Abstract
The in vitro activities of 10 families of antimicrobial agents alone and in combination with a synthetic polycationic polymer, polyethylenimine (PEI), against a resistant clinical isolate of Pseudomonas aeruginosa were investigated by MIC assays, checkerboard testing, and killing curve studies. At a concentration of 250 nM, PEI (10 kDa) was not directly bactericidal or bacteriostatic; but when it was used in combination with novobiocin, ceftazidime, ampicillin, ticarcillin, carbenicillin, piperacillin, cefotaxime, chloramphenicol, rifampin, or norfloxacin, it significantly reduced the MICs of these antibiotics by 1.5- to 56-fold. However, the MICs of aminoglycosides, polymyxins, and vancomycins were increased by 1.2- to 5-fold when these drugs were combined with PEI; and the MICs of tetracycline, erythromycin, ciprofloxacin, and ofloxacin were not affected when these drugs were combined with PEI. In the killing curve studies, combinations of PEI with novobiocin, ceftazidime, chloramphenicol, or rifampin resulted in 5- to 8-log10 CFU/ml reductions in bacterial counts when 25% of the MIC of each antibiotic was used. These results indicate that infections due to resistant Pseudomonas strains could be treated by the use of a synergistic combination of PEI and antimicrobial drugs.
Over the past two decades, Pseudomonas aeruginosa has attracted attention as an opportunistic pathogen in hospitalized, immunocompromised, and cystic fibrosis patients (6, 15, 20). Despite aggressive antibiotic therapy, P. aeruginosa is rarely eradicated owing to its high level of intrinsic resistance to many drugs (14, 19, 27). This resistance is due to the effective permeability of the outer membrane (OM) of gram-negative bacteria to both hydrophobic antibiotics and high-molecular-weight hydrophilic drugs (for reviews, see references 4, 25, 28, and 30). Unlike most cell types, gram-negative bacteria surround themselves with a double membrane. The outermost of these two membranes is asymmetric, with the inner leaflet composed of glycerophospholipids and the outer leaflet predominantly composed of lipopolysaccharide (LPS). Tight interactions between the highly negatively charged LPS molecules are believed to form an effective barrier against hydrophobic compounds. For these polyanionic molecules to form a stable “tiled roof” on the surface of the OM, adjacent LPS molecules are linked electrostatically by divalent cations (Ca2+, Mg2+) present in the OM.
Polyethylenimine (PEI) is a weakly basic, aliphatic, nontoxic synthetic polymer which is polycationic owing to the presence of primary, secondary, and tertiary amino groups. It is well known that certain polycationic agents such as polymyxin and its derivatives polylysines and protamine can increase the permeability of the gram-negative bacterial OM to solutes that are normally unable to penetrate (25). Helander et al. found that, when it was applied alone, PEI has a strong permeabilizing effect but no bactericidal effect on gram-negative bacteria (8). Escherichia coli, P. aeruginosa, and Salmonella enterica serovar Typhimurium were effectively sensitized to hydrophobic antibiotics (erythromycin, novobiocin, rifampin, clindamycin, and fucidin) by PEI in an agar diffusion assay (8). In the present study, we have investigated whether this permeabilizer compound could be used in combination with a large spectrum of antibiotics against a resistant clinical strain of P. aeruginosa. To assess the synergistic effects of PEI with various antibiotics, three currently used methods were used. First, we evaluated the MIC of each antibiotic with or without PEI. The synergism was also assessed by measuring the fractional inhibitory concentration (FIC) indices of each combination by checkerboard testing (3). Finally, synergism was measured by the killing curve test. Our results indicate that PEI enhances the bactericidal efficacies of both hydrophilic and hydrophobic antibiotics, with some exceptions. PEI reduces the uptake of polycationic antibiotics of the polymyxin group and the uptake of aminoglycosides, probably owing to competition for the cation-binding sites of bacterial LPS. All evidence indicates that the enhanced bactericidal activities of antibiotics in the presence of PEI is because of its strong binding to LPS, which leads to disorganization of the bacterial OM and facilitates antibiotic penetration.
MATERIALS AND METHODS
Reagents.
