Abstract
Oxazolidinone and pleuromutilin antibiotics are currently used in the treatment of staphylococcal infections. Although both antibiotics inhibit protein synthesis and have overlapping binding regions on 23S rRNA, the potential for cross-resistance between the two classes through target site mutations has not been thoroughly examined. Mutants of Staphylococcus aureus resistant to linezolid were selected and found to exhibit cross-resistance to tiamulin, a member of the pleuromutilin class of antibiotics. However, resistance was unidirectional because mutants of S. aureus selected for resistance to tiamulin did not exhibit cross-resistance to linezolid. This contrasts with the recently described PhLOPSA phenotype, which confers resistance to both oxazolidinones and pleuromutilins. The genotypes responsible for the phenotypes we observed were examined. Selection with tiamulin resulted in recovery of mutants with changes in the single-copy rplC gene (Gly155Arg, Ser158Leu, or Arg149Ser), whereas selection with linezolid led to recovery of mutants with changes (G2576U in 23S rRNA) in all five copies of the multicopy operon rrn. In contrast, cross-resistance to linezolid was exhibited by tiamulin-resistant mutants generated in a single-copy rrn knockout strains of Escherichia coli, illustrating that the copy number of 23S rRNA is the limiting factor in the selection of 23S rRNA tiamulin-resistant mutants. The interactions of linezolid and tiamulin with the ribosome were modeled to seek explanations for resistance to both classes in the 23S rRNA mutants and the lack of cross-resistance between tiamulin and linezolid following mutation in rplC.
Linezolid (LZD) is a synthetic antimicrobial agent belonging to the oxazolidinone class. It inhibits bacterial protein synthesis and has recently been introduced for the treatment of infections caused by gram-positive pathogens, including Staphylococcus aureus. Despite early predictions that LZD would display a low potential for the development of resistance, resistant clinical isolates of S. aureus with a G2576U mutation in all copies of the 23S rRNA gene have, unfortunately, emerged following prolonged therapy with the agent (13, 23, 24).
Pleuromutilins, such as tiamulin (TML) and valnemulin (VML), are used in veterinary medicine to treat infections caused by gram-positive pathogens. TML, the most widely used pleuromutilin, is a semisynthetic inhibitor of bacterial protein synthesis, most commonly used in the treatment of Brachyspira hyodysenteria infections in swine. Another pleuromutilin derivative, retapamulin, has recently been approved for the treatment of impetigo caused by methicillin-susceptible S. aureus and Streptococcus pyogenes (6, 19), and further members of the pleuromutilin class are in development as potential oral agents for systemic application (Nabriva, Vienna, Austria).
Pleuromutilin resistance has been identified in clinical isolates of Brachyspira spp. and in Escherichia coli, Streptococcus pyogenes, and S. aureus isolates selected in vitro (1, 2, 5, 8, 9, 18). Furthermore, chemical rRNA footprinting and mutational information show overlapping binding regions on the 23S rRNA, suggesting common loci for resistance to pleuromutilin and oxazolidinone antibiotics may occur (18). In view of the increasing interest in the application of pleuromutilins to control staphylococcal infections in humans and the recent observation that the PhLOPSA phenotype, caused by Cfr-mediated methylation of nucleotide A2503 of the 23S rRNA, confers resistance to both LZD and TML in S. aureus (12, 22), we have investigated whether other mechanisms of resistance to linezolid in clinical isolates of S. aureus might also confer cross-resistance to pleuromutilins.
(Portions of the work presented here were recently presented at the 46th Interscience Conference on Antimicrobial Agents and Chemotherapy [15].)
MATERIALS AND METHODS
Bacterial strains, plasmids, antibiotics, chemicals, and growth media.
