Abstract
Translational attenuation has been proposed to be the mechanism by which the erm(B) gene is induced. Here, we report genetic and biochemical evidence, obtained by using erythromycin as the inducing antibiotic, that supports this hypothesis. We also show that erythromycin increases the level of the erm(B) transcript by stalling the ribosome on the leader mRNA and thereby facilitating the stabilization and processing of the mRNA. Erythromycin-induced mRNA stabilization and processing were observed with an ochre stop at codons 11 to 13 of the leader but not with an ochre stop at codon 10. This suggests that erythromycin does not stall the ribosome before codon 11 of the leader reaches the aminoacyl site. Secondary structure analyses of the erm(B) transcripts by in vitro and in vivo chemical probing techniques identified conformational changes in the transcripts that result from induction by erythromycin. These findings demonstrate that stalling of erythromycin-bound ribosomes at leader codon 11 causes the refolding of mRNA into a conformation in which the translational initiation site for the structural gene is unmasked and renders erm(B) translationally active.
Dimethylation of a single adenine in 23S rRNA causes high-level cross-resistance to macrolide-lincosamide-streptogramin B (MLSB) antibiotics. The erm gene encodes the methylase responsible for this modification and remains the most widespread determinant of MLSB resistance. The expression of erm can be either constitutive or inducible. When the expression is inducible, there is variation in induction specificity. All of the MLSB antibiotics act as inducers of erm(B) to various degrees (18). This feature of erm(B) is different from the features of other classes of erm, the expression of which is induced by only certain specific MLSB antibiotics (1, 17, 30).
Translational attenuation is believed to control the expression of erm(A), erm(C), and erm(D) (14, 23, 31). For erm(C), the translational attenuation mechanism has been well elucidated (31). The structural gene of erm(C) is preceded by a leader peptide that comprises 19 amino acids encoded by a regulatory sequence. Without an inducer, the erm(C) mRNA is synthesized, but in a translationally unfavorable conformation. With the inducer, the inducer-bound ribosome stalls on the leader sequence with leader peptide codon 9 occupied by its aminoacyl site (A site), which leads to the conformational isomerization of the mRNA to the translationally active form. The ribosome stalling concomitantly brings about the stabilization of erm(C) mRNA owing to the blockade of its 5′ end from the access of a nuclease (3).
Despite its clinical importance, the induction mechanism of the erm(B) gene has not been elucidated in detail (18). A translational attenuation model has been proposed for erm(B) on the basis of sequence analysis (12), and enhancement of the transcript corresponding to erm(B) mRNA has also been reported (27). The erm(B) transcript contains a 5′ leader sequence of 259 nucleotides (nt) which has the potential to encode a leader peptide of 27 amino acids. In the current study, the induction mechanism of erm(B) by the 14-membered-ring macrolide erythromycin (Em) was investigated. We confirmed the translational attenuation as a model of erm(B) induction by Em and demonstrated the enhanced stabilization, processing, and conformational change of the mRNA in response to Em.
MATERIALS AND METHODS
Bacterial strains.
Strain BR151 (trpC2 lys3 metB10) was used as the host for all Bacillus subtilis strains except strain EL17. Strain Eryr Spo(Ts) was made from BR151 as described previously (29). Escherichia coli CSH26 [ara Δ(lac pro) thi] was used for the transformation of B. subtilis, and E. coli JM109 (Promega) was used for plasmid construction.
Plasmid constructions.
To make the erm(B)-lacZ translational fusion construct (strain EL100), PCR was performed with plasmid pAM225 (27) and primers FB (5′-CTTAGAATTCAACTTAAGAGTGTGT-3′) and FB1 (5′-GCCGGATCCGTCGTTAAATGCCCT-3′). The PCR product was digested with EcoRI and BamHI and ligated to plasmid pMM156 (4) digested with the same enzymes. The ligation product was electroporated to strain CSH26, and the resulting plasmid was introduced into strain BR151 as described above. In the same way, the erm(B) leader peptide-lacZ translational fusion construct (strain EL160) was obtained, except that primer BB3 (5′-TAAGGATCCGATTTATCTGCGTAATC-3′) instead of primer FB1 was used. The megaprimer PCR method was employed for in vitro mutagenesis (26). To replace the erm(B) promoter with the rapB promoter in EL100 (strain EL153), a BglII site was introduced into the transcription start site of erm(B). The EcoRI-BglII fragment containing the promoter was removed, and then the small EcoRI-BamHI fragment from pB1 (15) was inserted.
An E. coli-B. subtilis shuttle vector carrying the origins from pC194 (11) was constructed. NheI sites were introduced at each flanking region of the origin for B. subtilis in pHY300PLK (Takara) and the cat gene in pC194. Subsequently, the plasmids were each cut with NheI and the large fragments from each plasmid were mixed and ligated. The resulting vector, pAZA, is compatible with pMM156 in B. subtilis. To clone the expression unit for the erm(B) leader peptide to pAZA, a BamHI site was introduced downstream of the leader open reading frame (ORF) in EL153. The small EcoRI-BamHI fragment of the resulting plasmid was inserted in EcoRI-BamHI-cleaved pAZA. Then, a terminator (16) was inserted into the SalI and BglII sites of this vector in the correct orientation, yielding pAZA2. The strains of strain EL151 carrying pAZA or pAZA2 were designated EL210 and EL220, respectively.
