Abstract
The antigenicity, structural location, and function of the predicted lipoprotein TP0136 of Treponema pallidum subsp. pallidum were investigated based on previous screening studies indicating that anti-TP0136 antibodies are present in the sera of syphilis patients and experimentally infected rabbits. Recombinant TP0136 (rTP0136) protein was purified and shown to be strongly antigenic during human and experimental rabbit infection. The TP0136 protein was exposed on the surface of the bacterial outer membrane and bound to the host extracellular matrix glycoproteins fibronectin and laminin. In addition, the TP0136 open reading frame was shown to be highly polymorphic among T. pallidum subspecies and strains at the nucleotide and amino acid levels. Finally, the ability of rTP0136 protein to act as a protective antigen to subsequent challenge with infectious T. pallidum in the rabbit model of infection was assessed. Immunization with rTP0136 delayed ulceration but did not prevent infection or the formation of lesions. These results demonstrate that TP0136 is expressed on the outer membrane of the treponeme during infection and may be involved in attachment to host extracellular matrix components.
Treponema pallidum subsp. pallidum is a spiral-shaped bacterium that causes syphilis, a systemic, long-term infection that if untreated can damage the cardiovascular and nervous systems, ultimately leading to debilitation and death. Although syphilis is readily treated with penicillin, no vaccine is available, and the disease remains a significant public health problem in developing nations, with an estimated 12 million new cases occurring per year (54). Recent studies indicating that a syphilitic infection increases vulnerability to infection with human immunodeficiency virus (19, 44) reinforce the need for more-effective control of syphilis. The development of a vaccine would be a significant aid to the global effort for eradication (13), but progress has been hindered by the inability to continuously culture this organism in vitro (12).
Syphilis patients as well as animals experimentally infected with T. pallidum remain infected for years, often for the lifetime of the individual. The mechanism of this persistence is not well understood but is likely to involve the unusual properties of the T. pallidum outer membrane. The bacterial outer membrane displays few potential antigens to the host as it lacks lipopolysaccharide and contains few outer membrane proteins compared to other bacteria (24, 40). Humans with syphilis, as well as animals that have been experimentally infected for several months, fail to develop lesions after reinoculation with homologous strains, indicative of protective immunity. It is thought that a vaccine for syphilis is feasible because complete protective immunity to experimental infection was seen in the rabbit model of infection following multiple rounds of immunization with gamma-irradiated treponemes (33). This procedure is clearly impractical for routine use; therefore, attempts have been made to identify T. pallidum antigens that could be used in subunit vaccines (13). Although several T. pallidum antigens have been purified and tested for protection (13), thus far no single antigen or combination of antigens has been shown to provide substantial protection to subsequent infection with T. pallidum (13).
Similar to other invasive pathogens, T. pallidum uses components of the extracellular matrix (ECM) as targets for initial adherence and colonization (26). Common targets of bacterial adhesins include ECM proteins, such as fibronectin, laminin, collagen, fibrinogen, elastin, and vitronectin (26). T. pallidum has been shown to bind to fibronectin and laminin (18, 39), and recent studies suggest that the purified T. pallidum proteins TP0155 and TP0483 bind to fibronectin and that TP0751 binds to laminin (6, 7). However, as with other invasive bacteria (26), it is probable that the organism possesses multiple bacterial receptors responsible for attachment to host cells. Since the existence of functionally redundant adhesins is likely, it is crucial to identify these surface-localized proteins to assess their immunoprotective effects and to more fully understand the mechanisms of T. pallidum adherence.
Previously, genomic screens for identifying important antigens in the rabbit and human immune responses to T. pallidum infection were performed (4, 31). Several antigens of interest were identified, including TP0136, a 50-kDa protein of unknown function that was reactive with sera collected from patients with primary-stage syphilis. The amino acid sequence of TP0136 predicts that the protein contains a bacterial type II secretion signal sequence (36). Additionally, TP0136 mRNA transcript levels were shown to be significantly high relative to those in the rest of the transcriptome during experimental rabbit infection (46). The goal of this study was to analyze of the role of TP0136 in T. pallidum host interactions. This report shows that TP0136 is surface localized, reacts strongly with serum antibodies from syphilis patients and T. pallidum-infected rabbits, and binds to the host ECM glycoproteins fibronectin and laminin.
MATERIALS AND METHODS
General procedures.
Oligonucleotides were synthesized by Integrated DNA Technologies, Inc. Molecular biology procedures not described in detail below followed those in Sambrook et al. (42). Nucleotide sequencing was performed using an ABI3100 automated sequencer (Applied Biosystems). Protein purity was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis with Coomassie blue staining. Treponema pallidum subsp. pallidum (Nichols) was maintained by rabbit inoculation and, unless otherwise indicated, purified by Percoll gradient centrifugation as described previously (2, 23).
Cloning of TP0136.
The TP0136 gene without its predicted signal sequence (genome coordinates 156900 to 158276) was previously cloned into pUNI-D (32). The plasmid TP0136-UniD-PRSET-E, encoding TP0136-His6, was generated by Cre-mediated recombination of TP0136-UniD and PRSET-E (Novagen) (30).
Recombinant protein purification.
Isopropyl β-d-thiogalactoside (IPTG) concentrations (in the range of 0.1 mM to 1.0 mM), expression temperatures (25, 30, and 37°C), and culture media (Media Optimization Kit; AthenaES) were examined to determine optimal conditions for expression of both the His6 construct and a glutathione S-transferase-tagged construct according to the instructions in the Echo system manual (Novagen). No conditions tested produced soluble protein, and therefore, the conditions that produced the highest levels of expression of the insoluble His6-tagged protein were used. A single colony of Escherichia coli BL21(DE3) harboring TP0136-UniD-PRSET-E (recombinant TP0136 [rTP0136]) was used to inoculate 100 ml of Luria-Bertani broth (42) with kanamycin (25 μg/ml) and ampicillin (100 μg/ml) and grown aerobically overnight at 37°C with shaking. The entire culture was used to inoculate 2 liters of Superior Broth (AthenaES)-kanamycin-ampicillin, and growth with shaking was continued at 30°C overnight. Cells were harvested by centrifugation (8,000 × g, 10 min, 4°C) and lysed using a microfluidizer (Microfluidics, Newton, MA) with phosphate-buffered saline (PBS; pH 7.4) augmented with 0.5 mg/ml lysozyme (Sigma), 0.1 μl/ml Benzonase nuclease (Novagen), and one tablet of complete EDTA-free protease inhibitors (Roche). The cell lysate was fractionated by centrifugation (17,000 × g, 20 min, 4°C). The pellet was washed twice with 1% Triton X-100 in PBS. The pellet was solubilized overnight in 8 M urea-PBS with stirring at room temperature. Solubilized protein was loaded onto a Talon resin column (BD Biosciences) and eluted using 150 mM imidazole. Fractions containing rTP0136 (determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis) were pooled. Optimal conditions for protein refolding were screened using a QuickFold protein refolding kit (AthenaES). However, no conditions tested produced markedly more soluble protein than direct dialysis into PBS. Thus, following elution, protein was dialyzed into 4 liters of PBS for 6 to 8 h at room temperature. Protein was next transferred to a fresh 4-liter volume of PBS and allowed to dialyze overnight. Some precipitation occurred during dialysis, and this was removed by filtration prior to concentration of the protein in Amicon Ultra-15 concentration devices (Millipore) at room temperature. Concentrated protein was placed in 1.5-ml microcentrifuge tubes and spun at high speed for 1 to 2 min to pellet any remaining insoluble portions before transfer to a fresh 1.5-ml microcentrifuge tube and protein concentration determination. The protein samples used in all assays contained no visible precipitation or turbidity at the time of the assay. Approximately 1.5 mg of purified rTP0136 (quantitated by the Bio-Rad protein assay [Bio-Rad, Hercules, CA], using bovine serum albumin [BSA] as the standard) was obtained per liter of E. coli culture. Purified, soluble Shewanella oneidensis NarP-His6 was generously provided by Donna Pattison.
