Abstract
Resistance profiles were compared among 18 extended-spectrum-β-lactamase-producing (ESBL) and 27 acquired AmpC β-lactamase-producing Escherichia coli isolates collected from Canadian intensive care units from 2005 to 2006. ESBL-producing E. coli isolates were more likely to be gentamicin resistant (P < 0.03), fluoroquinolone resistant (P < 0.0001), and multidrug resistant (P < 0.0001) than AmpC-producing E. coli isolates.
Escherichia coli is the most commonly encountered pathogen within the Enterobacteriaceae family, causing significant infections worldwide. The spread of resistance to extended-spectrum cephalosporins in E. coli has been increasing in both hospital and community settings. Resistance to cephalosporins often is mediated by the production of extended-spectrum β-lactamases (ESBL). However, it also may be attributed to AmpC cephalosporinases, which can result either from the overexpression of the chromosomally encoded AmpC enzyme in E. coli or by the acquisition of a plasmid-mediated AmpC enzyme (13).
Generally, isolates with AmpC differ from ESBL producers phenotypically by yielding a negative ESBL disk test result, and also they are resistant to cefoxitin and usually are susceptible to cefepime/cefpirome. Resistance to other classes of antimicrobials among ESBL-producing E. coli isolates and, to a lesser extent, among AmpC-producing E. coli isolates has been escalating and limits therapeutic options (4, 11).
The purpose of this study was to compare the prevalence as well as the resistance patterns of ESBL-producing E. coli (ESBL-EC) isolates and acquired AmpC β-lactamase-producing E. coli (AmpC-EC) isolates from Canadian intensive care units (ICUs).
Study isolates were obtained as part of the Canadian ICU (CAN-ICU) surveillance study. CAN-ICU is a laboratory-based surveillance study coordinated by the Health Sciences Centre in Winnipeg, Canada. From 1 September 2005 to 1 August 2006, inclusive, 19 tertiary-care hospitals from across Canada with active ICUs voluntarily participated as study sites. Each center collected a maximum of 300 consecutive pathogens isolated from blood, urine, wound tissue, and respiratory specimens of ICU patients. As only pathogens causing infectious diseases were of interest, surveillance cultures and eye, ear, nose, and genital swabs were excluded. Duplicate isolates from the same site also were excluded. No anaerobes or fungi were assessed in the CAN-ICU study. All isolates were identified at participating sites by routine procedures performed at each laboratory. Isolates were shipped to the reference laboratory at the Winnipeg Health Sciences Centre on Amies charcoal swabs, subcultured onto blood agar, and stocked in skim milk at −80°C until MIC testing was carried out. The most common infectious sites and pathogens causing infection in Canadian ICUs have been reported previously (3) (www.can-r.ca). In this study, only the analysis of all unique genotypically confirmed ESBL- and AmpC β-lactamase-producing E. coli isolates obtained from CAN-ICU will be reported.
After two subcultures from frozen stock, the in vitro activities of ampicillin, piperacillin-tazobactam, ceftriaxone, cefepime, cefoxitin, gentamicin, ciprofloxacin, levofloxacin, meropenem, ertapenem, doxycycline, tigecycline, and trimethoprim-sulfamethoxazole were determined in triplicate by broth microdilution in accordance with CLSI guidelines (2).
Any E. coli isolate for which the ceftriaxone MIC was ≥1 μg/ml was identified as a putative ESBL or AmpC β-lactamase producer. All 50 E. coli isolates identified as putative ESBL or AmpC producers were tested genotypically for ESBL and AmpC genes by PCR. Universal primer sets were used to identify blaSHV, blaTEM, blaCTX-M, and blaOXA-1,-2,-10-like ESBL genes by PCR as previously described (1, 6, 12).
Genes encoding EBC (ACT-1/MIR-1-related genes), DHA (DHA-related genes), FOX (FOX-related genes), and CIT (CMY-2-related genes) phylogenetic groups of AmpC-acquired enzymes were detected using a previously described multiplex PCR (8). The PCR products were purified and sequenced. A BLAST search of the DNA sequence was conducted to determine the specific gene.