For in vitro transcription, the 5′-end region of erm(B) lacking the promoter was amplified with primers P456 (5′-ATAGGAATTCAAGTTAAATTAGATGC-3′) and FB1. The PCR product was cut with EcoRI and BamHI and ligated to pGEM-3Zf(−) (Promega), which had been digested with the same enzymes. E. coli JM109 was electroporated with the resulting plasmid (pEBL456).
β-Galactosidase assay.
The β-galactosidase assay was performed with 20 ml of mid-log-phase cultures, as described previously (22).
Measurement of mRNA half-lives.
B. subtilis cells carrying wild-type or mutant erm(B)-lacZ were grown to mid-log phase at 37°C in SPII medium (21), followed by incubation with or without Em (0.1 μg/ml) for 10 min. After the addition of rifampin at 150 μg/ml, samples were removed and mixed with a half volume of ice-cold quenching solution (50 mM NaN3) at the time intervals indicated in Fig. 1 to 3. Total RNA from the cultures was isolated, and a primer extension assay was carried out to quantify the amount of erm(B) mRNA, as described previously (20). A total of 5 μg of RNA was hybridized to 5 ng of end-labeled erm(B) probe P3 [complementary to nt 140 to 164 in the erm(B) mRNA].
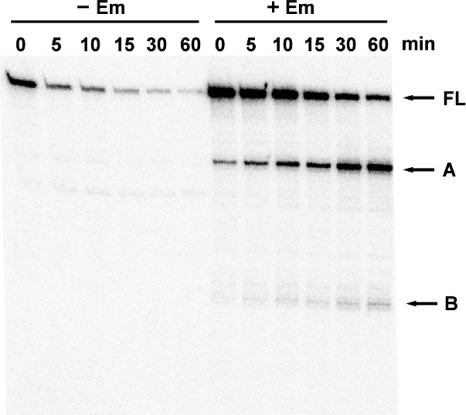
FIG. 1.
Decay of erm(B) mRNA analyzed by primer extension. B. subtilis EL100 harboring the wild-type erm(B)-lacZ fusion was grown exponentially in the absence (−) or presence (+) of 0.1 μg/ml Em for 10 min. Rifampin (150 μg/ml) was added to the cultures, and total RNA was isolated at the indicated times. Extension was performed with primer P3, which is specific for the erm(B) leader region. The bands corresponding to full-length (FL) and processed (A and B) transcripts are marked.
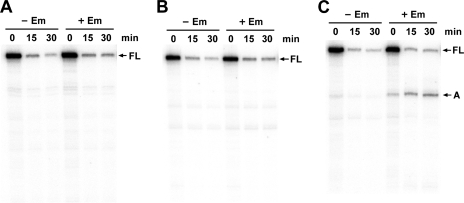
FIG. 3.
Primer extension analysis showing the differential decay of erm(B)-lacZ mRNA in various B. subtilis constructs. (A) EL151, EL100 with RBS1 deletion; (B) EL152, EL100 with a Val-to-Gly change at leader codon 9; (C) EL17, Eryr Spo(Ts) with wild-type erm(B)-lacZ. −Em, absence of Em; +Em, presence of Em; FL, full-length transcript.
In vitro chemical probing.
The erm(B) RNA was synthesized in vitro from BamHI-linearized pEBL456 with Riboprobe system T7 (Promega). Chemical modifications with dimethylsulfate (DMS) and 1-cyclohexyl-3-(2-morpholinoethyl)-carbodiimide metho-p-toluene sulfonate (CMCT) were performed as described previously (28). The modifications of 15 pmol of the RNA in the presence of 2 μg yeast RNA were carried out in 50-μl reaction volumes. Semidenaturing conditions were tested by the use of the same buffers but with the buffers containing 1 mM EDTA instead of MgCl2 and KCl. The modified RNAs (5 pmol) were annealed with 1 ng of end-labeled primers. Primer extension was conducted as described above with primers P1, P2, P3, and P4, which are complementary to nt 298 to 322, 219 to 249, 140 to 164, and 64 to 89 in the erm(B) mRNA, respectively.
In vivo chemical probing.
In vivo DMS modification, RNA isolation, and primer extension were carried out by the procedures described previously (20) and in the legend to Fig. 6.
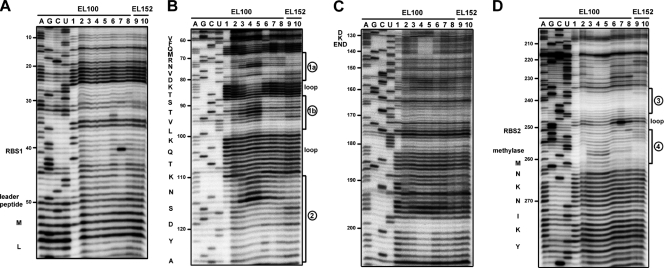
FIG. 6.
