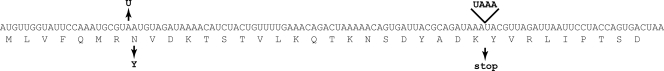Abstract
The methyltransferase genes erm(B) and cfr are adjacent to each other in the chromosome of methicillin-resistant Staphylococcus aureus strain CM05. Analyses of the transcriptional organization of the erm(B) and cfr genes in the chromosome of strain CM05 showed that the two genes are organized into an operon, designated mlr (for modification of the large ribosomal subunit), which is controlled by the erm(B) promoter. Analysis of the translation control and the inducibility of the erm(B) and cfr genes in the mlr operon showed that despite the presence of putative regulatory short open reading frames, both genes are expressed constitutively. The combined action of the two methyltransferases encoded in the mlr operon results in modification of two specific residues in 23S rRNA, A2058 and A2503, and renders cells resistant to all clinically useful antibiotics that target the large ribosomal subunit. Furthermore, simultaneous modification of both rRNA sites synergistically enhances resistance to 16-member-ring macrolides.
Antibiotic resistance genes are widespread in nature. Resistance mechanisms that originate in antibiotic-producing microorganisms often provide protection through modification of the drug target site or by active efflux of the drug from the cell (5, 21). The resistance genes can be hijacked by pathogenic bacteria and used as a defense against clinically useful antibiotics. Multiple resistance genes can be organized into clusters, allowing the spread of “resistance units,” such as those found on integrons, in which several resistance genes are controlled by a common promoter (1). Understanding of the origin, evolution, and mode of expression of resistance genes is critical for preventing the spread of resistance as well as for the development of new antibiotics.
Approximately half of all the known natural antibiotics, including many clinically useful drugs, act upon the ribosome, the central component of the cellular protein synthesis machinery and one of the key enzymes in the gene expression pathway (34). The binding sites of most of the ribosome-targeting antibiotics are composed exclusively or primarily of rRNA. It is therefore not surprising that many resistance mechanisms operate upon antibiotic bindings sites in rRNA. One of the most powerful mechanisms of this sort is the posttranscriptional modification of rRNA by methyltransferase enzymes. Methylation of distinct rRNA residues can efficiently prevent the binding of protein synthesis inhibitors. As different classes of antibiotics often bind to overlapping sites in the ribosome, the modification of one site can potentially render an organism resistant to a variety of drugs. In the large ribosomal subunit, all clinically relevant antibiotics act at or near the ribosome peptidyltransferase center. Therefore, rRNA methyltransferases which act at this site provide resistance to an exceptionally broad variety of drugs. For instance, the methylation of adenosine 2058 in the 23S rRNA of the large ribosomal subunit (the Escherichia coli numbering is used here and throughout) by Erm-type methyltransferases renders bacteria resistant to macrolides, lincosamides, and streptogramins B (27, 39). The recently described Cfr methyltransferase, which modifies A2503 in 23S rRNA, confers resistance to an even broader range of drugs, including phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramins A (15, 17, 20).
The erm genes account for one of the most common macrolide resistance mechanisms. They are found both on the chromosomes and on the plasmids of a wide range of gram-positive and gram-negative bacterial pathogens. The broad distribution of erm genes can be attributed in part to their frequent association with transposons that have a wide host range (3, 8). In addition, erm genes are often associated with other resistance genes (27).
The gene encoding the Cfr methyltransferase was originally found on plasmid pSCFS1 in a bovine isolate of Staphylococcus sciuri and was later found on plasmids pSCFS3 and pSCFS6 in staphylococcal isolates from other animal sources (15, 16, 31). Recently, we reported on the first case of the occurrence of cfr in a clinical strain (strain CM05) of methicillin-resistant Staphylococcus aureus (MRSA) isolated from a human patient (35). Despite the presence of a 35-kb plasmid in CM05, the cfr gene is located on the chromosome, where it is positioned immediately downstream of the erm(B) methyltransferase gene. The close association of erm(B) and cfr in the CM05 chromosome raised the possibility that the expression of these two genes is coordinated. The presence of short open reading frames (ORFs) upstream of the erm(B) and cfr cistrons suggested that a translation attenuation mechanism might control the expression of these genes. In this work we tested these hypotheses by exploring the transcriptional and the translational control of the erm(B)-cfr cluster from this clinical MRSA strain.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Clinical MRSA strain CM05 was isolated in 2005 in Colombia (35). S. aureus laboratory strain RN4220 was used as a plasmid host (24). E. coli XL-10 Gold cells (Stratagene) or E. coli TOP10 cells (Invitrogen) were used in the mutagenesis experiments and as alternative hosts for the shuttle plasmids.
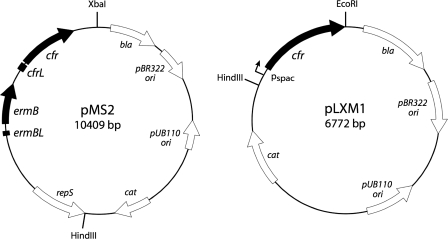
Plasmid pMS2 was constructed by excising a 4.9-kb HindIII-XbaI segment of the strain CM05 chromosomal DNA sequence containing the mlr operon from plasmid pMS1 (35) and cloning it into the corresponding sites of E. coli-S. aureus shuttle vector pL150 (18). Deletions were introduced into plasmid pMS2 by PCR amplification of the desired parts of the pMS2 sequence. The primers used in this study are listed in Table 1. For each deletion yielding plasmid pErmBΔ1, pErmBΔ2, or pErmBΔ3, one of the forward primers (primers permBdel1, pdel2, and pdel3, respectively) contained a HindIII site at the 3′ end and was used in conjunction with a reverse primer (primer RevpMS2) positioned immediately downstream of the original HindIII site. The amplification was carried out with highly accurate Accutaq LA DNA polymerase (Sigma) by using the following cycling conditions: 98°C for 30 s and 30 cycles of 94°C for 10 s, 58°C for 20 s, and 68°C for 12 min. Following PCR amplification, template plasmid pMS2 was removed by DpnI digestion and the PCR products were purified with a Wizard SV PCR purification kit (Promega). The PCR product was digested with HindIII, purified again with the same kit, ligated at 16°C overnight, and transformed into E. coli TOP10 competent cells (Invitrogen). The presence of the desired deletions in the resulting pErmBΔ plasmids and the lack of mutations in the mlr operon were verified by sequencing. Construction of plasmid pLXM1, which contains the cfr gene under the control of the Pspac promoter in the pLI50 vector, was described previously (35).
TABLE 1.