Methicillin-sensitive S. aureus strains RN4220, RN4220mutS, and 8325-4 (University of Leeds culture collection), LZD-resistant S. aureus clinical isolates NRS119-121 (Network on Antimicrobial Resistance in S. aureus, Herndon, VA), and E. coli SQ110 were used throughout the present study. E. coli SQ110 is a single-copy rrn operon construct kindly provided by Catherine Squires, Tufts University, Boston, MA. The pDG148-StuI gram-positive-E. coli shuttle vector (7) was used for cloning (Bacillus Genetic Stock Centre, Columbus, OH). Chemicals and antibiotics were from Sigma-Aldrich (Poole, United Kingdom), with the exception of LZD, which was a gift from Pfizer (Kalamazoo, MI), and TML, which was a gift from Sandoz (Vienna, Austria). Mueller-Hinton broth (MHB) and Mueller-Hinton agar were from Fisher (Loughborough, United Kingdom).
Determination of susceptibility to antimicrobial agents.
MICs were determined by agar dilution according to BSAC guidelines (3) in MHB. For E. coli, MHB was supplemented with 16 μg of polymyxin B nonapeptide (PMBN)/ml to permeabilize the gram-negative cell envelope (4).
Determination of mutation frequencies and timelines for emergence of resistance to antibiotics.
Spontaneous mutation frequency determinations were performed essentially as described by O'Neill et al. (17) using MH growth medium (supplemented with 16 μg of PMBN/ml for E. coli). In cases where the mutation frequencies were below the level of detection by direct plating (<10−9), a semiquantitative method for estimating mutation frequency was used in which mutants were selected by serial subculture in the presence of antibiotics (14), and the time to emergence of resistance was recorded.
PCR amplification and DNA sequencing.
The 23S rRNA alleles of S. aureus strains were amplified according to the method of Tsiodras et al. (23).
Oligonucleotide primers designed by using the Oligo 6.0 program (Molecular Biology Insights, Cascade, CO) were used to amplify the rplC and rplD genes from both E. coli and S. aureus (E. coli, rplC [5′-GAACCAGCGCTTCGTTG-3′ and 5′-TGATGCTCTGATGCGTCTG-3′] and rplD [5′-CGCTTTTTCAGAAACGTGC-3′ and 5′-AGCTGTGAAGGCGTAAGGA-3′]; S. aureus, rplC [5′-AACCTGATTTAGTTCCGTCTA-3′ and 5′-GTTGACGCTTTAATGGGCTTA-3′] and rplD [5′-TCGCTTACCTCCTTAATG-3′ and 5′-GGTGGAAACACTGTAACTG-3′]) and the resistance-defining region on the E. coli 23S rRNA allele (5′-ATAACGGTCCTAAGGTAGCGAAATTC-3′ and 5′-GCTTACACACCCGGCCTATCAACGTC-3′).
Confirmation of resistance loci.
Mutant rplC genes were amplified from KM164-168 (5′-AAGGAGGAAGCAGGTATGACCAAAGGAATCTTAGGAAGA-3′ and 5′-GACACGCACGAGTTATTTATTACCTTTTTTAATTGAAG-3′) and were cloned in a ligation independent manner described previously into the gram-positive E. coli shuttle vector pDG148-Stu. The construct was propagated in E. coli XL1-Blue after transformation and was then cloned back into electrocompetent, LZD- and TML-sensitive RN4220 cells.
Molecular modeling of mutated residues in the ribosome.
Molecular models of the mutated ribosome were constructed by using Maestro version 7.0110 (Schrödinger LLC, New York, NY) in conjunction with the structure file 1XBP.pdb, which contains TML cocrystallized within the 23S rRNA structure from Deinococcus radiodurans (20). We inserted LZD into our ribosomal model according to the conformation described by Leach et al. (10). The mutant L3 protein was constructed by using the BB modeling package (http://mccammon.ucsd.edu/∼adcock/bb.html) from the backbone coordinates present in the X-ray crystal structure. Residues within 7 Å of bound drug and 10 Å of L3 were used to create the drug-binding sites, extended to incorporate the relevant section of L3 that contained the mutated residues. The lowest energy conformations of the mutated ribosome were then subjected to full energy minimization by using the MMFFs forcefield within Macromodel (version 9.0014; Schrödinger LLC) with water selected as the solvent. During the minimization process, the outer atoms on the periphery of the model were frozen to maintain a compact structure.
RESULTS AND DISCUSSION
Selection of mutants resistant to LZD and TML.
