DMEM) and fetal bovine serum (FBS) were from HyClone (Logan, UT). Trypsin and isopropyl thio-β-D-galactopyranoside (IPTG) were obtained from Sigma-Aldrich. Oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA). Restriction endonucleases were purchased from either New England Biolabs (Ipswich, MA) or Invitrogen.
2.2. Cell cultures and treatments
Human hepatoma cell line, Huh-7 (Japan Health Research Resources Bank, Osaka, Japan) was maintained in DMEM supplemented with 100 units/mL penicillin, 100 µg/mL streptomycin, and 10% (v/v) FBS. ZnMP or SnMP was dissolved in DMSO and stored at -20°C. The addition of DMSO to the cultures did not exceed 1 µL of DMSO per mL of media.
2.3. Transfection and luciferase activity assay
Plasmid construct, phHOGL3/11.6, containing -11.6 kb (11.6 kb from transcriptional start site) 5’- flanking region of the human HO-1 promoter was a gift from Dr. A. Agarwal (University of Alabama at Birmingham, Birmingham, AL) [16]. Huh-7 cells were transfected with phHOGL3/11.6 using Lipofectamine™ 2000 from Invitrogen according to the manufacturer’s protocol. Cells lysates were collected and luciferase reporter gene expression were assessed by quantitation of luciferase activity, normalized to β-galactosidase activity and protein content.
2.4. Quantitative RT-PCR
Total RNA from treated cells was extracted and cDNA was synthesized as described before [12]. Real time quantitative RT-PCR was performed using a MyiQ™ Single Color Real-Time PCR Detection System from Bio-Rad (Hercules, CA) and iQ™ SYBR Green Supermix Real-Time PCR kit (Bio-Rad). We included samples without template and without reverse transcriptase as negative controls, which were expected to produce negligible signals (Ct value>35). Standard curves of Bach1, HO-1 and GAPDH were constructed with results of parallel PCR reactions performed on serial dilutions of a standard DNA (from one of the controls). Fold-change values were calculated by comparative Ct analysis after normalizing for the quantity of GAPDH in the same samples.
2.5. Western blots
Protein preparations and Western blots were carried out as described before [12]. In brief, total proteins (25–50 µg) were separated on 4–15% gradient SDS-PAGE gels (Bio-Rad). After electrophoretic transfer onto ImmunBlot PVDF membrane (Bio-Rad), membranes were blocked for 1 hour in PBS containing 5% nonfat dry milk and 0.1% Tween-20, and then incubated overnight with primary antibody at 4 °C. The dilutions of the primary antibodies were as follows: 1:2000 for anti-HO-1 antibody, and 1:1000 for anti-Bach1, and anti-GAPDH antibodies. The membranes were then incubated for 1 hour with horseradish peroxidase-conjugated secondary antibodies (dilution 1:10,000). Finally, the bound antibodies were visualized with the ECL-Plus chemiluminescence system according to the manufacturer’s protocol (Amersham). A Kodak 1DV3.6 computer-based imaging system (Rochester, NY) was used to measure the relative optical density of each specific band obtained after Western blotting. Data are expressed as percentages of the controls (corresponding to the value obtained with the vehicle-treated cells), which were assigned values of one.
2.6. Construction of prokaryotic expression vector pQE-Bach1C
pQE-30 vector was used to construct the prokaryotic expression vector pQE-Bach1C by subcloning the fragment containing the Bach1C gene [17]. The cDNA sequence encoding Bach1C was amplified by PCR from vector pCMV-Bach1 using a forward primer (P1: 5'-CGGGATCCTTATTTGAAAAGAAAGTGTATCTCTC-3') and reverse primer (P2: 5'-CCCGAGCTCATCAAATGAAGGGGCCGCACACTGAGG-3'), in which Bam HI and Hind III sites exist at the ends of Bach1C cDNA. PCR conditions consisted of 30 cycles of 94 °C for 0.5 min, 55 °C for 0.5 min, and 72 °C for 1.5 min. The PCR-amplified DNA fragment was separated on 1% agarose gel containing ethidium bromide and visualized by UV-light illumination. The amplified cDNA fragment was inserted into vector pQE-30 to construct prokaryotic expression vector pQE-Bach1C and this was verified by sequencing.
2.7. Expression and identification of recombinant Bach1C (417–739) in E. coli M15
E. coli M15 was cultured in LB medium for approximately 2 h until the cells reached an A600 of 0.6. Subsequently, recombinant Bach1C expression was induced by addition of IPTG to a final concentration of 1.0 mM and then incubating the cells for an additional 4 h at 37 °C [18]. Cells were finally harvested and washed with PBS (phosphate-buffered saline) buffer, and then resuspended in lysis buffer. The cell suspensions were sonicated on ice and centrifuged at 8000 g for 30 min. Soluble protein fractions were loaded onto a Ni2+-nitrilotriacetic acid (Ni-NTA) agarose column (Qiagen) equilibrated with lysis buffer. After washing the column with washing buffer, the recombinant Bach1C proteins were eluted with elution buffer. The expression and purification of Bach1C protein were monitored by SDS-PAGE and Western blot using goat polyclonal anti-Bach1 antibody.
2.8. Spectroscopic assays
Recombinant Bach1C was purified and immobilized on Ni-NTA beads. The resin with bound Bach1C was suspended in TBS (Tris-buffered saline), 0.1% Tween-20 (TBS-T), pH 8.0 and incubated with heme or ZnMP at room temperature for 1 h. The resulting mixtures were loaded on a column, and washed with sufficient TBS-T until free heme or ZnMP was completely removed. The column was subsequently washed with washing buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 8.0) and elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, pH 8.0). The absorption characteristics of elutes were scanned using a HT Synergy spectrophotometer from Bio-Tek Instruments (Winooski, VT) [8].
2.9. Statistical analyses of data
Experiments were repeated at least three times with similar results. All experiments included at least triplicate samples for each treatment group. Representative results from single experiments are presented. Statistical analyses were performed with JMP 4.0.4 software from SAS Institute (Cary, NC). Differences in mean values were assessed by analysis-of-variance techniques and Student’s ttest analysis. Values of P <0.05 were considered significant.
3. Results
3.1. ZnMP-mediated induction of HO-1 gene expression in human hepatoma Huh-7 cells
We examined whether ZnMP up-regulates the HO-1 promoter activity, mRNA and protein levels in human hepatoma Huh-7 cells. We found that ZnMP significantly up-regulates luciferase reporter activity by 1.7-fold (P<0.05). ZnMP induced HO-1 mRNA and protein levels in these cells in a dose-dependent fashion (Figs. 1B and 1C). A concentration of ZnMP as low as 1 µM significantly up-regulated HO-1 mRNA levels by 5.3-fold (P<0.05), the maximum effect (8.6-fold) was reached following 10 µM ZnMP treatment (Fig.1B), a concentration of 20 µM ZnMP was unable to dissolve completely in cell culture medium and the cells treated with 20 µM ZnMP failed to grow well. Therefore, 10 µM ZnMP was the highest concentrations used for the subsequent studies. To investigate whether these effects require the MePn macrocycle, we treated Huh-7 cells with 10 µM free mesoporphyrin or ZnCl2. As shown in Fig. 1C, HO-1 protein levels were not affected by free mesoporphyrin or ZnCl2.
Fig. 1. Effects of ZnMP on luciferase reporter gene expression, HO-1 mRNA and protein levels in Huh-7 cells.
