Abstract
Entecavir (ETV) was developed for the treatment of chronic hepatitis B virus (HBV) infection and is globally approved for that indication. Initial preclinical studies indicated that ETV had no significant activity against human immunodeficiency virus type 1 (HIV-1) in cultured cell lines at physiologically relevant ETV concentrations, using traditional anti-HIV assays. In response to recent clinical observations of anti-HIV activity of ETV in HIV/HBV-coinfected patients not receiving highly active antiretroviral therapy (HAART), additional investigative studies were conducted to expand upon earlier results. An extended panel of HIV-1 laboratory and clinical strains and cell types was tested against ETV, along with a comparison of assay methodologies and resistance profiling. These latest studies confirmed that ETV has only weak activity against HIV, using established assay systems. However, a >100-fold enhancement of antiviral activity (equivalent to the antiviral activity of lamivudine) could be obtained when assay conditions were modified to reduce the initial viral challenge. Also, the selection of a M184I virus variant during the passage of HIV-1 at high concentrations of ETV confirmed that ETV can exert inhibitory pressure on the virus. These findings may have a significant impact on how future assays are performed with compounds to be used in patients infected with HIV. These results support the recommendation that ETV therapy should be administered in concert with HAART for HIV/HBV-coinfected patients.
Patients coinfected with human immunodeficiency virus (HIV) and hepatitis B virus (HBV) are a unique and challenging treatment population, with increased morbidity and mortality due to the concurrent infections (19). Numerous nucleoside/nucleotide reverse transcriptase inhibitors (NRTIs) have been used as part of a highly active antiretroviral therapy (HAART) to successfully treat patients infected with HIV (2). The effectiveness of the combination therapy results in the creation of a high genetic barrier to resistance, requiring resistance substitutions to emerge against each of the individual therapies within the combination, under conditions of significant viral suppression. However, the selection of these viral resistance substitutions often causes complete or partial cross-resistance to other agents in the same drug class, increasing the importance of treatment combinations and sequences of therapies (22).
NRTIs are also used to treat chronic HBV infections. Lamivudine (LVD), adefovir, entecavir (ETV), and telbivudine are approved for anti-HBV therapy (16), and emtricitabine (FTC), tenofovir, and clevudine (2′-fluoro-5-methyl-β-l-arabinofuranosyluracil [l-FMAU]) are in late clinical development. Activity against both HIV and HBV has been noted for several NRTIs, including LVD, adefovir, FTC and tenofovir (13). ETV has previously been reported to exhibit highly potent and specific inhibition of HBV, with a 50% effective concentration (EC50) of 4 nM. In contrast, initial assays against HIV-1 yielded an EC50 of >10 μM, with a 50% cytotoxic concentration of 21 μM (10). The combination of weak in vitro activity and a maximum serum concentration (Cmax) of the drug in human plasma of 30 nM following the administration of the 1-mg dose of ETV (23) led to the conclusion that ETV did not exhibit clinically relevant activity against HIV-1. However, a recent report suggested that ETV may exhibit clinically relevant activity against HIV in some patients (15). Three patients coinfected with HIV and HBV showed a reduction of approximately 1 log10 in their circulating HIV-1 RNA levels, in addition to the reductions expected in their HBV levels of several log units while receiving ETV monotherapy. One of the three patients, who had received prior HAART that included LVD, showed an enrichment of an LVD-resistant (M184V) HIV variant over a 6-month treatment period with 1 mg of ETV. The report also noted that in vitro studies using a single-cycle, single-cell reporter assay showed a subnanomolar level of inhibition by ETV of a pseudotype virus carrying the reverse transcriptase (RT) gene from HIV laboratory and patient isolate strains. ETV was also shown to exhibit reduced susceptibility against an HIV-1 variant containing the M184V substitution. In order to gain greater insight into this observation and better understand the basis for the HIV activity in coinfected patients, we performed additional studies to reexamine the in vitro anti-HIV profile of ETV. The results confirm that ETV should not be used for HBV/HIV-coinfected patients without concomitant HAART.
MATERIALS AND METHODS
Cells.
MT-2, HEK-293, and 293T cells were obtained from the NIH AIDS Research and Reference Reagent Program. MOLT-4/IIIB cells, human T cells chronically infected with the HIV-1 IIIB virus, were a generous gift from Frederic Bushman. Both the MT-2 and the MOLT-4/IIIB cell lines were propagated in RPMI 1640 media supplemented with 10% heat-inactivated fetal bovine serum (FBS), 10 mM HEPES buffer (pH 7.55), 2 mM l-glutamine, 100 U/ml of penicillin, and 100 μg/ml of streptomycin, and subcultured twice a week. The HeLa-CD4-CXCR4 cell line was isolated at Bristol-Myers Squibb. The HeLa-CD4-CXCR4 and 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% heat-inactivated FBS and supplemented as above. Peripheral blood mononuclear cells (PBMCs) were isolated from the blood of HIV-seronegative individuals. Blood treated with sodium citrate to prevent clotting was fractionated by layering over a Ficoll (Histopaque 1077; Sigma) cushion in 50-ml tubes and by spinning in a Beckman GS6K centrifuge at 1,480 rpm for 45 min. The PBMCs at the interface were then washed three times with phosphate-buffered saline and resuspended at 2 × 106 cells/ml in RPMI 1640 medium or DMEM supplemented with 20% FBS and stimulated for 2 to 4 days with 4 μg/ml phytohemagglutinin (PHA; Sigma) and 10 U/ml interleukin 2 (IL-2; Invitrogen) prior to infection.
Viruses.
The HIV-1 B subtype viruses RF, HXB2, SF2, LAI, and IIIB and the proviral DNA clone of NL4-3 were obtained from the NIH AIDS Research and Reference Reagent Program. The NL4-3 M184V virus was constructed at Bristol-Myers Squibb. Titration of viral stocks was performed in the cell lines in which the viruses were originally amplified, using a virus infectivity assay (11), with supernatant RT activity or cytopathic effect (CPE) as endpoints. A recombinant NL-Rluc virus, in which a section of the nef gene from NL4-3 was replaced with the Renilla luciferase gene, was constructed at Bristol-Myers Squibb. NL-RLuc virus was prepared by the cotransfection of two plasmids, pNL-RLuc, containing NL-RLuc DNA, and pVSVenv, expressing the vesicular stomatitis virus (VSV) G envelope protein. Transfection of 293T cells was performed at a 1:3 ratio of pNL-RLuc to pVSVenv, using a Lipofectamine PLUS kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instruction, and the pseudotype virus generated was titrated in MT-2 cells. A pseudotype virus, HIV-1LAI Δenv-luc (LAI-Luc), was prepared at Bristol-Myers Squibb by cotransfecting HEK-293T cells with both a plasmid containing proviral DNA of HIV-1LAI Δenv-luc (firefly luciferase) and a plasmid expressing LAI-env, driven by the HIV long terminal repeat. This pseudotype virus was titrated in HeLa-CD4-CXCR4 cells, using a luciferase reporter gene assay (high sensitivity; Roche) as the endpoint.