Autoradiograms showing the results of in vivo chemical probing of the erm(B) leader region. B. subtilis BR151 carrying the wild-type erm(B) leader peptide (strain EL100) or the mutant erm(B) leader peptide (strain EL152) was incubated in the absence or presence of the antibiotics for 10 min, before it was subjected to DMS treatment. The positions of the base modifications were determined by the primer extension analysis with primers P4 (A), P3 (B), P2 (C), and P1 (D). Lanes A, G, C, and U, dideoxysequencing ladders with RNA without DMS treatment; lanes 1, in vitro transcript reacted with DMS under native conditions; lanes 2 to 5, wild-type erm(B) mRNA from EL100 upon exposure to 0, 0.01, 0.1, and 1 μg/ml Em, respectively; lanes 6 to 8, wild-type erm(B) mRNA treated with tylosin (0.3 μg/ml), clindamycin (0.1 μg/ml), and quinupristin (0.3 μg/ml), respectively; lanes 9 and 10, mutant mRNA from EL152 treated with 0 and 0.1 μg/ml of Em, respectively.
RESULTS
The erm(B) gene is regulated by translational attenuation.
To determine whether the erm(B) leader peptide is indeed translated, we constructed strain EL160, which is B. subtilis BR151 harboring the erm(B) leader peptide-lacZ translational fusion construct. For comparison, the same host harboring the erm(B)-lacZ translational fusion was also constructed (strain EL100). After 2 h exposure to Em (0.1 μg/ml), β-galactosidase activity was measured (Table 1). While the activity in the wild-type fusion was induced 15.2-fold, the activity in EL160 was both high and constitutive, which indicated that the leader peptide had been efficiently translated. Then, the ribosome binding site (RBS1) for the leader peptide was deleted in the erm(B)-lacZ construct (strain EL151). This resulted in the complete abolishment of induction. Moreover, we constructed various mutants with mutations at the leader region in the preliminary experiment and found that a Val-to-Gly change at codon 9 of the leader peptide (strain EL152) abolished the induction by Em. Therefore, the translation of the leader peptide is necessary for induction, and the amino acid sequence of the leader peptide is critical for the inducibility of Em.
TABLE 1.
β-Galactosidase activities of various B. subtilis strains
| Strain | Description | β-Galactosidase activitya
|
Fold induction | |
|---|---|---|---|---|
| Without Em | With Emb | |||
| EL100 | BR151 with erm(B)-lacZ | 212 ± 9 | 3,223 ± 315 | 15.2 |
| EL160 | BR151 with erm(B) leader peptide-lacZ | 3,511 ± 375 | 2,989 ± 225 | 0.9 |
| EL151 | EL100 with RBS1 (GGAGGG) deletion | 410 ± 26 | 400 ± 72 | 1.0 |
| EL152 | EL100 with GTA→GGA change at leader codon 9 | 289 ± 28 | 379 ± 3 | 1.3 |
| EL153 | EL100 with replacement by rapB promoter | 465 ± 49 | 2,540 ± 329 | 5.5 |
| EL17 | Eryr Spo(Ts) with erm(B)-lacZ | 209 ± 43 | 691 ± 89 | 3.3 |
| EL154 | EL100 with G→C mutation at nt 254c | 2,303 ± 163 | 4,378 ± 310 | 1.9 |
| EL220 | EL151 with trans expression of leader peptide | 370 ± 38 | 448 ± 53 | 1.2 |
| EL210 | Vector control of EL220 | 359 ± 21 | 473 ± 28 | 1.3 |
| EL110 | EL100 with ochre mutation at leader codon 10 | 3,876 ± 281 | 3,447 ± 350 | 0.9 |
| EL111 | EL100 with ochre mutation at leader codon 11 | 3,874 ± 309 | 3,533 ± 324 | 0.9 |
| EL112 | EL100 with ochre mutation at leader codon 12 | 2,612 ± 267 | 2,670 ± 287 | 1.0 |
| EL113 | EL100 with ochre mutation at leader codon 13 | 2,998 ± 310 | 3,146 ± 299 | 1.0 |
β-Galactosidase activity is expressed as Miller units.
Incubation at 0.1 μg/ml for 120 min.
The nucleotide number in erm(B) mRNA.