Both LZD and TML possessed low intrinsic resistance potentials against wild-type S. aureus strains exhibiting mutation frequencies of <10−9 (Table 1) . Mutants were also not recovered in a mutator strain of S. aureus (RN4220mutS) defective in the methyl-directed mismatch repair system (16) (data not shown). S. aureus mutants therefore had to be selected by continuous subculture. TML resistance emerged in the multicopy rrn operon S. aureus strains far more rapidly than LZD resistance (Table 1).
TABLE 1.
Mutation frequencies and timelines to resistance for LZD and TML in S. aureus strains and E. coli SQ110a
| Strain | Organism | Antibiotic | MIC (μg/ml) | Mutation frequency | Days of subculture to selection |
|---|---|---|---|---|---|
| RN4220 | S. aureus | LZD | 4 | <10−9 | 21 |
| S. aureus | TML | 1 | <10−9 | 2 | |
| RN4220mutS | S. aureus | LZD | 4 | <10−9 | ND† |
| S. aureus | TML | 2 | <10−9 | 1 | |
| 8325-4 | S. aureus | LZD | 4 | <10−9 | ND† |
| S. aureus | TML | 1 | <10−9 | 1 | |
| SQ110 | E. coli | LZD | 256 | ND* | ND* |
| E. coli | TML | 128 | ND* | ND* | |
| SQ110 (supplemented)b | E. coli | LZD | 16 | (2.86 ± 1.26) × 10−8 | ND† |
| E. coli | TML | 8 | (5.63 ± 2.36) × 10−7 | ND† |
ND*, not determinable; ND†, not determined.
That is, supplemented with 16 μg of PMBN/ml.
Cultures of E. coli strain SQ110 were insusceptible to LZD and TML (Table 1), most likely due to poor penetration of the antibiotics across the outer membrane. However, after exposure to PMBN the MIC of LZD was reduced from 256 to 16 μg/ml and the MIC of TML was reduced from 128 to 8 μg/ml (Table 1), thereby making mutant selection possible. When sensitized with PMBN, the single rrn operon E. coli strain SQ110 had measurable mutation frequencies to both antibiotics (Table 1), and mutants were therefore more readily selected than in the S. aureus strains. This suggests that a low rrn operon copy number favors selection of resistance to both agents.
Genetic analysis of mutants and cross-resistance studies.
LZD-resistant S. aureus mutants KM187 and KM198 derived from the five copy rrn operon strain RN4220 and clinical isolates NRS119-121, also with five copies of rrn, possessed G2576U mutations in all five copies of the 23S rRNA (Table 2). Such mutations are known to confer LZD resistance in S. aureus (13, 21, 23). LZD-resistant E. coli mutant KM55-68 had G2576U mutations on the single copy of the 23S rRNA, whereas KM69-73 had single G2447U 23S rRNA mutations and KM74 had a single U2500A mutation on the 23S rRNA (Table 2). All mutations have previously been implicated in LZD resistance (13, 21, 23). The LZD-resistant mutants listed above were also resistant to TML (Table 2).
TABLE 2.
Properties of parental strains and LZD/TML-resistant derivatives
| Straina | Speciesb | Antibiotic selection | MIC (μg/ml)
|
Mutation
|
||
|---|---|---|---|---|---|---|
| LZD | TML | 23S rRNAc | rplC | |||
| SQ110* | E. coli | 16 | 8 | |||
| KM55-68* | E. coli | LZD | 64 | 64 | G2576U | |
| KM69-73* | E. coli | LZD | 64 | 64 | G2447U | |
| KM74* | E. coli | LZD | 64 | 64 | U2500A | |
| KM75-84* | E. coli | TML | 16 | 32 | Gly153Arg | |
| KM85-86* | E. coli | TML | 16 | 32 | Lys157Gln | |
| KM87-88* | E. coli | TML | 16 | 64 | Arg141Ser | |
| KM89-94* | E. coli | TML | 64 | 64 | G2576U | |
| 8325-4 | S. aureus | 4 | 1 | |||
| KM164-165 | S. aureus | TML | 4 | 8 | Gly155Arg | |
| RN4220mutS | S. aureus | 4 | 2 | |||
| KM166-167 | S. aureus | TML | 4 | 16 | Ser158Leu | |
| RN4220 | S. aureus | 4 | 1 | |||
| KM168 | S. aureus | TML | 4 | 8 | Arg149Ser | |
| KM187+198 | S. aureus | LZD | 64 | 16 | G2576U* | |
| NRS119 | S. aureusT | LZD | 64 | 32 | G2576U* | |
| NRS120 | S. aureusT | LZD | 64 | 32 | G2576U* | |
| NRS121 | S. aureusT | LZD | 64 | 32 | G2576U* | |
*, Strain cultured in the presence of 16 μg of PMBN/ml.