Clinical isolates were obtained from the NIH AIDS Research and Reference Reagent Program and were grown in human PBMCs prepared as described above. Virus yields were determined for clinical isolates at 5 to 7 days postinfection by using α p24 enzyme-linked immunosorbent assay (PerkinElmer Life Sciences) per the manufacturer's instructions. The multiplicity of infection (MOI) was determined by the 50% tissue culture infectious dose method of endpoint dilution (6) with a p24 readout. The 50% tissue culture infectious dose of each viral stock was calculated by the Spearman-Karber statistical method, as described previously (6).
Reagents.
ETV and efavirenz (EFV) were synthesized at Bristol-Myers Squibb. LVD was extracted from the commercial formulation of the drug and purified at Bristol-Myers Squibb. Zidovudine (AZT) was purchased from Sigma.
Drug susceptibility and cytotoxicity assays.
The susceptibility of viruses to compounds was determined by incubation in the presence of serial dilutions of the compound. The EC50 value was calculated by using the one-ligand-binding-site model with XLFit software for Microsoft Excel, where the percentage of inhibition = 1/[1 + (EC50/drug concentration)m], and m is a parameter that reflects the slope of the concentration response curve. Cytotoxicity assays were performed in parallel for all experiments by exposing uninfected cells to serially diluted compounds and assaying them after 3 to 6 days for cell viability, using MTS [3-(4,5-dimethylthiazol-2yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt] (5) or XTT {2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-5[(phenylamino)carbonyl]-2H-tetrazolium hydroxide} (10) assays. The 50% cytotoxic concentration values were calculated using the same equation as described above for EC50 estimation.
Drug susceptibility assays using laboratory strains.
Drug susceptibility assays for laboratory isolates were carried out using two slightly different protocols. The chief differences are the volumes in which cells were initially infected with virus and the lengths of time cells were preincubated with virus prior to the addition of antivirals, with protocol 1 being 1 h (+1 h) and protocol 2 generally being 2 to 3 h (+2 to +3 h) with a smaller (more concentrated) virus inoculum. For ease of understanding, both protocols are listed below, and the protocol used in each experiment is also listed in the text.
Protocol 1.
Antiviral assays using various laboratory strains were conducted with MT-2 cells in RPMI 1640 medium supplemented with 10% heat-inactivated FBS, 10 mM HEPES buffer (pH 7.55), 2 mM l-glutamine, 100 units/ml penicillin G, and 100 μg/ml streptomycin. Cells diluted to 2.5 × 105/ml were infected with virus (MOI range, 0.003 to 0.014) for 1 h before the addition of an equal volume (100 μl) of compounds diluted in medium with 2% dimethyl sulfoxide. The extent of virus replication was determined 5 days postinfection by using an RT assay performed in a 96-well scintillation proximity assay (SPA) format. In the RT SPA, 20 μl of the culture supernatant was added to 40 μl of a reaction cocktail [62.5 mM Tris-HCl [pH 7.8], 100 mM KCl, 0.0625% NP-40, 2.5 mM dithiothreitol, 6.25 mM MgCl2, 1 mM EGTA, 6.25 μg/ml poly(rA), 2.5 μg/ml streptavidin SPA beads prebound with biotinylated-dT12-18, and 1 μCi [3H]thymidine triphosphate]. The reaction mixtures were incubated at 37°C for 1.5 h and quenched with 100 μl of 0.25 M EDTA per reaction. The SPA beads were allowed to settle for 2 h prior to counting with a Wallac Trilux, programmed for tritium counting with SPA beads in opaque plates.
Protocol 2.
This protocol used a longer preincubation period for virus and cells and a smaller volume than protocol 1. MT-2 cells were centrifuged at 1,200 rpm in a Beckman Allegra centrifuge for 10 min, and virus was added to the cell pellets in a volume of 0.5 ml. A MOI ranging from 0.005 to 0.05 with NL4-3 or NL4-3 M184V virus was used in these experiments. If a preincubation step was not used, the cell-virus mixtures were directly resuspended in culture medium and seeded into 96-well microtiter plates (final density, 1.0 × 105 cells/ml) containing serial dilutions of test compounds. In experiments designed to determine the effect of preincubation on antiviral activity, cell-virus mixtures were first incubated in a small volume (0.5 ml) for 2 to 3 h (+2, +3 h) and then diluted before the addition to plates containing drugs. Cultures were incubated at 37°C in 5% CO2 until CPEs were apparent (5 days), at which time virus yields were quantitated using 20 μl of supernatants in the RT assay, and the remaining suspensions of infected cells were used in a MTS cell viability assay.
Drug susceptibility assessment using clinical isolates.
Antiviral assays for clinical isolates were conducted with stimulated PBMCs. Cells were sedimented for 5 min at 1,200 rpm in a Beckman Allegra centrifuge and resuspended in 1 ml of RPMI 1640 medium supplemented as described above and containing 4 μg/ml PHA and 10 U/ml IL-2. An appropriate volume of virus was added and incubated at 37°C in 5% CO2 for 1.5 h. Infected cells were diluted to 1 × 106 cells/ml with medium supplemented with 2× PHA and 2× IL-2. The diluted cells (100 μl) were immediately added to compounds (100 μl) diluted in medium (plus 2% dimethyl sulfoxide) without PHA and IL-2. Virus yields were determined at 5 days postinfection, using a p24 enzyme-linked immunosorbent assay kit as described by the manufacturer (PerkinElmer Life Sciences). Typically, a 1:1,000 dilution of cell supernatant in medium was used in order to be in the dynamic range of the p24 assay.
Drug susceptibility assessment using pseudotype reporter viruses.
A single-cycle viral infection system was also utilized to determine drug susceptibility (11). For the recombinant NL-RLuc virus expressing the Renilla luciferase protein and pseudotyped with the VSV G protein, antiviral activity was evaluated by measuring the production of luciferase in infected cells at 5 days postinfection, using a Dual luciferase kit (Promega, Madison, WI) with a minor modification. Diluted passive lysis solution was premixed with both the resuspended luciferase assay substrate and the resuspended Stop & Glo substrate (2:1:1 ratio). Fifty microliters of the mixture was added to each aspirated cell monolayer of the assay plates, and luciferase activity was measured immediately on a Wallac TriLux (PerkinElmer Life Sciences). The HIVLAI Δenv-luc virus, pseudotyped with an LAI envelope, was also used to infect HeLa-CD4-CXCR4 and MT-2 cells and PBMCs containing serially diluted compounds. Firefly luciferase activity was monitored at 3 days postinfection, according to the manufacturer's specifications (Luciferase reporter gene assay, high sensitivity; Roche).
Monogram Biosciences pseudovirion (PhenoSense) assay.
An ETV stock solution was diluted in DMEM plus 10% FBS (17). Threefold dilutions were used in the range from 600 to 0.03 μM. HEK-293 cells were cotransfected in 10-cm dishes with an HIV-1 genomic vector that contains a luciferase indicator gene cassette in the envelope region and a plasmid that expresses the amphotropic murine leukemia virus envelope. For drug evaluation, two HIV-1 expression plasmids were used, NL4-3 wild type (a drug-sensitive virus) and NL4-3 M184V (an LVD- and FTC-resistant virus). Viruses were harvested from the transfected HEK-293 cells and were used to infect naïve HEK-293 cells. ETV was added to the HEK-293 cells at 24 h prior to viral infection of naïve cells. The ability of the pseudovirions to infect target cells was monitored by the production of luciferase in the infected cells. Just prior to the luciferase readout, cells were evaluated by visual inspection for evidence of toxicity (cell death) or morphological changes (cell growth affected) due to the presence of drug. From an analysis of the measured luciferase signal, the drug concentration required to inhibit viral infection by 50% was calculated for each virus tested. Data points that had visible cell death were omitted in calculating EC50 values.