PEI (mean molecular weight, 10 kDa) was purchased from Polysciences Inc. (Warrington, PA). A stock solution of PEI, in which the pH was adjusted to 7.4, was freshly prepared and filter sterilized through a 0.22-μm-pore-size membrane. Novobiocin, tobramycin, kanamycin, gentamicin, tetracycline, cefotaxime, chloramphenicol, erythromycin, polymyxin B, polymyxin E, rifampin, vancomycin, ofloxacin, and norfloxacin were obtained from Sigma (Oakville, Ontario, Canada). Ciprofloxacin was from Bayer (Kankakee, IL), and ceftazidime was from Glaxo Canada Inc. (Montréal, Quebec, Canada). The chemical structures of the antibiotics tested are shown in Fig. 1.
FIG. 1.
Chemical structures of the hydrophilic (A) and the hydrophobic (B) drugs tested in this study.
Bacterial strains and growth conditions.
The resistant clinical strains used for this experiment were P. aeruginosa 100609, which was isolated from the sputum of a cystic fibrosis patient, and Pseudomonas aeruginosa ATCC 25619, which was obtained from the American Type Culture Collection (ATCC; Rockville, MD). The microorganisms were stored at −70°C in brain heart infusion broth (Difco Laboratories, Detroit, MI) supplemented with 10% glycerol. The growth medium in all liquid cultures was tryptic soy broth (TSB; Difco Laboratories).
MIC determinations.
The MICs were determined by broth microdilution assay in accordance with the procedures recommended by the Clinical and Laboratory Standards Institute (formerly the National Committee for Clinical Laboratory Standards) (5). Briefly, a bacterial inoculum of 50 μl, corresponding to 5 × 105 CFU/ml (optical density at 660 nm, 0.02), with or without PEI (final concentration, 250 nM), was added to 50 μl of serial twofold dilutions of the antibiotics in the wells of microtiter plates (Sarstedt, Quebec, Canada). The plates were incubated under agitation at 37°C for 20 h. The MIC was defined as the lowest concentration of antibiotic that inhibited visible growth. The MICs were determined in triplicate.
Checkerboard testing.
Checkerboard tests were performed in triplicate for all combinations, as described previously (5). TSB with an inoculum of approximately 5 × 105 CFU/ml was used for all checkerboard assays. Synergy was measured by determination of the FIC, which is the ratio of the MIC of a drug in combination and MIC of the drug alone. For two interacting compounds A and B, the sum of the FICs (ΣFIC = FICA + FICB) indicates the extent of the interaction. When ΣFIC is ≤0.5, there is a synergistic effect. A ΣFIC of 0.5 to 2.0 is defined as indifference, and antagonism is defined as a ΣFIC of ≥4.0 (29).
Killing curve studies.
In vitro killing curve studies were performed as described previously (10, 12), with modifications. Briefly, TSB was inoculated with a 17-h preculture of P. aeruginosa 100609, giving a stock preparation containing a final inoculum of 5 × 107 CFU/ml, as determined by measurement of the optical density at 660 nm. This value allowed the control growth to reach a density of 108 to 1010 CFU/ml. PEI was added to give a final concentration of 250 nM. Studies were performed in the presence of sub-MICs (25% of the MIC) of each antibiotic and in the presence of controls comprising PEI alone, antibiotics alone, or bacteria alone. After 0, 1.5, 3, 6, and 9 h, samples were serially diluted and plated onto LB agar plates to obtain viable colony counts. Experiments were performed in triplicate. Synergy according to time-concentration killing curves was defined as a reduction in the mean log10 CFU/ml bacterial counts of ≥2.0 at any time during the 9-h experiment when the time-concentration killing curves generated with the combination of antibiotic and PEI were compared with those generated with the antibiotic alone. Indifference or antagonism in time-concentration killing curves was defined as 0 to 2 or ≤0 reductions in the mean log10 CFU/ml bacterial counts, respectively, at any time point during the 9-h experiments (10).
RESULTS
MIC and checkerboard synergy.
