Abstract
The inhibitory quotient (IQ) of human immunodeficiency virus (HIV) protease inhibitors (PIs), which is the ratio of drug concentration to viral susceptibility, is considered to be predictive of the virological response. We used several approaches to calculate the IQs of amprenavir and lopinavir in a subset of heavily pretreated patients participating in the French National Agency for AIDS Research (ANRS) 104 trial and then compared their potentials for predicting changes in the plasma HIV RNA level. Thirty-seven patients were randomly assigned to receive either amprenavir (600 mg twice a day [BID]) or lopinavir (400 mg BID) plus ritonavir (100 or 200 mg BID) for 2 weeks before combining the two PIs. The 90% inhibitory concentration (IC90) was measured using a recombinant assay without or with additional human serum (IC90+serum). Total and unbound PI concentrations in plasma were measured. Univariate linear regression was used to estimate the relation between the change in viral load and the IC90 or IQ values. The amprenavir phenotypic IQ values were very similar when measured with the standard and protein binding-adjusted IC90s. No relationship was found between the viral load decline and the lopinavir IQ. During combination therapy, the amprenavir and lopinavir genotypic IQ values were predictive of the viral response at week 6 (P = 0.03). The number of protease mutations (<5 or ≥5) was related to the virological response throughout the study. These findings suggest that the combined genotypic IQ and the number of protease mutations are the best predictors of virological response. High amprenavir and lopinavir concentrations in these patients might explain why plasma concentrations and the phenotypic IQ have poor predictive value.
The virological efficacy of protease inhibitors (PIs) in patients infected by the human immunodeficiency virus (HIV) is dependent on both their pharmacodynamic and their pharmacokinetic properties. In vitro high and sustained plasma (or cell) concentrations are needed to maximally suppress viral replication. The dose administered in HIV-infected patients should provide such concentrations.
The inhibitory quotient (IQ), defined as the ratio of the trough drug concentration in serum (Cmin) to viral susceptibility expressed as an inhibitory concentration (ideally, the 90% inhibitory concentration [IC90]), has been used to estimate the antiviral potency of PIs in vivo (24). This parameter has also been proposed to optimize the dosing regimen of treatment-experienced patients (12, 20). Although there is a strong theoretical rationale for using the IQ, the practical value of this parameter is controversial. First, there are few prospective studies of the relationship between IQ and virological response (22). Second, there is no consensus method for calculating this parameter (1, 24, 27). Standard calculations estimate IQ from both the plasma drug concentration and virus susceptibility. However, several pharmacokinetic and virological issues remain unsolved. In most pharmacokinetic studies, the total drug concentration in plasma is measured, whereas the active component is the free (protein-unbound) fraction. Furthermore, the addition of human albumin to the cell culture medium increases the IC90 in vitro. In summary, either the total concentration or the protein-adjusted concentration, and either the standard or the serum-adjusted IC90, can be used to calculate the IQ. More recently, a “genotypic IQ” (GIQ) was proposed as the ratio of the plasma concentration to the number of mutations on the viral protease gene. Indeed, genotypic resistance assays can be performed rapidly and are less costly than phenotypic resistance assays (11). Both genotypic and phenotypic IQs are predictive of changes in the HIV RNA level in treatment-experienced patients (5, 9, 11, 17, 22, 23, 31), but only one study of the predictive value of the IQ is available with data from patients treated with two ritonavir-boosted PIs (8).
We aimed at estimating the relationship between the IQs of amprenavir and lopinavir and the virological response after 2, 6, and 26 weeks of treatment in a group of heavily pretreated patients who participated in the French National Agency for AIDS Research (ANRS) 104 trial. Several methods for calculating IQs were compared with the viral phenotype and genotype for their ability to predict changes in plasma viral load.
(This study was presented in part at the 43rd Interscience Conference on Antimicrobial Agents and Chemotherapy, Chicago, IL, 14 to 17 September 2003 [2a].)
MATERIALS AND METHODS
Study design and study population.