Primers used in this study
| Primer | Sequence |
|---|---|
| atgx1 | AAATTGATTCTTAACTAGAGCAAATTGTGAAAGGCAGAAAGAAATGAATTTTAATAATAAAACAAAGTATG |
| atgx2 | CTTAACTAGAGCAAATTGTGAAAGGATGAAAGAACAGAATTTTAATAATAAAACAAAGTATGGTAAAATAC |
| c1 | TTCCTGTATTTTACCATACTTTG |
| c2 | ACATGATATAACTTCCCTG |
| c3 | CTATAATCAGGCTCATTATTACTT |
| e1 | GTCTGTTTCAAAACAGTAGATG |
| e2 | GATTGTTGAAGAAGGATTCTAC |
| e3 | GTAATTAAGAAGGAGGGATTCG |
| e4 | GTTATCTATTATTTAACGGGAGG |
| pdel2 | GCGCAAGCTTAGGTATAGGGCACCTCTAATA |
| pdel3 | GCGCAAGCTTGTATGTTTTGACTTTCGGCAC |
| permBdel1 | GGGCAAGCTTCGTCATGTTGGTATTCCAAAT |
| RevpMS2 | GGCTAATAGGGAATACATTACCA |
| SAL2060 | GTAAAGCTCCACGGGGTC |
| SAL2507 | CCAGGATGCGATGAGCCG |
| tRNADir | ATCCGGCCCCCGCAACC |
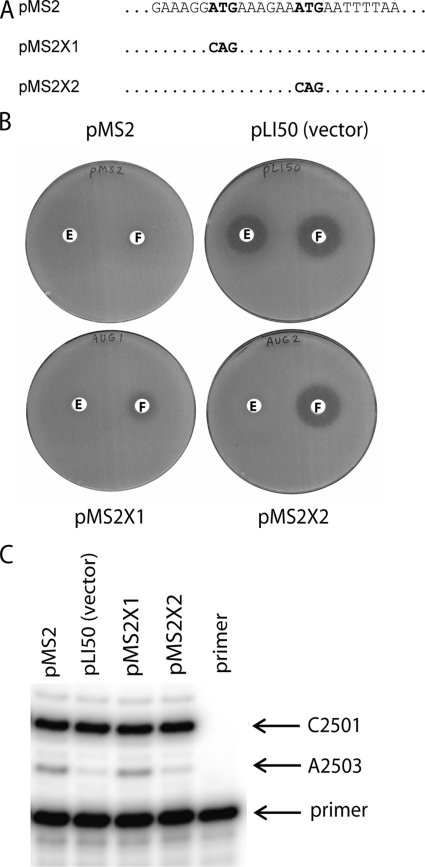
Site-directed mutagenesis of the first and second putative initiation AUG codons in the cfr gene (which yielded plasmids pMS2X1 and pMS2X2, respectively) was performed with a QuikChange multikit (Stratagene). Primer atgx1 was used to introduce the A-to-C mutation in the upstream AUG codon, and primer atgx2 was used for introduction of the A-to-C mutation in the second AUG codon. Mutagenized plasmids prepared according to the manufacturer's protocol were transformed into E. coli XL-10 Gold cells (Stratagene). Transformants were selected on LB agar plates supplemented with 100 μg/ml ampicillin.
All shuttle plasmids were isolated from E. coli XL-10 or TOP10 cells by using a GenElute plasmid mini kit (Sigma) and were introduced into S. aureus RN4220 cells by electroporation (29). S. aureus transformants were selected on LB agar plates supplemented with 10 μg/ml chloramphenicol.
Microbiological testing.
For disk diffusion testing, ca. 107 S. aureus RN4220 cells transformed with the appropriate plasmid were plated in 4 ml of soft (0.6%) agar on 1.5% LB agar plates supplemented with 10 μg/ml chloramphenicol. Whatman 3MM paper disks (diameter, 5 mm), containing 20 μg of erythromycin or 30 μg of florfenicol, were placed on top of the soft agar layer, and the plates were incubated overnight at 37°C and photographed. The MICs of the antibiotics were determined by the broth microdilution method by following the accepted recommendations (23).
Isolation of total RNA from S. aureus.
Overnight cultures of S. aureus RN4220 or CM05 cells were diluted 1:100 and grown to an optical density at 600 nm of 0.5 in LB broth. The culture medium was supplemented with 10 μg/ml chloramphenicol for RN4220 cells containing pLI50 and its derivatives. Cells from a 5-ml culture were pelleted, washed with 500 μl H2O, and resuspended in 200 μl of buffer (10 mM Tris-HCl pH 7.5, 30 mM MgCl2, 30 mM NH4Cl) containing 0.5 mg/ml of lysostaphin (Sigma). Lysis was carried out for 15 to 30 min at 37°C with shaking at 1,000 rpm on an Eppendorf thermomixer. Total RNA was subsequently isolated by using an RNeasy mini purification kit (Qiagen), according to the manufacturer's protocol.
Primer extension analysis.
To determine the transcription start site, 0.5 pmol of one of the primers (primer e1 or c1) labeled with 32P at the 5′ end was annealed to 10 μg of total S. aureus RNA and extended with avian myeloblastosis virus reverse transcriptase (RT; Seikagaku America) by using the protocol described previously (36). The 1,443-bp and 495-bp DNA markers for electrophoresis were prepared by PCR amplification of the pMS2 DNA template by using 32P-labeled primer c1 in conjunction with primer e3 and primer e4, respectively.
To determine the extent of rRNA modification, the general protocol of Sigmund et al. (33) was used, with some modifications described elsewhere (2). Primers SAL2060 and SAL2507 were used to assess the extent of modification of A2058 and A2503, respectively. The fraction of the extension product terminated at the modified nucleotide was calculated by subtracting the background and then dividing the intensity of the “stop” band by the sum of the stop band and the “top” band. Induction experiments with S. aureus RN4220 and CM05 pretreated with erythromycin and florfenicol were repeated at least twice.
RT-PCR.
Twenty picomoles of primer c2 was annealed to 5 μg of DNase-treated total RNA isolated from S. aureus RN4220 transformed with pLI50 or pMS2 and extended with Transcriptor RT (Roche), according to the manufacturer's protocol. The resulting cDNA was diluted 10-fold, and 2 μl was used as a template for PCR amplification in a 25-μl volume with primers e2 and c3. As a “no-RT” control, an equal amount of RNA was used as the template in a parallel PCR. Thermocycling was performed under the following conditions: 94°C for 1 min, followed by 30 cycles of 94°C for 30 s, 56°C for 30 s, and 68°C for 70 s. The PCR products were visualized on a 1% agarose gel.
RACE analysis.
The general protocol described previously (11) was used for rapid amplification of cDNA ends (RACE) analysis. Specifically, 12 μg DNase-treated total RNA isolated from S. aureus cells transformed with pMS2 was resuspended in 87.5 μl water, to which 10 μl 10× tobacco acid pyrophosphatase digestion buffer (Epicenter) and 0.5 μl RNase inhibitor (Roche) was added. The mixture was split into two tubes, to one of which 1 μl (10 U) of tobacco acid pyrophosphatase was added (Epicenter). Both samples were incubated at 37°C for 1 h. A total of 500 pmol (13.5 μg) of E. coli fMet-tRNA, which served as an RNA adapter, in 100 μl of H2O was then added to each tube. The mixtures were purified by phenol and phenol-chloroform extraction, followed by ethanol precipitation. The RNA pellets were dissolved in 14 μl H2O, heated to 90°C for 5 min, and placed on ice. To each tube, 2 μl 10× RNA ligation buffer, 2 μl dimethyl sulfoxide, 1.8 μl (36 U) T4 RNA ligase (New England Biolabs), and 0.2 μl (8 U) RNase inhibitor (Roche) were added. Ligation of the RNA was performed overnight at 15°C. Water (130 μl) was added to each ligation mixture, and the products were purified by phenol and phenol-chloroform extraction, followed by ethanol precipitation. The pellets were resuspended in 25 μl water and split into three tubes: 10 μl as a template for reverse transcription with the erm(B)-specific primer (primer e1), 10 μl for the cfr-specific primer (primer c1), and 5 μl for the no-RT control. Reverse transcription was performed with Transcriptor RT (Roche), according to the manufacturer's protocol. Two microliters of the reverse transcription reaction was used as a template for PCR with primers tRNADir and e1. The PCR products were purified with a Wizard SV DNA purification kit (Promega) and sequenced from primer e1.