Table 1 shows the MICs obtained with P. aeruginosa 100609 grown in TSB with 16 antibiotics both in the absence of PEI and in the presence of 250 nM PEI. This PEI concentration was subinhibitory; indeed, the MIC of PEI for P. aeruginosa 100609 was 1 μM. PEI at a final concentration of 250 nM reduced the MICs of novobiocin, ceftazidime, cefotaxime, chloramphenicol, rifampin, and norfloxacin by 1.5- to 40-fold. With the exception of norfloxacin, all these antibiotics had synergistic effects with PEI (FICs ≤ 0.5) by checkerboard testing. By contrast, with the aminoglycosides, polymyxins, and vancomycin, 250 nM PEI increased the MICs. Of these antibiotics, only tobramycin and vancomycin had FICs of ≥4.0 (antagonism); the others had an indifferent effect by checkerboard testing. With tetracycline, erythromycin, ciprofloxacin, and ofloxacin, PEI had no effect on the MIC. The synergistic effect of PEI in combination with antibiotics was not strain specific, as shown in Table 2; an additional strain, P. aeruginosa 25619, was tested for the synergistic effect of PEI with novobiocin and ceftazidime. Although the degree of synergistic values for the different strains were different, the positive effect was confirmed for both of the strains.
TABLE 1.
Effect of PEI on efficacies of different families of antibiotics against P. aeruginosa 100609
| Antibiotic class | Antibiotic | MIC (μg/ml) with:
|
Checkerboard FIC index (interpretationa) with PEI (250 μg/ml) | Checkerboard effect | Killing curve effect | |
|---|---|---|---|---|---|---|
| Antibiotic alone | Antibiotic and PEI (250 μg/ml) | |||||
| Novobiocin | Novobiocin | 1,600 | 140 | 70.10 (S) | Synergy | Synergy |
| Aminoglycosides | Tobramycin | 50 | 240 | 4.81 (I) | Antagonism | Indifference |
| Kanamycin A | 1,600 | 1,920 | 1.21 (I) | Indifference | NDb | |
| Gentamicin | 1,120 | 1,920 | 1.72 (I) | Indifference | ND | |
| Tetracyclines | Tetracycline | 20 | 20 | 1.00 (I) | Indifference | ND |
| Cephalosporins | Ceftazidime | 400 | 10 | 0.04 (S) | Synergy | Synergy |
| Cefotaxime | 960 | 120 | 0.14 (S) | Synergy | ND | |
| Chloramphenicol | Chloramphenicol | 100 | 40 | 0.41 (S) | Synergy | Synergy |
| Macrolides | Erythromycin | 140 | 140 | 1.00 (I) | Indifference | ND |
| Polymyxins | Polymyxin E | 5 | 10 | 2.01 (I) | Antagonism | ND |
| Polymyxin B | 5 | 10 | 2.01 (I) | Antagonism | Antagonism | |
| Rifamycins | Rifampin | 20 | 10 | 0.05 (S) | Synergy | Synergy |
| Cationic glycopeptides | Vancomycin | 1,120 | 3,800 | 3.40 (I) | Antagonism | Antagonism |
| Fluoroquinolones | Ciprofloxacin | 30 | 30 | 1.00 (I) | Indifference | Indifference |
| Ofloxacin | 10 | 10 | 1.00 (I) | Indifference | ND | |
| Norfloxacin | 15 | 10 | 0.68 (I) | Indifference | ND | |
| β-Lactams | Ampicillin | 2,240 | 100c | ND | ND | Synergy |
| Carbenicillin | 2,240 | 60c | ND | ND | Synergy | |
| Piperacillin | 40 | 7.5c | ND | ND | Synergy | |
| Ticarcillin | 1,120 | 20c | ND | ND | Synergy | |
S, synergy; I, indifference.
ND, not determined.
Synergy between β-lactams and PEI was investigated in a previous study in which the PEI concentration was 200 nM (11).
TABLE 2.
Synergistic effect of PEI in combination with novobiocin and ceftazidime against different P. aeruginosa strains
| Organism | Antibiotic | Checkerboard FIC index (interpretation) | Killing curve effecta |
|---|---|---|---|
| P. aeruginosa 100609 | Novobiocin | 0.10 (Sb) | Synergy |
| Ceftazidime | 0.04 (S) | Synergy | |
| P. aeruginosa 25619 | Novobiocin | 0.35 (S) | Synergy |
| Ceftazidime | 0.09 (S) | Synergy |
Combination of 250 μg/ml PEI and one-fourth the MIC of novobiocin or ceftazidime.
