Abstract
Specific inhibitors of hepatitis C virus (HCV) replication that target the NS3/4A protease (e.g., VX-950) or the NS5B polymerase (e.g., R1479/R1626, PSI-6130/R7128, NM107/NM283, and HCV-796) have advanced into clinical development. Treatment of patients with VX-950 or HCV-796 rapidly selected for drug-resistant variants after a 14-day monotherapy treatment period. However, no viral resistance was identified after monotherapy with R1626 (prodrug of R1479) or NM283 (prodrug of NM107) after 14 days of monotherapy. Based upon the rapid selection of resistance to the protease and nonnucleoside inhibitors during clinical trials and the lack of selection of resistance to the nucleoside inhibitors, we used the replicon system to determine whether nucleoside inhibitors demonstrate a higher genetic barrier to resistance than protease and nonnucleoside inhibitors. Treatment of replicon cells with nucleoside inhibitors at 10 and 15 times the 50% effective concentration resulted in clearance of the replicon, while treatment with a nonnucleoside or protease inhibitor selected resistant colonies. In combination, the presence of a nucleoside inhibitor reduced the frequency of colonies resistant to the other classes of inhibitors. These results indicate that the HCV replicon presents a higher barrier to the selection of resistance to nucleoside inhibitors than to nonnucleoside or protease inhibitors. Furthermore, the combination of a nonnucleoside or protease inhibitor with a nucleoside polymerase inhibitor could have a clear clinical benefit through the delay of resistance emergence.
Hepatitis C virus (HCV) is a positive-strand RNA virus that is a member of the Hepacivirus genus within the Flaviridae family. There are an estimated 170 million individuals chronically infected with HCV worldwide, which amounts to almost 3% of the global population (1). In the United States, an estimated 20,000 new HCV infections occurred in 2005, adding to the approximately 4 million individuals previously infected with HCV (2, 38). Liver cirrhosis, as a result of HCV infection, is currently the leading reason justifying liver transplantation; however, reinfection occurs immediately posttransplantation and can result in graft loss (39).
The current treatment of pegylated alpha interferon in combination with ribavirin results in a sustained viral response in approximately 50% of HCV patients infected with genotype (GT) 1 virus, the most prevalent GT worldwide. Therefore, a specific HCV antiviral therapy is highly desirable. Viral proteases and viral polymerases have been validated as clinically effective targets for a number of different viruses, including human immunodeficiency virus, hepatitis B virus, and herpesviruses (6, 7, 14, 15). Two potential drug targets encoded by HCV are the NS3/4A serine protease and the NS5B RNA-dependent RNA polymerase (5). Several anti-HCV compounds that inhibit the activity of either the NS3/4A protease or the NS5B RNA-dependent RNA polymerase have resulted in decreased viral loads when administered to HCV-infected patients (10, 29, 30, 32).
VX-950 is a peptidomimetic inhibitor of the NS3/4A serine protease that is currently undergoing clinical evaluation. In a phase 1b study, the viral load in HCV infected patients dosed with 750 mg of VX-950 every 8 h was reduced by greater than 4 log10 IU/ml (29). However, a number of patients administered VX-950 showed a subsequent viral load rebound or plateau during the 14-day dosing period. Population sequencing of the viral NS3 region identified a number of mutations near the NS3 protease catalytic domain (31). The changes at NS3 residues 36, 54, 155, and 156 were shown to confer a loss of sensitivity to the protease inhibitor VX-950 when tested using an enzyme or replicon assay (31).
The NS5B enzyme activity can be inhibited by different classes of compounds, including nucleoside inhibitors, which can act as alternative substrates for the viral polymerase, and nonnucleoside inhibitors, which act as allosteric inhibitors of the polymerase. A number of compounds that inhibit the RNA-dependent RNA polymerase activity of NS5B have also entered into clinical development. HCV-796 is a nonnucleoside inhibitor that resulted in a decrease in viral load when administered to HCV-infected patients. Patients treated with HCV-796 for 14 days had a maximum viral load reduction of 1.4 log10 on day 4, followed by a viral load rebound during the dosing period (4). Sequencing of the patient isolates identified the NS5B amino acid substitution C316Y, previously identified using the HCV replicon system and known to confer reduced sensitivity to HCV-796 (36, 37; A. Howe et al., presented at the 13th International Meeting on Hepatitis C Virus and Related Viruses, Cairns, Australia, 27 to 31 August 2006). Three nucleoside inhibitors have progressed into clinical development: NM283 (prodrug of NM107), R1626 (prodrug of R1479), and R7128 (prodrug of PSI-6130). Treatment of HCV-infected patients with R1626 resulted in a mean viral load reduction of 3.7 log10 at 4,500 mg twice a day (30). Unlike the case for VX-950- and HCV-796 treated patients, there was no evidence for the emergence of viral resistance to R1626 (30). However, mutations that confer reduced sensitivity to R1479 have been identified using the HCV replicon system (19). NM-283 has also shown clinical efficacy (1.2 log drop in viral load at 800 mg once a day) (10), and an NS5B resistance mutation, S282T, which confers resistance to NM107 (19) has been identified in vitro.
Within infected patients, HCV replicates at a high rate and exists as a population known as a quasispecies. When a selective pressure is applied to HCV (e.g., antiviral compounds or immunological response), minor variants within the population that are less susceptible to the pressure will have a replicative advantage. As a result, the dynamics of the genetically diverse population are altered, and those resistant variants will likely become the predominant species in the population. It is therefore important to understand how the inhibition of HCV replication, by various classes of compounds, may affect the selection of less sensitive variants. In addition, similar to the currently recommended human immunodeficiency virus therapy (12), the future therapy options for HCV-infected patients may include multiple specific anti-HCV inhibitors. In this study we characterized the selection of resistance, alone and in combination, to several clinically advanced anti-HCV compounds, including the protease inhibitor VX-950; the nonnucleoside inhibitor HCV-796; and the nucleoside inhibitors R1479, PSI-6130, and NM107.
MATERIALS AND METHODS
Plasmid construction.
The selectable Con1 HCV subgenomic replicon used in this study is based on the adapted dicistronic HCV subgenomic replicon construct previously described (18). The Con1 adapted transient replicon (rep PI-luc/ET) was obtained from R. Bartenschlager (24).
All site-directed mutations were introduced into rep PI-luc/ET using the QuikChange site-directed mutagenesis kit according to the manufacturer's instructions (Stratagene) and confirmed on both strands by DNA sequencing using ABI technology.
Compounds.