RESULTS
Transcription of cfr originates exclusively from the erm(B) promoter in MRSA CM05.
In the chromosome of clinical MRSA isolate CM05, the cfr gene, which encodes the A2503 methyltransferase, is located very close (426 bp downstream) to the erm(B) gene, which encodes the A2058 methyltransferase (Fig. 1B). The close proximity of the two resistance genes raised the possibility that they can be coexpressed.
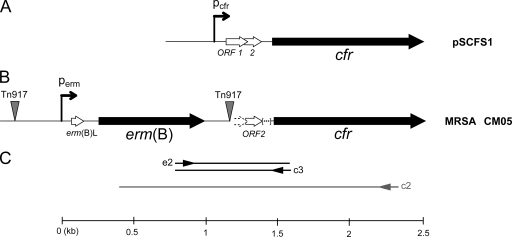
FIG. 1.
Organization of the monocistronic operon containing cfr from plasmid pSCFS1 (A) and organization of the mlr operon containing the erm(B) and the cfr genes in the chromosome of MRSA isolate CM05 (B). In pSCFS1, cfr is preceded by two overlapping ORFs (shown as ORF 1 and ORF 2) and a putative promoter. In CM05, the insertion of Tn917, which carries erm(B) and its regulatory elements, disrupts ORF 1 and the putative cfr promoter. An additional 35-nucleotide sequence present in pSCFS1 immediately after ORF 2 (indicated by brackets) is absent from CM05. (C) The scheme of the RT-PCR experiment for the detection of a transcript spanning the erm(B)-cfr intergenic spacer is shown. cDNA was synthesized from primer c2, which is complementary to the 5′ proximal region of the cfr gene, and was subsequently used as a template for PCR amplification with primers e2 and c3.
A strong promoter, Perm, which was previously found to control the transcription of erm(B) in transposon Tn917 (12), is present upstream from the erm(B) gene in the MRSA CM05 chromosome. Transcription from this promoter should initiate at the G residue located 258 bp upstream from the start codon of the erm(B) gene. The cfr gene was originally found in plasmids pSCFS1 and pSCFS3 residing in staphylococcal strains isolated from cattle (14, 15, 31). In these plasmids, a promoter-like sequence 366 bp upstream from the start of the cfr gene was proposed to control its transcription (31). However, the homology between the cfr 5′ proximal regions in plasmids pSCFS1 and pSCFS3 and in the chromosome of clinical isolate CM05 extends only 237 nucleotides upstream from the putative cfr start codon and, thus, does not include the cfr promoter sequence found in the plasmids. Analysis of the nucleotide sequence of the erm(B)-cfr intergenic spacer with the BPROM algorithm of the Softberry genome analysis suite (http://softberry.com/berry.phtml) did not reveal any strong promoter sequence. However, a sequence that exhibited weak promoter properties was identified and had a putative transcription start located 199 bp upstream from the start codon of the cfr gene, the −35′ box TTAAAG and the −10′ box TTTTAACT. It was unclear, however, whether S. aureus RNA polymerase does indeed recognize this sequence as a promoter and, thus, whether this DNA element contributes to the expression of cfr in the clinical strain. Therefore, we used a combination of several experimental approaches to locate the promoters that control the expression of the erm(B) and the cfr genes.
In order to facilitate subsequent experiments, the segment of the CM05 chromosome that carries the erm(B)-cfr cassette was cloned in a multicopy shuttle vector, pLI50 (18). The 5,788-bp EcoRI-HindIII segment of the CM05 chromosome, which contains the erm(B)-cfr region, was initially cloned in the pBR322 vector to produce plasmid pMS1 (35). Subsequently, the 4,933-bp HindIII-XbaI segment from pMS1 that included 2,303 bp of the sequence upstream from the erm(B) start codon, the erm(B)-cfr segment, and 418 bp of the sequence downstream from the cfr stop codon was cloned into the pLI50 shuttle vector. The resulting plasmid, pMS2 (Fig. 2), was introduced into laboratory S. aureus strain RN4220 (24). Transformation with pMS2 rendered S. aureus resistant to a broad range of antibiotics that target the large ribosomal subunit (Table 2). Judging from the known specificities of the Erm(B) and Cfr enzymes, resistance to lincosamides and streptogramins could be a result of the individual action or the combined actions of two methyltransferases; resistance to macrolides (known to be conferred exclusively by A2058 methyltransferase enzymes) was a signature of erm(B) expression, whereas resistance to phenicols, tiamulin, and linezolid (which depends on the hypermethylation of A2053) indicated faithful expression of the Cfr methyltransferase.
FIG. 2.
Maps of plasmids pMS2 and pLXM1 used in this study.
TABLE 2.
Antibiotic sensitivity profiles of clinical MRSA isolate CM05 and S. aureus laboratory strain RN4220 transformed with mlr operon-expressing plasmid pMS2 or empty vector pLI50
| Antibiotic | MIC (μg/ml)
|
||
|---|---|---|---|
| RN4220(pLI50) | RN4220(pMS2) | CM05 | |
| Chloramphenicol | NDa | ND | 64 |
| Clindamycin | <1.0 | >1,024 | >1,024 |
| Erythromycin | 0.5 | >1,024 | >1,024 |
| Florfenicol | 4 | 128 | ND |
| Linezolid | 2 | 4 | 8 |
| Quinupristin-dalfopristin | <0.05 | 0.5 | 2 |
| Tiamulin | 1 | 128 | 512 |
| Tylosin | 2 | >1,024 | ND |
ND, not determined.
Having established that both erm(B) and cfr were expressed in the transformed S. aureus RN4220 cells, we then used primer extension analysis to locate a promoter(s) that controls the transcription of the erm(B) and the cfr genes. Total RNA was isolated from RN4220 cells transformed with pMS2. Oligonucleotide primers complementary to positions 84 to 106 downstream from the erm(B) transcription start site or positions 24 to 47 of the cfr ORF were annealed to RNA and extended with RT (Fig. 3). Extension of the primer complementary to the 5′ proximal transcript of the erm(B) gene revealed the presence of mRNA initiated at the Perm promoter (Fig. 3, lane e1). However, no specific RT stops corresponding to a transcript initiated in the erm(B)-cfr intergenic spacer, including the computer-predicted promoter, were observed with the cfr-specific primer (Fig. 3, lane c1). The lack of a transcription start site within the erm(B)-cfr spacer indicated that cfr may be cotranscribed together with the erm(B) gene from the Perm promoter.