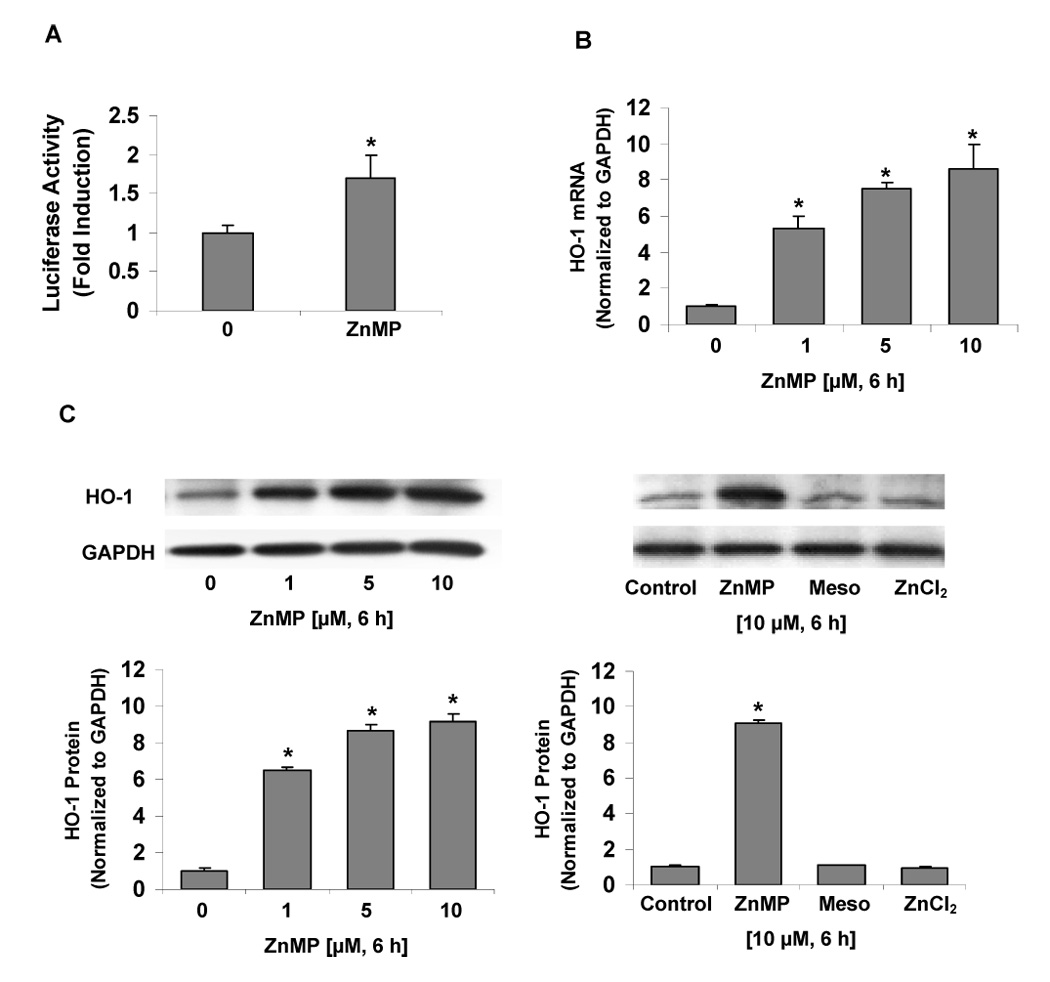
Huh-7 cells, transfected with phHOGL3/11.6, were treated with or without 10 µM ZnMP for 15 h. Cells were harvested and cell lysates were assayed for luciferase activity. For measurement of HO-1 mRNA and protein levels, Huh-7 cells on 12-well plates were treated with 0 (vehicle only), 1.0, 5.0 and 10 µM ZnMP, or with 10 µM free mesoporphyrin (Meso) and ZnCl2 for 6 hours, after which cells were harvested, and total RNA and protein were extracted. mRNA and protein levels were measured by quantitative RT-PCR and Western blot, respectively, as described in Materials and Methods. (A) Effect of ZnMP on hHOGL3/11.6-Luc reporter activity in Huh-7 cells. (B) Dose response of effects of ZnMP (0–10 µM) on HO-1 mRNA levels in Huh-7 cells. (C) Effects of ZnMP (0–10 µM), free Meso or ZnCl2 on HO-1 protein levels in Huh-7 cells. The amounts of HO-1 mRNA were normalized to GAPDH which did not vary with treatment. Values for cells treated with vehicle only were set equal to 1. Data are presented as means ± SD from triplicate samples. * differs from vehicle only, P<0.05.
3.2. Down-regulation of Bach1 protein levels by ZnMP in human hepatoma Huh-7 cells
To investigate whether ZnMP affects the Bach1 gene expression in human hepatoma Huh-7 cells, we measured Bach1 mRNA and protein levels in Huh-7 cells treated with indicated concentrations of ZnMP. ZnMP led to a rapid and profound decrease of Bach1 protein levels in Huh-7 cells, while Bach1 protein levels were not affected by 10 µM free mesoporphyrin or ZnCl2 (Fig. 2A). Bach1 protein band was undetectable 6 h after treatment with 10 µM ZnMP (Fig. 2A). In contrast, ZnMP did not affect mRNA levels of Bach1 (Fig. 2B).
Fig. 2. Effects of ZnMP on Bach1 protein levels in Huh-7 cells.
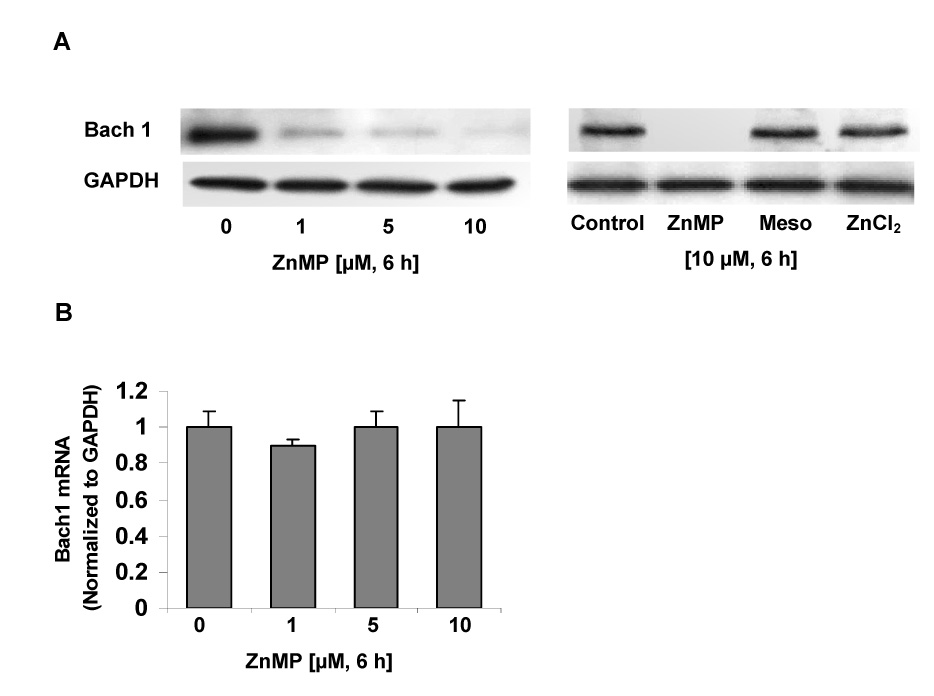
Huh-7 cells were treated with different concentrations of ZnMP (0, 1, 5, 10 µM) or with 10 µM mesoporphyrin (Meso) and ZnCl2 for 6 hours, after which cells were harvested and total protein and RNA were isolated. Proteins were separated on 4–15% SDS-polyacrylamide gel, transferred to a PVDF membrane, and probed with anti-human Bach1 and GAPDH specific antibodies. Bach1 mRNA levels were measured by quantitative RT-PCR as described in Materials and Methods. (A) Bach1 protein levels in Huh-7 cells exposed to ZnMP (0–10 µM), free Meso or ZnCl2 for 6 hours. (B) Bach1 mRNA levels in Huh-7 cells treated with ZnMP (0–10 µM) for 6 hours. The amounts of Bach1 mRNA were normalized to GAPDH which did not vary with treatment. Values for cells treated with vehicle only were set equal to 1. Data are presented as means ± SD from triplicate samples. * differs from vehicle only, P<0.05.