Analysis of activity in chronically infected cells.
MOLT-4/IIIB cells were extensively washed to remove extracellular virus before the compounds were added. Cells (30 ml) were sedimented for 5 min at 1,200 rpm in a Beckman Allegra centrifuge, decanted, and resuspended in 30 ml of RPMI 1640 medium supplemented as described above. This pelleting/washing process was repeated three times. The final cell pellet was resuspended to 2 × 105 cells/ml and was added to an equal volume (0.1 ml) of medium containing compound dilutions. The culture supernatant was assayed for RT activity after 4 days.
Selection of resistant variants.
MT-2 cells at a density of 2 × 105 cells/ml were infected with the NL4-3 strain at an MOI of 0.005. At the time of virus exposure, 0.1 μM or 0.4 μM ETV (∼two- or sevenfold the EC50 value) was added to the cultures. As a control, LVD resistance was also selected with a starting concentration of 0.2 μM (fivefold the EC50 value). The cultures were incubated until a CPE was observed, at which time the culture supernatant was harvested. Twenty-five microliters of this supernatant was used to infect a fresh culture of MT-2 cells, to which double the initial concentration of compound was added. Cultures were again incubated until a CPE was observed, at which time the entire process was repeated, with drug concentrations doubling at each dose. Consequently, at passage eight, the concentration of ETV was 12.8 μM or 51.2 μM. For selected passages, the NL4-3 RNA was extracted from the culture supernatants, using a viral RNA extraction kit (Qiagen). The viral RNA served as a template for reverse transcription-PCR (First-Strand Synthesis and Platinum high-fidelity DNA polymerase kits; Invitrogen) using primers that annealed to HIV-1 gag and vpr. The resulting DNA products were subjected to fluorescent dye-terminator nucleotide sequencing (ABI) using HIV-specific primers, and the resulting data were analyzed using DNASTAR version 6 software (Lasergene).
The ability of LVD and ETV to selectively enrich for the M184V HIV-1 virus from a mixture of M184V/wild-type viruses was also examined. The two NL4-3 viruses were mixed at a ratio of 1/100 (M184V/wild type) and used to infect 105 MT-2 cells/ml at a total MOI of 0.005 (with no preincubation step). Parallel cultures were then incubated in the presence of 2.5 and 10 μM ETV (45-fold and 179-fold the EC50 values) or 2.5 μM LVD (63-fold the EC50 value). After a 6-day incubation (when CPE was evident), supernatants were harvested, viral RNA was isolated, and sequence analyses were performed as described above.
RESULTS
Historical data for ETV activity against HIV-1.
Prior to 2007, the inhibitory activity of ETV against laboratory HIV-1 strains RF, BRU (LAI), and NL4-3 was examined and characterized, using various cell types in studies that used a 1- to 3-h preincubation of virus/cells prior to the addition of ETV (see Materials and Methods, protocols 1 and 2). The determination of virus inhibition was performed using either a cell survival endpoint (XTT or MTS) or by titration of RT activity in cell supernatants. The results of these various assays, conducted over a 10-year period, consistently indicated only weak activity, in the μM range, and in many instances, activity overlapped with cytotoxicity levels (10). In those assays where some degree of HIV inhibitory activity was observed with ETV, the levels required were orders of magnitude higher than that found in the plasma of healthy subjects treated with ETV (23).
Assessment of ETV activity using pseudotype HIV-1.
Routine screening for inhibitory activity and resistance has been improved through the use of pseudotype viruses (17). These recombinant viruses may contain the G glycoprotein from VSV within their envelope (the LAI pseudotype used here was pseudotyped with the LAI envelope protein). The G glycoprotein improves the stability and infectivity of the virus population, enabling greater ease of use and sensitivity of the assay. In addition, the pseudotype virus may contain a reporter gene, such as luciferase, the expression of which can be readily measured and quantitated. ETV activity was assayed with two such pseudotype virus assays derived from either the HIV-1 NL4-3 or LAI viral backbones. The NL-RLuc virus contains the Renilla luciferase gene, while the LAI-Luc virus contains the firefly luciferase gene, both in lieu of the nef gene. In addition, ETV was also tested with a PhenoSense assay (Monogram Biosciences) to determine the activity of ETV against that of their NL4-3 pseudotype virus stock, relative to other antiretrovirals. The results show EC50 values for ETV in the μM range regardless of the pseudotype strain, cell type, or assay used (Table 1). Examination of the NL4-3 pseudotypes against LVD, AZT, and EFV produced EC50 values within the range of expected values.
TABLE 1.
Activity of ETV against pseudotype HIV-1a
| Strain | Test site | Host cell | Inhibitor | EC50 (μΜ) ± SD |
|---|---|---|---|---|
| LAI-Luc | BMS | HeLa-CD4 | ETV | >4 |
| LAI-Luc | BMS | MT-2 | ETV | 1.2->10 |
| LAI-Luc | BMS | PBMC | ETV | >2.5 |
| NL-RLuc | BMS | MT-2 | ETV | 1.30 ± 0.02 |
| NL-Luc | Monogram | HEK-293 | ETV | 30.6b |
| NL-RLuc | BMS | MT-2 | LVD | 1.14 ± 0.95 |
| NL-RLuc | BMS | MT-2 | AZT | 0.016 ± 0.012 |
| NL-RLuc | BMS | MT-2 | EFV | 0.00034 ± 0.00000 |
Results are the averages of two or more independent experiments performed in duplicate ± standard deviations (SD).
SD not available.
Assessment of ETV activity using multicycle assay systems.
A panel of six HIV-1 subtype B laboratory viruses was assayed to evaluate ETV for anti-HIV activity, using cell culture protocol 1 and the RT readout in all cases. The MOI was determined for all experiments and varied over the range of 0.003 to 0.014. Results are summarized in Table 2 and demonstrate a wider-than-expected range of activity against this panel of laboratory viruses. ETV exhibited antiviral activity against HIV strains HXB2 (EC50, 1.030 μM), RF (EC50, 0.839 μM), and SF2 (EC50, 0.526 μM), with somewhat greater activity against the NL4-3 (EC50, 0.085 μM), IIIB (EC50, 0.081 μM), and LAI (EC50, 0.071 μM) strains. This activity level was comparable to that of LVD but much lower than that of AZT or EFV. It should be noted that the titration curves become somewhat flatter for the viruses showing the greatest susceptibility (discussed further below). The susceptibilities of the laboratory strains to LVD, AZT, and EFV did not vary substantially and were within the commonly accepted ranges of activity reported for these compounds. In addition, the HIV-1 NL4-3 strain was also examined by using cell culture protocol 2, which includes the modification incorporating a +2 h or +3 h virus/cell preincubation step in a small volume, prior to exposure to the compound. The inhibitory activity determined by using this method was substantially reduced in a preincubation time-dependent manner compared to the results obtained when virus infection preceded drug exposure by +1 h (Table 2).
TABLE 2.