DNA sequence comparison.
Treponema pallidum subsp. pallidum (Nichols and SS-14), Treponema pallidum subsp. pertenue (Samoa D and Samoa F), and Treponema paraluiscuniculi (Cuniculi A) were maintained by rabbit inoculation and purified by Hypaque gradient centrifugation as described previously (2, 20), and genomic DNA was prepared as described previously (20). The TP0136 gene and flanking chromosomal regions extending at least 150 bp on each side were amplified using PFU polymerase and oligonucleotide primers that were designed using GeneTool (53) software. The resulting PCR products were purified using a QIAquick PCR purification kit (Qiagen), and the DNA sequences were determined using the original amplification primers and internal primers where applicable. Sequences were aligned with the T-Coffee multiple sequence alignment program (38), and the resulting alignment was formatted with BOXSHADE. Percents similarity and identity of aligned sequences were calculated using MatGAT (8). The GenBank accession numbers for the sequence files reported in this study, which include primer sequences, are EF514691, EF514692, EF514693, EF137743, and EF514694 for TP0136 strains SS-14, Samoa D, Samoa F, Cuniculi A, and TP0462/3 Nichols, respectively.
Enzyme-linked immunosorbent assay (ELISA).
All human sera were collected under established guidelines with prior approval by the Committee for the Protection of Human Subjects, University of Texas Health Science Center at Houston. Anti-rTP0136 rabbit sera were obtained from the rabbit protection study described in detail below, and all sera were collected prior to inoculation with T. pallidum. The T. pallidum lysate was prepared by boiling Percoll-purified organisms for 10 min at 90°C. Purified rTP0136 (0.25 μg/μl) or recombinant NarP (0.25 μg/μl) or a T. pallidum lysate (2 × 104 organisms/μl) was diluted to the indicated concentration in carbonate buffer (pH 9.4), and Immulon 96-well plates (Thermo) were coated overnight with 100 μl of each, followed by blocking for 2 h at room temperature with 5% milk diluted in PBS Superblock (Pierce). Each serum sample (either individual rabbit sera or human sera pooled as described previously [4, 31]) was diluted (1:100 [rabbit] or 1:200 [human]) in Superblock and added in a 100-μl volume to the coated plates, followed by incubation at room temperature for 2 h. The plates were washed four times with PBS containing 0.05% Tween 20 before addition of a 1:1,500 dilution of anti-rabbit immunoglobulin G (IgG)-horseradish peroxidase (HRP; Amersham) and a 1:2,500 dilution of goat anti-human IgA-IgG-IgM-HRP (Pierce, Rockford, IL) to each well, followed by incubation for 1 h at room temperature. The plates were washed four times with PBS containing 0.05% Tween 20 before the addition of 1-Step ABTS colorimetric substrate (Pierce, Rockford, IL). Each well was read 40 min after peroxidase substrate addition with a Genios plate reader (Tecan, Durham, NC), and the experiments were performed in triplicate. Statistical significance (P < 0.005) was determined by comparison with negative controls (normal human sera [NHS]) or prechallenge sera by using the Student two-tailed t test.
Rabbit protection study.
Three New Zealand White rabbits (Myrtle's Rabbitry) were bled from the ear artery prior to immunization with rTP0136-monophosphoryl lipid A-trehalose dimycolate-cell wall skeleton adjuvant (Sigma) according to the protocol provided by the manufacturer. Bleeding and immunizations (1.5 mg rTP0136 per immunization) were repeated at 21-day intervals, for a total of four immunizations. On day 84, three immunized rabbits and three nonimmunized control rabbits were challenged intradermally on their shaved backs with live T. pallidum suspensions at eight sites per rabbit (104 organisms/site). The animals were observed daily for lesion appearance and development, and one representative site from each animal was examined by needle aspiration at day 20 postchallenge to verify the presence of treponemes by dark-field microscopy. The animals were observed until all lesions had healed.
Immunoelectron microscopy.
Samples of freshly isolated, motile Percoll-purified T. pallidum cells (4.5 × 107 organisms/10 μl) were applied to carbon-coated 400-mesh copper grids with a 2% Parlodion film for 10 min at room temperature in a humid chamber. Excess liquid was absorbed from the side of the grid with filter paper. The grids were exposed to 0.05% Triton X-100 for 1 min or processed without Triton X-100 treatment as indicated. After being washed five times with PBS, the grid was incubated with anti-rTP0136 (1:100 dilution; Melon Gel [Pierce, Rockford, IL] IgG purified) or preimmune sera (1:100 dilution; Melon Gel IgG purified) for 1 h. Following three washes with PBS, the grids were incubated for 1 h with anti-rabbit IgG-10-nm colloidal gold probe(s) (Sigma) (1:50 dilution) and then washed five times with PBS. Finally, the grids were negatively stained with 1% uranyl acetate (pH 5) for 60 seconds and excess liquid was blotted, followed by air drying for 15 min. The grids were examined with a JEOL JEM1230 electron microscope operating at 80 kV.
Immunofluorescence.
Samples of freshly isolated, motile, Percoll-purified T. pallidum cells (4 × 107 organisms/ml) were encapsulated in agarose microdroplets (11), and immunolocalization was performed as described previously (3). An aliquot of the encapsulated spirochetes was treated with 0.1% Triton X-100 for 30 min to solubilize the T. pallidum outer membrane. The agarose-embedded T. pallidum cells (with and without Triton X-100 treatment) were incubated for 2 h at 30°C with rabbit anti-rTP0136 and rat anti-T. pallidum periplasmic flagellar sheath protein (FlaA) (a gift from J. D. Radolf), followed by washing and a 2-h incubation with 2 μg of Alexa Fluor 595-labeled anti-rat IgG secondary antibody (Invitrogen) or Alexa Fluor 488-labeled anti-rabbit IgG secondary antibody (Invitrogen). After the wash, the treponemes were examined for the presence of fluorescence by using dark-field and fluorescence microscopy.