All ESBL-producing and AmpC-producing E. coli isolates were subtyped by pulsed-field gel electrophoresis by the standardized protocol for E. coli as previously described (6).
χ2 analysis or t tests were used to evaluate statistical significance, as appropriate, using Graphpad Quickcalcs.
A total of 4,092 clinical isolates were obtained from the CAN-ICU study. Of the 4,092 isolates, 493 (12.0%) were E. coli isolates. The ceftriaxone MIC for 50 of the 493 (10.1%) E. coli isolates was ≥1 μg/ml, and these isolates were identified as putative ESBLs. Eighteen (3.7%) and 27 (5.5%) of 50 E. coli isolates were identified genotypically as unique ESBL and acquired AmpC producers, respectively. The mechanism producing elevated ceftriaxone MICs for the remaining five E. coli isolates was not identified. No isolates carried both ESBL and acquired AmpC genes, as determined by PCR. All 18 (100%) ESBL-EC isolates were of the CTX-M genotype, with 13 (72.2%) being blaCTX-M-15, 2 (11.1%) being blaCTX-M-2, and 1 (5.6%) each being blaCTX-M-9, blaCTX-M-14, or blaCTX-M-1. Of the 27 AmpC-EC isolates, 26 (96.3%) were blaCMY-2 and 1 (3.7%) was blaACT-1. A comparison of the patient demographics for the 18 ESBL- and 27 AmpC-producing E. coli isolates is summarized in Table 1. AmpC-producing E. coli isolates were significantly more likely to be isolated from eastern Canada (P < 0.005) and from urine (P < 0.018) than ESBL producers, which were more likely to be isolated from central Canada (P < 0.002) and from a variety of specimen sources (Table 1).
TABLE 1.
Comparison of patient demographics, region of isolation, and specimen source of ESBL-EC and AmpC-EC isolates from Canadian ICUs
| Isolate type (n) | Patient age
|
Gender (%)
|
% Isolates from regiona:
|
% Isolates from specimen type:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean age | Range | Female | Male | W (n = 7) | C (n = 7) | E (n = 5) | Urine | Blood | Wound | RTb | |
| ESBL-EC (18) | 60 | 25-83 | 55.6 | 44.4 | 11.1 | 55.6 | 33.3 | 50.0 | 27.8 | 11.1 | 11.1 |
| AmpC-EC (27) | 66 | 5-96 | 77.8 | 22.2 | 11.1 | 11.1 | 77.8 | 85.2 | 7.4 | 3.7 | 3.7 |
| P value | NSc | NS | NS | NS | <0.002 | <0.005 | <0.018 | NS | NS | NS | |
The west region (W) encompasses British Columbia, Alberta, Saskatchewan, and Manitoba; the central region (C) encompasses Ontario and Quebec; the east region (E) encompasses New Brunswick and Nova Scotia. The numbers in parentheses indicate the number of sites from which isolates were obtained.
RT, respiratory tract.
NS, not significant (P > 0.05).
A comparison of antimicrobial susceptibilities of the AmpC- and ESBL-producing E. coli isolates is shown in Table 2. Susceptibility testing demonstrated that resistance to fluoroquinolones (ciprofloxacin and levofloxacin), trimethoprim-sulfamethoxazole, doxycycline, or gentamicin occurred in 83.3 and 11.1% (P < 0.0001), 61.1 and 26.6% (not significant; P < 0.064), 33.3 and 18.5% (not significant; P < 0.304), and 27.8 and 3.7% (P < 0.03) of ESBL-EC and AmpC-EC isolates, respectively (Table 2). The AmpC-EC isolates were more susceptible to all antimicrobials tested, with the exception of cefoxitin (by definition). All ESBL- and AmpC-producing isolates were susceptible to meropenem, ertapenem, and tigecycline (Table 2). Eighteen (100%) ESBL-EC isolates and nine (33.3%) AmpC-EC isolates were multidrug resistant (P < 0.0001) (i.e., resistant to three or more different antimicrobial classes).