Compounds R1479 (4′-azido-cytidine), NM107 (2′-C-methylcytidine), and HCV-796 (benzofuran carboxamide) were synthesized at Roche, Palo Alto. VX-950 was synthesized by Acme Bioscience, Inc. PSI-6130 (β-d-2′-deoxy-2′-fluoro-2′-C-methylcytidine) was synthesized by Pharmasset, Inc. Stocks of 10 mM were prepared in 100% dimethyl sulfoxide and stored at −20°C.
EC50 determinations and replication capacity of transient replicons.
The concentration of compound required to inhibit the expression of the HCV replicon reporter by 50% compared to untreated controls (50% effective concentration [EC50])was determined as described previously using 2209-23 stably transfected replicon cells (19), except that Huh7-Lunet cells were used for transient transfections. The Huh7-Lunet cells were kindly provided by V. Lohmann and have been described previously (17). The replication capacity of the transient replicons was determined as previously described (20).
In vitro combination studies.
The combination studies were performed as previously described (19). Briefly, HCV replicon 2209-23 cells were plated and inhibitors were added at different concentrations in threefold dilutions at a final dimethyl sulfoxide concentration of 1% (vol/vol) in a checkerboard fashion, including titrations of each of the compounds alone as reference controls. The concentrations of compound PSI-6130 ranged from 11 μM to 0.005 μM and from 1.23 μM to 0.005 μM for HCV-796 and from 11 μM to 0.05 μM for VX-950. The concentrations of compound R1479 ranged from 33.3 μM to 0.137 μM and from 0.41 μM to 0.0002 μM for HCV-796 and from 11 μM to 0.005 μM for VX-950. The degree of interaction between the two compounds was determined using a seven-parameter nonlinear model based on the Loewe additivity theory (13), which uses the concentration response surface approach as previously described (11). Drug interaction is interpreted using the parameter α as well as the percent synergy. The predicted additivity of the drugs combined is calculated by using all estimated parameters of the Greco model, except for α, which is fixed at 0. The deviance between the predicted response surface and the predicted additive surface is interpreted as percent synergy (positive percentages, if the response surface is above the additive surface) or percent antagonism (negative percentages, if the response surface is under the additive surface). Synergy is indicated when the parameter α is positive with a 95% confidence interval not including 0, and the maximum percent synergy is greater than 10%. Antagonism is indicated when α is negative with a 95% confidence interval not including 0, and the maximum percent antagonism is less than −10%. Loewe additivity or no interaction is indicated when the 95% confidence interval of α includes 0.
Colony formation assay.
For the analysis of resistance selection, Huh7 cells that stably maintain a GT 1b subgenomic replicon carrying the neoR gene, described previously (16), were plated at 1 × 105 cells/well in six-well plates in the presence of 0.5 mg/ml G418 and in the absence or presence of inhibitors. The cells were treated with either one or two compounds at 1, 10, or 15 times their respective EC50. The culture medium was changed twice per week, and cells were passaged at a 1:4 split when ∼95% confluence was reached. After 3 weeks in culture, cells were either fixed with 1% (vol/vol) formaldehyde and stained with crystal violet or the total cellular RNA was extracted using the RNeasy minikit (Qiagen). The NS5B and NS3 coding sequences were determined through direct sequencing of PCR products.
RESULTS
Lack of resistant colonies after treatment with the NS5B nucleoside inhibitor NM107, R1479, or PSI-6130.
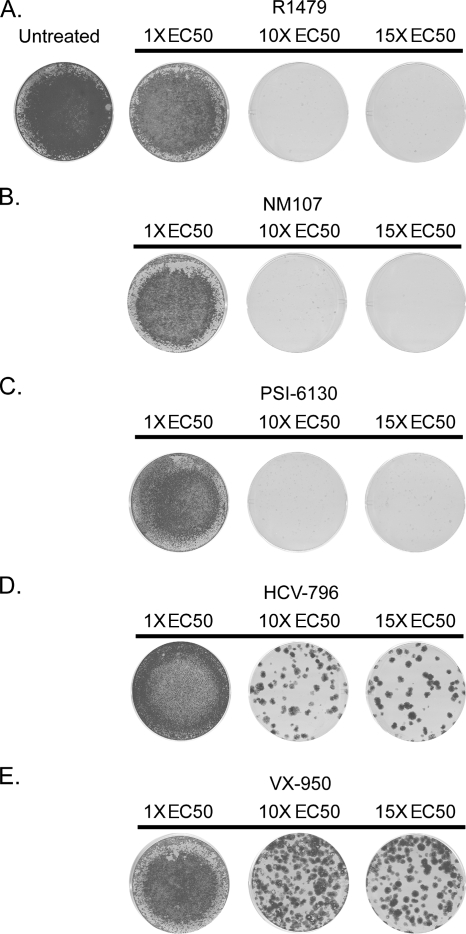
A stable GT 1b replicon cell line was treated with the NS5B nucleoside inhibitors NM107, R1479, and PSI-6130 for 3 weeks at 1, 10, or 15 times their respective EC50 (calculated using the stably transfected Con1 replicon cell line 2209-23 [Table 1] and comparable to previously reported values [16, 19, 23, 36]). The cells were selected with G418 in order to eliminate replicon cells susceptible to the inhibitor and to select for cells that contained replicon variants with reduced sensitivity to the inhibitor(s). After the 3-week selection period, the remaining cells were either stained with crystal violet or used for the extraction of total RNA in order to determine the sequence of the NS5B-coding region. As seen in Fig. 1A to C, incubation with 1 times the EC50 of any of the nucleoside inhibitors did not reduce the replicon replication to levels sufficient to render the cells sensitive to G418 selection, as seen by the intact monolayers in Fig. 1, whereas at higher concentrations of the inhibitor(s), the replicon was cleared.
TABLE 1.
Inhibitory activity and cytotoxicity against stable HCV 1b replicon cells (Gt 1b Con1)
| Compound | Mean ± SD
|
|
|---|---|---|
| EC50 (μM)a | CC50 (μM)b | |
| R1479 | 1.28 ± 0.85 | >100 |
| PSI-6130 | 0.61 ± 0.26 | >100 |
| NM107 | 1.23 ± 0.52 | >100 |
| HCV-796 | 0.017 ± 0.005 | >100 |
| VX-950 | 0.56 ± 0.11 | 26.7 ± 3.1 |
Inhibition of HCV replicon-encoded Renilla luciferase reporter activity after 3 days of incubation (n = 101 for R1479, n = 46 for PSI-6130, n = 567 for NM107, n = 18 for HCV-796, and n = 4 for VX-950).
CC50, 50% cytotoxic concentration. Cell viability was determined by either MTT or WST-1 assay with the same number of experiments as for the EC50 determinations.