ECM protein binding assays.
Collagen (IV), fibronectin, superfibronectin, laminin, and the negative controls BSA and fetuin were purchased from Sigma Chemical Co., resuspended where applicable according to the manufacturer's instructions, and diluted in carbonate buffer (pH 9.4; 1.5 μg/well). Immulon 96-well plates were coated with 100 μl of each protein overnight at 4°C. To determine relative coating efficiency, representative wells were washed three times with PBS, followed by the addition of 200 μl of EZQ protein detection reagent (Invitrogen), and fluorescent signal was visualized using UV illumination (Versadoc; Bio-Rad), followed by densitometry according to the instructions of the manufacturer. After being coated, the plates were blocked for 2 h at room temperature with 200 μl of histidine-blocking reagent (Qiagen). Twenty-five micrograms of either rTP0136 protein or the negative control NarP was added to each well, followed by incubation at room temperature for 2 h. The plates were then washed four times with histidine-blocking reagent. A 1:5,000 dilution of anti-His-HRP antibody (Qiagen) was added to each well, followed by incubation for 1 h at room temperature. The plates were washed eight times with PBS before the addition of SuperSignal ELISA Pico chemiluminescent substrate (Pierce, Rockford, IL). Light emission from each well was recorded 10 min after peroxidase substrate addition with a Genios plate reader. The experiments were performed in triplicate.
RESULTS
Functional genomic identification of TP0136.
The TP0136 protein from Treponema pallidum subsp. pallidum (Nichols) was previously identified from a screen of antigens reactive in both a profile of the rabbit immunoproteome (31) and a profile of the reactivity of primary-stage human syphilitic sera to T. pallidum proteins (4). Additionally, TP0136 mRNA levels were shown to be high relative to those in the rest of the transcriptome in a study of an experimental rabbit infection (46). With modification of the predicted translational start site from the original location (nucleotide [nt] 156792 in the Nichols strain genome [20]) to one 30 nt downstream (nt 156822), the TP0136 gene is predicted to encode a lipoprotein signal peptide and a signal peptidase II (SPaseII) cleavage site between amino acids 23 and 24 (SLLTT CDFTG) by several signal sequence prediction programs, including LipoP (27). Taken together, the above data suggested that the TP0136 protein is expressed during infection and recognized by the immune system. Therefore, it was of interest to study the antigenicity and possible host interactions of this protein in more detail.
Overexpression and purification of rTP0136.
To facilitate further study, it was necessary to purify the rTP0136 protein, and this required optimization of expression and purification conditions (see Materials and Methods). The protein was ultimately purified from the insoluble pellet as a histidine-tagged fusion protein in the presence of 8 M urea, followed by dialysis with PBS. The refolded protein preparation was soluble and reacted with anti-His6 tag antibodies in immunoblots (data not shown).
Analysis of TP0136 sequence heterogeneity among strains.
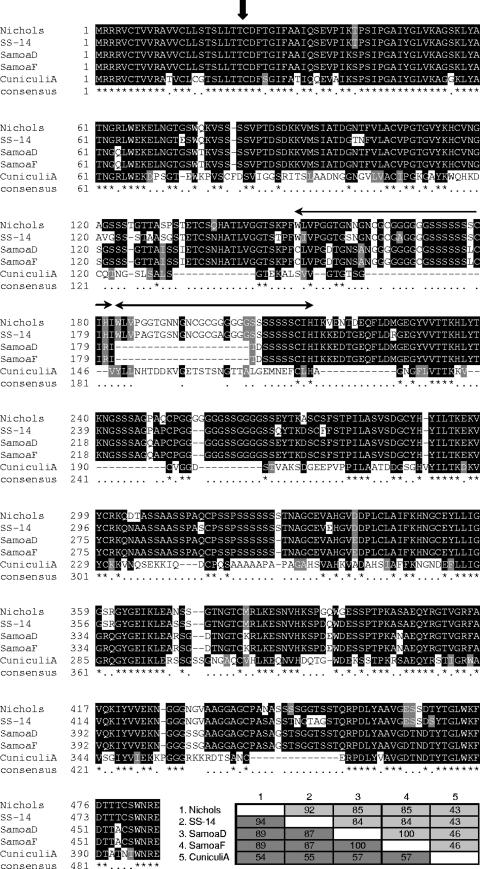
The DNA and protein sequences among T. pallidum subspecies and strains are generally highly conserved, with the exception of some of the well-characterized tpr genes, which are heterogeneous and may undergo intrastrain sequence variation (29, 48). This variation may represent immune evasion mechanisms. In addition, a recent DNA microarray and whole-genome amplification study indicates sequence variability between the TP0136 genes in T. pallidum subsp. pallidum (Nichols) and T. paraluiscuniculi Cuniculi A (47). To investigate further the levels of sequence heterogeneity in TP0136 among different strains, PCR amplification and DNA sequencing of the TP0136 gene were performed using genomic DNA from the syphilis-causing strain T. pallidum subsp. pallidum Street 14, two yaws strains (T. pallidum subsp. pertenue Samoa D and Samoa F), and the rabbit pathogen T. paraluiscuniculi Cuniculi A. The resulting sequences were compared with the published Nichols strain sequence (Fig. 1). Multiple nucleotide differences and small insertions and deletions were observed among the TP0136 genes of different strains, resulting in differences in the predicted amino acid sequences of TP0136 homologs, as shown in Fig. 1. The TP0136 gene thus contains regions that are highly variable among different strains while still maintaining an intact open reading frame and >50% similarity, suggesting that the TP0136 gene may be under selective pressure.
FIG. 1.
Amino acid sequence heterogeneity of TP0136 among strains of Treponema. Sequences are identified by strain name, and the strains included are two syphilis-causing strains (T. pallidum subsp. pallidum Nichols and T. pallidum subsp. pallidum Street strain 14), two yaws strains (T. pallidum subsp. pertenue Samoa D and T. pallidum subsp. pertenue Samoa F), and the rabbit pathogen T. paraluiscuniculi Cuniculi A. The vertical arrow indicates the predicted signal sequence cleavage site, and a repeat region is indicated by horizontal arrows. Black shading indicates 50% sequence identity, gray shading indicates >50% functionally similar amino acids, and no shading indicates <50% sequence similarity. Percent identity (light gray) and percent similarity (dark gray) for each sequence are indicated in the inset table at bottom right. -, gap introduced to optimize alignment. The consensus line contains the following annotations: *, identical amino acid residues; functionally similar amino acid residues. Numbers on the left indicate the positions of the amino acid residues.