If the expression of erm(B) gene is controlled by translational attenuation, the gene would be induced when the wild-type promoter is replaced with a heterologous promoter. We constructed strain EL153 in which the erm(B)-lacZ was under the control of the rapB promoter (15). This fusion was inducible by Em (Table 1). Then, we tested whether drug-bound ribosome is required for the induction of erm(B) gene expression by using the Eryr Spo(Ts) mutant of B. subtilis BR151. It was shown that the Eryr Spo(Ts) mutant of B. subtilis contained an alteration in ribosomal protein L17 (29). The L17 mutation in this strain probably causes the ribosome to have a weak affinity of binding to Em. Strain EL17 was constructed by transforming the Eryr Spo(Ts) strain with the erm(B)-lacZ gene. In strain EL17, Em induced the gene expression only approximately threefold (Table 1). Therefore, the binding of Em to the ribosome is a prerequisite to induction. For erm(B) a stem-loop spanning the translation initiation site of the methylase structural gene was predicted (12) (see Fig. 7A). To confirm that this structure was an attenuator of structural gene expression, we made erm(B)-lacZ with a change in sequence near RBS2 from GGAGTG to GGAGTC (strain EL154), which resulted in destabilization of the stem. This substitution increased the minimum free energy from −13.2 kcal/mol to −7.0 kcal/mol and as a result increased the basal (uninduced) level of the expression ∼11-fold (Table 1). Taken together, these results verify that the expression is regulated by translational attenuation.
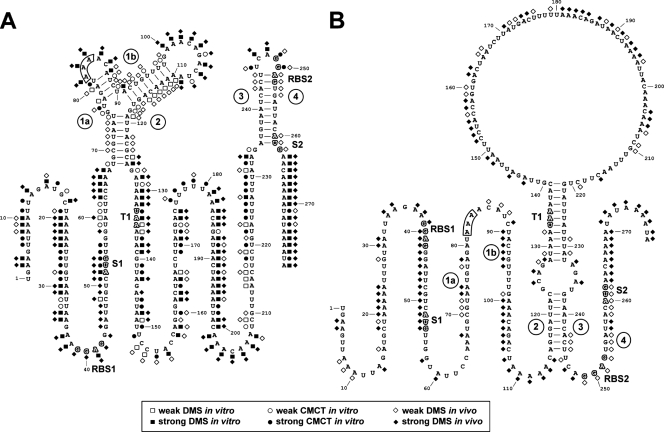
FIG. 7.
Secondary structure models of erm(B) mRNA showing the reactivity of the bases to chemical probes. (A) The inactive conformation is derived from the results of in vitro probing and in vivo probing without Em induction. (B) The active conformation is derived from the results of in vivo probing with Em induction. RBS1, S1, and T1, RBS, start codon, and termination codon of the leader peptide ORF, respectively; RBS2 and S2, RBS and start codon of the methylase gene, respectively; circled numbers 1a, 1b, 2, 3, and 4, segments of nt 68U to 81U, 87A to 98U, 110A to 126A, 235A to 245U, and 251A to 261U forming stems in the inactive conformation, respectively. The 11th codon of leader peptide, the ribosome stalling site, is boxed. The reactivities for the first two bases were not determined. These results were obtained from more than three independent experiments.
Em-induced stabilization and processing of erm(B) mRNA.
Shaw and Clewell observed a significant increase in the erm(B) transcript level by Em (27). This enhancement of the transcript level could be a consequence of increased transcription or the stabilization of mRNA. As shown above, the level of the erm(B)-lacZ fusion with a heterologous promoter (strain EL153) induced by Em decreased in comparison with that of the wild type. Thus, it is possible that Em somewhat induced the initiation of transcription. The mutant with the deletion of RBS1 (strain EL151) was, however, not inducible, even though it had the wild-type promoter. It is reasonable to hypothesize that the leader peptide is necessary for the induction of transcriptional initiation; i.e., the leader peptide might function as a trans-acting transcriptional activator. However, when the leader peptide was provided in trans by a plasmid carrying the leader peptide ORF in strain EL151 (strain EL220) to test this hypothesis, the phenotype was also not inducible (Table 1).
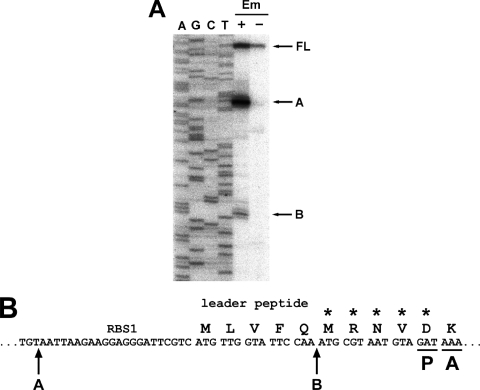
It has been reported that Em markedly stabilizes erm(C) and erm(A) mRNA (2, 24). Thus, we tested whether the increase in the erm(B) transcript level is due to the stabilization of the transcript. The half-life of the erm(B)-lacZ fusion mRNA was estimated by primer extension analysis (Fig. 1), since fusion with lacZ has little effect on the stability of erm(C) and erm(A) mRNA (7, 24). In the absence of Em, we detected a band corresponding to the full-length transcript that had a half-life of ∼3 min. In the presence of Em, the half-life of the full-length transcript increased to ∼27 min. Moreover, two additional bands (labeled A and B in Fig. 1) of different lengths were detected specifically in the cells induced with Em. The bands were 5′-end-truncated forms of the full-length transcript. They accumulated over time, which indicates that they are decay intermediates of the full-length transcript and are not from an alternative initiation of transcription. The 5′ ends of the truncated transcripts corresponding to bands A and B were mapped to 22 nt upstream from the start codon and before codon 6 of the leader peptide ORF, respectively (Fig. 2). It has been shown that in the presence of Em, transcripts of erm(A) and a deletion derivative of erm(C) are also cleaved in the sequences that encode leader peptides (9, 25).