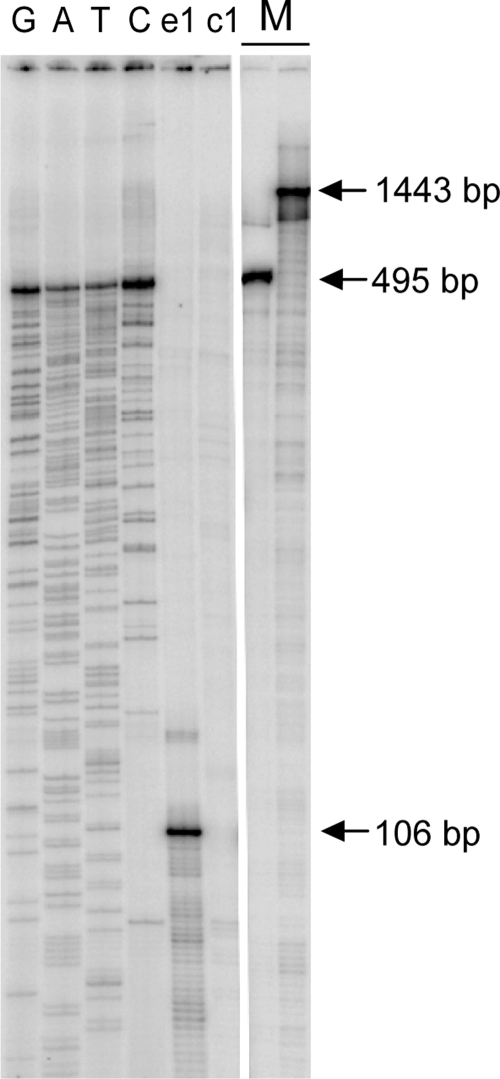
FIG. 3.
Primer extension analysis of the erm(B) and cfr transcription start sites. Total RNA isolated from S. aureus RN4220 cells transformed with plasmid pMS2 was used as the template. Primers e1 and c1 were used in the experiment and were complementary to the 5′ proximal region of the erm(B) ORF (lane e1) or the 5′ proximal region of cfr ORF (lane c1). Extension products carried on pMS2 DNA were separated on a 6% denaturing polyacrylamide gel along with sequencing reactions with primer e1 (lanes G, A, T, and C). The band in lane e1 that indicates the transcription start site from the Perm promoter corresponds to a 106-nucleotide-long cDNA product. 32P-labeled 1,443-bp and 495-bp DNA markers (lane M) indicate the approximate sizes of the putative cDNA products extending from primer c1 and terminating at the erm(B) transcription start site or a putative site within the erm(B)-cfr spacer, respectively.
If this scenario is correct, then transcripts initiated at Perm should span the erm(B)-cfr intergenic spacer. To detect the presence of such transcripts in the cell, we employed RT-PCR analysis using primers whose sequences are located within the coding sequences of the erm(B) and cfr genes (Fig. 1). A DNA product could be amplified by PCR from the cDNA that originated at primer c2, whose sequence is located within the cfr sequence, indicating the presence of mRNA that includes sequences of both the erm(B) and the cfr genes (data not shown).
In order to verify that transcription of cfr mRNA is indeed initiated at the Perm promoter, we used the RACE technique (7). Total RNA prepared from RN4220(pMS2) cells was treated with pyrophosphatase and ligated with an excess of tRNAfMet. cDNA was synthesized with a reverse primer c1 whose sequence is complementary to the cfr ORF and was then amplified by PCR with a forward tRNAfMet-specific primer (data not shown). Sequencing of the tRNA-transcript junction in the resulting RT-PCR product revealed that cfr transcription initiates at the Perm promoter.
The fact that in RN4220(pMS2) cells (and, thus, in clinical MRSA isolate CM05) cfr is under the transcriptional control of the erm(B) promoter does not completely rule out the possibility that, in addition to Perm, cfr may utilize another promoter located downstream from Perm. To determine whether cfr-dependent drug resistance depends exclusively on the Perm promoter, we introduced a series of deletions in plasmid pMS2 and analyzed the antibiotic resistance of S. aureus RN4220 cells transformed with the truncated plasmids (Fig. 4). Since cfr does not affect susceptibility to 14-member-ring macrolides (20), resistance to erythromycin was used as a measure of erm(B) expression. Conversely, cfr, but not erm(B), renders cells resistant to phenicols (15); therefore, the florfenicol resistance of the transformants was used to assess the expression of the cfr gene. The MICs in liquid cultures and the zones of inhibition around antibiotic disks on agar plates were determined for S. aureus RN4220 transformed with pMS2 [erm(B) positive, cfr positive], pLI50 [empty vector; erm(B) negative, cfr negative], or pErmBΔ1 [pMS2 in which the erm(B) promoter is deleted]. The identical antibiotic susceptibility profiles of cells transformed with pErmBΔ1 or the empty vector demonstrated that deletion of the erm(B) promoter abolished resistance to both erythromycin and florfenicol (Table 3 and Fig. 4). Deletions of more extended segments of pMS2, including the erm(B)-cfr spacer region (pErmBΔ2 and pErmBΔ3 in Fig. 4A), did not produce any additional changes in antibiotic sensitivity of the transformants (data not shown). To make sure that antibiotic resistance in S. aureus cells carrying pMS2 or its derivatives is determined by expression of the Erm(B) and the Cfr methyltransferases, primer extension analysis was used to assess the modification of A2058 [by Erm(B)] and A2503 (by Cfr) in 23S rRNA (17, 33, 38). Specific stops corresponding to the modification of A2058 and A2503 were observed in cells carrying plasmid pMS2 (Fig. 4C). However, in rRNA isolated from cells containing pErmBΔ1, the nucleotides at both position A2058 and position A2503 remained unmodified.
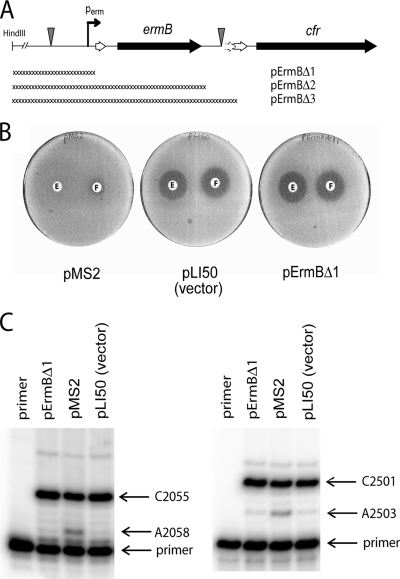
FIG. 4.