The ANRS 104 study was a prospective, randomized, open-label, multicenter trial involving patients with CD4 counts of <500/mm3 and plasma HIV RNA at >10,000 copies/ml after receiving successive antiretroviral treatments including at least two PIs and one nonnucleoside reverse transcriptase inhibitor. The main objective of this trial was to compare the clinical efficacies and tolerabilities of a combination of amprenavir and lopinavir-ritonavir in treatment-experienced patients (26). The study was divided into two periods. For the first 2 weeks (period 1), patients were randomized to receive, in addition to their ongoing nucleoside reverse transcriptase inhibitors (NRTI), (i) lopinavir-ritonavir (400/100 mg twice a day [BID]) or lopinavir-ritonavir (400/100 mg BID) plus ritonavir (100 mg BID) or (ii) amprenavir (600 mg BID) plus ritonavir (100 mg BID) or amprenavir (600 mg BID) plus ritonavir (200 mg BID). From week 3 to week 26 (period 2), all the patients received amprenavir plus lopinavir-ritonavir, with or without an additional boost of ritonavir (100 mg BID). The NRTIs were optimized on the basis of viral genotyping results and previous antiretroviral exposure. The genotype was interpreted for each inhibitor by using the 2001 update of the ANRS algorithm. Patients were recruited from 16 French clinical AIDS units. All patients signed an Ethics Committee-approved informed consent form. Patients were instructed to take their medication in the morning and evening, with a light meal. Physical examinations, CD4 and CD8 cell counts, and hematological and clinical chemistry measurements were performed at each study visit (weeks −2, 0, 2, 4, and 6 and then monthly for the subsequent 20 weeks). Blood samples were also drawn at weeks −2, 0, 2, 6, and 26 for plasma HIV type 1 (HIV-1) RNA, viral genotyping and phenotyping, and drug assays in plasma.
Laboratory measurements. (i) Virological parameters.
Plasma HIV-1 RNA was assayed locally at weeks −2, 0, 2, 4, 6, 14, and 26 by using the Roche Amplicor HIV-1 monitor kit (Roche, France; limit of quantitation, 200 copies/ml) or the Quantiplex HIV RNA 3.0 assay (Bayer Diagnostics, France; limit of quantitation, 50 copies/ml).
(ii) HIV-1 protease genotyping and phenotyping.
Viral genotyping was performed at weeks −2, 2, 6, and 26 based on direct sequencing of the HIV-1 protease-coding region and using the consensus technique of the ANRS AC11 resistance group or the TruGene HIV-1 genotyping kit (Visible Genetics, Bayer). The genotype was taken into account only if a complete protease sequence (amino acids 1 to 99) was obtained. Protease sequences from each patient were examined for the presence of mutations associated with protease resistance at the following 21 relevant codons (11): 10, 20, 24, 30, 32, 33, 36, 46, 47, 48, 50, 53, 54, 63, 71, 73, 77, 82, 84, 88, and 90 (the Stanford HIV drug resistance database, 2004; http://hivdb.stanford.edu/). The genotype for lopinavir-ritonavir and amprenavir-ritonavir was interpreted by using the 2006 update of the ANRS algorithm.
Resistance phenotyping was performed at screening (week −2) for all the patients and at week 26 for those patients in whom the antiretroviral drug regimen failed and had plasma HIV RNA levels above 50 copies/ml (20/37 patients) according to a recombinant virus assay (Phenoscript; Viralliance) (25). Results were expressed as the PI concentration inhibiting virus spread by 90% (IC90), in a standard method without added human serum (IC90). In a subgroup of 12 patients who started their study regimen with amprenavir, the amprenavir IC90 was also determined after adding 40% human serum to the growth medium which contains 10% fetal bovine serum in order to reach a total protein concentration close to that found in human plasma (IC90+serum).
(iii) Drug assays in plasma.
Blood samples were drawn at week 2 and week 6, just before the scheduled drug intake (Cmin). The total and unbound amprenavir and total lopinavir Cmin values were measured by high-pressure liquid chromatography with separation on a C18 column after liquid/liquid extraction of alkaline plasma and UV detection as described elsewhere (2, 26, 29). Bound amprenavir and unbound amprenavir were separated by ultrafiltration using Centrifree devices (Amicon, YM-300 filter system; Millipore Corp., Bedford, MA). Amprenavir was then measured in the ultrafiltrate as previously described. The overall day-to-day coefficient of variation was below 12% (2). All concentrations were expressed in ng/ml or μmol/liter for IQ calculations.
Calculation of IQs.
Phenotypic IQs were calculated as the ratio of the plasma Cmin of each PI to the IC90 measured at baseline. For amprenavir, the IC90 values were determined with or without added protein (IC90+serum or IC90), and both the total Cmin and unbound Cmin (Cminu) values were measured. Two methods to adjust for protein binding were tested, namely, Cminu and IC90+serum (24). Amprenavir IQs were therefore calculated as the ratios of Cminu/IC90 (IQu) and Cmin/IC90+serum (IQserum).
The GIQ of each PI was calculated as the ratio of the plasma Cmin corrected for protein binding (Cminu) to the baseline number of protease resistance mutations. The following mutations on the viral protease were considered: L10I/F/R/V, K20M/R, L24I, D30N, V32I, L33F, M36I, M46I/L, I47A/V, G48V, I50V, F53L, I54L/T/V, L63P, A71I/L/V/T, G73S, V77I, V82A/F/T/S, A84V, N88D/S, and L90M.
During the second treatment period, in which patients received two ritonavir-boosted PIs, the combined IQs were calculated as the sum of the phenotypic IQu for each PI. Cminu was not measured but was calculated as the total Cmin corrected by the average protein binding of amprenavir (10%) (2, 15) and of lopinavir and ritonavir (1%) (3, 4). The combined GIQ was calculated as the sum of the ratios of Cminu/number of protease resistance mutations.