3.3. ZnMP influences the stability of Bach1 protein in Huh-7 cells
To further understand how ZnMP down-regulates Bach1, we examined the stability of Bach1 protein. As shown in Fig. 3, Bach1 protein levels in Huh-7 cells treated with ZnMP and cycloheximide (CHX), an inhibitor of protein synthesis, were greatly and rapidly reduced. However, Bach1 protein levels in Huh-7 cells that were not treated with ZnMP were also decreased by CHX, but to a less extent. Specifically, ZnMP [10 µM] decreased the Bach1 protein half life (t1/2) from 19 h to 45 min (Fig. 3).
Fig. 3. Post-transcriptional regulation of Bach1 expression by ZnMP in Huh-7 cells.
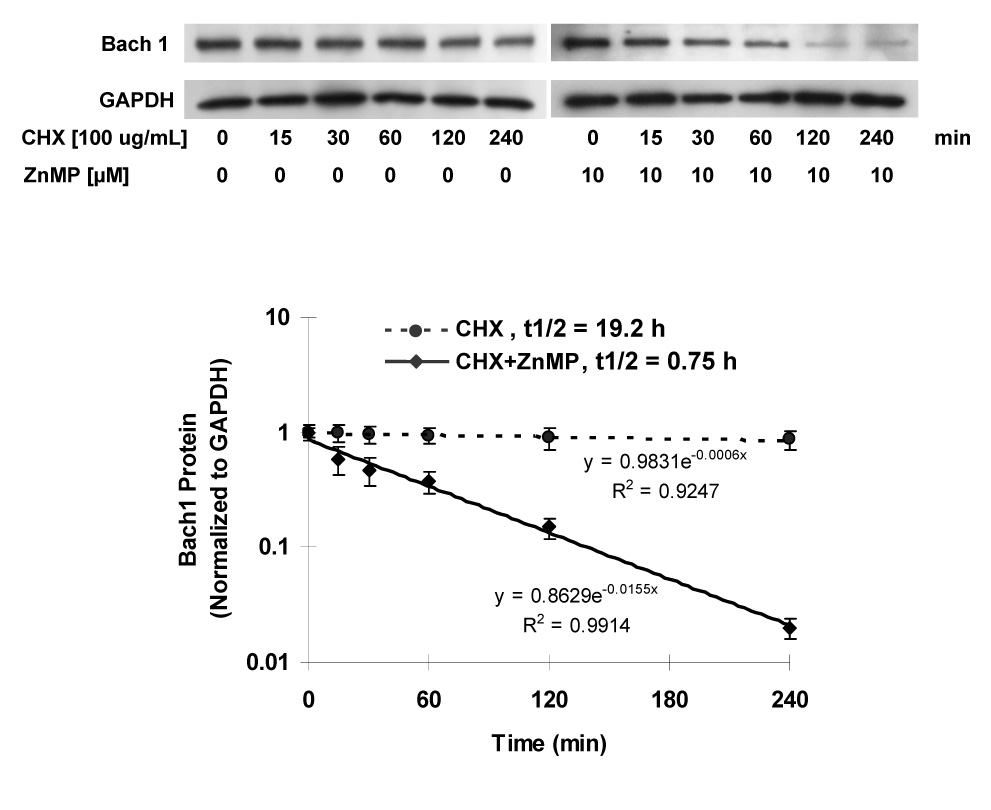
Huh-7 cells were treated with 100 ug/mL cycloheximide (CHX) and with or without 10 µM ZnMP for the indicated times, after which cells were harvested and total proteins were isolated, as described in Materials and Methods. Proteins were separated on 4–15% SDS-polyacrylamide gel, transferred to a PVDF membrane, and probed with anti-human Bach1- and GAPDH- specific antibodies. The relative amounts of Bach1 were normalized to GAPDH. Values for cells treated with vehicle only were set equal to 1. Representative results from one of three experiments are shown. Data are presented as means ± SD from triplicate samples.
3.4. Comparative studies of ZnMP vs SnMP on the down-regulation of Bach1 in Huh-7 cells
SnMP, another competitive HO inhibitor, has been reported recently to down-regulate Bach1 protein levels and induce the HO-1 gene expression in NIH 3T3 cells, demonstrating that up-regulation of expression of the HO-1 gene is mediated by a de-repression of Bach1 [19]. We performed comparative studies on ZnMP vs SnMP down-regulation of Bach1 in Huh-7 cells. As shown in Fig. 4, ZnMP markedly and rapidly decreased Bach1 protein levels after exposure to ZnMP for as little as 1 h and almost completely abrogated Bach1 protein expression by 8 h. In contrast, effects of SnMP were slower to occur and lesser in magnitude.
Fig. 4. ZnMP vs SnMP time-course of down-regulation Bach1 in Huh-7 cells.
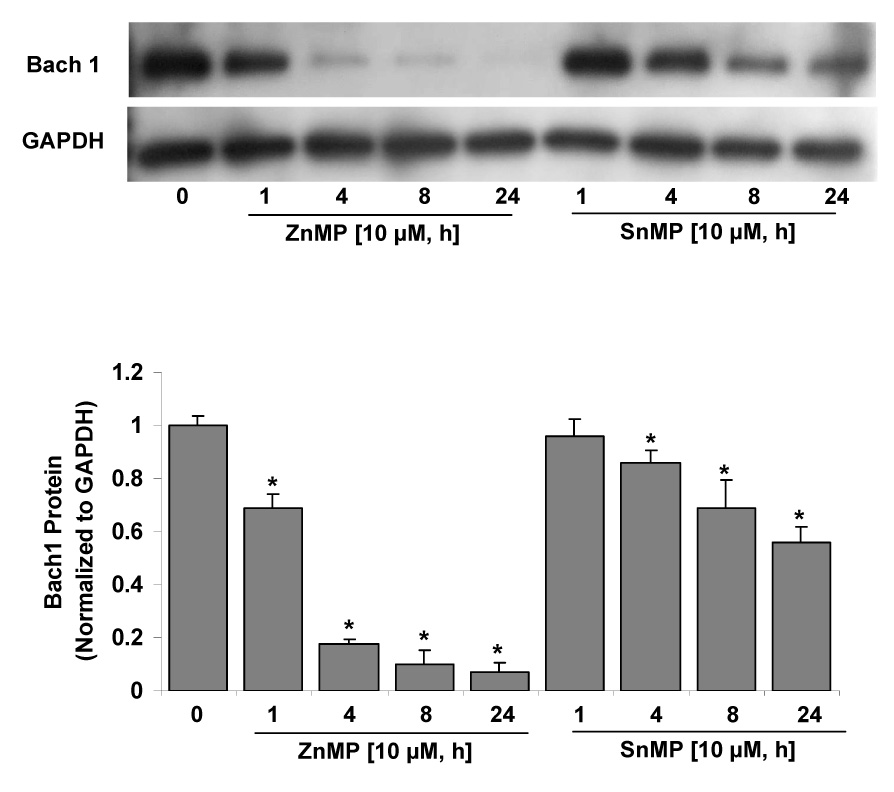
Huh-7 cells were treated with 10 µM ZnMP or SnMP for different times (0, 1, 4, 8, 24 h), and then the cells were harvested using harvest buffer containing the protease inhibitor cocktail. 25 µg of protein were loaded on a 4–15% SDS-polyacrylamide gel, transferred to a PVDF membrane, and probed with anti-Bach1, and anti-GAPDH specific antibodies, and then developed with ECL Plus reagent. The relative amounts of Bach1 protein were normalized to those for GAPDH, which did not vary with treatment. Values for cells treated with vehicle only were set equal to 1. Data are presented as means ± SD from triplicate samples. * differs from vehicle only, P<0.05.