Activity levels of NRTIs against multicycle HIV-1 replication
| Virus | EC50 value (μM) ± SD ofa:
|
Protocolb | |||
|---|---|---|---|---|---|
| ETV | LVD | AZT | EFV | ||
| HXB2 | 1.030 ± 0.356 | 0.202 ± 0.120 | 0.014 ± 0.006 | 0.0003 ± 0.0001 | 1 |
| RF | 0.839 ± 0.525 | 0.105 ± 0.084 | 0.0029 ± 0.0021 | 0.0003 ± 0.0001 | 1 |
| SF-2 | 0.526 ± 0.120 | 0.183 ± 0.042 | 0.0063 ± 0.0029 | 0.0004 ± 0.0001 | 1 |
| IIIB | 0.081 ± 0.028 | 0.047 ± 0.030 | 0.0012 ± 0.0007 | 0.0001 ± 0.0001 | 1 |
| LAI | 0.071 ± 0.029 | 0.182 ± 0.149 | 0.0038 ± 0.0021 | 0.0002 ± 0.0001 | 1 |
| NL4-3 | 0.085 ± 0.077 | 0.171 ± 0.025 | 0.0049 ± 0.0026 | 0.0003 ± 0.0001 | 1 |
| NL4-3 | 1.4 | ND | ND | ND | 2c |
| NL4-3 | >10 | ND | ND | ND | 2d |
| 93US143e | 1.753 ± 1.074 | 0.850 ± 0.453 | 0.034 ± 0.020 | 0.0007 ± 0.0006 | 1 |
| ASM 034e | 0.109 ± 0.105 | 0.071 ± 0.024 | 0.018 ± 0.010 | 0.0004 ± 0.0001 | 1 |
| 92US076e | 0.062 ± 0.036 | 0.081 ± 0.017 | 0.008 ± 0.002 | 0.0003 ± 0.0002 | 1 |
| ASM 044e | 0.026 ± 0.026 | 0.040 ± 0.042 | 0.016 ± 0.017 | 0.0005 ± 0.0006 | 1 |
The EC50 values ± standard deviations (SD) reported for the first six viruses are averages of four experiments using protocol 1 with 1-h preincubations of virus and cells. ND, not done.
The EC50 values reported for protocol 2 are averages of two experiments using various virus and cell preincubation times.
Time of drug addition at +2-h virus/cell incubation.
Time of drug addition +3-h virus/cell incubation.
EC50 values for viruses 92US076 and 93US143 are averages of four experiments, and EC50 values for viruses ASM 034 and ASM 044 are averages of three experiments.
ETV was also examined for its activity against four subtype B clinical isolates, with virus yields monitored and determined based on p24 production (Table 2). The MOI of each strain used ranged from 0.003 to 0.016. Again, a range of potencies was observed, with weak inhibitory activity against HIV-1 strain 93US143 and enhanced activity against other strains. HIV-1 strain ASM 044 was the most susceptible of the four isolates tested, with an EC50 value of 0.026 μM, while 93US143 was the least susceptible, with an EC50 value of 1.75 μM. Other NRTIs and EFV were again included as controls. All four clinical isolates exhibited susceptibilities similar to values expected for EFV (EC50 of 0.0003 to 0.0007 μM) and AZT (EC50 of 0.008 to 0.034 μM). For LVD, strain 93US143 exhibited susceptibility that was decreased compared to that of the other three viruses; however, genotyping failed to show evidence of a M184V resistance substitution (data not shown). The relatively potent activity observed against clinical isolates was again associated with flatter titration curves and appears to be related to the lower MOIs utilized for these viruses that do not achieve high titer stocks characteristic of laboratory strains.
Impact of assay conditions on observed potency of ETV.
The results described above suggest that ETV activity against HIV-1 is highly variable and is likely to be dependent on multiple factors. The wide range of susceptibilities to ETV observed with the panel of laboratory strains differed from the narrower range of activities observed with the approved HIV drugs included as controls. Also, only weak inhibitory activity was observed against pseudotype viruses (Table 1), while certain clinical isolates exhibited a high degree of susceptibility to ETV. In an attempt to better understand the underlying variables that contribute to the range of susceptibilities observed, a series of investigational studies was performed that further evaluated the conditions of infection, MOI, and assay readout methodologies.
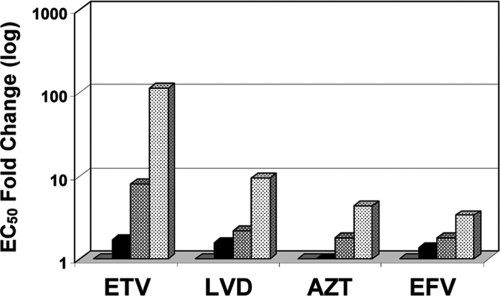
The effects of MOI and preincubation time of cells with virus prior to drug exposure were studied using the laboratory strain NL4-3. At various MOIs, ranging from 0.005 to 0.05, cells were infected in a small volume for 2 h prior to the addition of the drug (+2 h), or the drug was added at the time of virus inoculation (zero h). Parallel experiments were performed with ETV, LVD, AZT, and EFV, and results are summarized in Table 3 and presented in Fig. 1. MOI and the virus/cell preincubation step were key variables, significantly affecting the measured susceptibilities to all four drugs, although the observed effects were more dramatic with ETV than with the other control drugs. Increasing the virus inoculum from an MOI of 0.005 to 0.05 in the absence of a virus/cell preincubation step resulted in an increase in the EC50 values (decreased potency) for EFV, AZT, and LVD of 3.5-, 4.3-, and 9.3-fold, respectively (Fig. 1). For ETV, the EC50 at an MOI of 0.005 was 0.056 μM, rising to 6.45 μM at an MOI of 0.05, a 115-fold increase. The largest increases were observed between MOIs of 0.01 to 0.02, where the ETV EC50 increased ∼4.7-fold, and from 0.02 to 0.05, where the EC50 increased an additional 14.4-fold. The underlying mechanism responsible for this observation is unknown at this time, but clearly, decreased MOI has a greater effect on the potency of ETV than on the other established HIV RT inhibitors examined.
TABLE 3.
Relationship between MOI and virus/cell preincubation conditions and observed potencies
| MOI | Time of drug addition (h) | EC50 value (μM) ± SD ofa:
|
|||
|---|---|---|---|---|---|
| ETV | LVD | AZT | EFV | ||
| 0.005 | 0 | 0.056 ± 0.014 | 0.040 ± 0.002 | 0.010 ± 0.001 | 0.0002 ± 0.00005 |
| 0.01 | 0 | 0.095 ± 0.014 | 0.063 ± 0.041 | 0.008 ± 0.001 | 0.0003 ± 0.00005 |
| 0.02 | 0 | 0.449 ± 0.072 | 0.086 ± 0.009 | 0.018 ± 0.001 | 0.0004 ± 0.0001 |
| 0.05 | 0 | 6.450 ± 0.328 | 0.372 ± 0.078 | 0.043 ± 0.002 | 0.0007 ± 0.0002 |
| 0.005 | +2 | 3.620 ± 0.781 | 0.462 ± 0.180 | 0.051 ± 0.004 | 0.0009 ± 0.000004 |
| 0.01 | +2 | 9.573 ± 0.538 | 0.667 ± 0.011 | 0.075 ± 0.005 | 0.0009 ± 0.00002 |
| 0.02 | +2 | >10.000 | 1.985 ± 0.383 | 0.209 ± 0.029 | 0.0012 ± 0.00001 |
| 0.05 | +2 | >10.000 | 2.470 ± 0.015 | 0.249 ± 0.002 | 0.0030 ± 0.0005 |
Values ± standard deviations (SD) are the averages of two experiments, each performed in duplicate.
