Abstract
Burkholderia cenocepacia is an important opportunistic pathogen causing serious chronic infections in patients with cystic fibrosis (CF). Adaptation of B. cenocepacia to the CF airways may play an important role in the persistence of the infection. We have identified a sensor kinase-response regulator (BCAM0379) named AtsR in B. cenocepacia K56-2 that shares 19% amino acid identity with RetS from Pseudomonas aeruginosa. atsR inactivation led to increased biofilm production and a hyperadherent phenotype in both abiotic surfaces and lung epithelial cells. Also, the atsR mutant overexpressed and hypersecreted an Hcp-like protein known to be specifically secreted by the type VI secretion system (T6SS) in other gram-negative bacteria. Amoeba plaque assays demonstrated that the atsR mutant was more resistant to Dictyostelium predation than the wild-type strain and that this phenomenon was T6SS dependent. Macrophage infection assays also demonstrated that the atsR mutant induces the formation of actin-mediated protrusions from macrophages that require a functional Hcp-like protein, suggesting that the T6SS is involved in actin rearrangements. Three B. cenocepacia transposon mutants that were found in a previous study to be impaired for survival in chronic lung infection model were mapped to the T6SS gene cluster, indicating that the T6SS is required for infection in vivo. Together, our data show that AtsR is involved in the regulation of genes required for virulence in B. cenocepacia K56-2, including genes encoding a T6SS.
The Burkholderia cepacia complex (BCC) is a group of at least nine closely related gram-negative bacterial species. Ubiquitously present in the environment (3), BCC bacteria are also important opportunistic human pathogens that establish chronic lung infections in patients with chronic granulomatous disease and, most commonly, with cystic fibrosis (CF) (19). In some cases infection is further complicated by the “cepacia syndrome,” which is characterized by an often fatal rapid lung deterioration, acute necrotizing pneumonia, and septicemia (24). BCC infections are of great concern to the CF community due to the intrinsic resistance of these bacteria to most clinically relevant antimicrobial agents (19) and their ability to be transmitted from person to person (18, 46). In most CF centers worldwide and more particularly in Canada, B. cenocepacia is the most common BCC species recovered from patients (42, 48) and is frequently associated with the most severe infections (31).
Adaptation of B. cenocepacia to the environment of the CF airways probably plays an important role in the establishment of a chronic infection. Sensing and adaptation to new environmental conditions by bacteria is commonly governed by two-component regulatory systems leading to modification of gene expression patterns required for bacterial survival (50). Two-component systems usually consist of a membrane-associated sensor (histidine kinase protein) that monitors environmental signals and a response regulator (receiver) whose function is modulated by phosphotransfer from its cognate histidine kinase (2, 55). Sensor histidine kinases respond to a wide range of signals, including those encountered during infection.
A global virulence regulator, PA4856 (RetS), has recently been identified in Pseudomonas aeruginosa (16). The retS gene encodes an unusual two-component regulator, containing a periplasmic sensor domain linked by a seven-transmembrane-spanning region to a histidine domain fused with two response regulator domains. RetS inversely controls the expression of genes associated with acute and chronic infections (16). Deletion of retS results in overexpression of two exopolysaccharide biosynthetic gene clusters (pel and psl), leading to a hyperadhesive phenotype on eukaryotic cell surfaces and increased biofilm production. Despite demonstrating strong adherence to eukaryotic cells, P. aeruginosa retS mutants do not respond to host cell contact, which normally activates the expression of genes encoding the type III secretion system (TTSS). Thus, retS mutants are not cytotoxic for tissue culture cells and are severely attenuated for virulence in an acute murine pneumonia model (16). Microarray analyses revealed that RetS is also involved in the downregulation of a virulence locus encoding a type VI secretion system (T6SS) (34).
The newly described T6SS is conserved in numerous gram-negative pathogens that interact closely with eukaryotic cells (8, 9, 41) and it has been recognized as an important contributor to pathogenesis in many bacteria (34, 40, 44, 51, 57). Little is known about the structure and organization of the T6SS apparatus, but several proteins specifically secreted by the T6SS have been identified. One such protein shares sequence similarity to the hemolysin-coregulated protein (Hcp) from Vibrio cholerae (56). Hcp appears to be secreted by most of the T6SSs studied to date, and secretion of Hcp is the hallmark of a functional T6SS in many bacterial species (39). The function of Hcp-like proteins remains unknown. In Aeromonas hydrophila, Hcp is translocated into eukaryotic cells via the T6SS, and Hcp expression in transfected HeLa cells can induce apoptosis (51). In V. cholerae, Hcp is not cytotoxic by itself but is required for the extracellular secretion of three other proteins, namely, VgrG-1, VgrG-2, and VgrG-3 (40). The function of these proteins is also unknown, but VgrG-1 shares a subdomain with the V. cholerae RtxA toxin that mediates covalent actin cross-linking (39, 45). Recently, the crystal structure of the P. aeruginosa Hcp1 protein was determined (34). Hcp1 forms a hexameric ring with a large internal diameter, and it was hypothesized that Hcp1 may also participate as a structural scaffold in the secretion of other substrates. Protein structure search algorithms predicted that VgrG-related proteins likely assemble into a trimeric complex that is analogous to the baseplate “tail-spike” of bacteriophage T4 (39), differentiating the T6SS from other canonical secretion systems.
