Abstract
Nine proline-rich peptides ending with a proline-glutamine C terminus in a salivary peptidome were sequenced by matrix-assisted laser desorption ionization time of flight time of flight tandem mass spectrometry. A GPPPQGGRPQ peptide binds gram-positive Propionibacterium acnes and considerably inhibits bacterial growth. The peptide exhibiting innate immunity may be applied for treatment of various P. acnes-associated human diseases.
Saliva has been evaluated as a diagnostic and prognostic fluid. In this study, a human saliva peptidome was characterized by matrix-assisted laser desorption ionization time of flight time of flight (MALDI-TOF-TOF). Nine proline-rich peptides ending with a proline-glutamine sequence (PQ C terminus) were sequenced (Table 1). Previous studies have shown the release of a pentapeptide, RGRPQ, from salivary proline-rich proteins upon proteolysis by oral bacteria (6). The pentapeptide behaved as an innate-immunity-like peptide, since synthetic RGRPQ was found to desorb bound bacteria (6). In addition, a synthetic GGRPQ peptide showed activity equal to that of RGRPQ, which exhibited an excellent ability to inhibit the adhesion of oral bacteria to salivary proline-rich proteins with which hydroxyapatite beads were coated (6). By searching the nine peptides with PQ C termini found in the saliva peptidome (Table 1), we found that one peptide (GPPPQGGRPQ [m/z 990.60]) contained the GGRPQ C-terminal residues (underlined). Thus, this peptide was selected for investigation of its bacterial binding and antimicrobial activities.
TABLE 1.
Peptide PQ C termini detected in MALDI-TOF-TOF
| m/z | Sequence | Proteina | Accession No. | Mass (kDa) | Matched sequence positions |
|---|---|---|---|---|---|
| 971.53 | GPPPPPGKPQ | Basic salivary proline-rich protein 2 | P02812 | 37.3 | 221-230 |
| 990.60 | GPPPQGGRPQ | Salivary acidic proline-rich phosphoprotein 1/2 | P02810 | 17.0 | 148-157 |
| 1,068.66 | GPPPPPPGKPQ | Salivary acidic proline-rich phosphoprotein 1/2 | P02810 | 17.0 | 137-147 |
| 1,104.19 | GPPPQGGNRPQ | Basic proline-rich peptide P-E (IB-9) | P02811 | 6.0 | 29-39 |
| 1,333.77 | GRPQGPPQGQSPQ | Salivary acidic proline-rich phosphoprotein 1/2 | P02810 | 17.0 | 154-166 |
| 1,390.68b | GGRPQGPPQGQSPQ | Salivary acidic proline-rich phosphoprotein 1/2 | P02810 | 17.0 | 153-166 |
| 1,731.89 | GPPQQGGHPPPPQGRPQ | Salivary acidic proline-rich phosphoprotein 1/2 | P02810 | 17.0 | 93-109 |
| 1,957.14 | PQGPPQQGGHPPPPQGRPQ | Salivary acidic proline-rich phosphoprotein 1/2 | P02810 | 17.0 | 91-109 |
| 2,029.28 | GPPPPPGKPQGPPPQGGNKPQ | Basic proline-rich peptide P-E (IB-9) | P02811 | 6.0 | 18-39 |
Proteins identified by searching Mascot, a searching algorithm available at the Matrix Science home page. A human database obtained from ftp://ftp.ncbi.nih.gov/genomes/ containing 40,877 sequences was used. Ion scores greater than 70 were considered significant (P ≤ 0.05) (23) for protein identification.
A peptide is absent or has a low intensity in the MS spectra of MALDI-TOF-TOF.