3.6. Down-regulation of Bach1 protein levels by ZnMP is by proteasome-dependent degradation
Proteasome-dependent degradation is one of the major proteolytic pathways [20, 21]. To investigate if down-regulation of Bach1 protein is mediated by proteasome-dependent degradation, Huh-7 cells were treated with 5 µM ZnMP and selected concentrations of proteasome inhibitors [22, 23], epoxomicin (0, 1, 5, 10 µM) and MG132 (0, 1, 5, 10, 20 µM). Epoxomicin and MG132 did not affect Bach1 protein levels in Huh-7 cells without ZnMP treatment, but inhibited degradation of Bach1 by 5 µM ZnMP in a dose–dependent fashion. A concentration of epoxomicin and MG132 as low as 1 µM significantly inhibited degradation of Bach1 (Fig. 5A), and the maximum effect was reached with 10 µM epoxomicin treatment (Fig. 5A).The highest concentration of epoxomicin used in these experiments was 10 µM, because cells treated with 20 µM epoxomicin failed to grow well, suggesting that this concentration of epoxomicin was toxic to the cells.
Fig. 5. ZnMP down-regulates Bach1 by a proteasome-dependent process.
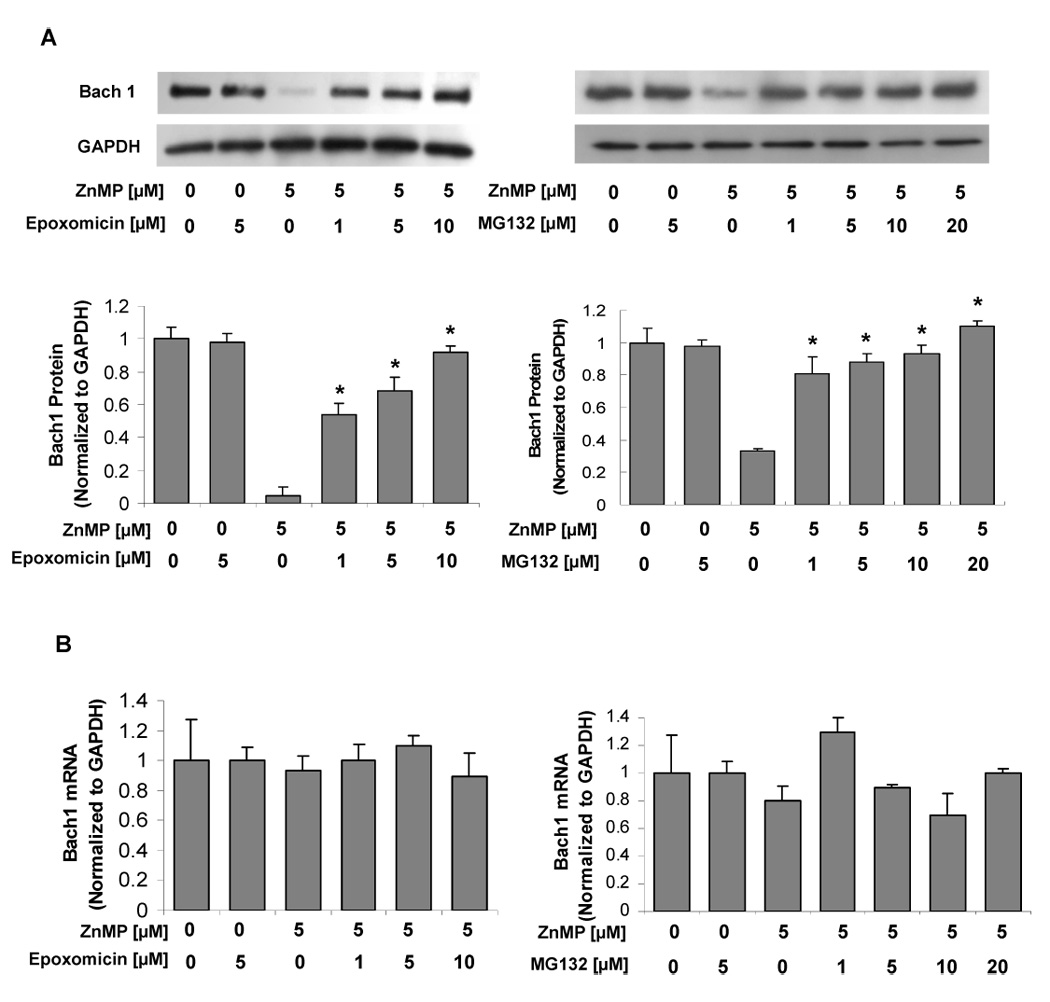
Huh-7 cells were treated with epoxomicin 20 min before ZnMP treatment, and then the cells were harvested 6 h after treatment with 5 µM ZnMP or with vehicle alone (DMSO) as control. Total RNA and protein were extracted. Bach1 and GAPDH mRNA and protein levels were measured by quantitative RT-PCR and Western blot as described in Materials and Methods. The amounts of Bach1 mRNA and protein were normalized to those for GAPDH, which did not vary with treatment. Values for cells treated with vehicle only were set equal to 1. (A) Bach1 protein levels in Huh-7 cells treated with 5 µM ZnMP and different concentrations of epoxomicin (0, 1, 5, 10 µM) or MG132 (0, 1, 5, 10, 20 µM). (B) Bach1 mRNA levels in Huh-7 cells treated with 5 µM ZnMP and different concentrations of epoxomicin (0, 1, 5, 10 µM) or MG132 (0, 1, 5, 10, 20 µM). Values for cells treated with vehicle only were set equal to 1. Data are presented as means ± SD from triplicate samples. * differs from 5 µM ZnMP treatment, P<0.05.