We report here the identification of AtsR (adherence and T6SS regulator), a putative sensor kinase hybrid homologous to RetS that controls the expression of hcp and biofilm formation in B. cenocepacia K56-2. Experiments employing amoeba plaque assays and macrophage infection assays demonstrate that an atsR mutant of B. cenocepacia K56-2 utilizes the T6SS to interact with eukaryotic cells. This study describes for the first time the presence of a functional T6SS in B. cenocepacia K56-2 and its importance for the virulence of this bacterium.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture media.
Bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were cultured in Luria broth (LB) (Difco) at 37°C with shaking. Escherichia coli cultures were supplemented as required with the following antibiotics (final concentrations): tetracycline (30 μg ml−1), kanamycin (30 μg ml−1), trimethoprim (50 μg ml−1), and gentamicin (50 μg ml−1). B. cenocepacia cultures were supplemented as required with trimethoprim (100 μg ml−1), tetracycline (100 μg ml−1), and gentamicin (50 μg ml−1). To assess the growth rates of parental and mutant strains of B. cenocepacia K56-2, overnight cultures were inoculated into fresh medium with a starting optical density at 600 nm (OD600) of 0.05. Growth rates were determined in 100-well microtiter plates by use of a Bioscreen C automated microbiology growth curve analysis system (MTX Lab Systems, Inc., Vienna, VA) under high-amplitude shaking conditions.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Source and/or reference |
|---|---|---|
| B. cenocepacia strains | ||
| K56-2 | ET12 clone related to J2315, CF clinical isolate | BCRRCb; 30 |
| DFA21 | K56-2 atsR::pDA27; Tpr | This study |
| DFA27 | K56-2 Δhcp | This study |
| DFA28 | K56-2 Δhcp atsR::pDA27; Tpr | This study |
| E. coli strains | ||
| DH5α | F− Φ80dlacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17(rK− mK+) supE44 thi-1 ΔgyrA96 relA1 | Laboratory stock |
| SY327 | araD Δ(lac pro) argE(Am) recA56 nalA λ pir; Rifr | 33 |
| K. pneumoniae strain | ||
| 18 | Laboratory stock | |
| S. aureus strain | ||
| RN6390 | D. E. Heinrichs | |
| Plasmids | ||
| pME6000 | oripBBR1, Tetr, lacZ, mob-+ | S. Heeb |
| pMLS7-eGFP | oripBBR1, Tpr, mob+, PS7 | 28 |
| pGPΩTp | oriR6K, ΩTpr cassette, mob+ | 14 |
| pGPISce-I | oriR6K, ΩTpr, mob+, containing the ISce-I restriction site | 15 |
| pAp2 | oripBBR1, Cmr, mob+, Pdhfr | S. Cardona |
| pRK2013 | oricolE1, RK2 derivative, Kanr, mob+, tra+ | 13 |
| pDA12 | Cloning vector, oripBBR1, Tetr, mob+, Pdhfr | This study |
| pDA27 | pGPΩTp; 272-bp internal fragment from atsR | This study |
| pAtsR | pDA12; 1.8-kbp atsR fragment | This study |
| pDA42 | pDA12; 0.7-kbp eGFP fragment | This study |
| pDA45 | pGPISce-I with fragments flanking hcp | This study |
| pHcp | pDA12; 0.5-kbp fragment encoding Hcp | This study |
| pDAISce-I | pDA12 encoding the ISce-I homing endonuclease | 15 |
Tpr, trimethoprim resistance; Cmr, chloramphenicol resistance; Kanr, kanamycin resistance; Tetr, tetracycline resistance; Rifr, rifampin resistance.
BCRRC, B. cepacia Research and Referral Repository for Canadian CF Clinics.
General molecular techniques.