Whole saliva from two males and one female between the ages of 20 and 40 was collected according to protocols described previously (9). After centrifugation (14,000 × g), the clear whole-saliva supernatants (0.4 μg/μl) collected from the three volunteers were pooled, digested immediately with trypsin (20 ng/μl) overnight (20), and then mixed 1:2 with alpha-cyano-4-hydroxycinnimic acid (7 mg/ml) for analysis by liquid chromatography-MALDI-TOF-TOF mass spectrometry (MS) (4800 TOF-TOF Analyzer; Applied Biosystems, Foster City, CA) (2, 21). Sixty-three saliva peptides corresponding to 22 proteins were identified from tryptic digests (data not shown). Nine saliva peptides ending with a PQ C terminus (Table 1) were derived from basic salivary proline-rich protein 2, salivary acidic proline-rich phosphoprotein (1/2), and basic proline-rich peptide P-E (IB-9).
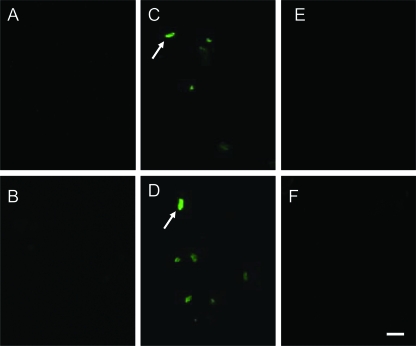
The GPPPQGGRPQ peptide was synthesized and labeled with fluorescein (GenScript Corp., Piscataway, NJ). The fluorescein-labeled peptide was obtained by coupling 5(6)-carboxyfluorescein to the amino terminus of GPPPQGGRPQ (10). Two gram-positive oral bacteria, Porphyromonas gingivalis (ATCC 33277) and Porphyromonas acnes (ATCC 6919) (11, 16), were chosen for interaction with GPPPQGGRPQ. Both bacteria (108 CFU) were incubated with fluorescein-labeled or unlabeled GPPPQGGRPQ (1 and 10 μM) for 1 h. The green fluorescence derived from the binding of peptides to the bacteria was detectable only when fluorescein-labeled peptide was incubated with P. acnes (Fig. 1), but not P. gingivalis (Fig. 1F), indicating that GPPPQGGRPQ selectively adhered to P. acnes. The green fluorescence was visible when P. acnes was incubated with 1 μM (Fig. 1C) or 10 μM (Fig. 1D) of fluorescein-labeled GPPPQGGRPQ, but not unlabeled GPPPQGGRPQ (Fig. 1A) and a fluorescein-labeled CGKRK (10 μM) (Fig. 1B) (a gift from Zhang Lianglin, Moores Cancer Center at the University of California, San Diego). Notably, the green fluorescence generated by 1 μM of fluorescein-labeled peptide was entirely quenched (Fig. 1E) when 1 mM of the unlabeled GPPPQGGRPQ was present in the reaction mixture of fluorescein-labeled peptide with P. acnes, indicating that the binding of GPPPQGGRPQ to P. acnes is peptide specific.
FIG. 1.
Fluorescence microscopic images of P. acnes upon incubation with a fluorescein-labeled GPPPQGGRPQ peptide. P. acnes was suspended in a phosphate buffer containing 1.1 mM NaH2PO4, 1 mM NaH2PO4, and 100 mM NaCl, pH 6.5, and incubated with GPPPQGGRPQ (10 μM) (A), fluorescein-labeled CGKRK (10 μM) (B), or fluorescein-labeled GPPPQGGRPQ (1 μM [C] and 10 μM [D and E]) for 1 h at 37°C under anaerobic conditions. Incubation of fluorescein-labeled CGKRK was performed to exclude nonspecific binding. (F) P. gingivalis incubated with fluorescein-labeled GPPPQGGRPQ (10 μM). The green fluorescence (arrows) indicated that fluorescein-labeled peptide bound to P. acnes. (E) The green fluorescence was entirely quenched when a high concentration (1 mM) of unlabeled GPPPQGGRPQ was added to the incubation of fluorescein-labeled peptide (1 μM) with bacteria. Bar, 1 μm.