S, synergy.
In vitro determination of antibacterial action over different time intervals.
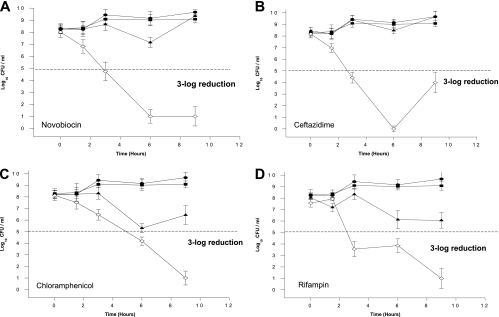
Eight antibiotics from different classes were chosen for time-kill curve studies. Of these, four had reductions in the MICs when they were administered in combination with PEI (Fig. 2), and the others had either an increase or no change in MIC (Fig. 3). For all killing curves, PEI alone (250 nM) had no effect on bacterial growth compared with the control growth. Figure 2A shows the effect of PEI on the bactericidal activity of novobiocin (25% of the MIC). When novobiocin and PEI were used in combination, bacterial growth was reduced more than 108 times after 9 h, whereas controls and novobiocin alone had no significant effect. The combination of PEI with ceftazidime (25% of the MIC) (Fig. 2B), chloramphenicol (25% of the MIC) (Fig. 2C), or rifampin (25% of the MIC) (Fig. 2D) increased the reductions in the numbers of P. aeruginosa CFU 105 times compared with the numbers of CFU obtained with each antibiotic alone after 9 h.
FIG. 2.
In vitro growth and killing curves of Pseudomonas aeruginosa 100609 exposed to four hydrophobic antibiotics whose activities are synergistic with PEI. •, control growth curve of the strain in the absence of antibiotics and PEI; ▪, growth curve with PEI (250 μg/ml); ▴, growth curve with antibiotic (25% of the MIC); and ⋄, growth curve with antibiotic (25% of MIC) and PEI (250 μg/ml). Data are the means ± standard deviations of an experiment performed in triplicate.
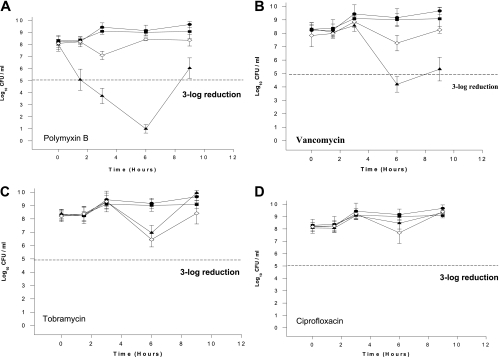
FIG. 3.
In vitro growth and killing curves of Pseudomonas aeruginosa 100609 exposed to three antibiotics antagonized or not affected by PEI. •, control growth curve of the strain in the absence of antibiotics and PEI; ▪, growth curve with PEI (250 μg/ml); ▴, growth curve with antibiotic (25% of the MIC); and ⋄, growth curve with antibiotic (25% of MIC) and PEI (250 μg/ml). Data are the means ± standard deviations of an experiment performed in triplicate.
By contrast, PEI reduced the bactericidal efficacies of polymyxin B and vancomycin. Bacterial growth was increased by 2 log10 after 9 h when PEI was combined with these antibiotics (Fig. 3A and B). Finally, when PEI was combined with tobramycin and ciprofloxacin, there were no visible differences between the efficacies of the combination, the antibiotic alone, and the controls (Fig. 3C and D).
DISCUSSION
Several agents have been used in attempts to alter the OM permeabilities of gram-negative bacteria (4); these include EDTA and lysozyme (2, 13), sodium hexametasulfonate (26), detergents like sodium dodecyl sulfate (1) and deoxycholate, as well as permeabilizing portions of antibiotics such as polymyxin B (18, 23) and other compounds containing positively charged amino groups, such as spermidine and lysine (24) and methylglyoxal bis-guanylhydrazone (16).