TABLE 2.
Comparison of antimicrobial susceptibilities of ESBL-EC and AmpC-EC isolates from Canadian ICUsa
| Antibiotic | ESBL-EC isolates (n = 18)
|
AmpC-EC isolates (n = 27)
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC50 | MIC90 | Range | % Sensitive | % Intermediate | % Resistant | MIC50 | MIC90 | Range | % Sensitive | % Intermediate | % Resistant | |
| Ampicillin | >256 | >256 | >256 | 100 | >256 | >256 | >256 | 100 | ||||
| Ceftriaxone | 128 | >256 | 8->256 | 5.6 | 11.1 | 83.3 | 16 | 64 | 2-128 | 44.4 | 44.4 | 11.1 |
| Cefepime | 8 | 32 | ≤1-256 | 50.0 | 38.9 | 11.1 | ≤1 | ≤1 | ≤1-2 | 100 | ||
| TZP | 2 | 8 | ≤1-64 | 94.4 | 5.6 | 2 | 8 | ≤1-16 | 100 | |||
| Cefoxitin | 8 | 32 | 4-32 | 61.1 | 22.2 | 16.7 | 256 | >256 | 32->256 | 100 | ||
| Ciprofloxacin | >16 | >16 | ≤0.06->16 | 16.7 | 83.3 | ≤0.06 | >16 | ≤0.06->16 | 85.2 | 3.7 | 11.1 | |
| Levofloxacin | 16 | 32 | ≤0.12->32 | 16.7 | 83.3 | ≤0.06 | 16 | ≤0.06->32 | 88.9 | 11.1 | ||
| SXT | >32 | >32 | 0.12->32 | 38.9 | 61.1 | 2 | 32 | ≤0.12-32 | 70.4 | 26.6 | ||
| Doxycycline | 4 | 64 | 1-128 | 61.1 | 5.6 | 33.3 | 2 | 64 | 0.5->256 | 66.7 | 14.8 | 18.5 |
| Gentamicin | 1 | 16 | ≤0.25-16 | 66.7 | 5.6 | 27.8 | ≤0.25 | 8 | ≤0.25-64 | 88.9 | 7.4 | 3.7 |
| Meropenem | ≤0.12 | ≤0.12 | ≤0.12 | 100 | ≤0.12 | ≤0.12 | ≤0.12 | 100 | ||||
| Ertapenem | 0.12 | 0.25 | 0.03-0.25 | 100 | 0.12 | 0.5 | 0.03-2 | 100 | ||||
| Tigecycline | 0.5 | 1 | 0.25-1 | 100 | 0.25 | 0.5 | 0.12-2 | 100 | ||||
MIC50, MIC at which 50% of the isolates tested are inhibited; MIC90, MIC at which 90% of the isolates tested are inhibited; TZP, piperacillin-tazobactam; SXT, trimethoprim-sulfamethoxazole.
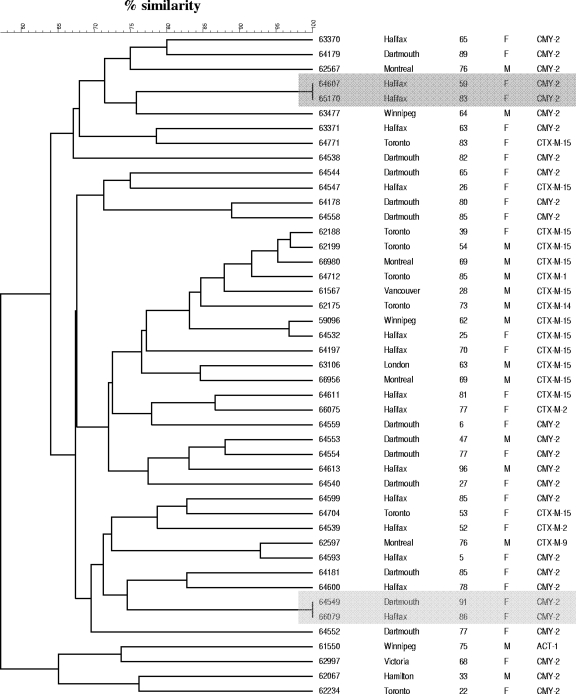
A dendrogram depicting the genetic relationships among ESBL-EC and AmpC-EC isolates from Canadian ICUs is shown in Fig. 1. Both genetically related (>80% homology) and unrelated clusters of E. coli isolates were observed. On the basis of macrorestriction patterns, intrahospital and interhospital spread may have occurred in eastern Canada and in each case that involved two AmpC-EC isolates.