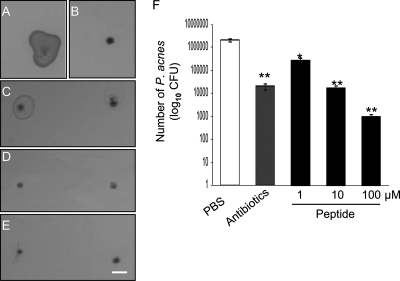
We next examined if the GPPPQGGRPQ peptide influences the growth of P. acnes. P. acnes (105 CFU) was preincubated with phosphate-buffered saline (PBS) (Fig. 2A) or GPPPQGGRPQ at concentrations of 1 μM (Fig. 2C), 10 μM (Fig. 2D), and 100 μM (Fig. 2E) for 3 h. Preincubation of P. acnes with an antibiotic mixture (500 units/ml of penicillin G and 0.5 mg/ml of streptomycin sulfate) for 3 h significantly inhibited the 2-day growth of P. acnes (2.1 × 103 ± 6.3 × 103 CFU) in comparison with that after incubation of P. acnes with PBS (20.5 × 105 ± 2.5 × 105 CFU) (Fig. 2B and F). Treatment of P. acnes with 1, 10, and 100 μM of GPPPQGGRPQ considerably attenuated the colonization (Fig. 2C to E) and the growth (26.5 × 104 ± 6.3 × 104, 18 × 103 ± 2.6 × 103, and 9.5 × 102 ± 2.1 × 102 CFU, respectively) of P. acnes. These results demonstrated that the GPPPQGGRPQ peptide exerts a bactericidal activity against P. acnes.
FIG. 2.
The GPPPQGGRPQ peptide suppresses the growth of P. acnes. P. acnes (1 × 105 CFU) was incubated with PBS (A), antibiotics (B), and GPPPQGGRPQ at final concentrations of 1 μM (C), 10 μM (D), and 100 μM (E) for 3 h. After a 2-day incubation, bacteria were spotted on agar plates for colonization. Bacteria incubated with peptide were spotted in duplicate (C to E). (F) The activity of the GPPPQGGRPQ peptide in bacterial growth was determined by plating serial dilutions (104 to 107) of bacteria on agar plates and quantifying the CFU. Bar, 0.5 mm. The bars represent means ± standard errors for three separate experiments. The numbers of P. acnes bacteria treated with antibiotics (hatched bar) and peptide (solid bars) were compared with that of P. acnes treated with PBS (open bar). Student's t test was conducted for comparison. P values of <0.01 (*) and <0.005 (**) were considered significant.
Although we failed to identify low-abundance proteins, such as cytokines and defensins, that normally are considerably elevated during oral inflammation (15), an advanced electrospray ionization tandem MS with higher sensitivity may make it possible to identify lower-abundance saliva proteins (7). Since the intensity of each peptide signal in an MS spectrum does not necessarily correlate with its biological abundance, quantitative MS using isotopic-tag labeling (19) will provide a means to determine the differential abundances of peptides under different biological conditions. Although it is unclear if the GPPPQGGRPQ peptide is derived from fragmentation of bacterium-bound proline-rich proteins, it has been documented that proline-rich proteins can be cleaved by proteases from oral Streptococcus and Actinomyces species and converted into peptides with bacterium-binding PQ C termini (13). Intriguingly, several peptides with PQ C termini were identified in undigested human parotid saliva (8), suggesting that peptides with PQ C termini may exist naturally in human whole saliva. When oral bacteria predominate, these peptides with PQ C termini may serve as innate-immunity-like peptides (5, 13) to bind and/or kill oral bacteria instantly. The PQ-rich repeats also exist in other proteins, such as diacylglycerol kinase, an enzyme involved in the regulation of signal transduction (18). It has also been reported that peptides with polyprolyl or polyglutamine sequences could contribute to amyloidogenic diseases (14) and display the activity of cellular permeation, as well as binding to heat shock proteins (1). Thus, peptides with PQ C termini in saliva may have biological functions other than acting as antimicrobials.