DNA manipulations were performed as described previously (43). T4 polynucleotide kinase and T4 DNA ligase (Roche Diagnostics, Laval, Quebec, Canada) were used as recommended by the manufacturers. Transformation of E. coli DH5α was done by the calcium chloride protocol (4). E. coli SY327 carrying the helper plasmid pRK2013 (13) was transformed by electroporation (11), and the transformants were used as donors for conjugation into B. cenocepacia K56-2. DNA amplification by PCR was performed using a PTC-221 DNA engine (MJ Research, Incline Village, NV) with either Taq DNA polymerase or proof start polymerase (Qiagen Inc., Mississauga, Ontario, Canada). DNA sequencing was performed by the Robarts Research Institute DNA sequencing facility at the University of Western Ontario, London, Canada.
RT-PCR and quantitative RT-PCR reactions.
Reverse transcriptase PCR (RT-PCR) was performed as described previously (36) to investigate the transcriptional organization of atsR (BCAM0379) and the neighboring genes BCAM0380 and BCAM0381. Total RNA was isolated from B. cenocepacia K56-2 by use of the RNeasy mini kit (Qiagen) following the manufacturer's protocol and treated with DNase (Qiagen) for 30 min at 37°C and at 75°C for 15 min. Reverse-transcribed cDNAs were amplified using primers 2798 and 2805 (Table 2). Each reverse transcription reaction was performed with RT and without RT (negative control). PCR amplifications with primer pairs 2798-2799 and 2805-2806 were performed using the cDNAs and B. cenocepacia K56-2 genomic DNA (positive control) as the template to amplify the intergenic regions of atsR-BCAM0380 and BCAM0380-BCAM0381, respectively.
TABLE 2.
Oligonucleotide primers
| Primer no. | 5′-3′ primer sequencea | Restriction enzymeb |
|---|---|---|
| 1548 | GAGCTCATCGATTTCGTTCCACTGA | ClaI |
| 1549 | TCATCGATCTGCACTTGAACGTGTGGCC | ClaI |
| 2143 | AGTCGAAGGCATATGTTACACATGCACTTGCAG | NdeI |
| 2165 | TAATCGATGGCTCTGCTGTAGTGAGTGG | ClaI |
| 2166 | CGATCGATGCGAAGAAGTTGTCCATATTG | ClaI |
| 2478 | TTTTTCTAGAGTCAACGAGGGCGTGCTC | XbaI |
| 2479 | CCGTCATGAATTCGCCCAGCGC | EcoRI |
| 2558 | TTTTCATATGCCGCTCGGCGAAGCCAAGT | NdeI |
| 2559 | GTTTTCTAGATCAGGCGAGCAGTGTCTCGA | XbaI |
| 2780 | GTGTTCTAGACCGTCGAATACGCGAAGTG | XbaI |
| 2781 | GCGGCTCGAGCAAACTGCAAGTGCAT | XhoI |
| 2782 | TTTTCTCGAGACGACAAGACCTACG | XhoI |
| 2783 | GGTGAATTCGGGGTAGTCGTC | EcoRI |
| 2798 | GCAGCCAGTATTGGGACACT | N/A |
| 2799 | TGCATCGACGAATACACGTT | N/A |
| 2805 | GACGTGACGATCGTATGCTG | N/A |
| 2806 | ATGGCTACATCCTCGACTGG | N/A |
| 2943 | ATGATCTAGATTAGACCGCGTAGGTCTTGTCGT | XbaI |
| 3143 | AGCTGGCAGTACACGGAAAG | N/A |
| 3136 | GCACGACCATCACGATCTC | N/A |
| 3315 | ACGTTCTCGCTGAAGTACGC | N/A |
| 3318 | CGCGTAGGTCTTGTCGTTCT | N/A |
Restriction endonuclease sites incorporated in the oligonucleotide sequences are underlined.
N/A indicates the absence of restriction site.