FIG. 1.
Dendrogram depicting the genetic relationships among ESBL- and AmpC-producing E. coli isolates from Canadian ICUs. The darkly shaded region highlights the intrahospital spread of two AmpC-EC isolates, whereas the lightly shaded region highlights the interhospital spread of two AmpC-EC isolates.
In 2005 and 2006, AmpC-EC isolates (5.5%) were found to be more prevalent than ESBL-EC isolates (3.7%) in Canadian ICUs. The increasing awareness and improved recognition of ESBL producers have led to improved infection control measures to minimize the spread of these emerging pathogens. Therefore, it is not surprising that the prevalence of ESBL-EC isolates is lower than that of AmpC producers, which generally are unidentified and not subjected to such infection control measures (4). However, the prevalence of ESBL-EC isolates in this study may be underestimated due to the use of a single extended-spectrum cephalosporin as an indicator for screening. This study, in agreement with other studies, found that CTX-M and CIT-type were the predominant enzymes being produced, with CTX-M-15 and CMY-2 being the predominant genotypes among ESBL-EC and AmpC-EC isolates, respectively (4, 5, 7, 9-11, 13). Since 1999 to 2000, CTX-M has replaced SHV as the most common ESBL in Canada; however, CIT-type remains the predominant acquired AmpC enzyme and has been increasing in prevalence (6, 7). No E. coli isolate carried both ESBL and acquired AmpC genes, an occurrence that has yet to be described in Canada. However, there were two ESBL-producing isolates that were resistant to cefoxitin. These isolates could have chromosomal promoter ampC mutations or porin mutations, or they may have acquired an unknown blaAmpC gene. In contrast to a previous study of cefoxitin-resistant E. coli isolates from Canadian hospitals, cefoxitin resistance in AmpC-EC isolates was attributed to the acquisition of plasmid-mediated AmpC genes rather than the overexpression of the chromosomally encoded AmpC gene (7). AmpC-EC isolates were significantly less likely to be multidrug resistant and more likely to be susceptible to fluoroquinolones and gentamicin than ESBL-EC isolates. Although AmpC-EC infections are, at the moment, more susceptible to many more antimicrobials than ESBL-EC infections, they are an increasing public health concern (10, 13). Like the ESBL genes, the AmpC genes have localized to multidrug-resistant plasmids and have further limited our therapeutic options (5). With no guidelines for proper identification and infection control measures, AmpC-producing E. coli infections may become a greater concern than ESBL-producing E. coli infections, as they are increasing in prevalence worldwide. In addition, multidrug-resistant plasmids bearing both ESBL and AmpC genes recently have been reported and will most likely spread among bacteria and become the new emerging threat (11, 13).
Acknowledgments
Patricia Baudry is supported by the Winnipeg Health Sciences Centre Department of Research and the Winnipeg Health Sciences Centre Foundation. This research was funded in part by Abbott, Bayer Canada, Bristol-Myers Squibb, Janssen-Ortho Inc., Merck Frosst, Sanofi-Aventis, and Wyeth.
We thank the participating centers, investigators, and laboratory staff for their continued support. The technical assistance of B. Weshnoweski and R. Vashisht, along with the secretarial support of M. Tarka, is appreciated.
Footnotes
Published ahead of print on 25 February 2008.