Although the target molecules of the GPPPQGGRPQ peptide in P. acnes are undetermined, it has been documented that an RGRPQ peptide could be similar to the ERGMT peptide signal that affects intra- or extracellular receptors and gene expression in Bacillus subtilis (12). Future studies will include determining the eukaryotic toxicity and MICs of the GPPPQGGRPQ peptide against P. acnes. The complete genome of P. acnes has been sequenced (3, 4). P. acnes is involved in many infectious diseases, including acne vulgaris and biofilm formation on implanted biomaterials (17). Therefore, the future applications of the GPPPQGGRPQ peptide may include monitoring of the distribution of P. acnes and/or treatment of P. acnes-associated diseases by inhibiting bacterial growth.
Acknowledgments
This work was supported by National Institutes of Health Grants (1-R01 AI067395, R21-R022754-01, and R21-I58002-01), a Dermatology Foundation Grant, and a SERCEB grant (5 U54 AI057157-02).
We thank M. Kirk and S. Barnes for their assistance with MS.
Footnotes
Published ahead of print on 19 February 2008.
REFERENCES
- 1.Bertoncini, C. W., R. M. Rasia, G. R. Lamberto, A. Binolfi, M. Zweckstetter, C. Griesinger, and C. O. Fernandez. 2007. Structural characterization of the intrinsically unfolded protein beta-synuclein, a natural negative regulator of alpha-synuclein aggregation. J. Mol. Biol. 372:708-722. [DOI] [PubMed] [Google Scholar]
- 2.Bodnar, W. M., P. K. Blackburn, J. M. Krise, and M. A. Moseley. 2003. Exploiting the complementary nature of LC/MALDI/MS/MS and LC/ESI/MS/MS for increased proteome coverage. J. Am. Soc. Mass Spectrom. 14:971-979. [DOI] [PubMed] [Google Scholar]
- 3.Bruggemann, H., A. Henne, F. Hoster, H. Liesegang, A. Wiezer, A. Strittmatter, S. Hujer, P. Durre, and G. Gottschalk. 2004. The complete genome sequence of Propionibacterium acnes, a commensal of human skin. Science 305:671-673. [DOI] [PubMed] [Google Scholar]
- 4.Bruggemann, H. 2005. Insights in the pathogenic potential of Propionibacterium acnes from its complete genome. Semin. Cutan. Med. Surg. 24:67-72. [DOI] [PubMed] [Google Scholar]
- 5.Davtyan, T. K., H. A. Manukyan, N. R. Mkrtchyan, S. A. Avetisyan, and A. A. Galoyan. 2005. Hypothalamic proline-rich polypeptide is a regulator of oxidative burst in human neutrophils and monocytes. Neuroimmunomodulation 12:270-284. [DOI] [PubMed] [Google Scholar]
- 6.Drobni, M., T. Li, C. Kruger, V. Loimaranta, M. Kilian, L. Hammarström, H. J. Örnvall, T. Bergman, and N. Strömberg. 2006. Host-derived pentapeptide affecting adhesion, proliferation, and local pH in biofilm communities composed of Streptococcus and Actinomyces species. Infect. Immun. 74:6293-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goebel, C., L. G. Mackay, E. R. Vickers, and L. E. Mather. 2000. Determination of defensin HNP-1, HNP-2, and HNP-3 in human saliva by using LC/MS. Peptides 21:757-765. [DOI] [PubMed] [Google Scholar]
- 8.Hardt, M., L. R. Thomas, S. E. Dixon, G. Newport, N. Agabian, A. Prakobphol, S. C. Hall, H. E. Witkowska, and S. J. Fisher. 2005. Toward defining the human parotid gland salivary proteome and peptidome: identification and characterization using 2D SDS-PAGE, ultrafiltration, HPLC, and mass spectrometry. Biochemistry 44:2885-2899. [DOI] [PubMed] [Google Scholar]