FIG. 1.
Replicon cells treated with a nucleoside inhibitor clear the replicon, while VX-950 or HCV-796 select for resistant colonies. Huh7 cells that stably maintain a GT 1b subgenomic replicon carrying the neoR gene were incubated with a specific HCV inhibitor at 1, 10, and 15 times the EC50 in the presence of G418. After selection for 3 weeks, the remaining cells were fixed and stained with crystal violet. The replicon cells were treated with the nucleoside inhibitor R1479 (A), NM107 (B), or PSI-6130 (C); the nonnucleoside inhibitor HCV-796 (D); or the protease inhibitor VX-950 (E). A representative example of experiments is shown (n = 6 for R1479, n = 4 for NM107 and PSI-6130, n = 7 for VX-950, and n = 10 for HCV-796).
Treatment with the NS3/4A protease inhibitor VX-950 or with the nonnucleoside NS5B polymerase inhibitor HCV-796 selects for resistant replicon colonies.
Similarly, in a parallel study, the stable GT 1b replicon cell line was treated with either the NS3/4A protease inhibitor VX-950 or the nonnucleoside NS5B inhibitor HCV-796. Unlike with the nucleoside inhibitors, resistant colonies were selected after 3 weeks of incubation under drug selective pressure of 10 or 15 times the EC50 with either VX-950 or HCV-796 (Fig. 1D and E). In the presence of 10 times the EC50 of VX-950, >200 colonies were obtained, whereas at 15 times the EC50 the number was 93 ± 15 (Table 2). In the presence of 10 times the EC50 of HCV-796, 60 ± 15 colonies were obtained, and 36 ± 12 colonies were obtained with 15 times the EC50 of HCV-796 (Table 2).
TABLE 2.
Frequency of resistant-colony formation after single or combination selection
| Selection (n)a | Mean no. of colonies ± SDb | Frequency (%)c |
|---|---|---|
| 10× HCV-796 (10) | 60 ± 15 | 0.060 |
| 10× HCV-796/1× R1479 (6) | 23 ± 9 | 0.023 |
| 10× HCV-796/1× PSI-6130 (4) | 6 ± 8 | 0.006 |
| 15× HCV-796 (10) | 36 ± 12 | 0.036 |
| 15× HCV-796/1× R1479 (6) | 12 ± 8 | 0.012 |
| 15× HCV-796/1× PSI-6130 (4) | 2 ± 3 | 0.002 |
| 15× VX-950 (7) | 93 ± 16 | 0.093 |
| 15× VX-950/1× R1479 (4) | 44 ± 30 | 0.044 |
| 15× VX-950/1× PSI-6130 (4) | 4 ± 4 | 0.004 |
Replicon cells were selected with 1, 10, or 15 times the EC50 of the indicated compounds.
Replicon cells were stained with crystal violet after undergoing selection for 3 weeks, and the number of colonies remaining were counted.
Frequency determined as number of colonies/initial number of cells × 100.
To determine whether the observed colonies were indeed resistant to the respective inhibitors, the total RNA was extracted and the sequence of the NS3- or NS5B-coding region was determined to identify any potential amino acid substitutions. Cells treated with any of the compounds at 1 times the EC50 either had the wild-type sequence, compared to the untreated sample, or had amino acid substitutions not known to confer resistance. Some of the latter were also observed in the untreated control or were observed after selection with different inhibitors (data not shown), suggesting that they are likely either random changes or adaptive mutations to the tissue culture conditions. Under inhibitor pressure of either 10 or 15 times the EC50 with VX-950 or HCV-796, replicons that contained known resistance mutations were selected (Table 3). Replicons selected under HCV-796 pressure contained NS5B amino acid substitutions C316Y and S365A, previously described to decrease the sensitivity to HCV-796 (Howe et al., presented at the 13th International Meeting on Hepatitis C Virus and Related Viruses). Similarly, replicons selected in the presence of VX-950 contained amino acid substitutions in the NS3-coding region, T54A and A156T/S, which are known to confer decreased sensitivity to VX-950 (22, 26, 35, 40).
TABLE 3.
Genotypic characterization of NS5B or NS3 from replicons after single or combination selection
| Selectiona | Mutation(s)b
|
|
|---|---|---|
| NS5B | NS3 | |
| Untreated | None | None |
| 1× R1479 | None | None |
| 1× PSI-6130 | None | NDg |
| 1× NM107 | None | ND |
| 1× HCV-796 | None | ND |
| 1× HCV-796/1× R1479 | None | ND |
| 10× HCV-796 | C316C/Yc, S365S/Ad | ND |
| 10× HCV-796/1× R1479 | C316Y | ND |
| 10× HCV-796/1× PSI-6130 | ND | ND |
| 15× HCV-796 | C316Y | ND |
| 15× HCV-796/1× R1479 | C316Y | ND |
| 15× HCV-796/1× PSI-6130 | C316Y | ND |
| 1× VX-950 | None | None |
| 1× VX-950/1× R1479 | None | None |
| 1× VX-950/1× PSI-6130 | None | None |
| 10× VX-950 | None | A156T/Se |
| 10× VX-950/1× R1479 | None | A156T/S |
| 10 × VX-950/1× PSI-6130 | None | T54 T/Af, A156A/S |
| 15× VX-950 | None | A156T/S |
| 15× VX-950/1× R1479 | None | A156T/S |
| 15× VX-950/1× PSI-6130 | None | A156S |
Replicon cells were either untreated or selected with 1, 10, or 15 times the EC50 of the indicated compounds.
Direct sequencing from PCR products, representing the major population (in one-letter amino acid code). Only amino acid substitutions known to confer resistance are reported.
The mutation C316Y is a known resistance mutation for HCV-796.
The mutation S365A is a known resistance mutation for HCV-796.
The mutations A156T and A156S are known resistance mutations for VX-950.
The mutation T54A is a known resistance mutation for VX-950.
ND, not determined.
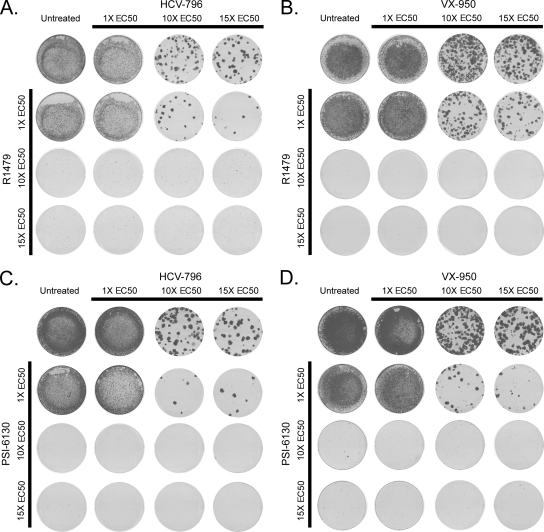
Combination using a nucleoside polymerase inhibitor reduces the frequency of drug resistance selection.