A superscript “T” indicates a clinical isolate.
*, In all five copies.
TML-resistant S. aureus mutants KM164-168, derived from strains with a single copy of rplC and five copies of rrn, all carried mutations in the L3 ribosomal protein encoded by rplC, and remained sensitive to LZD (Table 2). The L3 mutations were confirmed as TML resistance loci by cloning the mutant rplC into pDG148-Stu and introducing the construct into a TML-sensitive S. aureus RN4220 host. All mutations described conferred resistance to TML in the transformed host (data not shown). Fourteen of the twenty TML-resistant E. coli single copy rrn-operon SQ110 mutants also only had mutations in rplC and remained sensitive to LZD (Table 2). The remaining six TML-resistant mutants derived from SQ110 carried G2576U mutations on the 23S rRNA-encoding gene and were cross resistant to LZD (Table 2).
The relative ease with which rplC mutations are acquired, compared to multiple 23S rRNA mutations, explains why TML resistance through selection of 23S rRNA mutations is an unlikely event for strains possessing multiple copies of the rrn operon. Furthermore, the level of TML resistance conferred by mutations in rplC is similar to that mediated by 23S rRNA mutations, suggesting that there is no selective advantage in the accumulation of 23S rRNA mutations.
Homology models.
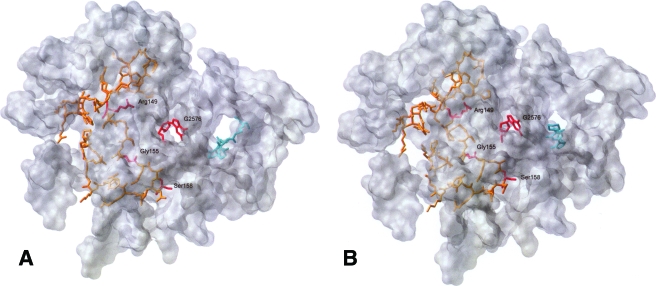
The TML- and LZD-bound staphylococcal ribosome models with the residues (highlighted) to be discussed here are illustrated in Fig. 1A and B, respectively.
FIG. 1.
(A) Model of TML (cyan) bound to the 23S subunit of S. aureus, with ribosomal protein L3 (orange) highlighted. The locations of the rRNA and L3 mutations are shown in red. (B) Model of LZD (cyan) bound to the 23S subunit of S. aureus, with ribosomal protein L3 (orange) highlighted. The locations of the rRNA and L3 mutations are shown in red.
Crystal structures are not available for the staphylococcal ribosome. Nevertheless, despite only 49% sequence homology between the L3 proteins of D. radiodurans and S. aureus, the homology between the 23S rRNA sequences in the active site is 74%, and only A2057G and U2611C nucleotide substitutions were necessary in the active site to form the S. aureus ribosome models. The location of TML was relatively unchanged in the S. aureus homology model (Fig. 2A) compared to the crystal structure of TML bound in the D. radiodurans ribosome (20).
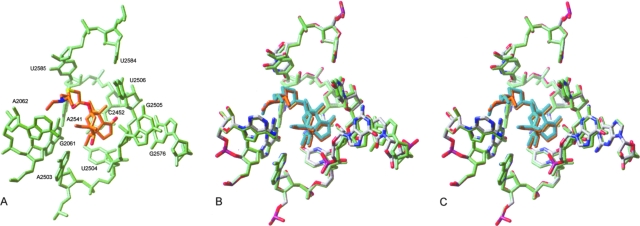
FIG. 2.