FIG. 1.
Drug susceptibility at different MOIs. MT2 cells were infected with the HIV-1 NL4-3 wild-type strain at MOIs of 0.005, 0.01, 0.02, and 0.05 according to protocol 2 (see Materials and Methods), with no virus/cell preincubation step (drug added at time zero). The changes in EC50 values are shown relative to that of the MOI of 0.005 (set at 1.0, gray bars) for MOIs of 0.01 (black bars), 0.02 (hatched bars), and 0.05 (dotted bars). Data are from Table 3, at zero-h conditions.
Further confirmation that the MOI or the amount of virus used to infect cells is an important variable was obtained from studies that examined the impact of the virus/cell preincubation time. Preincubation of virus with concentrated cells in small volumes at 2 h before the addition of drug (+2 h) was compared to conditions under which the cell/virus mixtures were added directly to the assay plates containing serially diluted compounds (zero h). The preincubation step used in protocol 2 is similar to using a higher MOI, since infection in a small volume likely increases the probability and efficiency of viral attachment and entry into cells. When the drug was added at 2 h after infection, EFV consistently exhibited an increase of only three- to fivefold in EC50 at all MOIs tested, while AZT gave a slightly higher, 5- to 12-fold increase (Table 3). LVD gave as much as a 23-fold increase. Interestingly, and consistent with the MOI experiments, ETV exhibited the greatest changes in potency at the 2-h incubation time of virus and cells prior to the ETV addition (+2 h). This is best illustrated at the lower MOIs of 0.005 and 0.01, as the differences between the +2-h and zero-h EC50s are 65- and 101-fold, respectively, compared with 12- and 11-fold, respectively, for LVD (Table 3). Once the MOI is increased to 0.02 or 0.05, the +2-h EC50 could not be determined, as it was above the highest drug concentration used. Together, these data suggest that the inhibitory activity of a weak anti-HIV compound such as ETV is highly sensitive to the virus inoculum used to infect cells.
These data demonstrate that the variables and underlying mechanisms of ETV inhibition of HIV-1 replication likely differ considerably from those of LVD, AZT, and EFV. Unlike these other drugs, ETV exhibits significant antiviral activity only under conditions of reduced viral challenge. Enhancing the infection by increasing the virus inoculum or by preincubating the cells with virus during the first cycle of a multicycle infection (+2 h) greatly reduces ETV potency. The +2-h virus/cell incubation step could also enable virus replication to proceed prior to the phosphorylation of ETV to the active triphosphate (ETV-TP), although the preincubation of cells with ETV prior to infection did not appear to substantially alter the EC50s of ETV (data not shown).
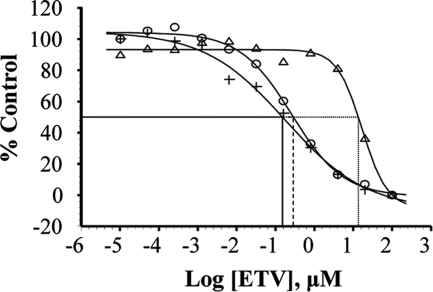
The conditions of infection also affect the shape of inhibition curves for ETV. Lowering the virus challenge appears to be associated with decreased EC50 values and flatter titration curves (Fig. 2). To enable a direct comparison of ETV potency levels under various assay conditions, the experiments described in Table 3, using the concentrated viral inoculum of protocol 2, with and without the 2-h virus/cell (zero h and +2 h), were repeated along with an arm utilizing a more diluted inoculum for protocol 1 with a 1-h preincubation step (+1 h). The ability of ETV to inhibit NL4-3 growth was examined at an MOI of 0.006, using RT as an endpoint. Figure 2 shows the inhibition curves for the three conditions tested in parallel. The zero-h sample produced the lowest EC50 of 0.151 μM (compared to 0.056 μM at an MOI of 0.005, as shown in Table 3 and Fig. 1), while the EC50 using the +1-h virus/cell incubation was 0.287 μM. As noted in Table 3, preincubation with cell pellets for a longer time (+2 h) resulted in much lower susceptibility to ETV inhibition, this time producing an EC50 of 13.5 μM. The shapes of the inhibition curves also differed between the conditions. The zero-h inhibition curve was much flatter than the +2-h curve (both protocol 2), while the shape of the +1-h curve with diluted virus (protocol 1) was somewhat intermediate (Fig. 2). The flat-shaped inhibition curves may indicate a plateau or threshold of inhibition.
FIG. 2.
Effect of virus preincubation on inhibition curves for ETV under different infection conditions. Inhibition curves for ETV inhibition of HIV NL4-3 in MT-2 cells were determined simultaneously with no preincubation of virus and cells (+), 1-h virus incubation with diluted cells (○), or 2-h virus incubation with concentrated cells (▵). MOI was 0.006 for each titration. Each point of the curves represents the mean value of triplicate RT activity. The percentage of control was calculated from the maximum RT activity under each condition.
Effect of assay endpoint on ETV potency.
An analysis of the earlier assay data suggested that increased ETV potency was observed when anti-HIV-1 activity was determined by using RT activity as a measure of virus replication. Other assays that measure the survival of cells, through the use of either XTT or MTS, did not exhibit any meaningful anti-HIV-1 activity for ETV (10). However, these experiments were all performed independently, so direct comparisons between endpoints of virus yield cannot be made. Therefore, additional studies were performed that compared the readout endpoint of RT activity to cell survival via an MTS assay on the same samples after a 5-day infection. The EC50 values using the RT assay are shown in Table 3, while Table 4 shows the EC50 values obtained for the same samples using the MTS assay and the changes between the two assays. Ratios were calculated only for those samples that gave measurable EC50 values with both assay methodologies. The most potent drugs, AZT and EFV, displayed little difference in EC50 values between RT and MTS endpoints, while LVD yielded a somewhat greater range of values, from as low as 1.2-fold to as high as 11.3-fold. The differences observed with ETV were more dramatic, although ratios could be calculated only for two sets of conditions (19- and >31-fold), as EC50 values under other conditions were above the highest ETV concentration tested. Thus, these studies suggest that only a more sensitive assay endpoint could reveal the activity of a weak HIV inhibitor such as ETV.
TABLE 4.