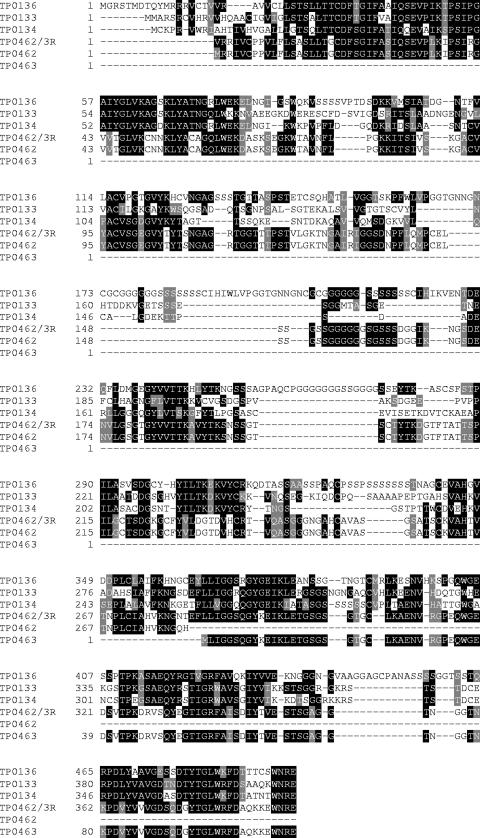
Although it has no significant similarity to proteins outside the Treponema genomes, TP0136 is a member of a family of paralogs including the TP0133, TP0134, TP0462, and TP0463 proteins (Fig. 2). Analysis of the paralog alignment indicated that the predicted TP0462 and TP0463 proteins are potentially a single protein. Dideoxy sequencing of genomic DNA in the intervening region indicated a single nucleotide difference from the published genome sequence which when corrected did indeed create a single protein from TP0462 and TP0463. All paralogs contain a signal sequence and are considered hypothetical proteins. In addition, the TP0133 and TP0463 proteins were significantly antigenic in immunoproteome assays (4, 31).
FIG. 2.
Sequence alignment of TP0136 paralogs indicated by genome sequence. Resequencing revealed a discrepancy with the published genome sequence that when translated yields the sequence TP0462/3R. Black shading indicates 100% sequence identity, gray shading indicates 75% sequence identity, and no shading indicates <50% sequence identity. -, gap introduced to optimize alignment. Numbers on the left indicate the positions of the amino acid residues.
Localization of endogenous TP0136 protein.
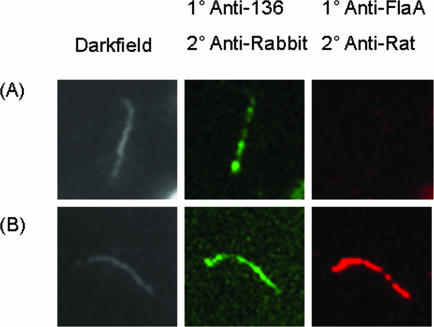
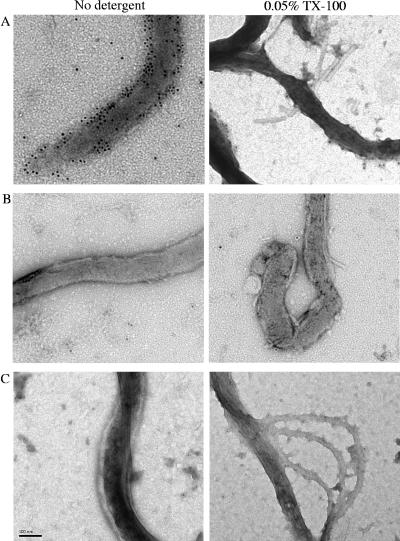
Because the TP0136 protein was predicted to contain a lipoprotein signal sequence, we examined whether it was localized on the outer surface of the treponeme. The localization of the protein was examined by both immunofluorescence and transmission electron microscopy, using rabbit anti-rTP0136 and rat anti-FlaA antibodies (Fig. 3 and 4). In the immunofluorescence assay, cellular localization was determined to be to the outer membrane if rTP0136 antibody binding, as indicated by green fluorescence, was observed in the absence of fluorescence of the known periplasmic protein FlaA, which was used as a control for outer membrane integrity (11). Cellular localization was considered to be to the periplasmic or inner membrane if antibody binding could be seen only with the addition of 0.1% Triton X-100, which solubilizes the outer membrane of T. pallidum. Green (Alexa Fluor 488) fluorescence was observed in the absence of detergent, indicating that the TP0136 protein was localized to the outer membrane (Fig. 3). As expected, red (Alexa Fluor 546) fluorescence, corresponding to binding of rat anti-FlaA antibodies, was observed only in the presence of detergent. No signal was observed when normal rabbit serum was used as a control (data not shown). These observations were confirmed using immunoelectron microscopy (22). Gold particles that were conjugated to anti-rTP0136 antibody were found to bind to the spirochete when the antibody was used in the absence of detergent, suggesting an outer membrane location for TP0136 protein (Fig. 4A). Additionally, few to no gold particles could be detected in association with treponemes by using preimmune sera in the presence or absence of detergent (Fig. 4B) or anti-FlaA antibody in the absence of detergent (Fig. 4C). Therefore, the TP0136 protein appears to be a surface-exposed protein.
FIG. 3.
Double label immunofluorescence microscopy following incubation with anti-rTP0136 and anti-FlaA antibodies with Percoll-purified T. pallidum in the absence (A) or presence (B) of 0.1% Triton X-100 detergent. Antibody binding of anti-rTP0136 antibody was detected using Alexa Fluor 488 goat anti-rabbit IgG (green), and antibody binding of anti-FlaA was detected using Alexa Fluor 546 goat anti-rat IgG (red).
FIG. 4.
Immunoelectron microscopy on T. pallidum cells following incubation with or without detergent and immunostaining with anti-rTP0136 antibody (A), the preimmune serum negative control (B), or anti-FlaA (C), followed by incubation with secondary antibody conjugated with gold particles. Bar length is 0.1 μm. TX-100, Triton X-100.
Detection of serum antibodies against TP0136 in infected rabbits and humans.
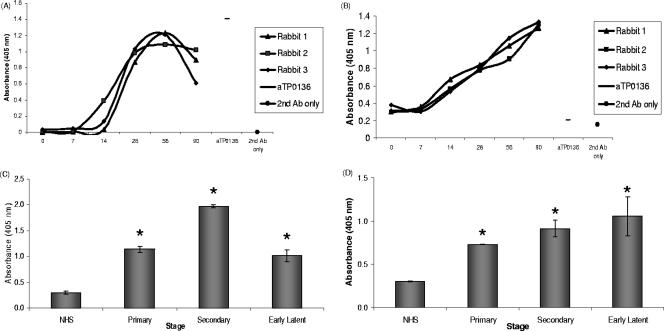
To determine if TP0136 protein is expressed in vivo and is a target of the immune response in infected hosts, the rabbit model of syphilitic infection was utilized. Three rabbits were infected with Treponema pallidum subsp. pallidum (Nichols strain), and sera were collected at days 0, 7, 14, 28, 56, and 90 postinoculation. ELISA analysis revealed that sera at days 14, 28, 56, and 90 postinoculation contained antibodies reactive with the rTP0136 protein and with the T. pallidum protein lysate (Fig. 5A and B), but no antibodies to rTP0136 were present in preinoculation (day 0) sera (Fig. 5A). There was a slight decrease in reactivity at day 90, which is consistent with our previous data; this result suggests that the antibody response to TP0136 is higher in the early stages of experimental infection.