REFERENCES
- 1.Boyd, D. A., S. Tyler, S. Christianson, A. McGeer, M. P. Muller, B. M. Willey, E. Bryce, M. Gardam, P. Nordmann, and M. R. Mulvey. 2004. Complete nucleotide sequence of a 92-kilobase plasmid harboring the CTX-M-15 extended-spectrum β-lactamase involved in an outbreak in long-term care facilities in Toronto, Canada. Antimicrob. Agents Chemother. 48:3758-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clinical and Laboratory Standards Institute. 2006. Methods for disk susceptibility tests for bacteria that grow aerobically, 7th ed. Approved standard M2-A7. Clinical and Laboratory Standards Institute, Wayne, PA.
- 3.DeCorby, M. M., B. Weshinoweski, J. A. Karlowsky, R. Vashisht, C. Siemens, M. McCracken, M. Mulvey, D. J. Hoban, and G. G. Zhanel. 2006. Characterization of MRSA, VRE, and ESBL isolated from Canadian intensive care units: results of the Canadian National Intensive Care Unit (CAN-ICU) study. Abstr. 46th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C2-1152.
- 4.Deshpande, L. M., R. N. Jones, T. R. Fritsche, and H. S. Sader. 2006. Occurrence of plasmidic AmpC type beta-lactamase-mediated resistance in Escherichia coli: report from the SENTRY Antimicrobial Surveillance Program (North America, 2004). Int. J. Antimicrob. Agents 28:578-581. [DOI] [PubMed] [Google Scholar]
- 5.Hopkins, K. L., M. J. Batchelor, E. Liebana, A. P. Deheer-Graham, and E. J. Threlfall. 2006. Characterisation of CTX-M and AmpC genes in human isolates of Escherichia coli identified between 1995 and 2003 in England and Wales. Int. J. Antimicrob. Agents 28:180-192. [DOI] [PubMed] [Google Scholar]
- 6.Mulvey, M. R., E. Bryce, D. Boyd, M. Ofner-Agostini, S. Christianson, A. E. Simor, and S. Paton. 2004. Ambler class A extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella spp. in Canadian hospitals. Antimicrob. Agents Chemother. 48:1204-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mulvey, M. R., E. Bryce, D. A. Boyd, M. Ofner-Agostini, A. M. Land, A. E. Simor, and S. Paton. 2005. Molecular characterization of cefoxitin-resistant Escherichia coli from Canadian hospitals. Antimicrob. Agents Chemother. 49:358-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pérez-Pérez, F. J., and N. D. Hanson. 2002. Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 40:2153-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pitout, J. D., D. L. Church, D. B. Gregson, B. L. Chow, M. McCracken, M. R. Mulvey, and K. B. Laupland. 2007. Molecular epidemiology of CTX-M-producing Escherichia coli in the Calgary health region: emergence of CTX-M-15-producing isolates. Antimicrob. Agents Chemother. 51:1281-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pitout, J. D., D. B. Gregson, D. L. Church, and K. B. Laupland. 2007. Population-based laboratory surveillance for AmpC beta-lactamase-producing Escherichia coli, Calgary. Emerg. Infect. Dis. 13:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Potz, N. A., R. Hope, M. Warner, A. P. Johnson, and D. M. Livermore. 2006. Prevalence and mechanisms of cephalosporin resistance in Enterobacteriaceae in London and south-east England. J. Antimicrob. Chemother. 58:320-326. [DOI] [PubMed] [Google Scholar]
- 12.Steward, C. D., J. K. Rasheed, S. K. Hubert, J. W. Biddle, et al. 2001. Characterization of clinical isolates of Klebsiella pneumoniae from 19 laboratories using National Committee for Clinical Laboratory Standards extended-spectrum β-lactamase detection methods. J. Clin. Microbiol. 39:2864-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woodford, N., S. Reddy, E. J. Fagan, R. L. Hill, K. L. Hopkins, M. E. Kaufmann, J. Kistler, M. F. Palepou, R. Pike, M. E. Ward, J. Cheesbrough, and D. M. Livermore. 2007. Wide geographic spread of diverse acquired AmpC beta-lactamases among Escherichia coli and Klebsiella spp. in the UK and Ireland. J. Antimicrob. Chemother. 59:102-105. [DOI] [PubMed] [Google Scholar]