Effect of Perm promoter deletion on expression of erm(B) and cfr genes. (A) Deletions engineered in plasmid pMS2, yielding plasmids pErmBΔ1, pErmBΔ2, and pErmBΔ3. (B) Disk diffusion assay. The plates carried lawns of S. aureus RN4220 cells transformed with plasmid pMS2, pLI50, or pErmBΔ1. Antibiotic disks contained 20 μg erythromycin (disks E) or 30 μg florfenicol (disks F). (C) Primer extension analysis of modification of A2058 and A2503 in 23S rRNA in cells transformed with plasmid pMS2, pLI50, or pErmBΔ1.
TABLE 3.
Sensitivity of S. aureus strain RN4220 transformed with various plasmids to erythromycin and florfenicol
| Strain | MIC (μg/ml)
|
|
|---|---|---|
| Erythromycin | Florfenicol | |
| RN4220(pMS2) | >1,024 | 128 |
| RN4220(pLI50)a | 0.5 | 4 |
| RN4220(pErmBΔ1)b | 0.5 | 4 |
| RN4220(pMS2X1)c | >1,024 | 128 |
| RN4220(pMS2X2)d | >1,024 | 4 |
Empty vector.
pMS2 with deletion of erm(B) promoter.
pMS2 with first AUG of cfr mutagenized to CAG.
pMS2 with second AUG of cfr mutagenized to CAG.
The conclusion that we drew from the microbiological and biochemical experiments is that Perm is the major (likely a sole) promoter that controls the expression of both the erm(B) and the cfr genes in the chromosome of clinical MRSA isolate CM05. The implication is that erm(B) and cfr are organized in an operon. We named this operon mlr, for modification of the large ribosomal subunit.
Identification of the cfr translation initiation codon.
In the genome of clinical strain CM05 and in plasmids pSCFS1 and pSCFS3, two closely spaced in-frame ATG codons are located near the 5′ end of the cfr ORF (Fig. 5A). The location of a putative Shine-Dalgarno sequence (AGGA) appears to favor the second (downstream) AUG as the translation initiation start site for cfr (31). However, since the presence of a Shine-Dalgarno sequence is not a universal requirement for translation initiation in bacteria (22), the first (upstream) AUG codon could potentially be used for the initiation of translation. Given the possibility that the expression of cfr may be under translation attenuation control (31), it was important to determine which of the two AUG codons is used as the cfr translation start codon. To this end, in plasmid pMS2 each of the two putative initiation AUG codons of cfr was individually mutated to CAG (Fig. 5A), and the resulting plasmids (pMS2X1 and pMS2X2, respectively) were introduced into S. aureus RN4220. The florfenicol sensitivities of the resulting strains, as assessed by the broth microdilution and antibiotic disk diffusion techniques, were used as a measure of cfr expression (Table 3 and Fig. 5B). Mutation of the first (upstream) AUG (pMS2X1) had only a minor effect on florfenicol resistance, whereas mutation of the second AUG (pMS2X2) led to the complete loss of resistance to florfenicol. The lack of florfenicol resistance correlated with the lack of hypermodification of A2503 in the strain transformed with pMS2X2 and the presence of this modification in cells carrying pMS2X1 (Fig. 5C). These results demonstrate that translation of cfr initiates at the second (downstream) AUG codon. A slight effect of mutation of the upstream AUG on florfenicol sensitivity (and, by inference, cfr expression) is possibly due to alteration in the cfr Shine-Dalgarno sequence.
FIG. 5.
Mapping of the translation start site of the cfr gene. (A) The 5′ terminal nucleotide sequence showing two putative initiator codons of the cfr gene and the mutations engineered in plasmids pMS2X1 and pMS2X2 plasmids; (B) antibiotic sensitivities of S. aureus RN4220 cells transformed with pMS2, empty vector pLI50, or plasmids pMS2X1 and pMS2X2; (C) primer extension analysis of the extent of Cfr-dependent modification of A2503 in S. aureus cells transformed with pMS2, pMS2X1, pMS2X2, or the empty vector.
Expression of erm(B) and cfr is constitutive in the clinical MRSA strain.
Wild-type erm(B) belongs to the class of inducible erm genes regulated posttranscriptionally by translation attenuation (12, 40). This mechanism involves stalling of the ribosome at the upstream regulatory ORF [leader ORF erm(B)L in Fig. 1]. Such stalling, which takes place in the presence of inducing concentrations of macrolide antibiotics, critically depends on the sequence of the leader peptide (Fig. 6) (40). By analogy with other inducible resistance genes, the presence of two short ORFs in front of the cfr gene in plasmids pSCFS1, pSCFS3, and pSCFS6 led to the hypothesis that the expression of cfr can also be inducible and a subject of translation attenuation control.
FIG. 6.
Differences in nucleotide sequences of the leader peptide ORF in transposon Tn917 [inducible erm(B)] (32) and the mlr operon [noninducible erm(B)]. The nucleotide sequence of the Tn917 erm(B) leader ORF is shown, and the nucleotide changes observed in mlr are indicated. The corresponding changes in the amino acid sequence of the encoded leader peptide are shown under the nucleotide sequence.
To determine whether the expression of erm(B) and cfr in the mlr operon is inducible, we monitored the levels of erm(B)- and cfr-specific modification of A2058 and A2503, respectively, in S. aureus 23S rRNA under induction conditions. CM05 cells were grown in the absence of antibiotics or in the presence of a subinhibitory concentration (256 μg/ml; approximately one-fourth the MIC for CM05) of erythromycin, an inducer of erm genes. Total RNA was isolated, and the extent of modification of A2058 and A2503 in 23S rRNA was assessed by primer extension. No substantial increase in the modification of either A2058 or A2503 in cells grown in the presence of erythromycin was observed, indicating that this classic erm(B) inducer does not affect the cellular level of activity of either Erm(B) or Cfr in strain CM05 (data not shown). Cfr-dependent methylation of A2503 renders cells resistant to several peptidyltransferase-targeting antibiotics. We therefore tested whether any of these drugs can possibly induce cfr expression. CM05 cells were preincubated in the presence of a subinhibitory concentration of chloramphenicol (8 μg/ml), florfenicol (15.6 μg/ml), clindamycin (256 μg/ml), linezolid (1.25 μg/ml), quinupristin-dalfopristin (Synercid; 0.4 μg/ml), or tiamulin (16 μg/ml). As a negative control, the small ribosomal subunit-targeting antibiotic tetracycline (0.0125 μg/ml) was included in the analysis. Primer extension analysis did not show any changes in A2503 modification upon exposure of the cells to chloramphenicol, florfenicol, clindamycin, linezolid, and tetracycline. Preincubation with either quinupristin-dalfopristin or tiamulin appeared to have a small negative effect on the extent of A2503 methylation by Cfr (data not shown). Thus, it appears that in the MRSA strain neither erm(B) nor cfr in the mlr operon is inducible and that translation attenuation does not contribute in any obvious way to the regulation of expression of these genes.