Replicon cells were treated with a nucleoside inhibitor (R1479 or PSI-6130) in combination with either VX-950 or HCV-796. Similar to the results shown in Fig. 1, single treatment with VX-950 or HCV-796 alone selected for resistant colonies, while the replicon was cleared in cells treated with a nucleoside inhibitor (Fig. 2A to D). The combination of 1 times the EC50 of R1479 with either VX-950 or HCV-796 reduced the number of colonies by approximately two- to threefold (Table 2). Higher concentrations of R1479, in combination with VX-950 or HCV-796, cleared the replicon. The combination of 1X PSI-6130 with either VX-950 or HCV-796 reduced the number of resistant colonies by at least 10-fold (Table 2), while higher concentrations of PSI-6130 cleared the replicon. Sequencing of the NS3- and NS5B-coding regions from replicon cells treated in combination identified known resistance mutations for HCV-796 and VX-950 but not for R1479 or PSI-6130 (Table 3).
FIG. 2.
Combination treatment with a nucleoside inhibitor reduces the emergence of resistant colonies. Huh7 cells that stably maintain a GT 1b subgenomic replicon carrying the neoR gene were incubated with one or two specific HCV inhibitors at 1, 10, and 15 times the EC50 in the presence of G418. After selection for 3 weeks, the remaining cells were fixed and stained with crystal violet. The replicon cells were treated with R1479 and HCV-796 (A), R1479 and VX-950 (B), PSI-6130 and HCV-796 (C), or PSI-6130 and VX-950 (D), and a representative example of at least four independent experiments is shown.
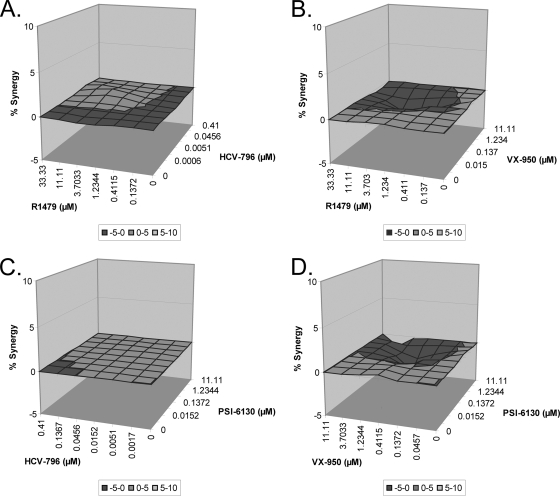
In vitro drug-drug interaction studies.
As shown above, the results of the combination colony formation assay indicated that there is a reduced emergence of resistant colonies as a result of combining a nucleoside inhibitor with either a protease or a nonnucleoside inhibitor. To specifically measure the in vitro drug-drug interaction, in vitro combination studies were performed to determine the inhibitory activity of VX-950 or HCV-796 in combination with R1479 or PSI-6130. All four of the combinations were analyzed using the Loewe additivity theory as specified in Materials and Methods, and the combinations resulted in an additive response; i.e., the percent synergy was approximately 0 and the 95% confidence interval of α included 0 (Fig. 3A to D).
FIG. 3.
Combination of a nucleoside inhibitor with either VX-950 or HCV-796 results in additive inhibitory activity against the replicon. Huh7 cells that stably maintain a GT 1b subgenomic replicon were treated with various concentrations of a nucleoside inhibitor, either R1479 (A and B) or PSI-6130 (C and D), and either HCV-796 (A and C) or VX-950 (B and D). After 72 h, the inhibitory activity of the compounds was determined by measuring the reduction in luciferase activity. The activity of the inhibitors in combination was determined from at least three experiments and is reported as percent synergy.
Lack of cross-resistance supports drug combination.
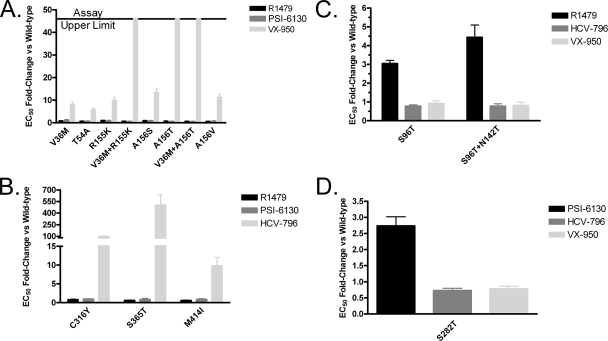
The reduced frequency of selection of drug-resistant replicon colonies during combination treatment suggests a lack of cross-resistance among the evaluated inhibitors. To confirm this, a panel of transient GT 1b replicons carrying resistance mutations for each inhibitor was generated (22, 26, 35, 40; Howe et al., presented at the 13th International Meeting on Hepatitis C Virus and Related Viruses). For the protease inhibitor VX-950, the NS3 amino acid substitutions A156S/T/V were originally identified in vitro. A subsequent phase 1b clinical trial confirmed the relevance of these mutations and identified additional amino acid substitutions in NS3 at residues 36, 54, and 155 (31). The introduction of the NS3 mutations (V36M, T54A, R155K, V36M-R155K, A156S, A156T, V36M-A156T, and A156V) into a transient replicon confirmed that these mutations resulted in decreased sensitivity (5- to >46-fold) to VX-950 (Fig. 4A), as previously described (31). The nucleoside inhibitors R1479 and PSI-6130 maintained potency against these VX-950-resistant transient replicons (Fig. 4A).
FIG. 4.