(A) Model of TML in its binding site. The nucleotides interacting with the drug are shown in green. The drug molecule is shown in orange (carbon atoms), red (oxygens), blue (nitrogens), and yellow (sulfur). (B) Model of TML (orange) in its binding site (green) overlapped with the positions of key nucleotides (atom colored) and TML (cyan) after G2576U mutation. (C) Model of TML (orange) in its binding site (green) overlapped with the positions of key nucleotides (atom colored) and TML (cyan) after Ser158Leu L3 mutation.
Comparison of our S. aureus model for LZD binding (Fig. 3A) with that reported previously (10) for E. coli revealed that LZD retains a similar conformation upon minimization, with the exception of the central aromatic ring, which is rotated ca. 45° relative to the plane of the oxazolidinone ring. In the reported model, the aromatic ring is directed toward the hydrophobic crevice formed between A2451 and C2452, whereas in our model this ring lies between C2452 and U2506. This alternative positioning of LDZ within the E. coli structure is likely explained by U2506 occupying a different position within the binding site that permits movement of the central aromatic ring to maximize hydrophobic interactions between this residue and C2452. The conformation of the oxazolidinone ring was similar in both E. coli and S. aureus models, which have this ring stacked with the uracil base of U2504, and also feature a hydrogen bond between the amide NH and the 5′ oxygen of G2505. Furthermore, the amide side chain maintained its folded position over the oxazolidinone ring in the S. aureus model (Fig. 3A).
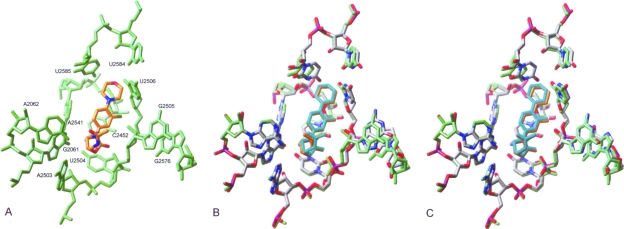
FIG. 3.
(A) Model of LZD in its binding site. The nucleotides interacting with the drug are shown in green. The drug molecule is shown in orange (carbon atoms), red (oxygens), and blue (nitrogens). (B) Model of LZD (orange) in its binding site (green) overlapped with the positions of key nucleotides (atom colored) and LZD (cyan) following G2576U mutation. (C) Model of LZD (orange) in its binding site (green) overlapped with the positions of key nucleotides (atom colored) and LZD (cyan) following the Ser158Leu L3 mutation.
TML resistance mutations.
Our model proposes that residues 2504 to 2506 form a key part of the binding pocket of the tricyclic mutilin core of TML (Fig. 2A); hence, modification of these nucleotides or conformational changes resulting from allosteric interactions would be expected to adversely affect TML binding. The guanine base of G2576 lies adjacent to the corresponding guanine base of G2505. Therefore, it is likely that mutation of this residue will have a direct effect on its neighbor. The spatial displacement of G2505 by the G2576U mutation in our model had a knock-on effect that resulted in a movement of up to 1.0 Å in the key sequence from positions 2504 to 2506. This in turn altered the position of TML within the binding cavity, which is expected to have an adverse effect on TML binding (Fig. 2B).
In addition, G2576 donates a hydrogen bond from N1 to the nonbridging oxygen atom of U2506. This interaction is expected to help stabilize the positioning of G2505 and U2506. Although it has previously been claimed that the G2576U mutation would be unable to form this hydrogen bond (10), this was not the case in our model, where N3 of U2576 is suitably positioned to fulfill this function. However, our model also suggests G2576 can form an additional hydrogen bond through N2 to the same oxygen atom of U2506. Since the pyrimidine ring of uracil would be incapable of donating a second hydrogen to U2506, we propose that the G2576U mutation weakens the interaction between this base and U2506 (and G2505). This decrease in the stability of key residues responsible for binding the mutilin core of TML is expected to reduce TML susceptibility. The increase in flexibility of U2506 results in the loss of a hydrogen bond between O4 of U2506 and N3 of U2584. As U2584 contributes hydrophobic interactions with the side chains of TML, conformational changes involving this residue are expected to further weaken drug binding.