Effect of assay endpoints on RTI potency using protocol 2
| MOI | Time of drug addition (h) | EC50 value (μM) ± SD ofa:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| ETV
|
LVD
|
AZT
|
EFV
|
||||||
| MTS | FC | MTS | FC | MTS | FC | MTS | FC | ||
| 0.005 | 0 | 1.364 ± 1.196 | 19 | 0.219 ± 0.108 | 5.5 | 0.027 ± 0.014 | 2.7 | 0.0004 ± 0.0001 | 2.0 |
| 0.01 | 0 | >2.901 | >31 | 0.554 ± 0.079 | 8.8 | 0.038 ± 0.009 | 4.8 | 0.0007 ± 0.0001 | 2.3 |
| 0.02 | 0 | >6.348 | 0.969 ± 0.003 | 11.3 | 0.054 ± 0.008 | 3.0 | 0.0008 ± 0.0002 | 2.0 | |
| 0.05 | 0 | >10 | 0.927 ± 0.046 | 2.5 | 0.087 ± 0.003 | 2.0 | 0.0010 ± 0.00008 | 1.4 | |
| 0.005 | +2 | >10 | 1.259 ± 0.046 | 2.7 | 0.117 ± 0.034 | 2.3 | 0.0011 ± 0.00001 | 1.2 | |
| 0.01 | +2 | >9.693 | 1.683 ± 0.205 | 2.5 | 0.144 ± 0.002 | 1.9 | 0.0012 ± 0.0001 | 1.3 | |
| 0.02 | +2 | >10 | 2.445 ± 0.332 | 1.2 | 0.223 ± 0.008 | 1.1 | 0.0013 ± 0.00008 | 1.1 | |
| 0.05 | +2 | >10 | 3.247 ± 0.277 | 1.3 | 0.302 ± 0.068 | 1.2 | 0.0017 ± 0.0002 | 0.5 | |
Values shown are the averages ± standard deviations (SD) of two experiments performed in duplicate. FC, fold change of MTS assay results relative to those of the RT assay of the sister sample (data from Table 3).
Lack of activity of ETV in chronically infected cells.
In order to rule out the possibility that the variability and MOI-dependent activity observed with ETV was due to an inhibitory mechanism unrelated to the inhibition of the viral RT, ETV was assayed with chronically infected cells, in which HIV yields are dependent only on the replication steps beyond RT activity. MOLT-4/IIIB cells have HIV-1 IIIB stably integrated into the chromosome and continually secrete virus. As a result, only drugs that affect postintegration steps should be able to inhibit virus production. However, since the cell is responsible for the transcription and translation of viral proteins, cytotoxic compounds also should appear to be active in this model. ETV, LVD, AZT, and the protease inhibitor nelfinavir were examined for activity levels against the production of HIV-1 in these chronically infected MOLT-4/IIIB cells. Supernatant was collected after 4 days of infection in the presence of various concentrations of drug, and the amount of mature virus released was measured by using the RT assay. As expected for a protease inhibitor, nelfinavir was active and able to inhibit mature virus release, with an EC50 of 0.029 μM. In contrast, the EC50s for ETV, LVD, and AZT were >38 μM, >100 μM, and >10 μM, respectively, indicating that the observed ETV activity in full cycle assays is not the result of inhibition of postintegration events or general cytotoxicity.
Activity of ETV against HIV-1 containing the M184V LVD resistance substitution.
Extensive studies and analyses of clinical samples from HBV-infected patients treated with ETV have demonstrated that HBV has decreased susceptibility to ETV when the M204V substitution is present and that clinical resistance requires the presence of M204V in HBV with additional ETV-specific substitutions (4, 18). The M204V substitution in the HBV RT is analogous to the M184V substitution in the HIV-RT, as they are both part of the YMDD motif (7). The recent case of an HIV/HBV-coinfected patient who exhibited an enrichment of HIV variants containing the M184V substitution while receiving ETV monotherapy (15) was of particular interest because of the overlapping resistance profiles of ETV and LVD when the equivalent M204V substitution was present in HBV (12). Therefore, the activity levels of ETV, LVD, AZT, and EFV against paired NL4-3 viruses with or without an M184V substitution were compared. Infection was performed at a low MOI (0.005) but without a virus/cell preincubation step in order to maximize ETV antiviral activity. The results, shown in Table 5, indicate that the presence of the M184V substitution reduced the virus' susceptibility to both ETV and LVD by >178-fold and >250-fold, respectively, while having no significant impact on AZT and EFV susceptibilities. A similar result was obtained with the PhenoSense assay performed at Monogram Biosciences, although the dynamic range of the assay was limited by the weak activity exhibited by ETV in the pseudotype assay. These data provide clear evidence that ETV can have antiviral activity against HIV-1 through the inhibition of HIV-1 RT and that the virus can escape via the LVD resistance pathway.
TABLE 5.
Anti-HIV activity of NRTIs on HIV-1 strains containing the M184V substitutiona
| NL4-3 virus | ETV
|
LVD
|
AZT
|
EFV
|
||||
|---|---|---|---|---|---|---|---|---|
| EC50 (μM) | FC | EC50 (μM) | FC | EC50 (μM) | FC | EC50 (μM) | FC | |
| Wild type | 0.056 | NA | 0.040 | NA | 0.010 | NA | 0.0002 | NA |
| M184V mutant | >10 | >178 | >10 | >250 | 0.005 | 0.5 | 0.0002 | 1.0 |
| Monogram wild type | 30.6 | NA | ND | ND | ND | ND | ND | ND |
| Monogram M184V mutant | >100 | >3.2 | ND | ND | ND | ND | ND | ND |
Values shown are the averages of two experiments performed in duplicate. FC, fold change (mutant versus wild type). NA, not applicable; ND, not done.
Selection/enrichment of drug-resistant HIV variants.
The observed activity of ETV against HIV-1 under conditions of reduced virus challenge and the decreased susceptibility of the M184V variant suggested that it may be possible to select for resistance in vitro. Therefore, HIV-1 NL4-3 was passaged in cell culture under conditions of low MOI and increased concentrations of ETV at each passage, starting at 0.1 μM or 0.4 μM (∼2- or 8-fold increase in EC50 value), or LVD, starting at 0.2 μM (5-fold increase in EC50 value). At each subsequent passage, the concentration of the drug was doubled. Population sequencing of the HIV RT gene was performed at selected passages to monitor for sequence changes. For the selection starting at 0.1 μM ETV, virus passaged for seven cycles (30 days) in the presence of ETV (final concentration, 6.4 μM) contained a G-to-A mutation at nucleotide 552 of the RT gene (which encodes the M184I substitution), representing ∼20% of the total population. The prevalence of this mutation increased to ∼50% by passage eight (34 days; final concentration, 12.8 μM). For the selection starting at 0.4 μM ETV, this nucleotide change appeared at passage six (27 days; final concentration, 12.8 μM) in ∼50% of the virus population. Consequently, this mutation progressed into ∼80 to 90% of the virus population at passage seven (30 days; final concentration, 25.6 μM) and 100% of the virus population at passage eight (34 days; final concentration, 51.2 μM). For virus passaged with LVD, this same nucleotide change was observed at passage six (34 days; final concentration, 3.2 μM) in ∼30% of the virus population. This mutation increased to 100% of the virus population at passage seven (39 days; final concentration, 6.4 μM). During the selection of LVD resistance in patients, the first substitution usually detected is M184I, which under continued selection is reported to progress to the M184V change (9, 21). Susceptibility assays were performed with the starting NL4-3 wild-type strain and passage eight (34 days) virus, at which time the ETV final concentration was 51.2 μM and the M184I substitution was detected in 100% of the virus population. The M184I substitution induced a change in EC50 value from 0.75 μM to >10 μM (average of four assays). While the M184V substitution was not detected in this selection experiment, it is evident that the M184I substitution alone provides sufficient escape from drug pressure so that further evolution to M184V is likely to be unnecessary in vitro (3). Nonetheless, this result further confirms that ETV can inhibit HIV RT and that at very high concentrations, it has the potential to select for resistance in a manner similar to that of LVD.