Statistical analysis.
The median (range) was used to describe the distribution of amprenavir, lopinavir, and ritonavir parameters and for the different IQ calculations. Univariate linear regression was used to estimate the relation between the decline in viral load (difference between baseline and week 2, 6, or 26) and the different Cmin, IC90, and IQ values. The higher was the proportion of explained variance (r2) of viral load, the better was the model. All statistical tests were run on SAS software (version 8.2; SAS Institute).
RESULTS
Overall, 37 patients were enrolled in the ANRS 104 study. Their baseline characteristics are shown in Table 1. Twenty-three patients started their study antiretroviral drug regimen with lopinavir-ritonavir (group 1) and 14 patients started with amprenavir (group 2).
TABLE 1.
Baseline characteristics of the patients participating in the ANRS 104 studya
| Parameter | Group 1, amprenavir/r (n = 14) | Group 2, lopinavir/r (n = 23) |
|---|---|---|
| Median age, yr (range) | 47 (32-53) | 41 (27-65) |
| Males, no. (%) | 12 (86) | 21 (91) |
| CDC clinical stage (no. [%]) | ||
| A | 6 (42) | 10 (43) |
| B | 4 (29) | 3 (13) |
| C | 4 (29) | 10 (43) |
| Median no. of CD4+ cells/mm3 (range) | 195 (65-385) | 185 (3-509) |
| Median HIV-1 RNA log10 copies/ml (range) | 4.9 (3.6-5.7) | 4.6 (3.8-5.6) |
| Median no. of previous antiretrovirals taken (range) | 7.5 (4-12) | 7.5 (4-12) |
| Median no. of antiretrovirals taken prior to inclusion and still in use at inclusion (range) | 9.5 (7-13) | 10 (8-13) |
| Genotypic resistance (no. [%]) | ||
| Amprenavir | 7 (50) | 9 (56) |
| Lopinavir | 7 (50) | 7 (30) |
| Median no. of protease mutations (range) | 7.0 (1.0-9.0) | 7.0 (1.0-10.0) |
| Median no. of reverse transcriptase mutations (range) | 6.5 (0-11.0) | 7.0 (0.0-11.0) |
| Median phenotypic resistance index (range) | ||
| Amprenavir | 2.8 (0.5-24.3) | 2.5 (0.5-19.5) |
| Lopinavir | 8.7 (0.3-84.0) | 10.7 (0.2-95.3) |
Patients from group 1 started with amprenavir-ritonavir (amprenavir/r) for the first 2 weeks and patients from group 2 started with lopinavir-ritonavir (lopinavir/r).
Amprenavir and lopinavir Cmin values are reported in detail elsewhere (26, 29) and were not related to the virological response. The median unbound amprenavir Cminu at week 2 was 177 ng/ml and was not predictive of the virological response at week 2.
In a subgroup of 12 patients who had phenotypic studies (group 2), the amprenavir IC90 was 57.8 ng/ml (8.7 to 150.9) and increased to 453 ng/ml (33 to 1,105) when measured in the presence of 50% human serum (IC90+serum). The IC90+serum was a good predictor of the early virological response (week 2, P = 0.006), whereas the IC90 was not. Amprenavir IQs adjusted for protein binding (IQu and IQserum) were rather similar (2.5 and 3.6, respectively). The relationship with viral load decline at week 2 was slightly better with IQu than with IQserum (r2 = 0.45, P = 0.02 versus r2 = 0.31, P = 0.06), although those results might be explained by reference to an outlier patient (r2 = 0.24, P = 0.12 versus r2 = 0.11, P = 0.31 without the outlier patient).
For the 21 patients treated with lopinavir during the first period, the lopinavir IC90 was 34.2 ng/ml (0.7 to 330.4) and was a good predictor of the virological response at weeks 2, 6, and 26 (week 2, r2 = 0.37, P = 0.003; week 6, r2 = 0.23, P = 0.03; week 26, r2 = 0.33, P = 0.006).
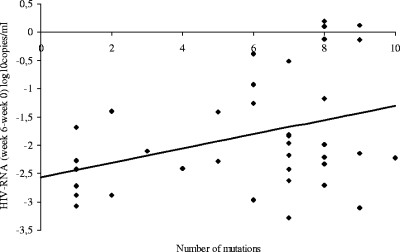
The number of protease mutations (<5 or ≥5) was related to the virological response throughout the study (at week 2, r2 = 0.18, P = 0.008; at week 6, r2 = 0.11, P = 0.046; and at week 26, r2 = 0.12, P = 0.034) (Fig. 1).
FIG. 1.
Relationship between viral load decline at week 6 and the number of protease mutations at screening (r2 = 0.11, P = 0.046).