Although none of the residues within L3, including those subject to mutation, make direct contact with TML, the mutations within L3 caused similar conformational changes in the environment around the drug as to those observed with the G2576U mutation (Fig. 2C). Our model suggests, for example, that all three L3 mutations described previously caused the displacement of residues 2504 to 2506, which form the cavity that binds the mutilin core of TML. Also, a hydrogen bond between U2506 and U2584 was again lost, thereby reducing the stability of the binding pocket further. Examination of the L3 mutations relative to the binding site of TML revealed Ser158Leu to be located closest to the peptidyl transferase center (PTC). The adjacent residue, Ala157, corresponds to Asn149 from E. coli, where an Asn149Asp mutation has been reported to confer resistance to TML (2). This resistance genotype was rationalized by the L3 mutation, causing a perturbation of the drug binding cavity, and a similar argument can be made for the Ser158Leu mutation in S. aureus. The mutated residue is located ca. 8 Å from U2504, which has been reported to be a key residue responsible for TML binding. Hence, changes in the conformation of U2504 as a result of indirect perturbation by Ser158Leu could possibly have a detrimental effect on TML binding (Fig. 2C).
However, it is more difficult to rationalize the resistance originating from the other two L3 mutations, Arg149Ser and Gly155Arg, in terms of their proximity to the PTC and TML. Nevertheless, despite being distant from the TML binding site, both mutations are located 7 to 8 Å from G2576. Since mutations of this nucleotide confer resistance to TML, it is plausible that indirect perturbation of the conformation of G2576 by L3 mutations located nearby will affect TML binding.
LZD resistance mutations.
In common with the TML S. aureus homology model, the LZD-containing complex also demonstrated that the guanine base of G2576 lies adjacent to that of G2505, whose ribose ring forms part of the binding pocket of the oxazolidinone ring of LZD (Fig. 3A). The G2576U mutation caused displacement of 1.6 Å in the position of the guanine group of G2505 relative to the unmodified model, which had a cumulative effect on the position of U2504, a key residue that binds the oxazolidinone ring of LZD. We predict that this movement of residues responsible for binding the key pharmacophore of LZD may have an adverse effect on drug binding. This view is reinforced by movement (up to 0.5 Å) of the oxazolidinone ring following the G2576U mutation (Fig. 3B).
In contrast, we predict that the L3 mutations had minimal effect on the conformation of the key residues, U2504 and G2505 (Fig. 3C). In addition, there was little apparent displacement of the oxazolidinone ring, reflecting the lack of perturbation of its binding residues. These predictions suggest that the modes of binding of LZD and TML are different such that LZD maintains contacts within the binding cavity that are able to resist conformational effects imposed by mutations within L3. However, all three L3 mutations did cause a movement of up to 2.6 Å of the uracil base of U2585 in our model. This movement caused reorientation of the adjacent residue U2584. In the case of TML, such a change in conformation is predicted to have a detrimental effect on drug binding since U2584 and U2585 form interactions with the side chain extensions that protrude from the TML mutilin core. However, since LZD is predicted to bind in the ribosomal A site, as opposed to TML, which binds across both A and P sites, these alterations are only expected to have a minor effect on LZD binding.
Interestingly, it has been reported that the Asn149Asp mutation that confers resistance to TML in E. coli does not confer cross-resistance to another pleuromutilin antibiotic, VML (11). The authors of that study explained this observation by suggesting that the extended side chain of VML can make additional interactions within the binding cavity, thereby overcoming local changes in conformation. Chemical footprinting data show increasing protection of U2585 going from TML to VML, confirming the importance of this base in binding these antibiotics. Possibly, the absence of LZD resistance observed after L3 mutation can be attributed to the fact that its main mode of binding does not involve significant interaction with residues U2584 and U2585. In contrast, the key oxazolidinone fragment is able to maintain its position and contacts close to the PTC.
Conclusions.
The potency, low potential for development of resistance, and favorable pharmacokinetics of oxazolidinones and pleuromutilins make them ideal antistaphylococcal drugs. However, the unidirectional cross-resistance between LZD and TML observed in LZD-resistant S. aureus could limit the systemic use of pleuromutilins in patients for which LZD treatment has been unsuccessful. Whether cross-resistance becomes an issue depends in part on the potency of the new pleuromutilins in development and whether they can overcome the 23S rRNA-mediated LZD resistance mechanism.