An additional study was conducted to determine if ETV could cause an enrichment of the M184V variant in a mixed population. A parallel study using LVD as the selective agent served as a control. In this experiment, the M184V variant and wild-type HIV-1 NL4-3 were mixed at a ratio of 1:100 (total MOI, 0.005) in the presence of 2.5 or 10 μM ETV (45-fold and 179-fold EC50, respectively) or 2.5 μM LVD (63-fold increase in the EC50) and incubated for 6 days, by which time CPE was observed. Sequence analyses of the harvested supernatants revealed that the treatment with the 2.5 μM dose of LVD resulted in an approximately 60-fold increase in the prevalence of the M184V variant in the population. Surprisingly, detectable levels of M184V were not observed for either of the ETV arms by population sequencing for reasons which remain unexplained at this time.
DISCUSSION
This report summarizes a comprehensive evaluation of the in vitro activity of ETV against HIV-1 strains. Previously unpublished studies conducted over the past decade have consistently yielded only very weak (EC50 >1 μM) activity against HIV-1 laboratory strains, using a variety of cell lines (10). These studies also included combination studies with known NRTIs in which ETV was added at concentrations approximately fourfold higher than the Cmax value observed with a patient's blood following the administration of the approved 1-mg dose (23). These studies again failed to show any significant activity against HIV, since the EC50 values of all the HIV NRTIs were unchanged when ETV was added.
These previously conducted studies conflict with a recent report by McMahon et al. (15), which indicated that ETV has potent activity (EC50, 0.1 nM) against HIV in a unique single-cycle, single-cell-based pseudovirus assay (24) with CD4+ lymphocytes using a green fluorescent protein reporter fluorescence-activated cell sorter assay as the endpoint. In their report, they also described that ETV monotherapy for HIV/HBV-coinfected patients resulted in a decrease in HIV levels of approximately 1 log10 in three patients and that enrichment of the M184V LVD resistance substitution occurred in one of these patients who had received prior HAART that included LVD. Our repeated assays utilizing both multicycle whole virus and pseudovirus single-cycle conventional methodologies failed to confirm such potent activity against HIV. These same assays and methodologies have accurately predicted the clinical activity of previously approved antiretroviral agents. Investigational studies into possible explanations for these differences identified MOI (the concentration of virus used to infect each cell) as a primary variable that could help explain the differences observed. While all of the HIV NRTIs show enhanced antiviral activity when lower MOIs are utilized, ETV appears to be particularly sensitive to this variable, giving a >100-fold increase in potency when the MOI is lowered by 10-fold, for reasons which remain unexplained (Table 3 and Fig. 1). The cell culture assay employed in the McMahon et al. study (15) involved measuring inhibition in individual cells, perhaps resulting in greater sensitivity to such a variable. Nonetheless, reducing the MOI, either directly or indirectly by eliminating the virus/cell preincubation step, clearly resulted in enhanced ETV activity. However, the titration curves become somewhat flatter, and results were more variable under these altered conditions. These flatter titration curves may indicate that ETV activity reaches a plateau or threshold. At this time, it is unclear what mechanism(s) is responsible for the unusual MOI-dependent boost in potency observed with ETV versus other HIV NRTIs, using a low viral inoculum.
Higher amounts of infecting virus are needed for the cell survival assays (XTT and MTS) than for the RT assay, likely reflecting the fact that XTT and MTS assays require significant cell death, while RT assays require only virus to be present in supernatants. This observation, along with the impact of virus/cell preincubation, likely provides a further basis for the reduced potencies observed in previous HIV assays using the cell survival endpoint. In addition, assays that used pseudotyped virus inoculum, including the commercial phenotyping assay (PhenoSense), also resulted in reduced ETV potency, presumably as a result of more efficient infection.
The current data confirmed that ETV has limited activity against HIV-1 (generally in the μM range), using traditional assay systems or a commercial phenotypic assay (PhenoSense). The observed ETV anti-HIV activity under reduced viral input appears to be directly related to the inhibition of the HIV RT, since assays using chronically infected (postintegration) cells failed to exhibit any antiretroviral activity. Importantly, ETV can select for the precursor M184I substitution in cell passage studies, and variants encoding the M184V substitution exhibited >178-fold reduced susceptibility to ETV. A recent study by Domaoal et al. (8) confirms that HIV-1 RT can utilize ETV-TP and that the M184V enzyme has lower affinity to ETV-TP than the wild-type enzyme.
The interpretation of these data is challenging, since standard assay methodologies predict that ETV should have no significant clinical activity against HIV under dosing conditions where the Cmax achieved in human blood is only 0.030 μM (23). However, the clinical data may tell a different story, as a patient taking the 1-mg ETV dose exhibited a selection/enrichment of the HIV-1 M184V variant. Treating HBV in HBV/HIV coinfection has been shown to reduce HIV-RNA levels, either by an indirect immunologic effect or potentially by diminishing some synergistic effect of coinfection such as the action of the HBV viral protein X, which has (at least in vitro) been shown to enhance the transactivation of HIV (1, 20). Similar reductions in HIV-1 RNA levels were noted in patients coinfected with HIV and herpes simplex virus (HSV) who received valacyclovir (14). However, the enhancement of the M184V substitution while a patient receives ETV therapy can occur only due to direct inhibitory pressure on HIV replication, and the enzyme study by Domaoal et al. supports this (8).
These results have important implications for drug discovery and profiling of antiviral compounds. The significant enhancement of ETV activity against HIV under conditions of reduced viral inoculum was not predicted using the standard assay conditions that have been optimized for robustness and reproducibility. In fact, little interest would be given to compounds that worked only under the modified conditions described here, since effective drugs need to demonstrate that they can work against high virus challenge. The lesson learned from the current studies is that non-HIV compounds need to be tested under reduced viral challenge to fully evaluate their degree of antiviral activity prior to being administered to HIV-infected patients.
ETV has been shown to be a potent agent for treating HBV in HIV/HBV-coinfected patients in whom HAART is currently being used to control their HIV infection (R. Colonno, R. Rose, C. Baldick, K. Pokornowski, B. Eggers, M. Plym, J. Fang, J. Yang, E. Ledesma, and D. Tenney, presented at the 13th Conference on Retroviruses and Opportunistic Infections, Denver, CO, 2006). The clinical relevance and the magnitude of the anti-HIV effect of ETV in coinfected patients cannot be assessed outside the context of a clinical study. However, due to its relatively weak activity against HIV, ETV should not be considered for use as an anti-HIV agent, and the use of ETV for the treatment of chronic HBV infection in HIV/HBV-coinfected patients in the absence of effective HAART is not recommended.
Acknowledgments
We thank Betsy Eggers, Zhufang Li, Susan Roberts, Louis Alexander, Kay Limoli, and Lan Trinh for their contributions to these studies, Bruce Kreter and Richard Wilber for a critical review of the manuscript, and John Leet for the preparation of antivirals.
Footnotes
Published ahead of print on 3 March 2008.