3.7. Preparation of Bach1C and studies on heme and ZnMP binding to Bach1C
Recent studies have indicated that heme up-regulates HO-1 by the mechanisms of binding of heme to Bach1 and degradation of Bach1 [5, 8, 12]. To investigate whether ZnMP binds to Bach1C, we constructed a prokaryotic expression vector, pQE-Bach1C, which highly expresses recombinant Bach1C with 6-His tag in E. coli strain M15. As a control, pQE-40 was introduced to E. coli M15 to express recombinant DHFR with 6-His tag in the presence of 1 mM IPTG. As expected, the cells with pQE-Bach1C transfection and 1 mM IPTG induction had a brownish color due to the binding of endogenous heme to the overexpressed Bach1C, whereas no color was seen in the cells transfected with pQE-Bach1C but without IPTG induction or in cells transfected with pQE-40 (Fig. 6A). We then purified Bach1C by Ni-NTA affinity chromatography as described in Materials and Methods. The expected size of recombinant Bach1C is 37.4 kDa (Fig. 6C). Western blot, using Bach1 antibody, was used to identify the recombinant Bach1C (Fig. 6D).
Fig. 6. Expression, purification and identification of mouse recombinant Bach1C.
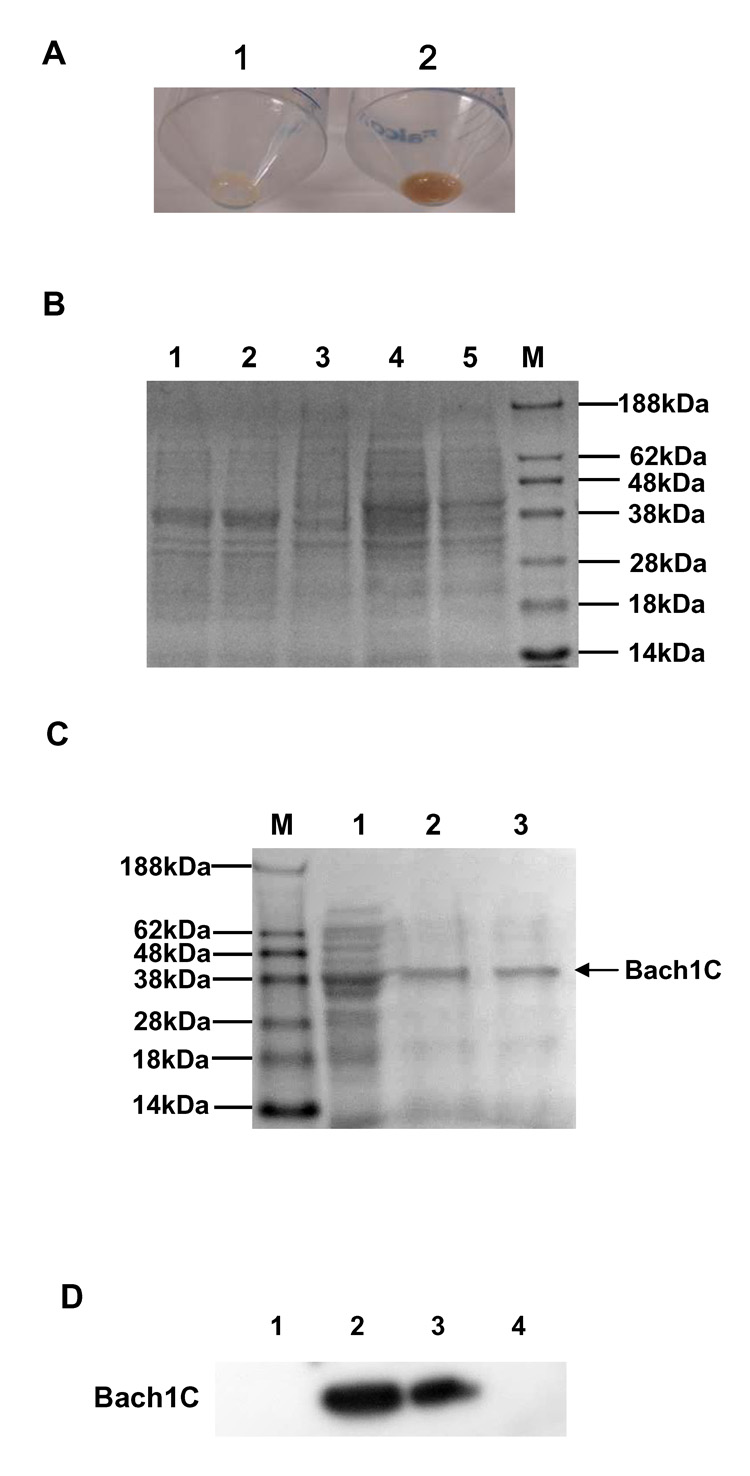
A plasmid that expresses mouse Bach1C was introduced into the E. coli strain M15, and the expression of the protein was induced with 1 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG). The protein was purified from the cell lysates by affinity column chromatography, stored on ice, and used within 3 weeks after preparation. (A) E. coli M15 with Bach1C and DHFR expression. 1: DHFR as control did not bind to endogenous heme from the E. coli and no color was seen; 2: Bach1C bound to endogenous heme which had brownish color. (B) SDS-PAGE analysis of Bach1C expression. Lane 1 and 2: pQE-Bach1C induced for 4 h by 1 mM IPTG; Lanes 3: pQE-Bach1C before IPTG induction; Lane 4: pQE-30 induced for 4 h with 1mM IPTG; Lanes 4: pQE-30 before IPTG induction; Lane M: protein marker. (C) SDS-PAGE analysis of Bach1C purification. Lane M: protein marker; Lane 1: E. coli M15 cells with pQE-Bach1C induction by 1 mM IPTG; Lanes 2 and 3: Bach1C purified by Ni-NTA agarose affinity chromatography. (D) Western blot identification of Bach1C expression in E. coli M15. Lane1 and 4: control; Lane 2 and 3: Bach1C.
We assayed binding of heme or ZnMP to Bach1 by immobilizing recombinant Bach1C on Ni-NTA agarose resin, as described in Materials and Methods. Resins with heme bound to Bach1 had a brownish color, whereas no color was seen in resins incubated with ZnMP or DMSO control (Fig. 7A). Only Bach1C exposed to heme showed a Soret band and a red shift of the maximum wavelength from 410 nm to 415 nm (Fig. 7B). Because the Bach1C that was overexpressed contained a 6-His tag to facilitate its purification on Ni-NTA columns, we were concerned that heme or ZnMP binding to the purified Bach1C might be due to the six histidine residues, rather than the CP heme binding motifs. Therefore, we tested whether 6-His tagged-DHFR that was similarly overexpressed and purified would show ZnMP binding. It did not, either by visual or spectrophotometric examination (data not shown). In addition, we cleaved the 6-His tag from some preparations of Bach1C, using SUMO protease and the methods of Zuo et al [24]. The preparations of Bach1C thus produced showed the same binding characteristics and spectra as those of 6-His tagged Bach1C (data not shown), further indicating that the 6-histidine residues were not involved in the binding. These findings demonstrated that ZnMP, unlike heme, does not bind to Bach1C.
Fig. 7. Recombinant Bach1C binds heme but not ZnMP.