- 9.Huang, C. M. 2004. Comparative proteomic analysis of human whole saliva. Arch. Oral Biol. 49:951-962. [DOI] [PubMed] [Google Scholar]
- 10.Huang, C. M., Y. T. Wu, and S. T. Chen. 2000. Targeting delivery of paclitaxel into tumor cells via somatostatin receptor endocytosis. Chem. Biol. 7:453-461. [DOI] [PubMed] [Google Scholar]
- 11.Kataoka, K., A. Amano, M. Kuboniwa, H. Horie, H. Nagata, and S. Shizukuishi. 1997. Active sites of salivary proline-rich protein for binding to Porphyromonas gingivalis fimbriae. Infect. Immun. 65:3159-3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lazazzera, B. A., and A. D. Grossman. 1998. The ins and outs of peptide signaling. Trends Microbiol. 6:288-294. [DOI] [PubMed] [Google Scholar]
- 13.Li, T., P. Bratt, A. P. Jonsson, M. Ryberg, I. Johansson, W. J. Griffiths, T. Bergman, and N. Stromberg. 2000. Possible release of an Arg Gly Arg Pro Gln pentapeptide with innate immunity properties from acidic proline-rich proteins by proteolytic activity in commensal Streptococcus and Actinomyces species. Infect. Immun. 68:5425-5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Markossian, K. A., A. A. Zamyatnin, and B. I. Kurganov. 2004. Antibacterial proline-rich oligopeptides and their target proteins. Biochemistry 69:1082-1091. [DOI] [PubMed] [Google Scholar]
- 15.Mizukawa, N., K. Sugiyama, T. Ueno, K. Mishima, S. Takagi, and T. Sugahara. 1999. Levels of human defensin-1, an antimicrobial peptide, in saliva of patients with oral inflammation. Oral. Surg. Oral. Med. Oral. Pathol. Oral. Radiol. Endod. 87:539-543. [DOI] [PubMed] [Google Scholar]
- 16.Oh, S., S. H. Kim, Y. Ko, J. H. Sim, K. S. Kim, S. H. Lee, S. Park, and Y. J. Kim. 2006. Effect of bacteriocin produced by Lactococcus sp. HY 449 on skin-inflammatory bacteria. Food Chem. Toxicol. 44:1184-1190. [DOI] [PubMed] [Google Scholar]
- 17.Perry, A. L., and P. A. Lambert. 2006. Propionibacterium acnes. Lett. Appl. Microbiol. 42:185-188. [DOI] [PubMed] [Google Scholar]
- 18.Sakane, F., and H. Kanoh. 1997. Molecules in focus: diacylglycerol kinase. Int. J. Biochem. Cell Biol. 29:1139-1143. [DOI] [PubMed] [Google Scholar]
- 19.Shi, Y., C. A. Elmets, J. W. Smith, Y. T. Liu, Y. R. Chen, C. P. Huang, W. Zhu, H. N. Ananthaswamy, R. L. Gallo, and C. M. Huang.. Quantitative proteomes and in vivo secretomes of progressive and regressive UV-induced fibrosarcoma tumor cells: mimicking tumor microenvironment using a dermis-based cell-trapped system linked to tissue chamber. Proteomics 27:4589-4600. [DOI] [PubMed]
- 20.Spahr, C. S., M. T. Davis, M. D. McGinley, J. H. Robinson, E. J. Bures, J. Beierle, J. Mort, P. L. Courchesne, K. Chen, R. C. Wahl, W. Yu, R. Luethy, and S. D. Patterson. 2001. Towards defining the urinary proteome using liquid chromatography-tandem mass spectrometry. I. Profiling an unfractionated tryptic digest. Proteomics 1:93-107. [DOI] [PubMed] [Google Scholar]
- 21.Vitorino, R., M. J. Lobo, A. J. Ferrer-Correira, J. R. Dubin, K. B. Tomer, P. M. Domingues, and F. M. Amado. 2004. Identification of human whole saliva protein components using proteomics. Proteomics 4:109-115. [DOI] [PubMed] [Google Scholar]