In the present study, we measured the effect of PEI on the bactericidal activities of 16 antibiotics of 10 different classes. PEI reduced the MICs of hydrophilic and hydrophobic compounds (with some exceptions) by 1.5- to 40-fold (Table 2) and increased their bactericidal effects in killing curve experiments (Fig. 2). We have previously demonstrated that PEI also reduced the MICs of four β-lactams by 5- to 50-fold.
An intact OM of gram-negative bacteria generally prevents the diffusion of hydrophobic solutes because its outer leaflet contains no glycerophospholipids but only highly ordered, quasicrystalline LPS (10, 12, 25). PEI has recently been reported to make gram-negative bacteria permeable to hydrophobic antibiotics (8). This polycationic polyamine has all the characteristics of a permeabilizer, as described by Vaara (25). Helander et al. (8) demonstrated that PEI (50 kDa) increased the bacterial uptake of 1-N-phenylnaphthylamine, a hydrophobic probe; sensitized the bacteria to hydrophobic antibiotics in an agar diffusion assay; and increased the susceptibility to lysozyme and the nonionic detergent Triton X-100. However, in contrast to other permeabilizers such as protamine and polylysines, PEI does not induce the release of LPS from the OM. Similar results were also reported for polymyxin B nonapetide (PMBN) (11).
Our results show that, depending on the chemical structures of the antibiotics tested, PEI can have a synergistic, an antagonistic, or no effect when it is used in combination with antibiotics. Synergistic effects were observed with both hydrophobic and hydrophilic antibiotics. Table 1 shows that a PEI concentration as low as 250 nM reduced the MICs of hydrophobic antibiotics such as chloramphenicol, novobiocin, and rifampin by 2- to 12-fold. This synergistic effect was also confirmed by checkerboard testing and killing curve studies (Fig. 2). All three hydrophobic antibiotics had FIC indices of <0.5, indicating synergy between PEI and the antibiotic. The molecular mechanism by which PEI increases the permeability of the OM has not been determined, but it is not due to any marked LPS release (9). The enhanced antibacterial activity is probably due to a redistribution of phospholipids from the inner to the outer layer of the OM owing to PEI. This would increase the bacterial membrane permeability, cause partitioning, and therefore allow the entry of hydrophobic antibacterial agents. The mechanisms may be similar to those of other agents known to be permeabilizers.
Two exceptions were observed among the hydrophobic drugs tested. First, the bactericidal efficacy of norfloxacin was only slightly increased by PEI, and the magnitude of the increase was not sufficient to decrease the FIC index to less than 0.5. This is due to a gyrase mutation, which results in fluoroquinolone resistance. Therefore, an increase in the uptake of norfloxacin does not significantly affect bacterial growth. The second exception was erythromycin: PEI had no effect on its bactericidal efficacy. This hydrophobic antibiotic is often used in experiments investigating the permeabilizing effect of E. coli but not P. aeruginosa membranes. This is because erythromycin has no effect on P. aeruginosa proliferation in vitro and only inhibits the production of virulence factors (22). These results are in agreement with those obtained by Helander et al. (8). Further studies with different bacterial strains will help show that PEI could have the same effect on erythromycin as PMBN, which increases the sensitivity of E. coli cells by 8- to 30-fold (25).
Among the hydrophilic drugs tested, we observed reductions in the MICs of 5- to 56-fold when ceftazidime, cefotaxime, ampicillin, carbenicillin, piperacillin, and ticarcillin were tested in combination with PEI (Table 1). Hydrophilic compounds usually cross the OM via porin channels (2). In contrast to PMBN, PEI has an effect on the MICs of antibiotics which are believed to traverse the OM through porin pores (25).
On the other hand, PEI reduced the bactericidal activities of polymyxins and aminoglycosides, and the intensity of the decrease was 1.2- to 4.8-fold (Table 1). These antibiotics act by binding to the structure of the OM of the bacteria at a site which is probably located in the deep core region and includes part of lipid A of LPSs (11). The interaction between PEI and these positively charged hydrophilic antibiotics seems to be a competition for the same binding site, because PEI has the highest positive charge density in comparison to those of other peptides, polymers, or antibiotics reported to have a positive charge; so the affinity between PEI and the OM should be greater than that between the antibiotics and the OM.