FIG. 5.
Reactivity of serum antibodies (Ab) from experimentally infected rabbits and human syphilis patients with purified rTP0136 protein, as determined by ELISA. (A) Reactivity of sera from individual rabbits (each indicated by a separate line) collected at sequential time points during an experimental T. pallidum infection with rTP0136 protein. Prechallenge rabbit sera (zero time) were used as a negative control, and anti-rTP0136 (aTP0136) antibody was used as a positive control for ELISA well coating. (B) Reactivity of individual rabbit sera to a protein lysate of T. pallidum was used as a control for the sensitivity of the rabbit sera used for panel A. As in panel A, prechallenge rabbit sera (zero time) were used as a negative control and anti-rTP0136 (aTP0136) was used as a positive control for plate coating. (C) Purified rTP0136 reactivity with immunoglobulin from sera pooled from patients with primary, secondary, and early latent syphilis. NHS were used as a negative control. (D) Reactivity of pooled human sera to the T. pallidum protein lysate was used as a positive control for the reactivity of the sera used for panel C. As in panel C, NHS were used as a negative control. Human serum samples were diluted 1:200, and rabbit samples were diluted 1:100. Statistical significance (P < 0.005) in comparison with negative controls (NHS) or prechallenge sera by the Student two-tailed t test is indicated by an asterisk.
Previously, a preliminary analysis was performed with sera from patients with syphilis for reactivity against the proteome of T. pallidum, including unpurified, glutathione S-transferase-tagged TP0136 protein (4). To extend this study, purified rTP0136 protein was immobilized and tested for binding to antibodies pooled from patients with primary, secondary, and early latent syphilis. As indicated by ELISA experiments, the purified rTP0136 protein and the T. pallidum lysate protein were recognized by pooled sera from patients at the primary, secondary, and early latent disease stages (Fig. 5C and D). An irrelevant His-tagged control protein, NarP, exhibited low reactivity for all sera tested, indicating that antibody binding is specific (data not shown). Taken together, the rabbit and human serologic data indicate that the TP0136 protein is expressed in vivo and is a target of the immune response in infected hosts.
Adherence of TP0136 to human ECM components.
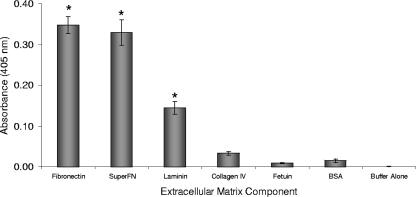
Preliminary genome-scale phage display experiments were carried out using an M13 phage expressing each of the T. pallidum proteins predicted to have a signal sequence linked to the gene III protein and testing for binding of the resulting phages to proteins of interest by ELISA. The results of this preliminary study suggested that the TP0136 protein may bind to the human ECM protein fibronectin (J. Petterson, unpublished data). Therefore, purified rTP0136 protein was also tested for binding to various ECM components previously identified as potentially involved in T. pallidum adherence (Fig. 6). For these experiments, serum fibronectin, superfibronectin (a combination of human plasma fibronectin and recombinant human fibronectin fragment III-C that is thought to result in multimers that better resemble matrix fibrils [35]), laminin, collagen IV, fetuin, and BSA were immobilized in microtiter wells and probed with purified rTP0136 protein. Binding of rTP0136 protein was detected with an anti-His6 tag antibody. rTP0136 protein bound significantly to fibronectin, superfibronectin, and laminin, as indicated by a P value of <0.005 in comparison with binding to the negative-control BSA protein by the Student two-tailed t test (Fig. 6). An irrelevant His-tagged control protein, NarP, was included as a control in this assay and exhibited low reactivity values, indicating that the binding is specific to the TP0136 portion of the protein (data not shown). The binding also appeared to be specific in that no significant binding to BSA, fetuin, or collagen IV was observed.
FIG. 6.
Binding of purified rTP0136 protein with purified ECM proteins as measured by ELISA. The non-ECM proteins fetuin and BSA were used as negative controls. Statistical significance (P < 0.005) in comparison with binding to the negative control BSA by the Student two-tailed t test is indicated by an asterisk.
Assessment of protection of rabbits against T. pallidum infection by immunization with TP0136.
Finally, because the TP0136 protein is an antigen localized to the outer membrane and a putative fibronectin-binding protein, it was of interest to assess the ability of rTP0136 protein to act as a protective antigen to subsequent challenge with infectious T. pallidum in the rabbit model of infection. New Zealand White rabbits were immunized with rTP0136/adjuvant and subsequently challenged with Treponema pallidum subsp. pallidum (Nichols strain). Rabbits immunized with rTP0136 showed a significant delay in time to ulceration (Table 1), but no significant difference in the time of lesion development was observed. Also, representative lesions from both immunized and nonimmunized animals were consistently positive for spirochetes, as determined by dark-field microscopy of needle aspirates. An adjuvant-alone control was not performed in this study. However, in other experiments, adjuvant alone has not had an effect on lesion development (M. McGill and S. Norris, unpublished results). These results indicated that immunization with rTP0136 protein did not prevent infection but altered the course of lesion development.
TABLE 1.
Syphilitic lesion development following intradermal challenge of rTP0136-immunized or control rabbits with 104 T. pallidum cells
| Immunization antigen | No. of lesions/no. of sites | Day (mean ± SD) of:
|
|
|---|---|---|---|
| Lesion appearance | Ulcerationa | ||
| None | 8/8 | 11.5 ± 0.5 | 18.0 ± 4.5 |
| None | 8/8 | 12.0 ± 1.3 | 18.4 ± 2.5 |
| None | 6/8 | 9.3 ± 1.4 | 18.2 ± 2.4 |
| rTP0136 | 8/8 | 11.3 ± 1.5 | 25.8 ± 4.1* |
| rTP0136 | 8/8 | 10.8 ± 0.9 | 21.8 ± 3.1* |
| rTP0136 | 8/8 | 11.5 ± 0.5 | 23.5 ± 0.9* |
Statistical significance (*) indicates a P value of <0.05 for comparison with nonimmunized negative controls by the Student two-tailed t test.