In RN4220 cells transformed with pMS2, preexposure of the cells to either erythromycin or florfenicol led to an apparent slight increase in modification of both A2058 and A2503 (Table 4). Since florfenicol is not an erm(B) inducer and it is hard to imagine that the possible induction of cfr expression by florfenicol would also increase the expression of the upstream erm(B) gene, the seemingly inducing effects of both antibiotics on the expression of the genes of the mlr operon in RN4220 cells transformed with pMS2 appear to be nonspecific. Given that in RN4220(pMS2) cells the mlr operon is expressed from a plasmid, we cannot rule out the possibility that the apparent increase in A2503 and A2058 modification may have arisen from the selection for a higher plasmid copy number during the pretreatment with sublethal concentrations of antibiotic.
TABLE 4.
Extent of modification of A2058 and A2503 in clinical MRSA isolate CM05 and S. aureus RN4220 transformed with pMS2 or empty vector pLI50 before and after incubation with erythromycin or florfenicol
| Strain | Site | % Modification after treatment witha:
|
||
|---|---|---|---|---|
| No treatment | Erythromycin | Florfenicol | ||
| RN4220(pLI50) | A2058 | 1 | NDb | ND |
| A2503 | 0 | ND | ND | |
| RN4220(pMS2) | A2058 | 12 | 23 | 20 |
| A2503 | 8 | 11 | 16 | |
| CM05 | A2058 | 34 | 35 | 32 |
| A2503 | 25 | 27 | 20 | |
The percentages shown were deduced from primer extension data and are the averages of at least two experiments.
ND, not determined.
Modification of A2503 renders cells less susceptible to 16-member-ring macrolides and synergistically enhances the resistance conferred by A2058 modification.
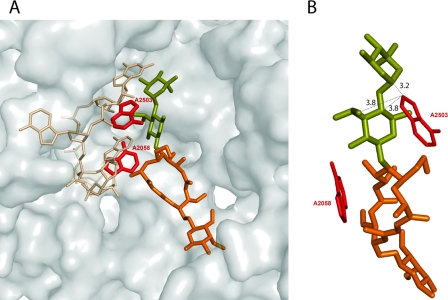
Methylation of either A2058 or A2503 perturbs the binding of individual antibiotics in their respective binding sites. However, since A2058 and A2503 are positioned in close proximity in the ribosome, some antibiotics that bind to the large ribosomal subunit simultaneously contact both of these nucleotides (Fig. 7). These include lincosamides, streptogramins, and some 16-member-ring macrolides (30, 37). Simultaneous modification of two closely positioned rRNA residues within an antibiotic binding site directed by the genes of the mlr operon may potentially synergistically increase antibiotic resistance (20). We indeed observed a synergistic resistance with respect to a 16-member-ring macrolide, tylosin. Although the effect of Cfr-dependent methylation of A2503 on binding of 16-member-ring macrolides was not reported previously, we found that RN4220 cells expressing Cfr alone from plasmid pLXM1 exhibited a twofold increased resistance to tylosin (Table 5). Dimethylation of A2058 by Erm(B) conferred a very high level of resistance to this drug (MIC, 2,000 μg/ml). However, the combined expression of Cfr and Erm(B) increased the level of resistance threefold (MIC, 6,000 μg/ml) compared to that attained with Erm(B) alone. This result demonstrates that modification of two nucleotides, A2058 and A2503, within an antibiotic binding site can have a synergistic effect on resistance to tylosin.
FIG. 7.
(A) Relative positions of A2058, A2503, and tylosin in the nascent peptide exit tunnel of the ribosome. The segment of 23S rRNA from positions 2058 to 2061 is shown as sticks (beige). The adenine bases of the residues 2058 and 2503 are highlighted in red. Tylosin is shown in orange, with the desosamine-mycarose side chain highlighted in olive. The position of the tRNA CCA 3′ end (cyan) and the attached amino acid (blue) in the P site of the peptidyltransferase center is shown as a landmark. The figure was prepared with the PyMol program (6) on the basis of the structures of antibiotic complexes of the Haloarcula marismortui large ribosomal subunits (PDB accession numbers 1K9M and 1YI2) (10, 37). (B) The structure shown in panel A was rotated by ca. 90 degrees clockwise around the y axis to illustrate the proximity of the desosamine-mycarose side chain of tylosin to A2503. The shortest distances (in Å) between the drug and the A2503 base are indicated.
TABLE 5.
Sensitivity of S. aureus RN4220 transformed with plasmids harboring erm(B) and/or cfr to clindamycin, quinupristin-dalfopristin, tylosin, josamycin, and spiramycin
| Plasmid (phenotype)a | MIC (μg/ml)b
|
||||
|---|---|---|---|---|---|
| CLI | Q-D | TYL | JOS | SPI | |
| pMS2 [erm(B)+cfr+] | >1,024 | 0.39 | 6,000 | >2,000 | >4,000 |
| pLI50 [erm(B)−cfr−] | <1 | <0.1 | 1 | 1 | 8 |
| pLXM1 [erm(B)−cfr+] | 1,024 | 0.39 | 2 | 64 | 128 |
| pMS2X2 [erm(B)+cfr−] | >1,024 | <0.1 | 2,000 | >2,000 | >4,000 |
erm(B)+, erm(B) positive; cfr+, cfr positive; erm(B)−, erm(B) negative; cfr−, cfr negative.
CLI clindamycin; Q-D, quinupristin-dalfopristin; TYL, tylosin; JOS, josamycin; SPI, spiramycin.
In accordance with the data obtained with tylosin, we found that the expression of Cfr alone notably decreased (16- to 64-fold) the susceptibility of S. aureus to josamycin and spiramycin, the other two 16-member-ring macrolides tested (Table 5). However, due to drug solubility limitations, we were unable to test the possible synergy achieved by modification of both the A2058 and the A2503 residues because the expression of Erm(B) alone conferred a very high level of resistance.
Even though lincosamides and streptogramins also contact both A2058 and A2503, no synergistic resistance could be observed when both of these nucleotides were modified. Modification of either of the two RNA residues (A2503 or A2058) provided an already very high level of resistance to the lincosamide clindamycin (Table 5), so that possible synergy could not be tested because of drug solubility limitations. Streptogramins are used clinically as a combination of two components (streptogramins A and B, which are composed of quinupristin and dalfopristin, respectively, in the clinical streptogramin quinupristin-dalfopristin). The dimethylation of A2058 conferred resistance to streptogramins of the B type but provided no resistance to quinupristin-dalfopristin (4) (Table 5). Modification of A2503 rendered Cfr-expressing cells resistant to streptogramin A and quinupristin-dalfopristin (20) (Table 4). However, the level of quinupristin-dalfopristin resistance was not increased when Cfr was coexpressed with Erm(B) (Table 5). Thus, resistance to quinupristin-dalfopristin in cells carrying the mlr operon is determined primarily by Cfr-dependent modification of A2503 rather than the combined actions of two methyltransferases.