FIG. 2.
Mapping of 5′ termini of the processed erm(B) transcripts. (A) Primer extension products from B. subtilis EL100 RNA with (+) or without (−) Em induction were run with the sequencing reactions by using the same primer. The 5′ termini of the transcripts corresponding to bands A and B are nt +30 and nt +67, respectively. (B) Location of the endonucleolytic cleavage in the leader region of erm(B). P and A, the codons occupied by the peptidyl and aminoacyl sites of the stalled ribosome, respectively; asterisks, the codons of the leader that are critical for induction.
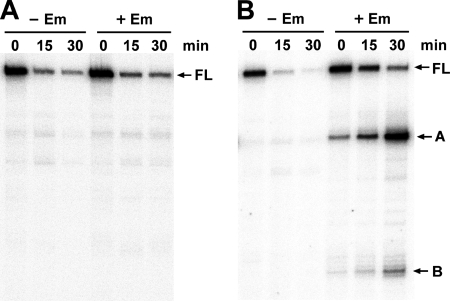
The ribosome stalling in the leader region appears to be essential for the induced stabilization and appearance of the decay intermediates of erm(B)-lacZ mRNA, as observed with erm(C) and its deletion derivative (2, 9). To confirm this, we analyzed the decay pattern in strain EL151, with the RBS1 deletion, and strain EL152, with a substitution at codon 9 of the leader region which was thought to prevent ribosome stalling by Em. These alterations resulted in the complete inhibition of stabilization of the full-length transcript and the loss of processed transcripts (Fig. 3A and B). Also, in the decay pattern of EL17, which possessed the ribosomal mutation, no stabilization of the full-length transcript was seen, and just a small amount of processed transcripts was detected (Fig. 3C).
Ribosome stalling site by Em.
To identify the specific Em-induced stalling site, leader codons 10 to 13 of erm(B)-lacZ were individually changed to the ochre nonsense codon (strains EL110 to EL113, respectively), and then the inducibility of the mutants was measured by the β-galactosidase assay. However, the basal expression levels of all mutants were approximately the same as the fully induced level of the wild type (Table 1), which was thought to result from a disruption of the mRNA secondary structure by the mutations, as discussed below. Because the induction of erm(C) mRNA stability has been reported to not be affected by the secondary structure of the leader region (13), we examined the decay profiles of these mutants. When leader codon 10 was replaced with the ochre codon, the full-length transcript was not stabilized by Em and no processed transcripts were seen in the presence of Em (Fig. 4A). On the other hand, when leader codon 11 was changed to the ochre codon, we could see the stabilization of both the full-length transcript and the processed transcripts (Fig. 4B). Replacement of leader codons 12 and 13 with the ochre codon also exhibited the same decay patterns as that of the wild type (data not shown). These results demonstrate that leader codon 11 is occupied by the A site of the ribosome stalled by Em.
FIG. 4.
Effects of ochre mutations in the leader peptide ORF on erm(B)-lacZ mRNA decay. Primer extension analysis was performed for the total RNAs from the ochre mutants at codons 10 (strain EL110) (A) and 11 (strain EL111) (B) of the leader peptide. −Em, absence of Em; +Em, presence of Em; FL, full-length transcript.
Structural probing of the erm(B) leader region in vitro and in vivo.
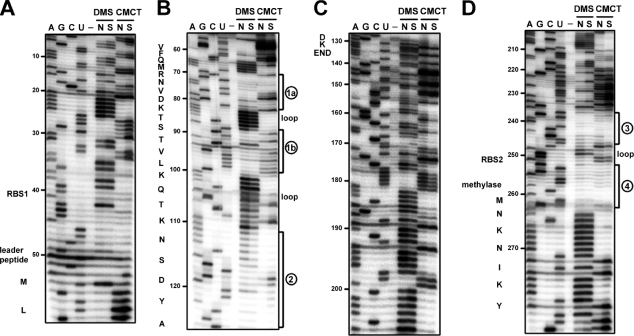
We performed chemical probing of the erm(B) leader region in vitro and in vivo. We used the experimental data with the help of a RNA secondary structure prediction program, Mfold (34), to derive the secondary structure models for the erm(B) leader. For in vitro probing, the transcript was synthesized in vitro by the use of T7 RNA polymerase. The transcript contains the entire leader sequence and the N-terminal 45 codons of the methylase gene. As shown in Fig. 5, little difference was seen between the native and the semidenaturing conditions. To examine the secondary structures in vivo, DMS probing of mRNA prepared from strain BR151 carrying erm(B)-lacZ (strain EL100) was conducted. The regions of nt 20A to 24A, 100A to 104A, 181A to 185A, and 263A to 270A served as the internal controls in Fig. 6A, B, C, and D, respectively.