DISCUSSION
The molecular mechanisms of T. pallidum pathogenesis are poorly understood (37). This is in large part because the organism is an obligate human pathogen that has not been cultured continuously in vitro (37). The inability to culture T. pallidum precludes the use of many experimental approaches, including gene inactivation studies. Therefore, different methods are needed to answer questions about the biology and pathogenesis of this organism. Studies that shed light on the cell surface of T. pallidum are particularly useful because surface proteins are likely to interface with the host immune response as well as potentially act as virulence factors. Several reports indicate that antibodies generated from a T. pallidum infection can block attachment to host cells (18) and promote macrophage-mediated phagocytosis (1). However, identifying outer membrane proteins responsible for these observations has been a challenge. This study has confirmed that the Treponema pallidum protein TP0136 induces an antibody response during human or rabbit infection (Fig. 5) and may play a role in adherence to ECM components (Fig. 6). Consistent with these findings, it was also observed that the TP0136 protein is localized to the outer membrane of the treponeme (Fig. 3 and 4).
T. pallidum is known to adhere to many cell types, although the mechanism by which T. pallidum attaches to and invades human tissue is only partially understood (16, 17, 25). T. pallidum has been shown to bind to the ECM proteins fibronectin and laminin (39, 49), analogous to related spirochetes Treponema denticola (51) and Borrelia burgdorferi (21). Recent studies suggest that the T. pallidum proteins TP0155 and TP0483 bind fibronectin (7) and that TP0751 binds laminin (6). Due to the likely possibility of redundant functions among T. pallidum adhesins, the identification of TP0136 protein as an additional putative adhesin is a step toward understanding T. pallidum adherence.
Bacterial lipoproteins contain an N-terminal signal peptide of approximately 19 amino acids (27), characterized at the extreme N terminus by 1 to 3 positively charged amino acids followed by a stretch of hydrophobic and neutral amino acids and then the consensus cleavage site for SPaseII ([L,V,I]−3[A,S,T,G]−2[G,A]−1C+1) (14). Determinants of lipoprotein localization are well understood for E. coli, where the presence of an aspartate at position +2 of the mature protein localizes the lipoprotein to the inner membrane, and seemingly any other amino acid at position +2 will lead to the lipoprotein localization to the bacterial outer membrane (55). While the membrane architecture and lipoprotein sorting of T. pallidum are markedly different from those of other gram-negative bacteria (5), TP0136 was predicted to contain a lipoprotein signal peptide and a SPaseII cleavage site between amino acids 23 and 24 (SLLTT CDFTG). TP0136 contains an aspartate at position +2, but nevertheless, the experimental results indicate that TP0136 is localized to the outer membrane of the spirochete. T. pallidum does not contain all of the components of the E. coli lipoprotein sorting system (5), however, and an examination of known spirochetal lipoprotein sorting did not reveal a correlation between localization and the amino acid at position +2 (43, 45). It has been shown that the disulfide environment around the sorting signal can influence lipoprotein localization (41), and Cullen et al. have speculated that conformation may direct localization more than amino acid sequence in spirochetes (14). The localization of TP0136 provides another example of the unusual properties of the outer membrane of T. pallidum.
As shown in Table 1, immunization with the rTP0136 protein resulted in a statistically significant delay in ulceration compared to what was found for control rabbits but did not lead to protection from infection with T. pallidum in the rabbit model system. The rTP0136 protein was shown to react more strongly to human sera than rabbit sera in ELISAs (Fig. 5), which may be due to inherent differences between human infection and the rabbit model system. It could also be that the single-challenge dose amount was too high in comparison to that in natural infection to allow for a sensitive assay. This is likely to be the case if immunization with TP0136 protein interferes with the initial step of attachment, since an unrealistically high challenge dose would allow some organisms to attach and proceed to the next stages of infection despite the presence of antibody. However, the fact that the immunized rabbits had a statistically significant delay in time to chancre formation suggests that immunization inhibited lesion development to some extent.
It has been shown that the tprK gene sequence is heterogeneous within and among T. pallidum isolates and that the tprK sequences change during infection and passage by an apparent gene conversion mechanism (9). Based on these results, it was proposed that T. pallidum has a mechanism for antigenic variation that may be important for immune evasion (9). Sequence heterogeneity is also observed in the TP0136 gene among different strains of T. pallidum. Interestingly, the TP0136 amino acid sequence contains multiple serine and glycine repeat tracts. These tracts are conserved among the different strains of T. pallidum but not among the paralogous genes of TP0136 (Fig. 2). Repeat sequence tracts are characteristic of proteins that are intrinsically unstructured or disordered (52), and it has been suggested that repetitive segments encode fundamental functions which are shaped by intense evolutionary pressure (50). Multiple runs of amino acids are uncommon in prokaryotic genomes (28), but a glycine repeat containing fibronectin-binding protein was described for Mycobacterium tuberculosis (15). The genome of the eukaryotic parasite P. falciparum contains several proteins rich in homorepeats (28), some of which are strong antigens and are being developed as a vaccine (10). Finally, it is of interest that the sequence of TP0136 from the rabbit pathogen T. paraluiscuniculi does not align well with the sequences from the human pathogens T. pallidum subsp. pallidum Street strain 14 and T. pallidum subsp. pallidum Nichols. The mechanism of species specificity that differentiates these similar pathogens is not yet known. Sequence heterogeneity present at the DNA and amino acid levels among proteins such as TP0136 may play a role in this mechanism.
The goal of syphilis eradication has been hindered by the lack of a vaccine. However, analysis of functional genomic data has led to the identification of potential candidate antigens that may allow for future vaccine development. Previous studies have found partial protection from challenge with Treponema pallidum by immunization with recombinant tprK protein (34). Similarly, we found that immunization with rTP0136 protein inhibited some aspects of lesion development in the rabbit model system. While the TP0136 gene is extremely variable, there are regions that are completely conserved among strains. Therefore, it is possible that further work with this protein may explain the lack of full protection and determine if this protein would be a good candidate in a multiple-subunit vaccine.
Understanding the mechanisms of pathogenesis of T. pallidum has been difficult due to the lack of an in vitro culture system. With the publication of the genome sequence, the development of new techniques became possible. The use of a T. pallidum clone set allowed the identification of TP0136 as an antigenic protein (4), and analysis of the location and ability to bind ECM proteins suggests that TP0136 is of interest with respect to host interactions.
Acknowledgments
This work was supported by NIH grants R01 AI45842 (T.P.) and R03 AI69107 (S.J.N.). This work was partly supported by grants from the Grant Agency of the Czech Republic (310/07/0321), the Ministry of Health of the Czech Republic (NR8967-4/2006), and the Ministry of Education of the Czech Republic (VZ MSM0021622415) to D..
Editor: R. P. Morrison
Footnotes
Published ahead of print on 10 March 2008.