Quantitative real-time PCR experiments were performed as follows. Overnight cultures grown at 37°C for 16 h were diluted to an OD600 of 0.05 in 30 ml of prewarmed LB. Diluted cultures of B. cenocepacia K56-2 and the K56-2 atsR::pDA27 mutant, DFA21, were grown at 37°C until exponential phase, harvested, and used for RNA extraction. Total RNA was isolated using a RiboPure-Bacteria kit (Ambion, Austin, TX), according to the manufacturer's instructions. RNA samples were treated with 8 U of DNaseI, which was inactivated using DNase inactivation reagent (Ambion, Austin, TX). The concentration of RNA samples was measured using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE). The histidinol dehydrogenase gene hisD (BCAL0312), which is involved in histidine biosynthesis, was used as a reference standard. The cDNAs from hisD and hcp were generated using the reverse gene-specific primers 3136 and 3318, respectively (Table 2). Reverse transcription reactions were performed in 20-μl mixtures (1× RT buffer, 2 μM hisD reverse primer [3136], 2 μM hcp reverse primer [3318], deoxynucleoside triphosphate at a concentration of 1 mM, 1 U/μl RNase inhibitor, 1 μg of total RNA template). The reverse transcription reactions were performed with 0.75 U/μl of RT and without RT (negative control). To construct standard curves for quantification, PCR products corresponding to internal fragments of hisD and hcp were amplified using primers 3143-3136 and 3315-3318, respectively (Table 2). The PCR products were purified and the concentration was measured using a NanoDrop ND-1000 spectrophotometer. The number of copies per microliter of each product was calculated and dilutions were performed to obtain solutions with a final concentration of 5.105 copies/μl. Then, 10-fold dilutions were performed up to a concentration of 5 copies/μl. Two microliters of each dilution or cDNA was subjected to real-time PCR using iQ SYBR green supermix (Bio-Rad) along with the primer pair 3143-3136 for hisD and the primer pair 3315-3318 for hcp. mRNA expression levels of hisD and hcp in the B. cenocepacia K56-2 and DFA21 strains were quantified on a Rotor-Gene 6000 quantitative multiplex PCR instrument (Corbett, Sydney, Australia) with the following parameters: 95°C for 3 min, followed by 40 PCR cycles consisting of 95°C for 10 s, 55°C for 15 s, and 72°C for 30 s. The concentrations of hcp mRNA in samples were normalized to concentrations of hisD mRNA. The experiment was repeated three times.
Construction of pDA12 and pDA42.
The plasmid pDA12 is a derivative of pAP2 (a pMLBAD derivative [28] containing the dhfr promoter and a cat gene; from S. Cardona, unpublished) in which the chloramphenicol resistance cassette has been replaced by a tetracycline resistance cassette as follows. The tetA and tetR genes were PCR amplified from pME6000 by use of the primers 2165 and 2166, which contain a ClaI restriction site at their 5′ ends (Table 2). The backbone of the pAP2 plasmid was PCR amplified using the Expand high-fidelity PCR system (Roche Diagnostics) and the primer pair 1548 and 1549, which also contained a ClaI restriction site. The resulting amplicons were digested with ClaI and ligated, giving rise to pDA12 (Table 1). The gene encoding the enhanced green fluorescent protein (eGFP) was excised from pMLS7-eGFP and subcloned into pDA12 by use of the restriction enzymes HindIII and EcoRI (Table 1). The resulting plasmid, pDA42, was introduced into B. cenocepacia K56-2 and DFA21 by conjugation, and expression of the eGFP was confirmed by fluorescence microscopy.
Mutagenesis of B. cenocepacia K56-2.
Insertional inactivation of B. cenocepacia K56-2 genes was performed using pGPΩTp, a suicide plasmid encoding resistance to trimethoprim (14). This plasmid is derived from the pGP704 backbone, which carries the Pir protein-dependent R6K origin of replication (25). The sensor kinase regulator gene atsR was mutated by first PCR amplifying a 272-bp internal fragment from B. cenocepacia K56-2 chromosomal DNA by use of the primer pair 2478 and 2479 (Table 2). The PCR cycling conditions used were as follows: 95°C for 5 min followed by 30 cycles of 95°C for 45 s, 60°C for 45 s, and 72°C for 30 s, with a final extension at 72°C for 5 min. The resulting amplicon was digested with the restriction enzymes EcoRI and XbaI and cloned into similarly digested pGPΩTp. The resulting plasmid, pDA27, was introduced into B. cenocepacia K56-2 by conjugation, and the resulting exconjugants were selected for resistance to trimethoprim. Candidate mutants were identified by PCR and confirmed by Southern blot hybridization using the internal fragment labeled with digoxigenin as a probe. The resulting mutant was named B. cenocepacia DFA21 (Table 1).