Table 2 shows the virological responses throughout the study as a function of the amprenavir and lopinavir phenotypic IQ and GIQ values measured at week 2 or week 6. The amprenavir GIQ (corrected amprenavir Cmin at week 2/number of protease mutations at week −2) was related to the virological response at week 2 (r2 = 0.66, P = 0.0004) and at week 6 (r2 = 0.38, P = 0.02). Patients who had a Cmin-corrected GIQ above 75 had a median decline in HIV RNA of 1.23 log10 copies/ml (−2.27; −0.97) versus 0.19 log10 copies/ml (−1.07; 0.15) in patients with a GIQ below 75 (P = 0.005). In contrast, none of the lopinavir IQs were predictive of antiretroviral activity.
TABLE 2.
Relationships between the viral load decline at weeks 2, 6, and 26 and the amprenavir, lopinavir, and ritonavir IQs (univariate analysis)
| Drug | IQ typea | Week | No. of patients | Median (range) |
P value at week:
|
||
|---|---|---|---|---|---|---|---|
| 2 | 6 | 26 | |||||
| Amprenavir | IQsd | 2 | 13 | 4.3 (0.62-25.9) | 0.04 | 0.24 | 0.70 |
| 6 | 13 | 1.8 (0.20-11.7) | 0.80 | 0.26 | |||
| GIQ | 2 | 14 | 81.2 (17.9-291.1) | 0.0004 | 0.02 | 0.16 | |
| 6 | 14 | 26.4 (9.8-69.1) | 0.56 | 0.59 | |||
| Lopinavir | IQsd | 2 | 21 | 4.0 (0.16-153.3) | 0.97 | 0.65 | 0.66 |
| 6 | 21 | 2.0 (0.15-157.5) | 0.70 | 0.80 | |||
| GIQ | 2 | 23 | 23.9 (6.6-177.6) | 0.37 | 0.20 | 0.20 | |
| 6 | 23 | 17.8 (3.6-182.5) | 0.15 | 0.36 | |||
| Ritonavir | IQsd | 2 | 34 | 0.01 (0.0004-0.5) | 0.55 | 0.18 | 0.28 |
| 6 | 34 | 0.005 (0.0004-0.8) | 0.34 | 0.87 | |||
| GIQ | 2 | 37 | 1.7 (0.12-19.1) | 0.18 | 0.10 | 0.12 | |
| 6 | 37 | 1.3 (0.12-20.4) | 0.16 | 0.45 | |||
| Amprenavir + lopinavir | IQsd | 6 | 34 | 4.9 (0.36-166.5) | 0.34 | 0.71 | |
| GIQ | 6 | 37 | 46.9 (15.9-543.4) | 0.03 | 0.15 | ||
| Amprenavir + lopinavir + ritonavir | IQsd | 6 | 34 | 4.9 (0.37-167.3) | 0.34 | 0.71 | |
| GIQ | 6 | 37 | 48.8 (16.0-549.3) | 0.03 | 0.15 | ||
Standard IQ (IQsd): (corrected Cmin [calculated protein-unbound Cminu] at week 2 or 6)/IC90 standard at week −2. GIQ: (corrected Cmin [calculated protein-unbound Cminu] at week 2 or 6)/number of protease mutations at week −2.
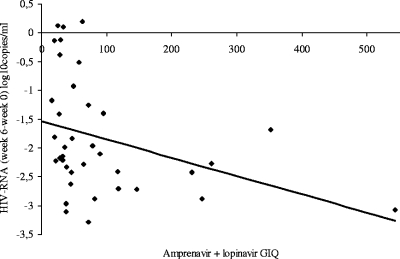
None of the phenotypic IQs or GIQs of amprenavir and lopinavir (determined with the Cmin corrected for protein binding and measured at week 6) were predictive of the viral load decline. However, a combination of the amprenavir and lopinavir GIQs, whether measured either with the standard equation or adjusted for protein binding, and with or without ritonavir, was predictive of the viral response at week 6 but not at the end of the study, as shown on Fig. 2. Patients who responded at week 6 (plasma HIV RNA <10,000 versus ≥10,000 copies/ml) had a higher median combined GIQ measured at week 6 (65 versus 29; P = 0.01).
FIG. 2.
Relationship between viral load decline at week 6 and combined (lopinavir and amprenavir) GIQ at week 6 (r2 = 0.12, P = 0.03).
DISCUSSION
A significant proportion of antiretroviral-experienced patients never have optimal viral suppression or experience a viral rebound shortly after starting a new treatment. Our population was heavily pretreated with a high proportion of baseline NRTI genotypic resistance; consequently, the efficacy of the combination results essentially from both PIs. Treatment thus needs to be optimized according to viral susceptibility and the plasma PI concentration. This study provides IQ data for two currently used ritonavir-boosted PIs, amprenavir and lopinavir, when administered alone (first 2 weeks of the study) or in combination, in heavily pretreated HIV-infected patients. This is one of the few studies to show that the combined IQ can be a useful predictor in patients who receive ritonavir-boosted dual-PI therapy (8).