REFERENCES
- 1.Barak, O., A. Aronheim, and Y. Shaul. 2001. HBV X protein targets HIV Tat-binding protein 1. Virology 283:110-120. [DOI] [PubMed] [Google Scholar]
- 2.Bartlett, J. A., M. J. Fath, R. Demasi, A. Hermes, J. Quinn, E. Mondou, and F. Rousseau. 2006. An updated systematic overview of triple combination therapy in antiretroviral-naive HIV-infected adults. AIDS 20:2051-2064. [DOI] [PubMed] [Google Scholar]
- 3.Boucher, C. A., N. Cammack, P. Schipper, R. Schuurman, P. Rouse, M. A. Wainberg, and J. M. Cameron. 1993. High-level resistance to (−) enantiomeric 2′-deoxy-3′-thiacytidine in vitro is due to one amino acid substitution in the catalytic site of human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 37:2231-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, T. T., R. G. Gish, R. de Man, A. Gadano, J. Sollano, Y. C. Chao, A. S. Lok, K. H. Han, Z. Goodman, J. Zhu, A. Cross, D. DeHertogh, R. Wilber, R. Colonno, and D. Apelian. 2006. A comparison of entecavir and lamivudine for HBeAg-positive chronic hepatitis B. N. Engl. J. Med. 354:1001-1010. [DOI] [PubMed] [Google Scholar]
- 5.Cory, A. H., T. C. Owen, J. A. Barltrop, and J. G. Cory. 1991. Use of an aqueous soluble tetrazolium/formazan assay for cell growth assays in culture. Cancer Commun. 3:207-212. [DOI] [PubMed] [Google Scholar]
- 6.DAIDS. 1997. Virology manual for HIV laboratories. Division of AIDS, National Institute of Allergy and Infectious Diseases, U.S. Department of Health and Human Services, Washington, DC. http://www.niaid.nih.gov/daids/vir_manual/full_vir_manual.pdf.
- 7.Das, K., X. Xiong, H. Yang, C. E. Westland, C. S. Gibbs, S. G. Sarafianos, and E. Arnold. 2001. Molecular modeling and biochemical characterization reveal the mechanism of hepatitis B virus polymerase resistance to lamivudine (3TC) and emtricitabine (FTC). J. Virol. 75:4771-4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Domaoal, R. A., M. McMahon, C. L. Thio, C. M. Bailey, J. Tirado-Rives, A. Obikhod, M. Detorio, K. L. Rapp, R. F. Siliciano, R. F. Schinazi, and K. S. Anderson. 2007. Pre-steady-state kinetic studies establish entecavir 5′-triphosphate as a substrate for HIV-1 reverse transcriptase. J. Biol. Chem. 283:5452-5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frost, S. D., M. Nijhuis, R. Schuurman, C. A. Boucher, and A. J. Brown. 2000. Evolution of lamivudine resistance in human immunodeficiency virus type 1-infected individuals: the relative roles of drift and selection. J. Virol. 74:6262-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Innaimo, S. F., M. Seifer, G. S. Bisacchi, D. N. Standring, R. Zahler, and R. J. Colonno. 1997. Identification of BMS-200475 as a potent and selective inhibitor of hepatitis B virus. Antimicrob. Agents Chemother. 41:1444-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson, V. A., and R. E. Byington. 1990. Infectivity assay (virus yield assay), p. 71-76. In A. Aldovani and B. D. Walker (ed.), Techniques in HIV research. Stockton Press, New York, NY.
- 12.Levine, S., D. Hernandez, G. Yamanaka, S. Zhang, R. Rose, S. Weinheimer, and R. J. Colonno. 2002. Efficacies of entecavir against lamivudine-resistant hepatitis B virus replication and recombinant polymerases in vitro. Antimicrob. Agents Chemother. 46:2525-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levy, V., and R. M. Grant. 2006. Antiretroviral therapy for hepatitis B virus-HIV-coinfected patients: promises and pitfalls. Clin. Infect. Dis. 43:904-910. [DOI] [PubMed] [Google Scholar]
- 14.Martinez, V., E. Caumes, M. Eisenhut, N. Nagot, H. Weiss, and P. Van de Perre. 2007. HSV Therapy and HIV-1 Reduction. N. Engl. J. Med. 356:2323-2324. [PubMed] [Google Scholar]
- 15.McMahon, M. A., B. L. Jilek, T. P. Brennan, L. Shen, Y. Zhou, M. Wind-Rotolo, S. Xing, S. Bhat, B. Hale, R. Hegarty, C. R. Chong, J. O. Liu, R. F. Siliciano, and C. L. Thio. 2007. The HBV drug entecavir: effects on HIV-1 replication and resistance. N. Engl. J. Med. 356:2614-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osborn, M. K., and A. S. Lok. 2006. Antiviral options for the treatment of chronic hepatitis B. J. Antimicrob. Chemother. 57:1030-1034. [DOI] [PubMed] [Google Scholar]
- 17.Petropoulos, C. J., N. T. Parkin, K. L. Limoli, Y. S. Lie, T. Wrin, W. Huang, H. Tian, D. Smith, G. A. Winslow, D. J. Capon, and J. M. Whitcomb. 2000. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44:920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sherman, M., C. Yurdaydin, J. Sollano, M. Silva, Y. F. Liaw, J. Cianciara, A. Boron-Kaczmarska, P. Martin, Z. Goodman, R. Colonno, A. Cross, G. Denisky, B. Kreter, and R. Hindes. 2006. Entecavir for treatment of lamivudine-refractory, HBeAg-positive chronic hepatitis B. Gastroenterology 130:2039-2049. [DOI] [PubMed] [Google Scholar]
- 19.Soriano, V., P. Barreiro, and M. Nunez. 2006. Management of chronic hepatitis B and C in HIV-coinfected patients. J. Antimicrob. Chemother. 57:815-818. [DOI] [PubMed] [Google Scholar]
- 20.Visvanathan, K., N. A. Skinner, A. J. Thompson, S. M. Riordan, V. Sozzi, R. Edwards, S. Rodgers, J. Kurtovic, J. Chang, S. Lewin, P. Desmond, and S. Locarnini. 2007. Regulation of Toll-like receptor-2 expression in chronic hepatitis B by the precore protein. Hepatology 45:102-110. [DOI] [PubMed] [Google Scholar]
- 21.Wainberg, M. A., H. Salomon, Z. Gu, J. S. Montaner, T. P. Cooley, R. McCaffrey, J. Ruedy, H. M. Hirst, N. Cammack, J. Cameron, et al. 1995. Development of HIV-1 resistance to (-)2′-deoxy-3′-thiacytidine in patients with AIDS or advanced AIDS-related complex. AIDS 9:351-357. [PubMed] [Google Scholar]
- 22.Wainberg, M. A., J. P. Sawyer, J. S. Montaner, R. L. Murphy, D. R. Kuritzkes, and F. Raffi. 2005. Challenges for the clinical development of new nucleoside reverse transcriptase inhibitors for HIV infection. Antivir. Ther. 10:13-28. [PubMed] [Google Scholar]
- 23.Yan, J. H., M. Bifano, S. Olsen, R. A. Smith, D. Zhang, D. M. Grasela, and F. LaCreta. 2006. Entecavir pharmacokinetics, safety, and tolerability after multiple ascending doses in healthy subjects. J. Clin. Pharmacol. 46:1250-1258. [DOI] [PubMed] [Google Scholar]
- 24.Zhang, H., Y. Zhou, C. Alcock, T. Kiefer, D. Monie, J. Siliciano, Q. Li, P. Pham, J. Cofrancesco, D. Persaud, and R. F. Siliciano. 2004. Novel single-cell-level phenotypic assay for residual drug susceptibility and reduced replication capacity of drug-resistant human immunodeficiency virus type 1. J. Virol. 78:1718-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]