REFERENCES
- 1.Baker-Zander, S. A., E. W. Hook III, P. Bonin, H. H. Handsfield, and S. A. Lukehart. 1985. Antigens of Treponema pallidum recognized by IgG and IgM antibodies during syphilis in humans. J. Infect. Dis. 151264-272. [DOI] [PubMed] [Google Scholar]
- 2.Baseman, J. B., J. C. Nichols, J. W. Rumpp, and N. S. Hayes. 1974. Purification of Treponema pallidum from infected rabbit tissue: resolution into two treponemal populations. Infect. Immun. 101062-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanco, D. R., C. I. Champion, A. Dooley, D. L. Cox, J. P. Whitelegge, K. Faull, and M. A. Lovett. 2005. A monoclonal antibody that conveys in vitro killing and partial protection in experimental syphilis binds a phosphorylcholine surface epitope of Treponema pallidum. Infect. Immun. 733083-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brinkman, M. B., M. McKevitt, M. McLoughlin, C. Perez, J. Howell, G. M. Weinstock, S. J. Norris, and T. Palzkill. 2006. Reactivity of antibodies from syphilis patients to a protein array representing the Treponema pallidum proteome. J. Clin. Microbiol. 44888-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cameron, C. 2006. The T. pallidum outer membrane and outer membrane proteins, p. 237-266. In J. D. Radolf and S. Lukehart (ed.), Pathogenic Treponema. Caister Academic Press, Norfolk, VA.
- 6.Cameron, C. E., N. L. Brouwer, L. M. Tisch, and J. M. Kuroiwa. 2005. Defining the interaction of the Treponema pallidum adhesin Tp0751 with laminin. Infect. Immun. 737485-7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cameron, C. E., E. L. Brown, J. M. Kuroiwa, L. M. Schnapp, and N. L. Brouwer. 2004. Treponema pallidum fibronectin-binding proteins. J. Bacteriol. 1867019-7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campanella, J. J., L. Bitincka, and J. Smalley. 2003. MatGAT: an application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinformatics 429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Centurion-Lara, A., R. E. LaFond, K. Hevner, C. Godornes, B. J. Molini, W. C. Van Voorhis, and S. A. Lukehart. 2004. Gene conversion: a mechanism for generation of heterogeneity in the tprK gene of Treponema pallidum during infection. Mol. Microbiol. 521579-1596. [DOI] [PubMed] [Google Scholar]
- 10.Chiang, P. K., J. M. Bujnicki, X. Su, and D. E. Lanar. 2006. Malaria: therapy, genes and vaccines. Curr. Mol. Med. 6309-326. [DOI] [PubMed] [Google Scholar]
- 11.Cox, D. L., D. R. Akins, S. F. Porcella, M. V. Norgard, and J. D. Radolf. 1995. Treponema pallidum in gel microdroplets: a novel strategy for investigation of treponemal molecular architecture. Mol. Microbiol. 151151-1164. [DOI] [PubMed] [Google Scholar]
- 12.Cox, D. L., and J. D. Radolf. 2006. Metabolism of the Treponema, p. 61-100. In J. D. Radolf and S. Lukehart (ed.), Pathogenic Treponema. Caister Academic Press, Norfolk, VA.
- 13.Cullen, P. A., and C. E. Cameron. 2006. Progress towards an effective syphilis vaccine: the past, present and future. Expert Rev. Vaccines 567-80. [DOI] [PubMed] [Google Scholar]
- 14.Cullen, P. A., D. A. Haake, and B. Adler. 2004. Outer membrane proteins of pathogenic spirochetes. FEMS Microbiol. Rev. 28291-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Espitia, C., J. P. Laclette, M. Mondragon-Palomino, A. Amador, J. Campuzano, A. Martens, M. Singh, R. Cicero, Y. Zhang, and C. Moreno. 1999. The PE-PGRS glycine-rich proteins of Mycobacterium tuberculosis: a new family of fibronectin-binding proteins? Microbiology 1453487-3495. [DOI] [PubMed] [Google Scholar]
- 16.Fitzgerald, T. J., R. C. Johnson, J. N. Miller, and J. A. Sykes. 1977. Characterization of the attachment of Treponema pallidum (Nichols strain) to cultured mammalian cells and the potential relationship of attachment to pathogenicity. Infect. Immun. 18467-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fitzgerald, T. J., J. N. Miller, and J. A. Sykes. 1975. Treponema pallidum (Nichols strain) in tissue cultures: cellular attachment, entry, and survival. Infect. Immun. 111133-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fitzgerald, T. J., L. A. Repesh, D. R. Blanco, and J. N. Miller. 1984. Attachment of Treponema pallidum to fibronectin, laminin, collagen IV, and collagen I, and blockage of attachment by immune rabbit IgG. Br. J. Vener. Dis. 60357-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fleming, D. T., and J. N. Wasserheit. 1999. From epidemiological synergy to public health policy and practice: the contribution of other sexually transmitted diseases to sexual transmission of HIV infection. Sex. Transm. Infect. 753-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fraser, C. M., S. J. Norris, G. M. Weinstock, O. White, G. G. Sutton, R. Dodson, M. Gwinn, E. K. Hickey, R. Clayton, K. A. Ketchum, E. Sodergren, J. M. Hardham, M. P. McLeod, S. Salzberg, J. Peterson, H. Khalak, D. Richardson, J. K. Howell, M. Chidambaram, T. Utterback, L. McDonald, P. Artiach, C. Bowman, M. D. Cotton, C. Fujii, S. Garland, B. Hatch, K. Horst, K. Roberts, M. Sandusky, J. Weidman, H. O. Smith, and J. C. Venter. 1998. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science 281375-388. [DOI] [PubMed] [Google Scholar]
- 21.Grab, D. J., C. Givens, and R. Kennedy. 1998. Fibronectin-binding activity in Borrelia burgdorferi. Biochim. Biophys. Acta 1407135-145. [DOI] [PubMed] [Google Scholar]
- 22.Haapasalo, M., K. H. Muller, V. J. Uitto, W. K. Leung, and B. C. McBride. 1992. Characterization, cloning, and binding properties of the major 53-kilodalton Treponema denticola surface antigen. Infect. Immun. 602058-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanff, P. A., S. J. Norris, M. A. Lovett, and J. N. Miller. 1984. Purification of Treponema pallidum, Nichols strain, by Percoll density gradient centrifugation. Sex. Transm. Dis. 11275-286. [DOI] [PubMed] [Google Scholar]
- 24.Hardy, P. H., Jr., and J. Levin. 1983. Lack of endotoxin in Borrelia hispanica and Treponema pallidum. Proc. Soc. Exp. Biol. Med. 17447-52. [DOI] [PubMed] [Google Scholar]
- 25.Hayes, N. S., K. E. Muse, A. M. Collier, and J. B. Baseman. 1977. Parasitism by virulent Treponema pallidum of host cell surfaces. Infect. Immun. 17174-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joh, D., E. R. Wann, B. Kreikemeyer, P. Speziale, and M. Hook. 1999. Role of fibronectin-binding MSCRAMMs in bacterial adherence and entry into mammalian cells. Matrix Biol. 18211-223. [DOI] [PubMed] [Google Scholar]