Deletion of hcp (BCAL0343) encoding an Hcp-like protein (B. cenocepacia K56-2 Δhcp; strain DFA27) was performed using a recently developed system based on the I-SceI homing endonuclease (15). Briefly, this system allows the creation of unmarked and nonpolar mutations and comprises two plasmids. One plasmid, pGPISce-I, serves to clone the regions flanking the gene to be deleted and contains a restriction site for a homing endonuclease. Once introduced by conjugation, the mutagenic plasmid integrates into the B. cenocepacia K56-2 chromosome, giving rise to trimethoprim-resistant mutants. A second plasmid, pDAISce-I (pDA12 derivative encoding the I-SceI endonuclease), is introduced by conjugation. Homing endonucleases catalyze site-specific double-strand breaks in the chromosome at the recognition site. As DNA double-strand breaks are lethal, only mutants undergoing second homologous recombination events, including those with a deletion of the gene of interest, can be recovered. PCR amplifications of regions flanking hcp were performed using 2780-2781 and 2782-2783 primer pairs (Table 2). The amplicons were digested with the restriction enzymes XbaI-XhoI and XhoI-EcoRI, respectively, and cloned into pGPISce-I digested with EcoRI and XbaI, giving rise to pDA45. The resulting mutant was designated DFA27 (Table 1). Deletion of hcp was also performed in combination with atsR inactivation, giving rise to B. cenocepacia DFA28 (the atsR::pDA27 Δhcp mutant). The deletion of hcp was first analyzed by PCR and then confirmed by Southern blot hybridization.
Complementation experiments.
To complement B. cenocepacia DFA21, wild-type atsR was PCR amplified from B. cenocepacia K56-2 with the primer pair 2558 and 2559 with the following thermal cycling conditions: 95°C for 5 min followed by 30 cycles of 95°C for 45 s, 60°C for 45 s, and 72°C for 2 min, with a final extension at 72°C for 10 min. The resulting amplicon was digested with the restriction enzymes NdeI and XbaI and ligated into similarly digested pDA12. The resulting plasmid, pAtsR, was introduced into the mutant by conjugation, and complementation was assessed by the biofilm assay. To complement DFA27 (Δhcp) and DFA28, hcp was PCR amplified using primers 2143 and 2943 cloned as an NdeI and XbaI fragment in pDA12 as indicated above, giving rise to pHcp.
Biofilm formation.
In a modification of the biofilm ring assay (37), overnight cultures grown at 37°C for 16 h were diluted to an OD600 of 0.005 in LB, and triplicate 500-μl aliquots were dispended into polystyrene tubes. Following 24 h of static incubation at 37°C, the medium was removed and the tubes were washed gently with distilled water. Adherent bacteria were stained with 1% (wt/vol) crystal violet and washed three times with distilled water. The bound crystal violet was dissolved in 1 ml of 100% methanol and quantified by measuring the absorbance at 540 nm. The experiment was repeated independently three times.
Adherence assays.
Abiotic adherence assays were performed as follows. Bacteria grown in LB at 37°C for 21 h were washed twice and resuspended in phosphate-buffered saline (PBS; Wisent). Approximately 107 CFU were added in duplicate to the wells of a six-well polystyrene plate (BD Biosciences, Mississauga, Ontario, Canada) containing 2 ml of PBS. Additionally, serial 10-fold dilutions were performed on the starting inoculum, and 100 μl of each dilution was plated on LB agar plates to determine precisely the number of bacteria added to each well (input). The six-well plates were centrifuged for 2 min at 300 × g and incubated at 37°C for 10 min. Each well was washed five times with 2 ml of PBS, and bound bacteria were detached from the surface with the addition of 500 μl of 10 mM EDTA and 1.5 ml of 1% (vol/vol) Triton X-100. Bacteria were then centrifuged at 4,500 × g for 4 min, and the pellet was resuspended in 1 ml PBS. Serial 10-fold dilutions were performed and 100 μl of each dilution was plated on LB agar plates and incubated at 37°C. The colony enumeration was performed the following day to determine the number of bacteria that remained adherent to the six-well plate after the PBS washes (output). The adherence of each strain tested was calculated by dividing the output number by the input number and expressed relative to the adherence of B. cenocepacia K56-2 wild type, which was set as 1. The experiment was repeated independently three times.
A549 lung epithelial cell adherence assays were performed with B. cenocepacia K56-2 and DFA21 carrying pDA42 (expressing eGFP) to facilitate bacterial numeration as follows. Bacterial cultures grown at 37°C for 21 h were washed twice and resuspended in RPMI 1640 (Wisent). Bacteria were added at a starting multiplicity of infection (MOI) of 100:1 in triplicate to A549 cells grown on glass coverslips in six-well tissue culture plates containing 2 ml of Dulbecco's modified Eagle medium (DMEM) with 10% (vol/vol) fetal bovine serum (FBS). No centrifugation was performed, and the plates were incubated at 37°C in a humidified atmosphere of 95% and 5% carbon dioxide (CO2). After 1 h, wells were gently washed five times with 2 ml of RPMI 1640 to remove nonadherent bacteria, and 4′,6′-diamidino-2-phenylindole dihydrochloride (DAPI) (final concentration, 25 μg/ml; Molecular Probes, Invitrogen, Eugene, OR) was added to visualize eukaryotic cell nuclei. The total number of cell-attached bacteria and the total number of cells were counted in 21 fields of view. The experiment was repeated independently three times.