Transient replicons show lack of cross-resistance to other classes of HCV inhibitors. Huh7-Lunet cells were transfected with either a wild-type GT 1b replicon or one engineered to contain mutations resulting in reduced sensitivity to VX-950 (A), HCV-796 (B), R1479 (C), or PSI-6130 (D). The transfected cells were incubated with various HCV inhibitors for 72 h, and then the replicon luciferase activity was measured. The EC50 determinations were obtained from at least three independent experiments: Con1, n = 6 for R1479, n = 8 for VX-950, n = 5 for PSI-6130, and n = 6 for HCV-796; V36M, n = 5 for R1479, n = 6 for VX-950, and n = 4 for PSI-6130; T54A, n = 4 for R1479 and VX-950 and n = 3 for PSI-6130; R155K, n = 3 for R1479 and PSI-6130 and n = 4 for VX-950; V36M+R155K, n = 5 for R1479 and n = 3 for VX-950 and PSI-6130; A156S, n = 5 for R1479 and PSI-6130 and n = 6 for VX-950; A156T, n = 4 for R1479 and n = 3 for VX-950 and PSI-6130; V36M+A156T, n = 3 for R1479, VX-950, and PSI-6130; A156V, n = 6 for R1479, n = 4 for VX-950, and n = 3 for PSI-6130; S96T, n = 6 for R1479, n = 8 for VX-950, and n = 4 for HCV-796; S96T+N142T, n = 6 for R1479, n = 7 for VX-950, and n = 4 for HCV-796; C316Y, n = 4 for R1479, n = 5 for PSI-6130, and n = 6 for HCV-796; S365T, n = 3 for R1479 and PSI-6130 and n = 5 for HCV-796; M414I, n = 4 for R1479 and n = 3 for PSI-6130 and HCV-796; and S282T, n = 6 for VX-950, n = 4 for PSI-6130, and HCV-796. The fold change of the inhibitor was determined as the EC50 of the inhibitor against the mutant replicon compared to the wild-type replicon and is presented as mean ± standard error of the mean.
The nonnucleoside inhibitor HCV-796 has been reported to bind to a site in the palm domain of the HCV polymerase (Howe et al., presented at the 13th International Meeting on Hepatitis C Virus and Related Viruses). In vitro replicon resistance studies identified substitutions at NS5B residues C316Y/F/S, S365T/A/L, and M414I, which are located near the inhibitor binding site and confer a loss of sensitivity to HCV-796 (Howe et al., presented at the 13th International Meeting on Hepatitis C Virus and Related Viruses). In addition, the amino acid substitution C316Y was observed in patients who experienced viral load rebound while on monotherapy with HCV-796 (36). Transient GT 1b replicons engineered to contain HCV-796 resistance amino acid substitutions (C316Y, S365T, and M414I) displayed a 10- to 500-fold decrease in sensitivity to HCV-796 (Fig. 4B). The HCV-796 resistance mutations did not alter the potency of R1479 or PSI-6130 (Fig. 4B).
Finally, the mutations that confer reduced sensitivity to NM107, R1479, or PSI-6130 were examined. NS5B amino acid substitutions S96T and S96T-N142T were previously reported to confer reduced sensitivity to R1479 (19). Transient replicons containing those mutations had approximately a three- to fivefold-reduced sensitivity to R1479, while VX-950 and HCV-796 maintained their potency (Fig. 4C). Similarly, amino acid substitution S282T in NS5B resulted in approximately 3-fold reduced sensitivity to PSI-6130 (33) or 30-fold reduced sensitivity to NM107 (19), while VX-950 and HCV-796 maintained their potency (Fig. 4D). These results indicate that during combination treatment, the potency of the compounds tested is not affected by the emergence of mutations conferring resistance to the other classes of HCV inhibitors.
Effect of resistance mutations on replication capacity.
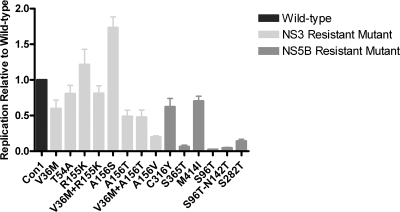
In addition to studying the impact of the NS3 and NS5B amino acid substitutions on compound potency, we also determined how the evaluated resistance mutations may affect the replicon replication capacity. The replication capacity for the wild-type GT 1b replicon was set at 1, and the relative replication capacities of the mutant replicons were compared against it. The replication capacity of the VX-950-resistant mutants ranged from 20% relative to wild type (A156V) to levels 70% higher than wild type (A156S) (Fig. 5). For the HCV-796-resistant mutants, the most common substitution observed in clinical trials was C316Y, which has an in vitro replication capacity of 70% compared to wild type and similar to that of M414I. However, the replication capacity of the S365T mutant replicon was only 10% compared to the wild type (Fig. 5). The mutations in NS5B that confer reduced sensitivity to R1479 or PSI-6130 have replication capacities of 4% for S96T, 5% for S96T-N142T, and 15% for S282T compared to the wild-type replicon (Fig. 5), similar to previously reported values (19, 28).
FIG. 5.
Replication capacity of replicons containing NS3 or NS5B mutations. Huh7-Lunet cells were transfected with either a wild-type GT 1b replicon or one engineered to contain mutations resulting in reduced sensitivity to VX-950, HCV-796, R1479, or PSI-6130. The replication capacity of the mutants was determined from at least four independent experiments (n = 8 for Con1 and S282T; n = 6 for T54A, R155K, A156S, and V36M+R155K; n = 5 for V36M+A156T, V201A, C316Y, S365T; and n = 4 for S156T, A156V, V36M, S96T, N142T, S96T+N142T, C316Y, and M414I) and is expressed as their normalized replication efficiency (mean ± standard error of the mean) compared to that of the wild-type, which was set at 1.
DISCUSSION
The current standard of care provides good clinical efficacy in patients infected with HCV GTs 2 and 3 but is less efficacious in patients infected with the most prevalent GT 1, thereby emphasizing the urgent need for more effective HCV-specific antiviral therapies (9, 27). This has triggered considerable drug discovery efforts that have resulted in the evaluation of a number of HCV inhibitors, both NS3/4A protease and NS5B polymerase inhibitors, in clinical trials. However, the selection of variants that are resistant to HCV inhibitors is likely to occur due to the high genetic heterogeneity of HCV, the error-prone nature of its RNA-dependent RNA polymerase, and the high virus production and turnover, resulting in every possible mutant likely present in a patient's quasispecies. After monotherapy treatment of HCV-infected patients with either the NS3/4A protease inhibitor VX-950 (31) or the nonnucleoside polymerase inhibitor HCV-796 (36, 37), HCV variants resistant to either inhibitor were selected. No viral resistance has been observed, however, after treatment with the nucleoside inhibitor R1626 (30). This suggests a potential difference in the selection of resistance to nucleoside inhibitors compared to nonnucleoside and protease inhibitors. In this study we investigated the ease with which selection of resistance to different classes of HCV inhibitors occurs, using the HCV replicon system.