A possible criticism of the procedure used here is that the permeabilizer might sequester ions from the medium, which could result in the production of either weakened cells or nutritionally defective cells, or both. However, this would not explain the effect of PEI on polymyxins and aminoglycosides, nor would it explain the increased uptake of 1-N-phenylnaphthylamine observed by Helander et al. (8).
P. aeruginosa infections are difficult to overcome owing to the high intrinsic resistance of P. aeruginosa to antibiotics. The primary cause of such resistance is attributed to reduced OM permeability (4, 7, 17). It has been estimated that the rate of permeation of β-lactam antibiotics across the P. aeruginosa OM is about 12-fold less than that across E. coli OMs (21). One possible therapeutic strategy against P. aeruginosa, therefore, might be to attempt to minimize the reduction in OM permeability by cotreatment with permeabilizers such as PEI, thus allowing the increased uptake of antibiotics across the OM. Further work is required to investigate the ability of PEI to increase the susceptibilities of other strains, such as Burkholderia cepacia and E. coli, to antibacterial agents.
Footnotes
Published ahead of print on 19 February 2008.
REFERENCES
- 1.Ayres, H. M., J. R. Furr, and A. D. Russell. 1998. Effect of divalent cations on permeabilizer-induced lysozyme lysis of Pseudomonas aeruginosa. Lett. Appl. Microbiol. 27:372-374. [DOI] [PubMed] [Google Scholar]
- 2.Bell, A., M. Brains, and R. E. W. Hancock. 1990. Pseudomonas aeruginosa outer membrane OprH: expression from the cloned gene and function in EDTA and gentamicin resistance. J. Bacteriol. 173:6657-6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bereenbaum, M. C. 1978. A method for testing for synergy with any number of agents. J. Infect. Dis. 137:122-130. [DOI] [PubMed] [Google Scholar]
- 4.Champlin, F. R., M. L. Ellison, J. W. Bullard, and R. S. Conrad. 2005. Effect of outer membrane permeabilisation on intrinsic resistance to low triclosan levels in Pseudomonas aeruginosa. Int. J. Antimicrob. Agents 26:159-164. [DOI] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 1993. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A4. Clinical and Laboratory Standards Institute, Villanova, PA.
- 6.Davis, P. B., M. Drumm, and M. W. Konstan. 1996. Cystic fibrosis. Am. J. Respir. Crit. Care Med. 154:1229-1256. [DOI] [PubMed] [Google Scholar]
- 7.Gotoh, N. 2001. Antibiotic resistance caused by membrane impermeability and multidrug efflux systems. Nippon Rinsho 59:712-718. [PubMed] [Google Scholar]
- 8.Helander, I. M., H. Alakomi, K. Latva-Kala, and P. Koski. 1997. Polyethylenimine is an effective permeabilizer of gram-negative bacteria. Microbiology 143:3193-3199. [DOI] [PubMed] [Google Scholar]
- 9.Helander, I. M., K. Latva-Kala, and K. Lounatmaa. 1998. Permeabilizing action of polyethylenimine on Salmonella typhimurium involves disruption of the outer membrane and interactions with lipopolysaccharide. Microbiology 144:385-390. [DOI] [PubMed] [Google Scholar]
- 10.Karlowsky J. A., G. A. Harding, S. A. Zelenitsky, D. J. Hoban, A. Kabani, T. V. Balko, M. Turik, and G. G. Zhanel. 1997. In vitro kill curves of a new semisynthetic echinocandin, LY-303366, against fluconazole-sensitive and -resistant Candida species. Antimicrob. Agents Chemother. 41:2576-2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwon, D. H., and C. D. Lu. 2006. Polyamines increase antibiotic susceptibility in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 50:1623-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Labischinski, H., G. Barnickel, H. Bradaczek, D. Naumann, E. T. Rietschel, and P. Giesbrecht. 1985. High state of order of isolated bacterial lipopolysaccharide and its possible contribution to the permeation barrier property of the outer membrane. J. Bacteriol. 162:9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lambert, R. J., G. W. Hanlon, and S. P. Denyer. 2004. The synergistic effect of EDTA/antimicrobial combinations on Pseudomonas aeruginosa. J. Appl. Microbiol. 96:244-253. [DOI] [PubMed] [Google Scholar]