DISCUSSION
In plasmids pSCFS1, pSCFS3, and pSCFS6, in which the gene encoding the Cfr methyltransferase was originally found, cfr comprises a monocistronic operon transcribed from its own promoter (15, 16, 31). In hospital MRSA isolate CM05, which bears the cfr gene, its genetic organization is drastically different (35). The nucleotide sequence of the region upstream from the cfr ORF reveals the presence of the Tn917 transposon segment containing the erm(B) cistron with its control elements, the promoter Perm and the leader peptide ORF (32). This insertion eliminated the putative cfr promoter and put it under the control of the Perm promoter, thereby combining erm(B) and cfr into a single operon, mlr. Although it is difficult to unequivocally rule out the existence of a weak promoter in front of the cfr gene in the CM05 chromosome, none of our experiments showed the presence of cfr transcripts initiated within the erm(B)-cfr spacer. Instead, all the results consistently led to the conclusion that the erm(B) and cfr ORFs are cotranscribed from a single promoter, Perm, located upstream of the erm(B) gene.
The expression of both the erm(B) and the cfr genes in the mlr operon was thought to be inducible. However, neither preincubation of CM05 MRSA cells with erythromycin, a classic erm inducer, nor exposure to peptidyltransferase-targeting inhibitors increased the level of posttranscriptional modification of A2058 and A2503, the targets of the Erm(B) and Cfr methyltransferases, respectively. Thus, our results argue that the two genes in the mlr operon are expressed constitutively.
It is generally assumed that the induction of erm(B) expression requires drug-dependent and nascent peptide-dependent stalling of the ribosome during the translation of the leader peptide ORF (12). A similar mechanism of induction was also proposed for cfr in plasmid pSCFS1 (31). The lack of inducibility of erm(B) and cfr in the mlr operon could be due to the mutations in the leader ORFs. However, since the molecular mechanisms of Cfr and Erm(B) induction have not been characterized, it is unclear how these alterations specifically contribute to the lack of inducibility. In comparison with transposon Tn917, which carries an inducible erm(B), the leader ORF upstream from the erm(B) gene in the mlr operon contains a duplication of the TAAA sequence, which introduces a premature stop codon that truncates the leader peptide by 9 amino acids (Fig. 6). Although a similar mutation was found in erm(B) variants that retained their inducibility (28), other reports linked the same mutation with a lack of induction (25, 41). In addition, the erm(B) leader in the mlr operon has a missense mutation in the eighth codon (Asn to Tyr), which may possibly lead to the constitutive expression of erm(B) due to the alteration in the nascent peptide sequence. Furthermore, in clinical MRSA isolate CM05 (but not in pMS2-transformed RN4220 cells), the constitutively high level of dimethylation of A2058 may also be affected by the presence of erm(A) in the CM05 chromosome (1a; S. Toh and A. S. Mankin, unpublished data). erm(A) encodes a methyltransferase similar to Erm(B) and targets A2058 in the 23S rRNA.
Preexposure of an animal isolate of a Staphylococcus sciuri strain carrying plasmid pSCFS1 to low concentrations of phenicols notably increased the levels of resistance to these antibiotics, indicating that expression of the cfr gene in this plasmid may be inducible (31). However, we observed no such effect in clinical strain CM05 or in RN4220 cells transformed with plasmid pMS2. Elimination of the first leader ORF due to the transposon insertion could be one of the causes for the constitutive expression of the cfr gene in the mlr operon (Fig. 1). In addition, a spacer separating the intact leader ORF and the cfr cistron in the mlr operon lacks a 35-nucleotide-long segment that is present in plasmid pSCFS1. This deletion alters the secondary structure of mRNA and might lead to the constitutive expression of cfr (16).
The mlr operon encodes two rRNA methyltransferase enzymes which target two adenine residues in bacterial 23S rRNA. The combined action of the two enzymes encoded by the mlr operon protects cells from all the clinically relevant antibiotics that target the large ribosomal subunit (Table 3). Furthermore, we observed previously unreported resistance to 16-member-ring macrolides resulting from the modification of A2503 by Cfr. The greatest effect of the A2503 modification was observed with spiramycin and josamycin, with 16- and 64-fold increases in the MICs, respectively, while the MIC of tylosin increased 2-fold. This difference could be due to the presence of the mycinose sugar at position 14 of the lactone ring of tylosin (which is absent from josamycin and spiramycin). This mycinose residue makes an additional contact with helix 35 in domain II of 23S rRNA (9) and may help tylosin bind to the ribosome, despite the A2503 modification.
Modification of two nucleotides within an antibiotic binding site can have a synergistic effect on antibiotic resistance (13, 19). We found that the combined expression of Erm(B) and Cfr confers enhanced resistance to tylosin. The extended desosamine-mycarose disaccharide at position 5 of the lactone ring of 16-member-ring macrolides contributes significantly to the binding energy of the antibiotics (9, 26, 30, 37). The disaccharide side chain bridges the distance between A2058 and A2503 and establishes contacts with both nucleotides (Fig. 7), which explains the synergy observed. Although we could not observe a similar synergy for other drugs that interact with both A2058 and A2503 because of the high level of resistance resulting from the individual action of Cfr or Erm(B), the synergistic action of both methyltransferases may affect future drugs designed to overcome Erm- or Cfr-based mechanisms of resistance.
Acknowledgments
We thank Seok-Ming Toh for providing plasmids pMS2 and pLXM1, Liqun Xiong for guidance with some experiments, Karen Lolans for quinupristin-dalfopristin, Cesar Arias for sharing unpublished data, and Nora Vazquez-Laslop for critical reading of the manuscript.
This work was supported by grant AI072445 from the National Institute of Allergy and Infectious Diseases, NIH.
Footnotes
Published ahead of print on 25 February 2008.
REFERENCES
- 1.Alekshun, M. N., and S. B. Levy. 2007. Molecular mechanisms of antibacterial multidrug resistance. Cell 128:1037-1050. [DOI] [PubMed] [Google Scholar]
- 1a.Arias, C. A., M. Vallejo, J. Reyes, D. Panesso, J. Moreno, E. Castañeda, M. V. Villegas, B. E. Murray, and J. P. Quinn. 2008. Clinical and microbiological aspects of linezolid resistance mediated by the cfr gene encoding a 23S rRNA methyl transferase. J. Clin. Microbiol. 46:892-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey, M., T. Chettiath, and A. S. Mankin. 2008. Induction of ermC expression by “noninducing” antibiotics. Antimicrob. Agents Chemother. [DOI] [PMC free article] [PubMed]
- 3.Clewell, D. B., S. E. Flannagan, D. D. Jaworski, and D. B. Clewell. 1995. Unconstrained bacterial promiscuity: the Tn916-Tn1545 family of conjugative transposons. Trends Microbiol. 3:229-236. [DOI] [PubMed] [Google Scholar]
- 4.Cocito, C., M. Di Giambattista, E. Nyssen, and P. Vannuffel. 1997. Inhibition of protein synthesis by streptogramins and related antibiotics. J. Antimicrob. Chemother. 39:7-13. [DOI] [PubMed] [Google Scholar]
- 5.Cundliffe, E. 1989. How antibiotic-producing organisms avoid suicide. Annu. Rev. Microbiol. 43:207-233. [DOI] [PubMed] [Google Scholar]
- 6.DeLano, W. L. 2002. The PyMol user's manual. DeLano Scientific, San Carlos, CA.