The emergence of variants with reduced sensitivity to antiviral compounds depends on several factors, including the prevalence of the variant within the quasispecies, its replication capacity, the level of resistance that it confers to the virus, and the number of mutations needed to introduce the required coding change(s). The primary amino acid substitutions that confer reduced sensitivity to each compound tested in this study (NS3 A156T for VX-950, NS5B C316Y or S365T for HCV-796, NS5B S282T for NM107 and PSI-6130, and NS5B S96T or S96T/N142T for R1479) require only a single nucleotide change. As a consequence, the frequency of generation of each of these mutants during a single viral replication cycle should be equivalent, but the viability of the resulting variants may differ. However, the data presented here demonstrate that the mutations that confer reduced sensitivity to VX-950 and to HCV-796 were observed after 3 weeks of selection, whereas no resistance mutations were observed in the case of the nucleoside inhibitors. This may be explained by a number of factors, such as the magnitude of the decrease of drug sensitivity in relation to the replication capacity of the drug-resistant mutant replicon. The NS5B mutations that confer reduced sensitivity to R1479 or PSI-6130 result in only a three- to fivefold loss in sensitivity to the inhibitors but also result in a greater than 85% reduction in replication capacity. This may explain the lack of emergence of nucleoside inhibitor-resistant variants over the wild-type replicon. In order for a resistance mutation to emerge from a population upon the introduction of a selection pressure, the mutation must confer reduced sensitivity to the selection agent with minimal compromise of the viral replication capacity. One example shown in this study is the selection of the HCV-796 resistance NS5B amino acid substitution C316Y instead of S365T. As indicated in Fig. 4, the C316Y mutation confers approximately 100-fold-reduced sensitivity to HCV-796, while S365T confers >500-fold-reduced sensitivity. However, the S365T mutation has a 10-fold-lower replication capacity than C316Y. The S365T mutant may not have a replicative advantage over C316Y in the presence of HCV-796 and thus was not selected during the 3-week assay. The S365A amino acid substitution was identified during a selection experiment with HCV-796 in this study. However, the S365A substitution has been reported to replicate at an approximately 10-fold-higher level than S365T (Howe et al., presented at the 13th International Meeting on Hepatitis C Virus and Related Viruses), which may provide further evidence that the replication capacity of S365T was not sufficient to provide a replicative advantage. It is also worth noting that the replication capacity determined for a particular mutation may vary depending on the experimental assay conditions. For example, we measured the replication capacity for the VX-950 resistant mutant A156T using a transient GT 1b replicon over a 96-h period. We determined that the replication capacity of the A156T mutant was approximately twofold lower than that of the wild type, while other groups using different assay systems (i.e., different replicon/adaptive mutations or a stable replicon assay) have reported a lower replication capacity for the A156T mutant (21, 26, 34, 40). Another important factor that can affect how quickly resistant variants emerge in patients upon exposure to a selection pressure is the prevalence of preexisting resistance mutations within the quasispecies. Mutations in NS3 that confer reduced sensitivity to VX-950 have been detected within the quasispecies of an HCV-infected patient who had not been treated with VX-950 (3, 8). Similarly, mutations in NS5B that confer reduced sensitivity to a number of different nonnucleoside inhibitors, including the HCV-796 resistance mutation C316Y, have also been identified within the quasispecies of HCV-infected patients (25; S. Le Pogam et al., submitted for publication). In contrast, the NS5B amino acid substitutions S282T and S96T, which confer reduced sensitivity to the nucleoside inhibitors NM107/PSI-6130 and R1479, respectively, have not been identified within the quasispecies of HCV-infected patients. These studies, with the exception of that of Lu et al. (25), could only detect variants whose frequency was approximately 1% of the total population. Future studies that utilize more sensitive assays and a larger number of clinical samples will need to be performed in order to truly understand the prevalence of preexisting resistance mutations in the HCV genetic population.
In summary, we have examined the development of resistance to three classes of anti-HCV compounds. We found that resistance mutations were selected when replicon cells were treated for 3 weeks with either a protease inhibitor or a nonnucleoside inhibitor, but not with a nucleoside inhibitor. Our results highlight the potentially important role of nucleoside inhibitors in any future HCV therapies. R1479 and PSI-6130 remain potent against both VX-950- and HCV-796-resistant mutants. In addition, selection of replicon cells with R1479 or PSI-6130 in combination with a protease inhibitor or nonnucleoside inhibitor reduced the emergence of resistant colonies suggesting a clear benefit of combination treatment. The NS5B mutations that confer reduced sensitivity to R1479 or PSI-6130 result in a small loss in antiviral activity but also result in a large reduction in the replication capacity. The VX-950- and HCV-796-resistant mutants confer a larger loss in compound activity, as much as 500-fold, and generally have higher replication capacities than the nucleoside inhibitor mutants. These data indicate that the HCV replicon presents a higher genetic barrier to resistance for nucleoside inhibitors than for either protease inhibitors or nonnucleoside inhibitors.
Acknowledgments
We thank Volker Lohmann for kindly supplying Huh 7 cells and Phillip Furman at Pharmasset, Inc., for providing PSI-6130. We acknowledge Nixy Zutshi and her group for cell culture support and Wen-Rong Jiang for advice on drug-drug interaction studies.
Footnotes
Published ahead of print on 19 February 2008.