Preparation of culture supernatant proteins and mass spectrometry.
Overnight cultures were diluted to an OD600 of 0.003 in 50 ml of prewarmed LB. After 4 h of incubation at 37°C, the cultures were centrifuged for 15 min at 10,000 × g at 4°C. Culture supernatants were sterilized through a 0.22-μm filter (Millipore), and proteins were precipitated overnight at 4°C with 10% (vol/vol) of trichloroacetic acid (final concentration). The precipitates were isolated by centrifugation at 10,000 × g for 20 min at 4°C and the pellets were washed with ice-cold acetone. Another centrifugation was performed at 10,000 × g for 20 min. The pellets were air dried and then resolubilized by the addition of 0.1 M sodium phosphate buffer, pH 7.0. The protein concentration of each sample was determined by Bradford assay (Bio-Rad), and aliquots containing 10 μg of protein were loaded on a 16% sodium dodecyl sulfate-polyacrylamide electrophoresis (SDS-PAGE) gel. Detection was performed with a Brilliant Blue G-Colloidal staining according to the manufacturer's recommendations (Sigma, St. Louis, MO).
The apparent 21-kDa protein band was excised from a Brilliant Blue G-Colloidal-stained one-dimensional gel loaded with 20 μg of secreted proteins from B. cenocepacia DFA21 by use of an Ettan spot picker (Amersham). In-gel trypsin digestion was performed by the Functional Proteomics Facility of the University of Western Ontario using the Waters MassPREP automated station. Mass spectrometry analysis was performed by the Biological Mass Spectrometry Laboratory of the University of Western Ontario on a Micromass Q-Tof global mass spectrometer equipped with a Z-spray source operating in the positive-ion mode. A 1-hour gradient was used on a C18 column fitted with a trapping column by use of the Waters Nano Acquity UPLC instrument. Data were acquired using MassLynx 4.1 and were submitted for screening via the MASCOT search engine (Matrix Science, London, United Kingdom).
Dictyostelium discoideum plaque assay.
D. discoideum axenic strain AX3 was obtained from the Dicty Stock Center (Columbia University) and grown in HL5 liquid medium at 22°C according to the Dictybase guidelines (http://www.dictybase.org/). D. discoideum cells from mid-log-phase cultures were collected by centrifugation (100 × g, 3 min), washed once, and resuspended in SorC (16.7 mM Na2H-KH2PO4-50 μM CaCl2, pH 6.0) (52). Bacteria were grown in LB for 16 h, pelleted by centrifugation, washed once, and resuspended in SorC. The growth of D. discoideum on a lawn of B. cenocepacia K56-2 cells was determined by plating 100 μl of a suspension containing ∼100 amoebae and 107 CFU on SM agar or on low-nutrient plates, SM/50 and SM/100, which contained 1/50 and 1/100 of the SM stock solution, respectively (52). Quantitative measurements of D. discoideum growth on a lawn of B. cenocepacia cells were obtained by first plating 200 μl of a bacterial suspension at 109 CFU/ml on SM agar. Then, the bacterial lawn was spotted with 5-μl droplets containing serial dilutions of D. discoideum. Plates were incubated at 22°C for 3 days and examined for plaque formation by D. discoideum. Klebsiella pneumoniae 18 was used as a positive control for predation by D. discoideum.
D. discoideum cytotoxicity assay.
D. discoideum cells were prepared as described above. K. pneumoniae 18, Staphylococcus aureus RN6390, and K56-2(pDA12) and DFA21(pDA12) cultures were grown in 5-ml portions of HL5 medium at 37°C for 16 h. The bacteria were pelleted by centrifugation, and culture supernatants were collected and filtered (0.22-μm filter pore size). One milliliter of supernatant was incubated with approximately 500 amoebae and mixed by inversion at room temperature for 1 h. The effect of bacterial culture supernatants on D. discoideum growth was determined by plating 200 μl of the amoebal suspension on SM agar plates on which 200 μl of a bacterial suspension at 109 CFU/ml of K. pneumoniae was previously dispersed. Plates were incubated at 22°C for 5 days and examined for plaque formation by D. discoideum. HL5 medium and S. aureus RN6390 were used as negative and positive controls of cytotoxicity toward D. discoideum, respectively.