FIG. 5.
Autoradiograms showing the results of in vitro chemical probing of the erm(B) leader region. The purified transcript was renatured and reacted with DMS and CMCT under native conditions (lanes N) or semidenaturing conditions (lanes S). The positions of base modification were determined by primer extension analysis with primers P4 (A), P3 (B), P2 (C), and P1 (D). Lanes A, G, C, and U, dideoxysequencing ladders; lanes −, incubation control in the buffer used for DMS. The RBSs and amino acid sequences of the leader peptide and methylase are indicated to the left of each gel. The positions of the stem-loops are indicated to the right of each gel (see Fig. 7).
Two structural elements were observed in a model derived from in vitro and in vivo studies without Em (Fig. 7A). One consists of the sequences from the 6th to the 25th codons of the leader peptide and contains three stems and two loops. We designated nt 68U to 81U, 87A to 98U, and 110A to 126A stem segments 1a, 1b, and 2, respectively. The other is a stem-loop in the translational initiation site of the methylase gene, where we designated two strands of the stem, nt 235A to 245U and 251A to 261U, stem segments 3 and 4, respectively. This structure may block the translational initiation of the structural gene.
Upon exposure to Em for 10 min, the in vivo methylation patterns were altered (Fig. 6, lanes 3 to 5). As a negative control, we used the mutant at leader codon 9 (strain EL152), and as expected, there was little change of the conformation (Fig. 6, lanes 9 and 10). As the concentration of Em increased, the change of the wild-type mRNA became more quantitatively distinct. The methylation was inhibited in the region of nt 79G to 85A in the transcript that corresponds successively to leader codons 10, 11, and the first position of codon 12, which was probably due to masking by the stalled ribosome. The stalled ribosome is thought to disrupt the association of stem segments 1a, 1b, and 2. Induction with Em increased the availability of the bases for methylation in the region of nt 71G to 78A of segment 1a, 88U to 93U of segment 1b, and 110A to 114C of segment 2. However, nt 115A to 123C of stem segment 2 remained protected. nt 130A to 139C, predicted to be single stranded, were also protected after Em induction. nt 130A to 139C are complementary to nt 221G to 230U, the accessibility of which was reduced after induction with Em. Therefore, these two segments are predicted to be duplexed. nt 115A to 123C of stem segment 2 are also complementary to nt 237G to 245U of stem segment 3, the accessibility of which remained inhibited. Thus, these two regions are also predicted to be associated (Fig. 7B). Ultimately, stem segment 4 could not associate with stem segment 3, which was demonstrated by the increased methylation in nt 255A to 261U of segment 4.
All MLSB antibiotics are known to be inducers of erm(B) (18). Therefore, for comparison, the effects of the 16-membered-ring macrolide tylosin, the lincosamide clindamycin, and the streptogramin B quinupristin were also tested (Fig. 6, lanes 6 to 8). The cells were treated at the optimal induction concentrations, determined by a preliminary β-galactosidase assay (tylosin, 0.3 μg/ml; clindamycin, 0.1 μg/ml; quinupristin, 0.3 μg/ml) for 10 min. Only weak conformational alterations were observed, in contrast to the result for Em, while these drugs as well as Em induced the expression of erm(B)-lacZ under the same induction condition (Em, 5.5-fold; tylosin, 4.1-fold; clindamycin, 1.5-fold; quinupristin, 1.9-fold).
DISCUSSION
A previous Northern blot analysis had shown that only the full-length transcript and no truncated form of erm(B) mRNA was detectable in both uninduced and induced cells (27). There is also no possible transcription terminator structure in the leader region, based on our chemical probing results. These findings rule out the possibility of transcriptional attenuation as a control model and strongly support the conclusion that Em-induced expression of erm(B) is regulated by translational attenuation.
In B. subtilis, mRNA decay has been thought to initiate with the endonucleolytic cleavage effected by a decay-initiating nuclease (probably RNase J1/J2) that requires a free 5′ end (5, 6). Recently, it was found that RNase J1/J2 also has 5′-to-3′ exonuclease activity (19). Thus, mRNA decay can be initiated by endonucleolytic or exonucleolytic cleavage at the 5′ end. We surmise that the stabilization of the full-length transcript is due to the shielding of the endonuclease target site near the 5′ end by the stalled ribosome, as suggested in the cases of erm(C) and cat mRNA (3, 8, 25). The stabilization of erm(B) mRNA also results from the accumulation of stable processed transcripts. In the presence of Em, an endonucleolytic activity associated with the stalled ribosome presumably cleaves the sequences near the stall site. Otherwise, the processed transcripts may arise from a blocking of the 5′-to-3′ migration of the exonuclease by the stalled ribosome. The stalled ribosome at the 5′ end of the processed transcripts restricts the access of endonuclease and exonuclease and thereby protects the downstream mRNA from degradation.