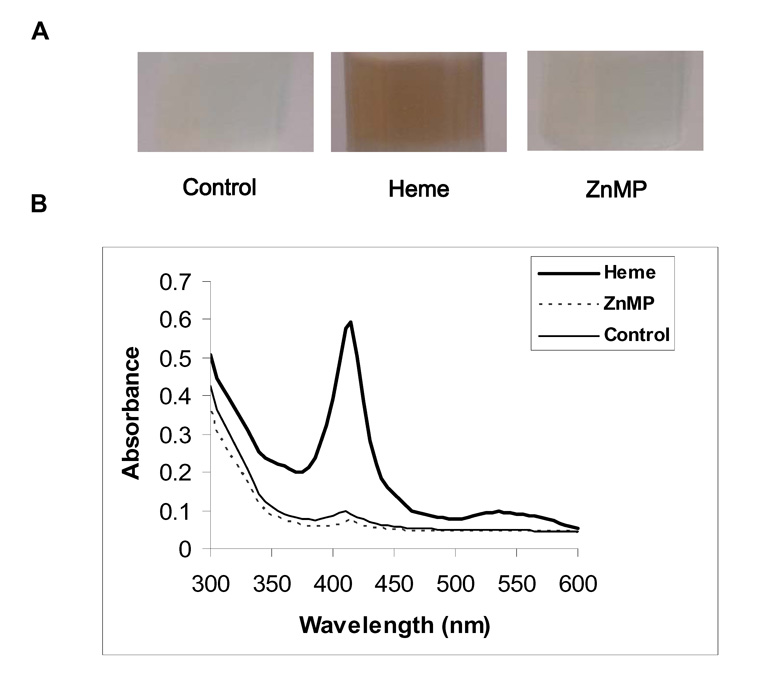
Recombinant Bach1C, produced in E. coli as described in Fig.7, was purified by affinity chromatography. The resin with bound Bach1C was suspended in TBS, 0.1%Tween-20, pH 8.0 and incubated 1 h with 20 µM heme, ZnMP, or DMSO (Control), followed by washing the column with 8 mL TBS-T. (A) shows photographs of the resin. The brownish color indicates high affinity binding of heme to Bach1C, whereas the much diminished color with ZnMP indicates little or no binding of ZnMP. (B) shows absorption spectra of the elutions. The Soret peaks for heme but not for control or ZnMP, confirm lack of ZnMP binding to Bach1C.
4. Discussion
Our major findings are: 1) ZnMP up-regulates HO-1 mRNA and protein levels in a dose–dependent fashion in human hepatoma Huh-7 cells (Fig. 1); 2) ZnMP induces profound down-regulation of Bach1 proteins in human hepatoma Huh-7 cells (Fig. 2A), but has no effect on Bach1 mRNA levels (Fig. 2B); 3) down-regulation of Bach1 protein by ZnMP is much greater and faster than SnMP (Fig. 4); 4) epoxomicin and MG132, proteasome inhibitors, block the degradation of Bach1 in Huh-7 cells (Fig. 5) in a dose-dependent manner, suggesting that down-regulation of Bach1 by ZnMP is mediated by a proteasome-dependent degradation; and 5) ZnMP does not bind to the heme-binding motifs of Bach1 (Fig. 7).
Bach1, a member of the basic leucine zipper family of proteins, has been recently shown to be a transcriptional repressor of HO-1, and to play a critical role in heme-, CoPP- and SnMP-dependent upregulation of the HO-1 gene [5, 9, 11–13, 19, 25]. Upon exposure to heme, heme binds to Bach1 and forms antagonizing heterodimers with the Maf-related oncogene family. These heterodimers bind to MAREs, also known as anti-oxidant responsive elements (AREs), and suppress expression of genes that respond to Maf-containing heterodimers and other positive transcriptional factors. After binding heme, Bach1 loses its DNA binding activity and then it is exported out of nuclei by triggering a Crm1- dependent mechanism.
It is well-known that ZnMP is a competitive inhibitor of HO activity. Earlier studies reported that zinc protoporphyrin (ZnPP) is a strong up-regulator of HO-1 transcription in liver cell cultures, in comparison to zinc bis glycol porphyrin (ZnBG), which showed minimal effects [26, 27]. In our studies, ZnMP up-regulated the HO-1 gene expression at both mRNA and protein levels in human hepatoma Huh-7 cells. ZnMP produced a profound and rapid degradation of Bach1 in human hepatoma Huh-7 cells, and also in human stellate LX-2 and NIH 3T3 cells (data not shown), suggesting that ZnMP-dependent degradation of Bach1 exists in many types of cells. Bach1 degradation by SnMP and heme have recently been reported [19, 28]. We performed studies comparing effects of SnMP and ZnMP (10 µM) in Huh-7 cells for various times (0–24 h). The degradation of Bach1 by ZnMP was both faster and more profound than that produced by SnMP (Fig. 4).
So far, two distinct systems for protein degradation have been found in mammals: the lysosome system and the ubiquitin-proteasome system [29–32]. In the ubiquitin-proteasome pathway, substrates are first marked for degradation by covalent linkage to multiple molecules of ubiquitin and then are hydrolyzed by the 26 S proteasome, a 2000 kDa ATP-dependent proteolytic complex [31, 32]. Since 1994, pharmacological inhibitors of the proteasome have become available. Several types of proteasome inhibitors have been identified that can readily enter cells and selectively inhibit this degradation pathway. The most widely used are hydrophobic peptide aldehydes, such as carbobenzoxyl-leucinyl-leucinyl-leucinal (MG132), carbobenzoxyl-leucinyl-leucinyl-norvalinal (MG115), and N-acetyl-leucinyl-leucinyl-norleucinal (MG101), which reversibly inhibit the proteasome dependent degradation of proteins. These agents are substrate analogues and potent transition-state inhibitors, primarily of the chymotrypsin-like activity of the proteasome [20, 33]. MG132 (50 µM) can block the rapid breakdown of ubiquitin-proline-β-galactosidase (UB-P-β-gal) by the ubiquitin pathway in 30 min without affecting protein synthesis [20]. One of the critical functions of this pathway is the rapid degradation of highly abnormal proteins or critical regulatory proteins, such as cyclins, p53, and IκB, that must be rapidly eliminated for control of growth and metabolism [32]. Epoxomicin, a natural product isolated from Actinomyces sp., is a cell-permeable, potent, selective and irreversible proteasome inhibitor. In the present study, epoxomicin and MG132 were used to investigate the mechanism by which ZnMP degrades Bach1 protein. Both epoxomicin and MG132 significantly blocked the ZnMP-dependent degradation of Bach1 in a dose-dependent manner, suggesting that down-regulation of Bach1 protein in the nucleus by proteosomal degradation is a highly likely mechanism. Further studies on how ZnMP is able to trigger proteasome-dependent degradation of Bach1 are underway in our laboratory.
Recent data of Igarashi et al indicated that the four C-terminal cysteine-proline (CP) motifs in Bach1 are essential for heme binding, and that this binding is one of the mechanisms by which heme upregulates HO-1 [8]. More recent data showed that heme binds to Bach1 in two modes located in Bach1ΔBTB (179–739 aa) [34]. CP motifs have been found in several heme regulatory motifs (HRMs) and play an important role in heme binding in various proteins. Native mouse Bach1 contains a total of six CP motifs. Two CP motifs are downstream of the BTB (Bric-a-brac/Tramtrack/Broad complex) domain (CP1 and CP2), three CP motifs are upstream of the bZip domain (CP3, CP4 and CP5) and one CP motif is downstream of the bZip domain (CP6). Among these CP motifs, CP3–CP6 has been demonstrated to be the most important for heme-binding [8]. Bach1C (417–739 aa) used in our studies showed heme, but not ZnMP, binding.