Macrophage infection assays and immunofluorescence microscopy.
The C57BL/6 murine bone marrow-derived macrophage cell line ANA-1 was obtained from the Department of Human Genetics, Montreal General Hospital Research Institute, McGill University, Montreal, Quebec, Canada (7). Cells were maintained in DMEM with 10% FBS and grown at 37°C in a humidified atmosphere with 5% CO2. One milliliter of bacterial cultures grown at 37°C for 16 h was washed twice and resuspended in DMEM-10% FBS. Heat-inactivated bacteria were obtained by incubation at 60°C for 25 min prior to infection. Bacteria were added to ANA-1 cells grown on glass coverslips at a MOI of 50:1 and centrifuged for 1 min at 300 × g. After 4 h of incubation at 37°C, infected macrophages were washed three times with 2 ml of DMEM and the coverslips were analyzed immediately or used for immunostaining as follows. Cells were fixed in 4% paraformaldehyde (vol/vol) for 30 min at room temperature and incubated for 10 min with 100 mM glycine in PBS (1×). Cells were then permeabilized with 0.1% Triton X-100 (vol/vol) and blocked with 5% milk powder (wt/vol) for 1 h at room temperature. Permeabilized cells were incubated with primary antibodies followed by secondary antibodies in 5% milk powder for 1 h each at room temperature. Coverslips were mounted onto glass slides by use of fluorescent mounting medium (DakoCytomation). Rabbit anti-B. cenocepacia K56-2 primary antibody was used at a dilution of 1:500, and the Alexa Fluor 647 goat anti-rabbit immunoglobulin G secondary antibody (Molecular Probes, Invitrogen) was used at a dilution of 1:1,000. For fluorescence labeling of actin, macrophages were incubated with one unit of Alexa Fluor 488 phalloidin (Molecular Probes, Invitrogen). Fluorescence and phase-contrast images were acquired using a Qimaging (Burnaby, British Columbia, Canada) cooled charge-coupled-device camera on an Axioscope 2 (Carl Zeiss, Thornwood, NY) microscope with an X100/1.3 numerical aperture Plan-Neofluor objective and a 50-W mercury arc lamp. Images were digitally processed using the Northern Eclipse version 6.0 imaging analysis software (Empix Imaging, Mississauga, Ontario, Canada). Confocal microscopy was performed using a Zeiss LSM 510 META/ConfoCor2 laser scanning confocal microscope and a 100× oil immersion objective. Bacterially induced cell death or damage was verified by using trypan blue staining (Sigma) (final concentration, 1 mg/ml) for 10 min.
RESULTS
AtsR regulates biofilm formation in B. cenocepacia K56-2.
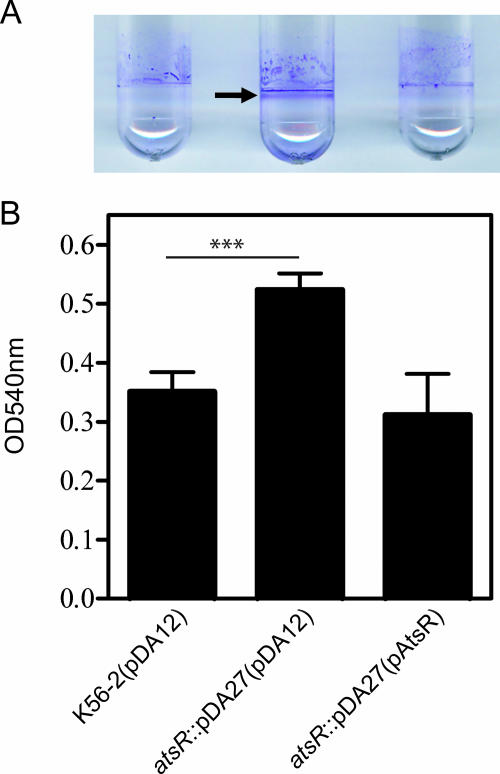
We examined the sequenced genome of B. cenocepacia J2315, which is clonally related to K56-2 (30), to identify genes encoding a homolog of P. aeruginosa RetS (PA4856) (16). BLASTP analysis (http://www.ncbi.nlm.nih.gov/BLAST/) allowed the identification of several genes encoding RetS-like putative response regulators: BCAM0227, BCAM0379, BCAM1505, and BCAM2757. Mutations in each of these putative response regulators were generated by insertional inactivation using the plasmid pGPΩTp, a mutagenesis system leading to the targeted insertion of a mutagenesis plasmid (14, 29). Since retS inactivation in P. aeruginosa leads to increased biofilm production (16), our insertional mutants were screened for biofilm formation. BCAM0227, BCAM1505, and BCAM2757 insertional mutants did not have an apparent biofilm defect in vitro (data not shown). The BCAM0379 insertional mutant formed a strong biofilm ring after 24 h of static incubation (Fig. 1A). For reasons discussed below, we have assigned to BCAM0379 the name atsR (adherence and T6SS regulator). Quantitation of the biofilm mass demonstrated that the atsR::pDA27 mutant, DFA21, produced 50% more biofilm than the wild-type strain (P = 0.0001) (Fig. 1B). Growth curves of wild-type K56-2 and DFA21 in liquid media were similar, suggesting that increased biofilm formation by DFA21 was not due to faster growth (data not shown). Trans-complementation of DFA21 with pAtsR, encoding only AtsR, restored biofilm formation to parental levels (Fig. 1A and B) and also confirmed that the insertion into atsR did not affect the neighboring genes. Moreover, the overexpression of atsR in the wild-type strain, K56-2, led to decreased biofilm formation (data not shown). Together, these results suggest that AtsR acts as a negative regulator of biofilm formation.