Acquisition of the PhLOPSA phenotype confers cross-resistance between LZD and pleuromutilins due to methylation of residue A2503 in 23S rRNA. In contrast, as reported here, the emergence of pleuromutilin-resistant isolates by point mutation should have little impact on susceptibility to LZD. This is because resistance to LZD is not observed with TML-resistant S. aureus mutants due to the preferential selection of single-copy rplC mutations rather than 23S rRNA mutations on the multicopy operon rrn. Cross-resistance to LZD is observed with TML-resistant mutants derived from a single-copy rrn knockout strain of E. coli, illustrating that the copy number of 23S rRNA genes is the limiting factor in the selection of 23S rRNA TML-resistant mutants. Finally, the molecular modeling of TML and LZD resistance mutations proposes a rational hypothesis for explaining the resistance phenotypes we have observed.
Acknowledgments
This study was supported by project grant 0410009 from the United Kingdom Department of Health awarded to I.C.
Footnotes
Published ahead of print on 7 January 2008.
REFERENCES
- 1.Bock, A., F. Turnowsky, and G. Hogenauer. 1982. Tiamulin resistance mutations in Escherichia coli. J. Bacteriol. 151:1253-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bosling, J., S. M. Poulsen, B. Vester, and K. S. Long. 2003. Resistance to the peptidyl transferase inhibitor tiamulin caused by mutation of ribosomal protein L3. Antimicrob. Agents Chemother. 47:2892-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.BSAC. 1991. A guide to sensitivity testing; Report of the working party on antibiotic sensitivity testing of the British Society for Antimicrobial Chemotherapy. J. Antimicrob. Chemother. 27(Suppl. D):1-50. [PubMed] [Google Scholar]
- 4.Chopra, I., and K. Hacker. 1992. Uptake of minocycline by Escherichia coli. J. Antimicrob. Chemother. 29:19-25. [DOI] [PubMed] [Google Scholar]
- 5.Gentry, D. R., S. F. Rittenhouse, L. McCloskey, and D. J. Holmes. 2007. Stepwise exposure of Staphylococcus aureus to pleuromutilins is associated with stepwise acquisition of mutations in rplC and minimally affects susceptibility to retapamulin. Antimicrob. Agents Chemother. 51:2048-2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones, R. N., T. R. Fritsche, H. S. Sader, and J. E. Ross. 2006. Activity of retapamulin (SB-275833), a novel pleuromutilin, against selected resistant gram-positive cocci. Antimicrob. Agents Chemother. 50:2583-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joseph, P., J. R. Fantino, M. L. Herbaud, and F. Denizot. 2001. Rapid orientated cloning in a shuttle vector allowing modulated gene expression in Bacillus subtilis. FEMS Microbiol. Lett. 205:91-97. [DOI] [PubMed] [Google Scholar]
- 8.Kloss, P., L. Xiong, D. L. Shinabarger, and A. S. Mankin. 1999. Resistance mutations in 23S rRNA identify the site of action of the protein synthesis inhibitor linezolid in the ribosomal peptidyl transferase center. J. Mol. Biol. 294:93-101. [DOI] [PubMed] [Google Scholar]
- 9.Kosowska-Shick, K., C. Clark, K. Credito, P. McGhee, B. Dewasse, T. Bogdanovich, and P. C. Appelbaum. 2006. Single- and multistep resistance selection studies on the activity of retapamulin compared to other agents against Staphylococcus aureus and Streptococcus pyogenes. Antimicrob. Agents Chemother. 50:765-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leach, K. L., S. M. Swaney, J. R. Colca, W. G. McDonald, J. R. Blinn, L. M. Thomasco, R. C. Gadwood, D. Shinabarger, L. Xiong, and A. S. Mankin. 2007. The site of action of oxazolidinone antibiotics in living bacteria and in human mitochondria. Mol. Cell 26:393. [DOI] [PubMed] [Google Scholar]