- 7.Frohman, M. A., M. K. Dush, and G. R. Martin. 1988. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 85:8998-9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grohmann, E., G. Muth, and M. Espinosa. 2003. Conjugative plasmid transfer in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67:277-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen, J. L., J. A. Ippolito, N. Ban, P. Nissen, P. B. Moore, and T. A. Steitz. 2002. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol. Cell 10:117-128. [DOI] [PubMed] [Google Scholar]
- 10.Hansen, J. L., P. B. Moore, and T. A. Steitz. 2003. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J. Mol. Biol. 330:1061-1075. [DOI] [PubMed] [Google Scholar]
- 11.Hartmann, R. K., A. Bindereif, A. Schön, and E. Westhof (ed.). 2005. Handbook of RNA biochemistry, vol. 2. Wiley-VCH, Weinheim, Germany.
- 12.Horinouchi, S., W. H. Byeon, and B. Weisblum. 1983. A complex attenuator regulates inducible resistance to macrolides, lincosamides, and streptogramin type B antibiotics in Streptococcus sanguis. J. Bacteriol. 154:1252-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johansen, S. K., C. E. Maus, B. B. Plikaytis, and S. Douthwaite. 2006. Capreomycin binds across the ribosomal subunit interface using tlyA-encoded 2′-O-methylations in 16S and 23S rRNAs. Mol. Cell 23:173-182. [DOI] [PubMed] [Google Scholar]
- 14.Kehrenberg, C., F. M. Aarestrup, and S. Schwarz. 2007. IS21-558 insertion sequences are involved in the mobility of the multiresistance gene cfr. Antimicrob. Agents Chemother. 51:483-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kehrenberg, C., K. K. Ojo, and S. Schwarz. 2004. Nucleotide sequence and organization of the multiresistance plasmid pSCFS1 from Staphylococcus sciuri. J. Antimicrob. Chemother. 54:936-939. [DOI] [PubMed] [Google Scholar]
- 16.Kehrenberg, C., and S. Schwarz. 2006. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob. Agents Chemother. 50:1156-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kehrenberg, C., S. Schwarz, L. Jacobsen, L. H. Hansen, and B. Vester. 2005. A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: methylation of 23S ribosomal RNA at A2503. Mol. Microbiol. 57:1064-1073. [DOI] [PubMed] [Google Scholar]
- 18.Lee, C. Y., S. L. Buranen, and Z. H. Ye. 1991. Construction of single-copy integration vectors for Staphylococcus aureus. Gene 103:101-105. [DOI] [PubMed] [Google Scholar]
- 19.Liu, M., and S. Douthwaite. 2002. Resistance to the macrolide antibiotic tylosin is conferred by single methylations at 23S rRNA nucleotides G748 and A2058 acting in synergy. Proc. Natl. Acad. Sci. USA 99:14658-14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Long, K. S., J. Poehlsgaard, C. Kehrenberg, S. Schwarz, and B. Vester. 2006. The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob. Agents Chemother. 50:2500-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuoka, M., and T. Sasaki. 2004. Inactivation of macrolides by producers and pathogens. Curr. Drug Targets Infect. Dis. 4:217-240. [DOI] [PubMed] [Google Scholar]
- 22.Moll, I., G. Hirokawa, M. C. Kiel, A. Kaji, and U. Blasi. 2004. Translation initiation with 70S ribosomes: an alternative pathway for leaderless mRNAs. Nucleic Acids Res. 32:3354-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard M7-A6. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 24.Novick, R. P. 1991. Genetic systems in staphylococci. Methods Enzymol. 204:587-636. [DOI] [PubMed] [Google Scholar]
- 25.Oh, T.-G., A.-R. Kwon, and E.-C. Choi. 1998. Induction of ermAMR from a clinical strain of Enterococcus faecalis by 16-membered-ring macrolide antibiotics. J. Bacteriol. 180:5788-5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poulsen, S. M., C. Kofoed, and B. Vester. 2000. Inhibition of the ribosomal peptidyl transferase reaction by the mycarose moiety of the antibiotics carbomycin, spiramycin and tylosin. J. Mol. Biol. 304:471-481. [DOI] [PubMed] [Google Scholar]
- 27.Roberts, M. C., J. Sutcliffe, P. Courvalin, L. B. Jensen, J. Rood, and H. Seppala. 1999. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob. Agents Chemother. 43:2823-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosato, A., H. Vicarini, and R. Leclercq. 1999. Inducible or constitutive expression of resistance in clinical isolates of streptococci and enterococci cross-resistant to erythromycin and lincomycin. J. Antimicrob. Chemother. 43:559-562. [DOI] [PubMed] [Google Scholar]
- 29.Schenk, S., and R. A. Laddaga. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 94:133-138. [DOI] [PubMed] [Google Scholar]
- 30.Schlünzen, F., R. Zarivach, J. Harms, A. Bashan, A. Tocilj, R. Albrecht, A. Yonath, and F. Franceschi. 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413:814-821. [DOI] [PubMed] [Google Scholar]
- 31.Schwarz, S., C. Werckenthin, and C. Kehrenberg. 2000. Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob. Agents Chemother. 44:2530-2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaw, J. H., and D. B. Clewell. 1985. Complete nucleotide sequence of macrolide-lincosamide-streptogramin B-resistance transposon Tn917 in Streptococcus faecalis. J. Bacteriol. 164:782-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sigmund, C. D., M. Ettayebi, A. Borden, and E. A. Morgan. 1988. Antibiotic resistance mutations in ribosomal RNA genes of Escherichia coli. Methods Enzymol. 164:673-690. [DOI] [PubMed] [Google Scholar]
- 34.Tenson, T., and A. Mankin. 2006. Antibiotics and the ribosome. Mol. Microbiol. 59:1664-1677. [DOI] [PubMed] [Google Scholar]
- 35.Toh, S.-M., L. Xiong, C. A. Arias, M. V. Villegas, K. Lolans, J. Quinn, and A. S. Mankin. 2007. Acquisition of a natural resistance gene renders a clinical strain of methicillin-resistant Staphylococcus aureus resistant to the synthetic antibiotic linezolid. Mol. Microbiol. 64:1506-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toh, S.-M., L. Xiong, T. Bae, and A. S. Mankin. 2008. The methyltransferase YfgB/RlmN is responsible for modification of adenosine 2503 in 23S rRNA. RNA 14:98-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tu, D., G. Blaha, P. B. Moore, and T. A. Steitz. 2005. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell 121:257-270. [DOI] [PubMed] [Google Scholar]
- 38.Vester, B., and S. Douthwaite. 1994. Domain V of 23S rRNA contains all the structural elements necessary for recognition by the ErmE methyltransferase. J. Bacteriol. 176:6999-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weisblum, B. 1995. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39:577-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weisblum, B. 1995. Insights into erythromycin action from studies of its activity as inducer of resistance. Antimicrob. Agents Chemother. 39:797-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Werckenthin, C., S. Schwarz, and K. Dyke. 1996. Macrolide-lincosamide-streptogramin B resistance in Staphylococcus lentus results from the integration of part of a transposon into a small plasmid. Antimicrob. Agents Chemother. 40:2224-2225. [DOI] [PMC free article] [PubMed] [Google Scholar]