The 5′ ends of the processed transcripts detected in the decay of the erm(A) transcript were mapped in codon 5 of the first leader peptide and in codon 5 and after codon 7 of the second leader peptide (25). Similarly, the 5′ end of the processed transcript of the erm(C) deletion mutant was mapped before leader codon 5 (9). Because a stalled ribosome has its A site occupied by leader codon 9 in erm(C) induction, the processing site is at codon 4 upstream from the A-site codon. Our results revealed that the A site is stalled at leader codon 11 of erm(B) and that the downstream cleavage site is before codon 6. Thus, on the erm(B) transcript, the distance and direction of the cleavage site from the ribosome stalling site are similar to those of erm(A) and erm(C). This confirms that in the presence of Em the cleavage of erm(B) mRNA is linked with the stalled ribosome and that the induction of the erm(B) gene demands that the A site of the stalled ribosome be occupied by leader codon 11.
Our data suggest that the critical erm(B) leader sequence where the Em-ribosome complex stalls consists of codons 6 to 10, which encode the amino acids MRNVD (Fig. 2B). Fragment B encodes the upstream boundary of the stall site, as determined by reverse transcription mapping, while the downstream extent was determined by substitution of an ochre triplet for codons 10 to 13. We previously found four kinds of mutations of the erm(B) leader region that cause alterations in the induction specificity for Em and 16-membered-ring macrolides such as tylosin (22). Three of the four mutations are consistent with the deduction: an Arg-to-Cys change at codon 7 [erm(Bv)], the duplication of four amino acids (Gln, Met, Arg, Tyr) before codon 9 of the wild-type leader peptide [erm(Bv1)], and a Gln-to-Lys change at leader codon 9 of erm(Bv1) [erm(Bv2)].
A previous X-ray crystallographic study showed that approximately 18 nt upstream and 9 nt downstream from the A-site codon of mRNA are in contact with the ribosome of Thermus thermophilus (32). Hence, although the ribosome stalling site (leader codon 11) is in a single-stranded loop region, the stalled ribosome possibly hinders stem segments 1a and 1b from associating with stem segment 2 and, instead, an association between stem segments 2 and 3 is favored (Fig. 7). Freeing the sequestered translational initiation site of the methylase gene leads to an increased rate of methylase translation. When we converted respective leader codons 10 to 13 to ochre codons to find the ribosome stalling site, the basal levels of expression were as high as the fully induced level (Table 1). The reason may be that the pausing of ribosomes at codons 10 to 13 prevents stem segment 2 from associating with segments 1a and 1b.
In the proposed model, the distance between segment 2 and segment 3 is over 100 nt, while the distance between segment 3 and segment 4 is 5 nt. Although segment 2 and segment 3 are duplexed after induction, as determined by the chemical probing results, this conformational change is thought to be kinetically unfavorable. However, long-distance base pairing could take place with the help of a protein. Examples of protein mediation of RNA base pairing, such as E. coli host factor I, Hfq (33), and trp RNA-binding attenuation protein (10), have been reported. In the case of erm(B), the leader peptide or any host protein might facilitate this long-distance interaction.
Acknowledgments
We thank D. B. Clewell for plasmid pAM225 and H. Jarmer for plasmid pB1.
This study was supported by a grant from the South Korea Centers for Disease Control and Prevention.
Footnotes
Published ahead of print on 25 February 2008.
REFERENCES
- 1.Allen, N. E. 1977. Macrolide resistance in Staphylococcus aureus: inducers of macrolide resistance. Antimicrob. Agents Chemother. 11:669-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bechhofer, D. H., and D. Dubnau. 1987. Induced mRNA stability in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 84:498-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bechhofer, D. H., and K. H. Zen. 1989. Mechanism of erythromycin-induced ermC mRNA stability in Bacillus subtilis. J. Bacteriol. 171:5803-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byeon, W. H., and B. Weisblum. 1990. Replication genes of plasmid pE194-cop and repF: transcripts and encoded proteins. J. Bacteriol. 172:5892-5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Condon, C. 2007. Maturation and degradation of RNA in bacteria. Curr. Opin. Microbiol. 10:271-278. [DOI] [PubMed] [Google Scholar]
- 6.Deana, A., and J. G. Belasco. 2005. Lost in translation: the influence of ribosomes on bacterial mRNA decay. Genes Dev. 19:2526-2533. [DOI] [PubMed] [Google Scholar]
- 7.DiMari, J. F., and D. H. Bechhofer. 1993. Initiation of mRNA decay in Bacillus subtilis. Mol. Microbiol. 7:705-717. [DOI] [PubMed] [Google Scholar]
- 8.Dreher, J., and H. Matzura. 1991. Chloramphenicol-induced stabilization of cat messenger RNA in Bacillus subtilis. Mol. Microbiol. 5:3025-3034. [DOI] [PubMed] [Google Scholar]