- 11.Long, K. S., L. H. Hansen, L. Jakobsen, and B. Vester. 2006. Interaction of pleuromutilin derivatives with the ribosomal peptidyl transferase center. Antimicrob. Agents Chemother. 50:1458-1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Long, K. S., J. Poehlsgaard, C. Kehrenberg, S. Schwarz, and B. Vester. 2006. The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob. Agents Chemother. 50:2500-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meka, V. G., S. K. Pillai, G. Sakoulas, C. Wennersten, L. Venkataraman, P. C. DeGirolami, G. M. Eliopoulos, R. C. Moellering, and H. S. Gold. 2004. Linezolid resistance in sequential Staphylococcus aureus isolates associated with a T2500A mutation in the 23S rRNA gene and loss of a single copy of rRNA. J. Infect. Dis. 190:311-317. [DOI] [PubMed] [Google Scholar]
- 14.Miller, K., A. J. O'Neill, and I. Chopra. 2002. Response of Escherichia coli hypermutators to selection pressure with antimicrobial agents from different classes. J. Antimicrob. Chemother. 49:925-934. [DOI] [PubMed] [Google Scholar]
- 15.Miller, K., M. H. Wilcox, and I. Chopra. 2006. Linezolid and tiamulin cross-resistance in Staphylococcus aureus mediated by overlapping binding regions in 23S rRNA. Abstr. 46th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C1-0944. [DOI] [PMC free article] [PubMed]
- 16.O'Neill, A. J., and I. Chopra. 2001. Use of mutator strains for characterization of novel antimicrobial agents. Antimicrob. Agents Chemother. 45:1599-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Neill, A. J., J. H. Cove, and I. Chopra. 2001. Mutation frequencies for resistance to fusidic acid and rifampicin in Staphylococcus aureus. J. Antimicrob. Chemother. 47:647-650. [DOI] [PubMed] [Google Scholar]
- 18.Pringle, M., J. Poehlsgaard, B. Vester, and K. S. Long. 2004. Mutations in ribosomal protein L3 and 23S rRNA at the peptidyl transferase centre are associated with reduced susceptibility to tiamulin in Brachyspira spp. isolates. Mol. Microbiol. 54:1295-1306. [DOI] [PubMed] [Google Scholar]
- 19.Rittenhouse, S., S. Biswas, J. Broskey, L. McCloskey, T. Moore, S. Vasey, J. West, M. Zalacain, R. Zonis, and D. Payne. 2006. Selection of retapamulin, a novel pleuromutilin for topical use. Antimicrob. Agents Chemother. 50:3882-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlunzen, F., E. Pyetan, P. Fucini, A. Yonath, and J. M. Harms. 2004. Inhibition of peptide bond formation by pleuromutilins: the structure of the 50S ribosomal subunit from Deinococcus radiodurans in complex with tiamulin. Mol. Microbiol. 54:1287-1294. [DOI] [PubMed] [Google Scholar]
- 21.Swaney, S. M., D. L. Shinabarger, R. D. Schaadt, J. H. Bock, J. L. Slightom, and G. E. Zurenko. 1998. Oxazolidinone resistance is associated with a mutation in the peptidyl-transferase region of 23S rRNA. Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C-104.
- 22.Toh, S.-M., L. Xiong, C. A. Arias, M. V. Villegas, K. Lolans, J. Quinn, and A. S. Mankin. 2007. Acquisition of a natural resistance gene renders a clinical strain of methicillin-resistant Staphylococcus aureus resistant to the synthetic antibiotic linezolid. Mol. Microbiol. 64:1506-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsiodras, S., H. S. Gold, G. Sakoulas, G. M. Eliopoulos, C. Wennersten, L. Venkataraman, R. C. Moellering, and M. J. Ferraro. 2001. Linezolid resistance in a clinical isolate of Staphylococcus aureus. Lancet 358:207-208. [DOI] [PubMed] [Google Scholar]
- 24.Wilson, P., J. A. Andrews, R. Charlesworth, R. Walesby, M. Singer, D. J. Farrell, and M. Robbins. 2003. Linezolid resistance in clinical isolates of Staphylococcus aureus. J. Antimicrob. Chemother. 51:186-187. [DOI] [PubMed] [Google Scholar]