We compared two methods for calculating the phenotypic IQ, which incorporates protein binding. Amprenavir and lopinavir are highly bound to plasma proteins and especially alpha1 acid glycoprotein (90% and 98 to 99%, respectively). As only free drug inhibits viral replication, protein binding affects the potency of these two PIs. ICs measured in vitro must be adjusted for protein binding before extrapolation to the clinical setting. Moreover, ICs suffer from variability due to a lack of standardization of phenotyping methods (14). There are three methods for adjusting the in vitro IC for protein binding, namely, multiplication of the IC by the free fraction measured in vivo, measurement of the IC in the presence of 50% human serum albumin, and multiplication of the IC by a constant to adjust for assay variations (20), but none of these methods has been clinically validated. Our results show that, whatever the equation used to calculate IQ (Cminu/standard IC90 or Cmin/IC90+serum), the relationship with viral load decline is very similar. The best correlation was obtained with the IQu (Cminu/IC90). It has previously been demonstrated that measuring and calculating the Cminu gives similar results (29). However, it remains to be determined whether these findings can be extrapolated to PIs other than amprenavir.
We found no relationship between lopinavir or amprenavir exposure and the decline in viral load, in agreement with previous studies (6, 7, 13, 16, 30). Amprenavir and lopinavir Cmin values obtained with the ritonavir boost were far higher than the IC50 reported for wild-type virus (28) and the IC90 measured at screening. This might explain why plasma concentrations were poorly predictive of the decline in plasma HIV RNA. Compliance with medication was not measured in our study; the amprenavir and lopinavir Cmin values suggested that it was maximal at week 2 and week 6 but declined thereafter. Thus, none of the phenotypic IQ values based on concentrations measured at week 6 were predictive of antiviral efficacy.
The GIQ is simpler to measure than the phenotypic IQ. We found that the GIQ of amprenavir was associated with the virological response at week 2 and week 6 but not at week 26. Furthermore the amprenavir GIQ cutoff of 75 could be a useful tool in clinical practice, as previously demonstrated by Marcelin et al. (18). Our findings confirm that the GIQ, which incorporates both baseline viral resistance and the level of drug exposure in plasma, is superior to drug exposure alone in predicting the virological response to a salvage regimen (18). However, lopinavir IQs did not correlate with virological efficacy, and our data do not support those reported elsewhere (5, 9, 16). This is probably because the lopinavir Cmin values for our patients were sufficiently high not to restrict efficacy (13), whereas lopinavir concentrations were lower in other studies (5, 9). In these latter studies, the Cmin was determined as part of routine therapeutic drug monitoring or observational studies, settings in which compliance is important for overall exposure (5, 9, 16). One limitation of therapeutic drug monitoring is the wide intrapatient variability of trough concentrations, as recently demonstrated by Nettles et al. (21) and Goujard et al. (10). Poor compliance and an effect of food may be involved in this variability. We acknowledge that one limitation of our study is that compliance was not measured. However, comparison of trough concentrations measured at weeks 6, 14, and 26 clearly shows that compliance is decreasing from week 6, where concentrations of lopinavir and amprenavir were in the expected range in all patients, in contrast to weeks 14 and 26, where 7 and 6 patients, respectively, had concentrations below the limit of quantification of the assay (26).
Interestingly, the combined GIQ was predictive of the virological response at week 6, as in the GigHAART trial (8). This suggests that a combination of two PIs has a strong antiviral effect that might overcome resistance in these strongly pretreated patients. As expected, ritonavir did not participate in the drug effect, as the concentrations used to boost PIs are too low (even though some patients received 200 mg BID). The combined IQ of amprenavir plus lopinavir plus ritonavir was close to the combined IQ of amprenavir plus lopinavir and had the same predictive potential. However, the predictive value of this parameter disappeared at the end of the study, for several possible reasons. In particular, the Cmin tends to fall, as a negative initial interaction between the two PIs and a decreased compliance tends to reduce their concentrations and exposure levels; additionally, the viral resistance profile is continually evolving (19). Further studies are needed to determine which viral mutations have the biggest impact on the GIQ and how these mutations can be taken into account in the calculations (30).
The combined GIQ did not have better predictive value than the number of mutations. In these highly pretreated patients with high PI Cmin values, the number of PI resistance mutations is a major determinant of virological outcome (5, 13, 16) and in our study was the only factor predictive of the virological response at week 26. Finally, as previously demonstrated (26), patients with more than five protease resistance mutations or a lopinavir mutation score (13, 14) of higher than 5 at baseline had a significantly poorer virological response than other patients (P = 0.04 and P = 0.006, respectively).