REFERENCES
- 1.Anonymous. 1997. Hepatitis C: global prevalence. Wkly. Epidemiol. Rec. 72:341-344. [PubMed] [Google Scholar]
- 2.Armstrong, G. L., A. Wasley, E. P. Simard, G. M. McQuillan, W. L. Kuhnert, and M. J. Alter. 2006. The prevalence of hepatitis C virus infection in the United States, 1999 through 2002. Ann. Intern. Med. 144:705-714. [DOI] [PubMed] [Google Scholar]
- 3.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, and D. L. Wheeler. 2007. GenBank. Nucleic Acids Res. 35:D21-D25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chandra, P., D. Raible, D. Harper, J. Speth, S. Villano, and G. Bichier. 2006. Antiviral activity of the non nucleoside polymerase inhibitor, HCV-796, in patients with chronic hepatitis C virus: preliminary results from a randomized, double-blind, placebo-controlled, ascending multiple dose study. Gastroenterology 130:A-748. [Google Scholar]
- 5.De Francesco, R., and G. Migliaccio. 2005. Challenges and successes in developing new therapies for hepatitis C. Nature 436:953-960. [DOI] [PubMed] [Google Scholar]
- 6.Dienstag, J. L., R. P. Perrillo, E. R. Schiff, M. Bartholomew, C. Vicary, and M. Rubin. 1995. A preliminary trial of lamivudine for chronic hepatitis B infection. N. Engl. J. Med. 333:1657-1661. [DOI] [PubMed] [Google Scholar]
- 7.Fischl, M. A., D. D. Richman, M. H. Grieco, M. S. Gottlieb, P. A. Volberding, O. L. Laskin, J. M. Leedom, J. E. Groopman, D. Mildvan, and R. T. Schooley. 1987. The efficacy of azidothymidine (AZT) in the treatment of patients with AIDS and AIDS-related complex. A double-blind, placebo-controlled trial. N. Engl. J. Med. 317:185-191. [DOI] [PubMed] [Google Scholar]
- 8.Franco, S., M. Parera, E. Aparicio, B. Clotet, and M. A. Martinez. 2007. Genetic and catalytic efficiency structure of an HCV protease quasispecies. Hepatology 45:899-910. [DOI] [PubMed] [Google Scholar]
- 9.Fried, M. W., M. L. Shiffman, K. R. Reddy, C. Smith, G. Marinos, F. L. Goncales, Jr., D. Haussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, A. Lin, J. Hoffman, and J. Yu. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975-982. [DOI] [PubMed] [Google Scholar]
- 10.Godofsky, E. W., N. Afdhal, V. Rustgi, L. Shick, L. Duncan, X. L. Zhou, G. Chao, C. Fang, B. Fielman, M. Myers, and N. A. Brown. 2004. First clinical results for a novel antiviral treatment for hepatitis C: a phase I/II dose escalation trial assessing tolerance, pharmacokinetics and antiviral activity on NM283, a novel antiviral treatment for hepatitis C. Gastroenterology 126:A681. [Google Scholar]
- 11.Greco, W. R., H. S. Park, and Y. M. Rustum. 1990. Application of a new approach for the quantitation of drug synergism to the combination of cis-diamminedichloroplatinum and 1-beta-d-arabinofuranosylcytosine. Cancer Res. 50:5318-5327. [PubMed] [Google Scholar]
- 12.Hammer, S. M., M. S. Saag, M. Schechter, J. S. Montaner, R. T. Schooley, D. M. Jacobsen, M. A. Thompson, C. C. Carpenter, M. A. Fischl, B. G. Gazzard, J. M. Gatell, M. S. Hirsch, D. A. Katzenstein, D. D. Richman, S. Vella, P. G. Yeni, and P. A. Volberding. 2006. Treatment for adult HIV infection: 2006 recommendations of the International AIDS Society—USA panel. Top. HIV Med. 14:827-843. [PubMed] [Google Scholar]
- 13.Ji, C., J. Zhang, M. Dioszegi, S. Chiu, E. Rao, A. Derosier, N. Cammack, M. Brandt, and S. Sankuratri. 2007. Ccr5 small molecule antagonists and monoclonal antibodies exert potent synergistic antiviral effects by co-binding to the receptor. Mol. Pharmacol. 72:18-28. [DOI] [PubMed] [Google Scholar]
- 14.Jones, B. R., D. J. Coster, P. N. Fison, G. M. Thompson, L. M. Cobo, and M. G. Falcon. 1979. Efficacy of acycloguanosine (Wellcome 248U) against herpes-simplex corneal ulcers. Lancet i:243-244. [DOI] [PubMed] [Google Scholar]
- 15.Kitchen, V. S., C. Skinner, K. Ariyoshi, E. A. Lane, I. B. Duncan, J. Burckhardt, H. U. Burger, K. Bragman, A. J. Pinching, and J. N. Weber. 1995. Safety and activity of saquinavir in HIV infection. Lancet 345:952-955. [DOI] [PubMed] [Google Scholar]
- 16.Klumpp, K., V. Leveque, S. Le Pogam, H. Ma, W. Jiang, H. Kang, C. Granycome, M. Singer, C. Laxton, J. Q. Hang, K. Sarma, D. B. Smith, D. Heindl, C. J. Hobbs, J. H. Merrett, J. Symons, N. Cammack, J. A. Martin, R. Devos, and I. Najera. 2006. The novel nucleoside analog R1479 (4′-azidocytidine) is a potent inhibitor of NS5B-dependent RNA synthesis and hepatitis C virus replication in cell culture. J. Biol. Chem. 281:3793-3799. [DOI] [PubMed] [Google Scholar]
- 17.Koutsoudakis, G., E. Herrmann, S. Kallis, R. Bartenschlager, and T. Pietschmann. 2007. The level of CD81 cell surface expression is a key determinant for productive entry of hepatitis C virus into host cells. J. Virol. 81:588-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Pogam, S., W. Jiang, V. Leveque, S. Rajyaguru, H. Ma, H. Kang, S. Jiang, M. Singer, S. Ali, K. Klumpp, D. B. Smith, J. Symons, N. Cammack, and I. Najera. 2006. In vitro selected Con1 subgenomic replicons resistant to 2′-C-methyl-cytidine or to R1479 show lack of cross resistance. Virology 351:349-359. [DOI] [PubMed] [Google Scholar]
- 20.Le Pogam, S., H. Kang, S. F. Harris, V. Leveque, A. M. Giannetti, S. Ali, W. R. Jiang, S. Rajyaguru, G. Tavares, C. Oshiro, T. Hendricks, K. Klumpp, J. Symons, M. F. Browner, N. Cammack, and I. Najera. 2006. Selection and characterization of replicon variants dually resistant to thumb- and palm-binding nonnucleoside polymerase inhibitors of the hepatitis C virus. J. Virol. 80:6146-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin, C., C. A. Gates, B. G. Rao, D. L. Brennan, J. R. Fulghum, Y. P. Luong, J. D. Frantz, K. Lin, S. Ma, Y. Y. Wei, R. B. Perni, and A. D. Kwong. 2005. In vitro studies of cross-resistance mutations against two hepatitis C virus serine protease inhibitors, VX-950 and BILN 2061. J. Biol. Chem. 280:36784-36791. [DOI] [PubMed] [Google Scholar]