Chromium, tin, and zinc porphyrins are potent inhibitors of HO-1 and -2 [1, 4, 35–37] and have been proposed as therapeutic agents for severe unconjugated hyperbilirubinemia (e.g., Crigler-Najjar syndrome type II) and for prolonging effects of heme in therapy of acute porphyric syndromes [38, 39]. Problems in their clinical application have included concerns about the carcinogenicity of chromium [40], the photosensitivity caused by tin porphyrins [37, 39, 41], and evidence of possible bone marrow toxicity due to zinc porphyrins [42, 43]. In addition, levels of HO-1 protein have been found to be increased in mammalian, although not avian, cells exposed to tin and zinc porphyrins [6, 44] . The findings of the present paper suggest that some or all of the up-regulation of HO-1 protein levels produced by tin or zinc porphyrins is due to their abilities to degrade Bach1, thereby lifting the tonic repression that Bach1 exerts on HO-1 gene expression [5, 8, 11–13]. The greater degree of inhibition of red cell development by zinc vs tin porphyrins previously described [42, 43] may similarly be related to the more rapid and dramatic effect of zinc vs tin porphyrins on Bach1 degradation.
In summary, the results of our and other recent studies indicate that certain non-heme MePns (CoPP, SnMP, ZnMP), but not the free porphyrins nor the metallic ions themselves, produce post-translational down-regulation of Bach1 proteins by increasing proteasomal degradation. The effect of ZnMP is particularly rapid and dramatic. Our present results further suggest that the Bach1 protein is broken down by the proteasome, but that the rapid and dramatic effect of ZnMP is not dependent upon binding to Bach1. Further studies are underway in our laboratory, designed to elucidate how ZnMP acts on Bach1 and the proteasome, as well as to search for other potential protein targets of these effects of ZnMP.
Acknowledgements
This work was supported by United States Public Health Service Grants RO1-DK38825 and contracts NO-1 DK92326 and UO-1 DK065193.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Elbirt KK, Bonkovsky HL. Heme oxygenase: recent advances in understanding its regulation and role. Proc Assoc Am Physicians. 1999;111:438–447. [PubMed] [Google Scholar]
- 2.Lambrecht RW, Fernandez M, Shan Y, Bonkovsky HL. Heme oxygenase and carbon monoxide in cirrhosis and portal hypertension, Ascites and Renal Dysfunction in Liver Disease. Blackwell Science, Oxford. 2005 [Google Scholar]
- 3.Hill-Kapturczak N, Chang SH, Agarwal A. Heme oxygenase and the kidney. DNA Cell Biol. 2002;21:307–321. doi: 10.1089/104454902753759726. [DOI] [PubMed] [Google Scholar]
- 4.Bonkovsky HL, Elbirt KK. Heme Oxygenase: Its Regulation and Role. In: Cutler RG, Rodriguez H, editors. Oxidative Stress and Aging. River Edge, NJ: World Scientific; 2002. pp. 699–706. [Google Scholar]
- 5.Shan Y, Lambrecht RW, Donohue SE, Bonkovsky HL. Role of Bach1 and Nrf2 in up-regulation of the heme oxygenase-1 gene by cobalt protoporphyrin. Faseb J. 2006;20:2651–2653. doi: 10.1096/fj.06-6346fje. [DOI] [PubMed] [Google Scholar]
- 6.Shan Y, Pepe J, Lu TH, Elbirt KK, Lambrecht RW, Bonkovsky HL. Induction of the heme oxygenase-1 gene by metalloporphyrins. Arch Biochem Biophys. 2000;380:219–227. doi: 10.1006/abbi.2000.1921. [DOI] [PubMed] [Google Scholar]
- 7.Oyake T, Itoh K, Motohashi H, Hayashi N, Hoshino H, Nishizawa M, Yamamoto M, Igarashi K. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol Cell Biol. 1996;16:6083–6095. doi: 10.1128/mcb.16.11.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogawa K, Sun J, Taketani S, Nakajima O, Nishitani C, Sassa S, Hayashi N, Yamamoto M, Shibahara S, Fujita H, Igarashi K. Heme mediates derepression of Maf recognition element through direct binding to transcription repressor Bach1. Embo J. 2001;20:2835–2843. doi: 10.1093/emboj/20.11.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun J, Hoshino H, Takaku K, Nakajima O, Muto A, Suzuki H, Tashiro S, Takahashi S, Shibahara S, Alam J, Taketo MM, Yamamoto M, Igarashi K. Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. Embo J. 2002;21:5216–5224. doi: 10.1093/emboj/cdf516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun J, Brand M, Zenke Y, Tashiro S, Groudine M, Igarashi K. Heme regulates the dynamic exchange of Bach1 and NF-E2-related factors in the Maf transcription factor network. Proc Natl Acad Sci U S A. 2004;101:1461–1466. doi: 10.1073/pnas.0308083100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki H, Tashiro S, Hira S, Sun J, Yamazaki C, Zenke Y, Ikeda-Saito M, Yoshida M, Igarashi K. Heme regulates gene expression by triggering Crm1-dependent nuclear export of Batch1. Embo J. 2004;23:2544–2553. doi: 10.1038/sj.emboj.7600248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shan Y, Lambrecht RW, Ghaziani T, Donohue SE, Bonkovsky HL. Role of Bach-1 in regulation of heme oxygenase-1 in human liver cells: insights from studies with small interfering RNAS. The Journal of biological chemistry. 2004;279:51769–51774. doi: 10.1074/jbc.M409463200. [DOI] [PubMed] [Google Scholar]
- 13.Igarashi K, Sun J. The heme-Bach1 pathway in the regulation of oxidative stress response and erythroid differentiation. Antioxid Redox Signal. 2006;8:107–118. doi: 10.1089/ars.2006.8.107. [DOI] [PubMed] [Google Scholar]
- 14.Schuurmans MM, Hoffmann F, Lindberg RL, Meyer UA. Zinc mesoporphyrin represses induced hepatic 5-aminolevulinic acid synthase and reduces heme oxygenase activity in a mouse model of acute hepatic porphyria. Hepatology (Baltimore, Md. 2001;33:1217–1222. doi: 10.1053/jhep.2001.24170. [DOI] [PubMed] [Google Scholar]
- 15.Greenbaum NL, Kappas A. Comparative photoactivity of tin and zinc porphyrin inhibitors of heme oxygenase: pronounced photolability of the zinc compounds. Photochem Photobiol. 1991;54:183–192. doi: 10.1111/j.1751-1097.1991.tb02005.x. [DOI] [PubMed] [Google Scholar]
- 16.Hill-Kapturczak N, Voakes C, Garcia J, Visner G, Nick HS, Agarwal A. A cis-acting region regulates oxidized lipid-mediated induction of the human heme oxygenase-1 gene in endothelial cells. Arteriosclerosis, thrombosis, and vascular biology. 2003;23:1416–1422. doi: 10.1161/01.ATV.0000081656.76378.A7. [DOI] [PubMed] [Google Scholar]