Thus, this study suggests that when treatment compliance is optimal and high PI concentrations are achieved, the viral genotype is the best predictor of virological outcome.
Acknowledgments
This work was supported by ANRS, Paris, France.
We thank Céline Boucherie, who assisted us in the statistical analysis and in the preparation of the revised manuscript.
The following institutions and investigators participated in the Puzzle-1 ANRS 104 study: W. Rozenbaum, L. Naït-Ighil, T. H. Nguyen, L. Slama, P. M. Girard, Hôpital Rothschild, Paris; J. M. Molina, D. Sereni, N. Colin de Verdière, C. Lascoux-Combes, C. Pintado, D. Ponscarme, F. Prevoteau de Clary, M. Tourneur, Hôpital Saint-Louis, Paris; M. Bentata, L. Guillevin, O. Launay, R. Mansouri, F. Rouges, Hôpital Avicenne, Bobigny; M. Kazatchkine, A. Aouba, M. Azizi, J. N. Fiessinger, P. Le Houssine, Hôpital Européen Georges Pompidou, Paris; D. Sicard, C. Bernasconi, D. Salmon, B. Silbermann, Hôpital Cochin, Paris; J. P. Cassuto, C. Ceppi, M. Poiree, Hôpital de l'Archet, Nice; G. Raguin, M. Merad, Hôpital de la Croix Saint Simon, Paris; J. F. Delfraissy, C. Goujard, Y. Quertainmont, Hôpital de Bicêtre, Le Kremlin-Bicêtre; C. Perronne, P. de Truchis, Hôpital Raymond Poincaré, Garches; B. Dupont, J. L. Bresson, I. Calatroni, Hôpital Necker, Paris; and F. Raffi, J. L. Esnault, S. Leautez, Hôtel Dieu, Nantes.
Footnotes
Published ahead of print on 19 February 2008.
REFERENCES
- 1.Aarnoutse, R. E., J. M. Schapiro, C. A. Boucher, Y. A. Hekster, and D. M. Burger. 2003. Therapeutic drug monitoring: an aid to optimising response to antiretroviral drugs? Drugs 63:741-753. [DOI] [PubMed] [Google Scholar]
- 2.Barrail, A., C. Le Tiec, S. Paci-Bonaventure, V. Furlan, I. Vincent, and A. M. Taburet. 2006. Determination of amprenavir total and unbound concentrations in plasma by high-performance liquid chromatography and ultrafiltration. Ther. Drug Monit. 28:89-94. [DOI] [PubMed] [Google Scholar]
- 2a.Barrail, A., C. Droz, L. Morand-Joubert, E. Dam, C. Le Tiec, F. Clavel, G. Chêne, G. Raguin, P. M. Giraard, A. M. Taburet, and the Puzzle 1 Study Group. 2003. Inhibitory quotients as predictors of the virological response to amprenavir in heavily pretreated patients, abstr. H-1998. Abstr. 43rd Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC.
- 3.Boffito, M., D. J. Back, T. F. Blaschke, M. Rowland, R. J. Bertz, J. G. Gerber, and V. Miller. 2003. Protein binding in antiretroviral therapies. AIDS Res. Hum. Retrovir. 19:825-835. [DOI] [PubMed] [Google Scholar]
- 4.Boffito, M., P. G. Hoggard, W. E. Lindup, S. Bonora, A. Sinicco, S. H. Khoo, G. Di Perri, and D. J. Back. 2004. Lopinavir protein binding in vivo through the 12-hour dosing interval. Ther. Drug Monit. 26:35-39. [DOI] [PubMed] [Google Scholar]
- 5.Breilh, D., I. Pellegrin, A. Rouzes, K. Berthoin, F. Xuereb, H. Budzinski, M. Munck, H. J. Fleury, M. C. Saux, and J. L. Pellegrin. 2004. Virological, intracellular and plasma pharmacological parameters predicting response to lopinavir/ritonavir (KALEPHAR study). AIDS 18:1305-1310. [DOI] [PubMed] [Google Scholar]
- 6.Clevenbergh, P., R. Boulme, M. Kirstetter, and P. Dellamonica. 2004. Efficacy, safety and predictive factors of virological success of a boosted amprenavir-based salvage regimen in heavily antiretroviral-experienced HIV-1-infected patients. HIV Med. 5:284-288. [DOI] [PubMed] [Google Scholar]
- 7.De Luca, A., F. Baldini, A. Cingolani, S. Di Giambenedetto, R. M. Hoetelmans, and R. Cauda. 2004. Deep salvage with amprenavir and lopinavir/ritonavir: correlation of pharmacokinetics and drug resistance with pharmacodynamics. J. Acquir. Immune Defic. Syndr. 35:359-366. [DOI] [PubMed] [Google Scholar]