- 22.Lin, C., K. Lin, Y. P. Luong, B. G. Rao, Y. Y. Wei, D. L. Brennan, J. R. Fulghum, H. M. Hsiao, S. Ma, J. P. Maxwell, K. M. Cottrell, R. B. Perni, C. A. Gates, and A. D. Kwong. 2004. In vitro resistance studies of hepatitis C virus serine protease inhibitors, VX-950 and BILN 2061: structural analysis indicates different resistance mechanisms. J. Biol. Chem. 279:17508-17514. [DOI] [PubMed] [Google Scholar]
- 23.Lin, K., R. B. Perni, A. D. Kwong, and C. Lin. 2006. VX-950, a novel hepatitis C virus (HCV) NS3-4A protease inhibitor, exhibits potent antiviral activities in HCV replicon cells. Antimicrob. Agents Chemother. 50:1813-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lohmann, V., S. Hoffmann, U. Herian, F. Penin, and R. Bartenschlager. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu, L., H. Mo, T. J. Pilot-Matias, and A. Molla. 2007. Evolution of resistant M414T mutants among hepatitis C virus replicon cells treated with polymerase inhibitor A-782759. Antimicrob. Agents Chemother. 51:1889-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu, L., T. J. Pilot-Matias, K. D. Stewart, J. T. Randolph, R. Pithawalla, W. He, P. P. Huang, L. L. Klein, H. Mo, and A. Molla. 2004. Mutations conferring resistance to a potent hepatitis C virus serine protease inhibitor in vitro. Antimicrob Agents Chemother. 48:2260-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manns, M. P., J. G. McHutchison, S. C. Gordon, V. K. Rustgi, M. Shiffman, R. Reindollar, Z. D. Goodman, K. Koury, M. Ling, and J. K. Albrecht. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958-965. [DOI] [PubMed] [Google Scholar]
- 28.Migliaccio, G., J. E. Tomassini, S. S. Carroll, L. Tomei, S. Altamura, B. Bhat, L. Bartholomew, M. R. Bosserman, A. Ceccacci, L. F. Colwell, R. Cortese, R. De Francesco, A. B. Eldrup, K. L. Getty, X. S. Hou, R. L. LaFemina, S. W. Ludmerer, M. MacCoss, D. R. McMasters, M. W. Stahlhut, D. B. Olsen, D. J. Hazuda, and O. A. Flores. 2003. Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. J. Biol. Chem. 278:49164-49170. [DOI] [PubMed] [Google Scholar]
- 29.Reesink, H. W., S. Zeuzem, C. J. Weegink, N. Forestier, A. van Vliet, J. van de Wetering de Rooij, L. McNair, S. Purdy, R. Kauffman, J. Alam, and P. L. Jansen. 2006. Rapid decline of viral RNA in hepatitis C patients treated with VX-950: a phase Ib, placebo-controlled, randomized study. Gastroenterology 131:997-1002. [DOI] [PubMed] [Google Scholar]
- 30.Roberts, S., G. Cooksley, G. Dore, R. Robson, D. Shaw, H. Berns, M. Brandl, S. Fettner, G. Hill, D. Ipe, K. Klumpp, M. Mannino, I. Najera, E. O'Mara, Y. Tu, and C. Washington. 2006. Results of a phase 1B, multiple dose study of R1626, a novel nucleoside analog targeting HCV polymerase in chronic HCV genotype 1 patients. Hepatology 44:692A. [Google Scholar]
- 31.Sarrazin, C., T. L. Kieffer, D. Bartels, B. Hanzelhalka, U. Muh, M. Welker, D. Wincheringer, Y. Zhou, H.-M. Chu, C. Lin, C. Weegink, H. Reesink, S. Zeuzem, and A. D. Kwong. 2007. Dynamic hepatitis C virus genotypic and phenotypic changes in patients treated with the protease inhibitor telaprevir. Gastroenterology 132:1767-1777. [DOI] [PubMed] [Google Scholar]
- 32.Sarrazin, C., R. Rouzier, F. Wagner, N. Forestier, D. Larrey, S. K. Gupta, M. Hussain, A. Shah, D. Cutler, J. Zhang, and S. Zeuzem. 2007. SCH 503034, a novel hepatitis C virus protease inhibitor, plus pegylated interferon alpha-2b for genotype 1 nonresponders. Gastroenterology 132:1270-1278. [DOI] [PubMed] [Google Scholar]
- 33.Stuyver, L. J., T. R. McBrayer, P. M. Tharnish, J. Clark, L. Hollecker, S. Lostia, T. Nachman, J. Grier, M. A. Bennett, M. Y. Xie, R. F. Schinazi, J. D. Morrey, J. L. Julander, P. A. Furman, and M. J. Otto. 2006. Inhibition of hepatitis C replicon RNA synthesis by beta-d-2′-deoxy-2′-fluoro-2′-C-methylcytidine: a specific inhibitor of hepatitis C virus replication. Antiviral Chem. Chemother. 17:79-87. [DOI] [PubMed] [Google Scholar]
- 34.Tong, X., R. Chase, A. Skelton, T. Chen, J. Wright-Minogue, and B. A. Malcolm. 2006. Identification and analysis of fitness of resistance mutations against the HCV protease inhibitor SCH 503034. Antiviral Res. 70:28-38. [DOI] [PubMed] [Google Scholar]
- 35.Trozzi, C., L. Bartholomew, A. Ceccacci, G. Biasiol, L. Pacini, S. Altamura, F. Narjes, E. Muraglia, G. Paonessa, U. Koch, R. De Francesco, C. Steinkuhler, and G. Migliaccio. 2003. In vitro selection and characterization of hepatitis C virus serine protease variants resistant to an active-site peptide inhibitor. J. Virol. 77:3669-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Villano, S., A. Howe, D. Raible, D. Harper, J. Speth, and G. Bichier. 2006. Analysis of HCV NS5B genetic variants following monotherapy with HCV-796, a non-nucleoside polymerase inhibitor, in treatment-naive HCV-infected patients. Hepatology 44:607A. [Google Scholar]
- 37.Villano, S., D. Raible, D. Harper, J. Speth, P. Chandra, P. Shaw, and G. Bichier. 2007. Antiviral activity of the non-nucleoside polymerase inhibitor, HCV-796, in combination with pegylated interferon alfa-2b in treatment-naive patients with chronic hepatitis C virus. J. Hepatol. 46:S24. [Google Scholar]
- 38.Wasley, A., J. T. Miller, and L. Finelli. 2007. Surveillance for acute viral hepatitis—United States, 2005. MMWR Surveill Summ. 56:1-24. [PubMed] [Google Scholar]
- 39.Wiesner, R. H., M. Sorrell, and F. Villamil. 2003. Report of the first International Liver Transplantation Society expert panel consensus conference on liver transplantation and hepatitis C. Liver Transpl. 9:S1-S9. [DOI] [PubMed] [Google Scholar]
- 40.Yi, M., X. Tong, A. Skelton, R. Chase, T. Chen, A. Prongay, S. L. Bogen, A. K. Saksena, F. G. Njoroge, R. L. Veselenak, R. B. Pyles, N. Bourne, B. A. Malcolm, and S. M. Lemon. 2006. Mutations conferring resistance to SCH6, a novel hepatitis C virus NS3/4A protease inhibitor. Reduced RNA replication fitness and partial rescue by second-site mutations. J. Biol. Chem. 281:8205-8215. [DOI] [PubMed] [Google Scholar]