- 17.Hou WH, Fang T, Chai YR, Wang TY, Wang JM, Xue LX. Expression of recombinant kringle 1-5 domains of human plasminogen by a prokaryote expression system. Protein Expr Purif. 2006;47:93–98. doi: 10.1016/j.pep.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 18.Hou WH, Wang TY, Yuan BM, Chai YR, Jia YL, Tian F, Wang JM, Xue LX. Recombinant mouse canstatin inhibits chicken embryo chorioallantoic membrane angiogenesis and endothelial cell proliferation. Acta Biochim Biophys Sin (Shanghai) 2004;36:845–850. doi: 10.1093/abbs/36.12.845. [DOI] [PubMed] [Google Scholar]
- 19.Abate A, Zhao H, Wong RJ, Stevenson DK. The role of Bach1 in the induction of heme oxygenase by tin mesoporphyrin. Biochemical and biophysical research communications. 2007;354:757–763. doi: 10.1016/j.bbrc.2007.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee DH, Goldberg AL. Selective inhibitors of the proteasome-dependent and vacuolar pathways of protein degradation in Saccharomyces cerevisiae. The Journal of biological chemistry. 1996;271:27280–27284. doi: 10.1074/jbc.271.44.27280. [DOI] [PubMed] [Google Scholar]
- 21.Reed SI. The ubiquitin-proteasome pathway in cell cycle control. Results Probl Cell Differ. 2006;42:147–181. doi: 10.1007/b136681. [DOI] [PubMed] [Google Scholar]
- 22.Lee DH, Goldberg AL. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 1998;8:397–403. doi: 10.1016/s0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- 23.Tsubuki S, Saito Y, Tomioka M, Ito H, Kawashima S. Differential inhibition of calpain and proteasome activities by peptidyl aldehydes of di-leucine and tri-leucine. J Biochem (Tokyo) 1996;119:572–576. doi: 10.1093/oxfordjournals.jbchem.a021280. [DOI] [PubMed] [Google Scholar]
- 24.Zuo X, Mattern MR, Tan R, Li S, Hall J, Sterner DE, Shoo J, Tran H, Lim P, Sarafianos SG, Kazi L, Navas-Martin S, Weiss SR, Butt TR. Expression and purification of SARS coronavirus proteins using SUMO-fusions. Protein Expr Purif. 2005;42:100–110. doi: 10.1016/j.pep.2005.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Igarashi K, Sun H. Oxidative stress protection by heme. Masui. 2002;51 Suppl:S16–S25. [PubMed] [Google Scholar]
- 26.Zhang W, Contag PR, Hardy J, Zhao H, Vreman HJ, Hajdena-Dawson M, Wong RJ, Stevenson DK, Contag CH. Selection of potential therapeutics based on in vivo spatiotemporal transcription patterns of heme oxygenase-1. J Mol Med. 2002;80:655–664. doi: 10.1007/s00109-002-0375-x. [DOI] [PubMed] [Google Scholar]
- 27.Hajdena-Dawson M, Zhang W, Contag PR, Wong RJ, Vreman HJ, Stevenson DK, Contag CH. Effects of metalloporphyrins on heme oxygenase-1 transcription: correlative cell culture assays guide in vivo imaging. Mol Imaging. 2003;2:138–149. doi: 10.1162/15353500200303139. [DOI] [PubMed] [Google Scholar]
- 28.Zenke-Kawasaki Y, Dohi Y, Katoh Y, Ikura T, Ikura M, Asahara T, Tokunaga F, Iwai K, Igarashi K. Heme induces ubiquitination and degradation of the transcription factor Bach1. Mol Cell Biol. 2007;27:6962–6971. doi: 10.1128/MCB.02415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dice JF. Molecular determinants of protein half-lives in eukaryotic cells. Faseb J. 1987;1:349–357. doi: 10.1096/fasebj.1.5.2824267. [DOI] [PubMed] [Google Scholar]
- 30.Hochstrasser M. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr Opin Cell Biol. 1995;7:215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- 31.Ciechanover A. The ubiquitin-proteasome proteolytic pathway. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 32.Finley D, Ciechanover A, Varshavsky A. Ubiquitin as a central cellular regulator. Cell. 2004;116:S29–S32. doi: 10.1016/s0092-8674(03)00971-1. 22 p following S32. [DOI] [PubMed] [Google Scholar]
- 33.Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 34.Hira S, Tomita T, Matsui T, Igarashi K, Ikeda-Saito M. Bach1, a heme-dependent transcription factor, reveals presence of multiple heme binding sites with distinct coordination structure. IUBMB Life. 2007;59:542–551. doi: 10.1080/15216540701225941. [DOI] [PubMed] [Google Scholar]
- 35.Vreman HJ, Ekstrand BC, Stevenson DK. Selection of metalloporphyrin heme oxygenase inhibitors based on potency and photoreactivity. Pediatr Res. 1993;33:195–200. doi: 10.1203/00006450-199302000-00021. [DOI] [PubMed] [Google Scholar]
- 36.Russo SM, Pepe JA, Donohue S, Cable EE, Lambrecht RW, Bonkovsky HL. Tissue distribution of zinc-mesoporphyrin in rats: relationship to inhibition of heme oxygenase. J Pharmacol Exp Ther. 1995;272:766–774. [PubMed] [Google Scholar]
- 37.Drummond GS, Kappas A. Sn-protoporphyrin inhibition of fetal and neonatal brain heme oxygenase. Transplacental passage of the metalloporphyrin and prenatal suppression of hyperbilirubinemia in the newborn animal. J Clin Invest. 1986;77:971–976. doi: 10.1172/JCI112398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russo SM, Pepe JA, Cable EE, Lambrecht RW, Bonkovsky HL. Repression of ALA synthase by heme and zinc-mesoporphyrin in a chick embryo liver cell culture model of acute porphyria. Eur J Clin Invest. 1994;24:406–415. doi: 10.1111/j.1365-2362.1994.tb02184.x. [DOI] [PubMed] [Google Scholar]
- 39.Dover SB, Moore MR, Fitzsimmons EJ, Graham A, McColl KE. Tin protoporphyrin prolongs the biochemical remission produced by heme arginate in acute hepatic porphyria. Gastroenterology. 1993;105:500–506. doi: 10.1016/0016-5085(93)90726-s. [DOI] [PubMed] [Google Scholar]
- 40.Juturu V, Komorowski JR. Chromium compounds: cytotoxicity and carcinogenesis. Toxicology. 2003;186:171–173. doi: 10.1016/s0300-483x(02)00707-2. author reply 175–177. [DOI] [PubMed] [Google Scholar]
- 41.Dover SB, Graham A, Fitzsimons E, Moore MR, McColl KE. Haem-arginate plus tin-protoporphyrin for acute hepatic porphyria. Lancet. 1991;338:263. doi: 10.1016/0140-6736(91)90411-h. [DOI] [PubMed] [Google Scholar]
- 42.Lutton JD, Abraham NG, Drummond GS, Levere RD, Kappas A. Zinc porphyrins: potent inhibitors of hematopoieses in animal and human bone marrow. Proc Natl Acad Sci U S A. 1997;94:1432–1436. doi: 10.1073/pnas.94.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lutton JD, Jiang S, Drummond GS, Abraham NG, Kappas A. Comparative pharmacology of zinc mesoporphyrin and tin mesoporphyrin: toxic actions of zinc mesoporphyrin on hematopoiesis and progenitor cell mobilization. Pharmacology. 1999;58:44–50. doi: 10.1159/000028267. [DOI] [PubMed] [Google Scholar]
- 44.Sardana MK, Kappas A. Dual control mechanism for heme oxygenase: tin(IV)-protoporphyrin potently inhibits enzyme activity while markedly increasing content of enzyme protein in liver. Proc Natl Acad Sci U S A. 1987;84:2464–2468. doi: 10.1073/pnas.84.8.2464. [DOI] [PMC free article] [PubMed] [Google Scholar]