- 8.Delaugerre, C., G. Peytavin, S. Dominguez, A. G. Marcelin, C. Duvivier, K. Gourlain, B. Amellal, M. Legrand, F. Raffi, D. Costagliola, C. Katlama, and V. Calvez. 2005. Virological and pharmacological factors associated with virological response to salvage therapy after an 8-week of treatment interruption in a context of very advanced HIV disease (GigHAART ANRS 097). J. Med. Virol. 77:345-350. [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez de Requena, D., O. Gallego, L. Valer, I. Jimenez-Nacher, and V. Soriano. 2004. Prediction of virological response to lopinavir/ritonavir using the genotypic inhibitory quotient. AIDS Res. Hum. Retrovir. 20:275-278. [DOI] [PubMed] [Google Scholar]
- 10.Goujard, C., M. Legrand, X. Panhard, B. Diquet, X. Duval, G. Peytavin, I. Vincent, C. Katlama, C. Leport, B. Bonnet, D. Salmon-Ceron, F. Mentre, and A. M. Taburet. 2005. High variability of indinavir and nelfinavir pharmacokinetics in HIV-infected patients with a sustained virological response on highly active antiretroviral therapy. Clin. Pharmacokinet. 44:1267-1278. [DOI] [PubMed] [Google Scholar]
- 11.Hirsch, M. S., F. Brun-Vezinet, B. Clotet, B. Conway, D. R. Kuritzkes, R. T. D'Aquila, L. M. Demeter, S. M. Hammer, V. A. Johnson, C. Loveday, J. W. Mellors, D. M. Jacobsen, and D. D. Richman. 2003. Antiretroviral drug resistance testing in adults infected with human immunodeficiency virus type 1: 2003 recommendations of an International AIDS Society-USA panel. Clin. Infect. Dis. 37:113-128. [DOI] [PubMed] [Google Scholar]
- 12.Hoefnagel, J. G., P. P. Koopmans, D. M. Burger, R. Schuurman, and J. M. Galama. 2005. Role of the inhibitory quotient in HIV therapy. Antivir. Ther. 10:879-892. [PubMed] [Google Scholar]
- 13.Hoefnagel, J. G., M. J. van der Lee, P. P. Koopmans, R. Schuurman, S. Jurriaans, A. I. van Sighem, L. Gras, F. de Wolf, J. M. Galama, and D. M. Burger. 2006. The genotypic inhibitory quotient and the (cumulative) number of mutations predict the response to lopinavir therapy. AIDS 20:1069-1071. [DOI] [PubMed] [Google Scholar]
- 14.Kappelhoff, B. S., K. M. Crommentuyn, M. M. de Maat, J. W. Mulder, A. D. Huitema, and J. H. Beijnen. 2004. Practical guidelines to interpret plasma concentrations of antiretroviral drugs. Clin. Pharmacokinet. 43:845-853. [DOI] [PubMed] [Google Scholar]
- 15.Livington, D. J., S. Pazhanisamy, D. J. Porter, J. A. Partaledis, R. D. Tung, and G. R. Painter. 1995. Weak binding of VX-478 to human plasma proteins and implications for anti-human immunodeficiency virus therapy. J. Infect. Dis. 172:1238-1245. [DOI] [PubMed] [Google Scholar]
- 16.Marcelin, A. G., I. Cohen-Codar, M. S. King, P. Colson, E. Guillevic, D. Descamps, C. Lamotte, V. Schneider, J. Ritter, M. Segondy, H. Peigue-Lafeuille, L. Morand-Joubert, A. Schmuck, A. Ruffault, P. Palmer, M. L. Chaix, V. Mackiewicz, V. Brodard, J. Izopet, J. Cottalorda, E. Kohli, J. P. Chauvin, D. J. Kempf, G. Peytavin, and V. Calvez. 2005. Virological and pharmacological parameters predicting the response to lopinavir-ritonavir in heavily protease inhibitor-experienced patients. Antimicrob. Agents Chemother. 49:1720-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marcelin, A. G., C. Dalban, G. Peytavin, C. Lamotte, R. Agher, C. Delaugerre, M. Wirden, F. Conan, S. Dantin, C. Katlama, D. Costagliola, and V. Calvez. 2004. Clinically relevant interpretation of genotype and relationship to plasma drug concentrations for resistance to saquinavir-ritonavir in human immunodeficiency virus type 1 protease inhibitor-experienced patients. Antimicrob. Agents Chemother. 48:4687-4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marcelin, A. G., C. Lamotte, C. Delaugerre, N. Ktorza, H. Ait Mohand, R. Cacace, M. Bonmarchand, M. Wirden, A. Simon, P. Bossi, F. Bricaire, D. Costagliola, C. Katlama, G. Peytavin, and V. Calvez. 2003. Genotypic inhibitory quotient as predictor of virological response to ritonavir-amprenavir in human immunodeficiency virus type 1 protease inhibitor-experienced patients. Antimicrob. Agents Chemother. 47:594-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morand-Joubert, L., C. Charpentier, G. Poizat, G. Chene, E. Dam, G. Raguin, A. M. Taburet, P. M. Girard, A. J. Hance, and F. Clavel. 2006. Low genetic barrier to large increases in HIV-1 cross-resistance to protease inhibitors during salvage therapy. Antivir. Ther. 11:143-154. [PubMed] [Google Scholar]