- 27.Juncker, A. S., H. Willenbrock, G. Von Heijne, S. Brunak, H. Nielsen, and A. Krogh. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 121652-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalita, M. K., G. Ramasamy, S. Duraisamy, V. S. Chauhan, and D. Gupta. 2006. ProtRepeatsDB: a database of amino acid repeats in genomes. BMC Bioinformatics 7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LaFond, R. E., A. Centurion-Lara, C. Godornes, W. C. Van Voorhis, and S. A. Lukehart. 2006. TprK sequence diversity accumulates during infection of rabbits with Treponema pallidum subsp. pallidum Nichols strain. Infect. Immun. 741896-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, Q., M. Z. Li, D. Leibham, D. Cortez, and S. J. Elledge. 1998. The univector plasmid-fusion system, a method for rapid construction of recombinant DNA without restriction enzymes. Curr. Biol. 81300-1309. [DOI] [PubMed] [Google Scholar]
- 31.McKevitt, M., M. B. Brinkman, M. McLoughlin, C. Perez, J. K. Howell, G. M. Weinstock, S. J. Norris, and T. Palzkill. 2005. Genome scale identification of Treponema pallidum antigens. Infect. Immun. 734445-4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKevitt, M., K. Patel, D. Smajs, M. Marsh, M. McLoughlin, S. J. Norris, G. M. Weinstock, and T. Palzkill. 2003. Systematic cloning of Treponema pallidum open reading frames for protein expression and antigen discovery. Genome Res. 131665-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, J. N. 1973. Immunity in experimental syphilis. VI. Successful vaccination of rabbits with Treponema pallidum, Nichols strain, attenuated by gamma-irradiation. J. Immunol. 1101206-1215. [PubMed] [Google Scholar]
- 34.Morgan, C. A., S. A. Lukehart, and W. C. Van Voorhis. 2003. Protection against syphilis correlates with specificity of antibodies to the variable regions of Treponema pallidum repeat protein K. Infect. Immun. 715605-5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morla, A., Z. Zhang, and E. Ruoslahti. 1994. Superfibronectin is a functionally distinct form of fibronectin. Nature 367193-196. [DOI] [PubMed] [Google Scholar]
- 36.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 101-6. [DOI] [PubMed] [Google Scholar]
- 37.Norris, S. J., D. L. Cox, and G. M. Weinstock. 2001. Biology of Treponema pallidum: correlation of functional activities with genome sequence data. J. Mol. Microbiol. Biotechnol. 337-62. [PubMed] [Google Scholar]
- 38.Notredame, C., D. G. Higgins, and J. Heringa. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302205-217. [DOI] [PubMed] [Google Scholar]
- 39.Peterson, K. M., J. B. Baseman, and J. F. Alderete. 1983. Treponema pallidum receptor binding proteins interact with fibronectin. J. Exp. Med. 1571958-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Radolf, J. D., M. V. Norgard, and W. W. Schulz. 1989. Outer membrane ultrastructure explains the limited antigenicity of virulent Treponema pallidum. Proc. Natl. Acad. Sci. 862051-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robichon, C., M. Bonhivers, and A. P. Pugsley. 2003. An intramolecular disulphide bond reduces the efficacy of a lipoprotein plasma membrane sorting signal. Mol. Microbiol. 491145-1154. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 43.Schulze, R. J., and W. R. Zuckert. 2006. Borrelia burgdorferi lipoproteins are secreted to the outer surface by default. Mol. Microbiol. 591473-1484. [DOI] [PubMed] [Google Scholar]
- 44.Sellati, T. J., D. A. Wilkinson, J. S. Sheffield, R. A. Koup, J. D. Radolf, and M. V. Norgard. 2000. Virulent Treponema pallidum, lipoprotein, and synthetic lipopeptides induce CCR5 on human monocytes and enhance their susceptibility to infection by human immunodeficiency virus type 1. J. Infect. Dis. 181283-293. [DOI] [PubMed] [Google Scholar]
- 45.Setubal, J. C., M. Reis, J. Matsunaga, and D. A. Haake. 2006. Lipoprotein computational prediction in spirochaetal genomes. Microbiology 152113-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smajs, D., M. McKevitt, J. K. Howell, S. J. Norris, W. W. Cai, T. Palzkill, and G. M. Weinstock. 2005. Transcriptome of Treponema pallidum: gene expression profile during experimental rabbit infection. J. Bacteriol. 1871866-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strouhal, M., D. Smajs, P. Matejkova, E. Sodergren, A. G. Amin, J. K. Howell, S. J. Norris, and G. M. Weinstock. 2007. Genome differences between Treponema pallidum subsp. pallidum strain Nichols and T. paraluiscuniculi strain Cuniculi A. Infect. Immun. 755859-5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun, E. S., B. J. Molini, L. K. Barrett, A. Centurion-Lara, S. A. Lukehart, and W. C. Van Voorhis. 2004. Subfamily I Treponema pallidum repeat protein family: sequence variation and immunity. Microbes Infect. 6725-737. [DOI] [PubMed] [Google Scholar]
- 49.Thomas, D. D., J. B. Baseman, and J. F. Alderete. 1985. Putative Treponema pallidum cytadhesins share a common functional domain. Infect. Immun. 49833-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tompa, P. 2003. Intrinsically unstructured proteins evolve by repeat expansion. Bioessays 25847-855. [DOI] [PubMed] [Google Scholar]
- 51.Umemoto, T., Y. Nakatani, Y. Nakamura, and I. Namikawa. 1993. Fibronectin-binding proteins of a human oral spirochete Treponema denticola. Microbiol. Immunol. 3775-78. [DOI] [PubMed] [Google Scholar]
- 52.Weathers, E. A., M. E. Paulaitis, T. B. Woolf, and J. H. Hoh. 2004. Reduced amino acid alphabet is sufficient to accurately recognize intrinsically disordered protein. FEBS Lett. 576348-352. [DOI] [PubMed] [Google Scholar]
- 53.Wishart, D. S., P. Stothard, and G. H. Van Domselaar. 2000. PepTool and GeneTool: platform-independent tools for biological sequence analysis. Methods Mol. Biol. 13293-113. [DOI] [PubMed] [Google Scholar]
- 54.World Health Organization. 1999. Global prevalence and incidence of selected curable sexually transmitted diseases: overview and estimates. World Health Organization, Geneva, Switzerland.
- 55.Yamaguchi, K., F. Yu, and M. Inouye. 1988. A single amino acid determinant of the membrane localization of lipoproteins in E. coli. Cell 53423-432. [DOI] [PubMed] [Google Scholar]