- 9.Drider, D., J. M. DiChiara, J. Wei, J. S. Sharp, and D. H. Bechhofer. 2002. Endonuclease cleavage of messenger RNA in Bacillus subtilis. Mol. Microbiol. 43:1319-1329. [DOI] [PubMed] [Google Scholar]
- 10.Du, H., and P. Babitzke. 1998. trp-RNA binding attenuation protein-mediated long-distance RNA refolding regulates translation of trpE in Bacillus subtilis. J. Biol. Chem. 273:20494-20503. [DOI] [PubMed] [Google Scholar]
- 11.Horinouchi, S., and B. Weisblum. 1982. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J. Bacteriol. 150:815-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horinouchi, S., W. H. Byeon, and B. Weisblum. 1983. A complex attenuator regulates inducible resistance to macrolides, lincosamides, and streptogramin type B antibiotics in Streptococcus sanguis. J. Bacteriol. 154:1252-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hue, K. K., and D. H. Bechhofer. 1991. Effect of ermC leader region mutations on induced mRNA stability. J. Bacteriol. 173:3732-3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hue, K. K., and D. H. Bechhofer. 1992. Regulation of the macrolide-lincosamide-streptogramin B resistance gene ermD. J. Bacteriol. 174:5860-5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarmer, H., T. S. Larsen, A. Krogh, H. H. Saxild, S. Brunak, and S. Knudsen. 2001. Sigma A recognition sites in the Bacillus subtilis genome. Microbiology 147:2417-2424. [DOI] [PubMed] [Google Scholar]
- 16.Kaltwasser, M., T. Wiegert, and W. Schumann. 2002. Construction and application of epitope- and green fluorescent protein-tagging integration vectors for Bacillus subtilis. Appl. Environ. Microbiol. 68:2624-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamimiya, S., and B. Weisblum. 1997. Induction of ermSV by 16-membered-ring macrolide antibiotics. Antimicrob. Agents Chemother. 41:530-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leclercq, R., and P. Courvalin. 2002. Resistance to macrolides and related antibiotics in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:2727-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathy, N., L. Bénard, O. Pellegrini, R. Daou, T. Wen, and C. Condon. 2007. 5′-to-3′ exoribonuclease activity in bacteria: role of RNase J1 in rRNA maturation and 5′ stability of mRNA. Cell 129:681-692. [DOI] [PubMed] [Google Scholar]
- 20.Mayford, M., and B. Weisblum. 1989. Conformational alterations in the ermC transcript in vivo during induction. EMBO J. 8:4307-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 22.Min, Y.-H., J.-H. Jeong, Y.-J. Choi, H.-J. Yun, K. Lee, M.-J. Shim, J.-H. Kwak, and E.-C. Choi. 2003. Heterogeneity of macrolide-lincosamide-streptogramin B resistance phenotypes in enterococci. Antimicrob. Agents Chemother. 47:3415-3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy, E. 1985. Nucleotide sequence of ermA, a macrolide-lincosamide-streptogramin B determinant in Staphylococcus aureus. J. Bacteriol. 162:633-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sandler, P., and B. Weisblum. 1988. Erythromycin-induced stabilization of ermA messenger RNA in Staphylococcus aureus and Bacillus subtilis. J. Mol. Biol. 203:905-915. [DOI] [PubMed] [Google Scholar]
- 25.Sandler, P., and B. Weisblum. 1989. Erythromycin-induced ribosome stall in the ermA leader: a barricade to 5′-to-3′ nucleolytic cleavage of the ermA transcript. J. Bacteriol. 171:6680-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarkar, G., and S. S. Sommer. 1990. The “megaprimer” method of site-directed mutagenesis. BioTechniques 8:404-407. [PubMed] [Google Scholar]
- 27.Shaw, J. H., and D. B. Clewell. 1985. Complete nucleotide sequence of macrolide-lincosamide-streptogramin B-resistance transposon Tn917 in Streptococcus faecalis. J. Bacteriol. 164:782-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang, T. H., T. S. Rozhdestvensky, B. C. d'Orval, M.-L. Bortolin, H. Huber, B. Charpentier, C. Branlant, J.-P. Bachellerie, J. Brosius, and A. Hüttenhofer. 2002. RNomics in Archaea reveals a further link between splicing of archaeal introns and rRNA processing. Nucleic Acids Res. 30:921-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tipper, D. J., C. W. Johnson, C. L. Ginther, T. Leighton, and G. Wittmann. 1977. Erythromycin resistant mutations in Bacillus subtilis cause temperature sensitive sporulation. Mol. Gen. Genet. 150:147-159. [DOI] [PubMed] [Google Scholar]
- 30.Weisblum, B. 1995. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39:577-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weisblum, B. 1995. Insights into erythromycin action from studies of its activity as inducer of resistance. Antimicrob. Agents Chemother. 39:797-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yusupova, G. Z., M. M. Yusupov, J. H. Cate, and H. F. Noller. 2001. The path of messenger RNA through the ribosome. Cell 106:233-241. [DOI] [PubMed] [Google Scholar]
- 33.Zhang, A., K. M. Wassarman, J. Ortega, A. C. Steven, and G. Storz. 2002. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell 9:11-22. [DOI] [PubMed] [Google Scholar]
- 34.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]