- 14.Leonard, A., T. Leal, and P. Lebecque. 2007. Cystic fibrosis and Pseudomonas aeruginosa: current and future strategies. J. Pharm. Belg. 62:25-28. [PubMed] [Google Scholar]
- 15.Marelich, G. P., and C. E. Cross. 1996. Cystic fibrosis in adults. From researcher to practitioner. West. J. Med. 164:321-334. [PMC free article] [PubMed] [Google Scholar]
- 16.Modha, J. L., K. J. Barrett-Bee, and R. J. Rowbury. 1990. Enhancement by cationic compounds of the growth inhibitory effect of novobiocin on Escherichia coli. Lett. Appl. Microbiol. 8:219-222. [Google Scholar]
- 17.Nikas, T. I., and R. E. W. Hancock. 1983. Pseudomonas aeruginosa outer membrane permeability: isolation of a porin protein F-deficient mutant. J. Bacteriol. 153:281-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ostronoff, M., F. Ostronoff, A. Sucupira, A. P. Souto Maior, M. Caniza, and R. Florencio. 2006. Multidrug-resistant Pseudomonas aeruginosa infection in neutropenic patients successfully treated with a combination of polymyxin B and rifampin. Int. J. Infect. Dis. 10:339-340. [DOI] [PubMed] [Google Scholar]
- 19.Pier, G. B. 1986. Pulmonary disease associated with Pseudomonas aeruginosa in cystic fibrosis: current status of the host-bacterium interaction. J. Infect. Dis. 151:575-580. [DOI] [PubMed] [Google Scholar]
- 20.Ramsey, B. W. 1996. Management of pulmonary disease in patients with cystic fibrosis. N. Engl. J. Med. 335:179-188. [DOI] [PubMed] [Google Scholar]
- 21.Russell, A. D. 1982. Modificatrion of the bacterial cell envelope and enhancement of antibiotic susceptibility, p. 119-168. In C. H. Stuart-Harris and D. M. Harris (ed.), The control of antibiotic-resistant bacteria. Academic Press, Inc., New York, NY.
- 22.Sakata, K., H. Yajima, K. Tanaka, Y. Sakamoto, K. Yamamoto, A. Yoshida, and Y. Dohi. 1993. Erythromycin inhibits the production of elastase by Pseudomonas aeruginosa without affecting its proliferation in vitro. Am. Rev. Respir. Dis. 148:1061-1065. [DOI] [PubMed] [Google Scholar]
- 23.Tam, V. H., A. N. Schilling, G. Vo, S. Kabbara, A. L. Kwa, and N. P. Wiederhold. 2005. Pharmacodynamics of polymyxin B against Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 49:3624-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaara, M. 1990. The effect of oligolysines Lys-3, Lys-4 and Lys-5 on the outer membrane permeability of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 55:15-19. [DOI] [PubMed] [Google Scholar]
- 25.Vaara, M. 1992. Agents that increase the permeability of the outer membrane. Microbiol. Rev. 56:395-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaara, M., and J. Jaakkola. 1989. Sodium hexametaphosphate sensitizes Pseudomonas aeruginosa, several other species of Pseudomonas, and Escherichia coli to hydrophobic drugs. Antimicrob. Agents Chemother. 33:1741-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vasil, M. L. 1986. Pseudomonas aeruginosa mechanism of virulence epidemiology. J. Pediatr. 108:800-805. [DOI] [PubMed] [Google Scholar]
- 28.Waters, V., and F. Ratjen. 2006. Multidrug-resistant organisms in cystic fibrosis: management and infection-control issues. Expert Rev. Anti-Infect. Ther. 4:807-819. [DOI] [PubMed] [Google Scholar]
- 29.White, R. L., D. S. Burgess, M. Manduru, and J. A. Bosso. 1996. Comparison of three different in vitro methods of detecting synergy: time-kill, checkerboard, and E test. Antimicrob. Agents Chemother. 40:1914-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yokoyama, T., M. Sugai, M. Ohara, and Y. Imamura. 2007. Multi-drug resistant Pseudomonas aeruginosa infection. J. Nippon Rinsho 65(Suppl. 3):418-422. [PubMed] [Google Scholar]