- 20.Morse, G. D., L. M. Catanzaro, and E. P. Acosta. 2006. Clinical pharmacodynamics of HIV-1 protease inhibitors: use of inhibitory quotients to optimise pharmacotherapy. Lancet Infect. Dis. 6:215-225. [DOI] [PubMed] [Google Scholar]
- 21.Nettles, R. E., T. L. Kieffer, T. Parsons, J. Johnson, J. Cofrancesco, Jr., J. E. Gallant, K. A. Carson, R. F. Siliciano, and C. Flexner. 2006. Marked intraindividual variability in antiretroviral concentrations may limit the utility of therapeutic drug monitoring. Clin. Infect. Dis. 42:1189-1196. [DOI] [PubMed] [Google Scholar]
- 22.Pellegrin, I., D. Breilh, G. Coureau, S. Boucher, D. Neau, P. Merel, D. Lacoste, H. Fleury, M. C. Saux, J. L. Pellegrin, E. Lazaro, F. Dabis, and R. Thiebaut. 2007. Interpretation of genotype and pharmacokinetics for resistance to fosamprenavir-ritonavir-based regimens in antiretroviral-experienced patients. Antimicrob. Agents Chemother. 51:1473-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pellegrin, I., D. Breilh, J. M. Ragnaud, S. Boucher, D. Neau, H. Fleury, M. H. Schrive, M. C. Saux, J. L. Pellegrin, E. Lazaro, and M. Vray. 2006. Virological responses to atazanavir-ritonavir-based regimens: resistance-substitutions score and pharmacokinetic parameters (Reyaphar study). Antivir. Ther. 11:421-429. [PubMed] [Google Scholar]
- 24.Piliero, P. J. 2002. The utility of inhibitory quotients in determining the relative potency of protease inhibitors. AIDS 16:799-800. [DOI] [PubMed] [Google Scholar]
- 25.Race, E., E. Dam, V. Obry, S. Paulous, and F. Clavel. 1999. Analysis of HIV cross-resistance to protease inhibitors using a rapid single-cycle recombinant virus assay for patients failing on combination therapies. AIDS 13:2061-2068. [DOI] [PubMed] [Google Scholar]
- 26.Raguin, G., G. Chene, L. Morand-Joubert, A. M. Taburet, C. Droz, C. Le Tiec, F. Clavel, and P. M. Girard. 2004. Salvage therapy with amprenavir, lopinavir and ritonavir 200 mg/d or 400 mg/d in HIV-infected patients in virological failure. Antivir. Ther. 9:615-625. [PubMed] [Google Scholar]
- 27.Ribera, E., L. F. Lopez-Cortes, V. Soriano, J. L. Casado, and J. Mallolas. 2005. Therapeutic drug monitoring and the inhibitory quotient of antiretroviral drugs: can they be applied to the current situation? Enferm. Infecc. Microbiol. Clin. 23:55-67. [PubMed] [Google Scholar]
- 28.Sadler, B. M., C. Gillotin, Y. Lou, and D. S. Stein. 2001. Pharmacokinetic and pharmacodynamic study of the human immunodeficiency virus protease inhibitor amprenavir after multiple oral dosing. Antimicrob. Agents Chemother. 45:30-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taburet, A. M., G. Raguin, C. Le Tiec, C. Droz, A. Barrail, I. Vincent, L. Morand-Joubert, G. Chene, F. Clavel, and P. M. Girard. 2004. Interactions between amprenavir and the lopinavir-ritonavir combination in heavily pretreated patients infected with human immunodeficiency virus. Clin. Pharmacol. Ther. 75:310-323. [DOI] [PubMed] [Google Scholar]
- 30.Torti, C., M. C. Uccelli, E. Quiros-Roldan, F. Gargiulo, V. Tirelli, G. Lapadula, M. Regazzi, P. Pierotti, C. Tinelli, A. De Luca, A. Patroni, N. Manca, and G. Carosi. 2006. Prediction of early and confirmed virological response by genotypic inhibitory quotients for lopinavir in patients naive for lopinavir with limited exposure to previous protease inhibitors. J. Clin. Virol. 35:414-419. [DOI] [PubMed] [Google Scholar]
- 31.Valer, L., C. de Mendoza, and V. Soriano. 2005. Predictive value of drug levels, HIV genotyping, and the genotypic inhibitory quotient (GIQ) on response to saquinavir/ritonavir in antiretroviral-experienced HIV-infected patients. J. Med. Virol. 77:460-464. [DOI] [PubMed] [Google Scholar]