FIG. 1.
Biofilm assays performed with B. cenocepacia K56-2 (wild type [WT]) and DFA21 (the atsR::pDA27 mutant) containing the control plasmid pDA12 or pAtsR, as indicated in parentheses. Biofilms were quantified by crystal violet staining after 24 h of static incubation at 37°C. (A) The ring corresponding to robust biofilm formation characteristic of the atsR mutant is indicated by an arrow. (B) The extent of crystal violet retention by adherent bacteria was measured by OD540. Data shown are the means of three independent experiments done in triplicate. Error bars represent the standard deviations. Significant differences were determined using unpaired t tests. The three asterisks indicate that the P value is 0.0001.
The atsR gene is on chromosome 2 and encodes a predicted 606-amino-acid protein that belongs to the sensor kinase-response regulator hybrid family but shares only 19% identity at the primary amino acid sequence with RetS (16). Analysis using SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/) and TMHMM (http://www.cbs.dtu.dk/services/TMHMM-2.0/) predicted a signal peptide sequence and one transmembrane domain at the N terminus, suggesting that AtsR is inserted in the inner membrane. Predicted histidine kinase and ATPase domains following the transmembrane domain were detected using Pfam (http://pfam.sanger.ac.uk/) (1). Unlike RetS, the C terminus of AtsR contains only one predicted response regulator-like receiver domain. However, similar to RetS, AtsR lacks the DNA binding domain characteristic of conventional response regulator proteins, suggesting that AtsR is a component of a multipart signaling pathway. Comparisons with sequences deposited in GenBank showed that AtsR is conserved among the Burkholderia genus. Homologs sharing 95%, 86%, 84%, 66%, and 65% amino acid identity with AtsR from B. cenocepacia are found in B. ambifaria, B. cepacia, B. vietnamiensis, B. thailandensis, and B. pseudomallei species, respectively.
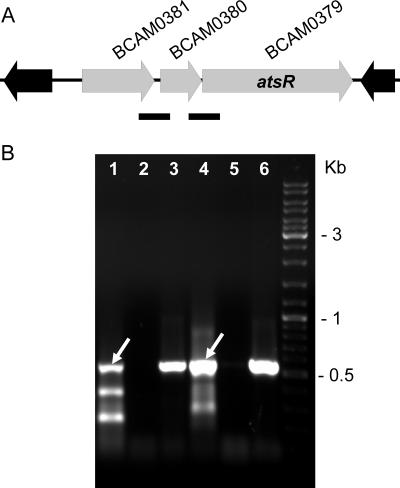
Two genes are located upstream of atsR and in the same transcriptional orientation (Fig. 2A): (i) BCAM0380, encoding a putative periplasmic protein of unknown function containing an oxidoreductase molybdopterin binding domain; and (ii) BCAM0381, encoding a putative cytoplasmic transcriptional regulator containing an N-terminal BetR helix-turn-helix domain and a response regulator domain at the C terminus. RT-PCR experiments performed on the intergenic regions of BCAM0381-BCAM0380 and BCAM0380-atsR revealed that these three genes are cotranscribed (Fig. 2B).
FIG. 2.
Genetic organization and analysis of the gene cluster encoding AtsR in B. cenocepacia J2315 and K56-2. The direction of transcription of each gene is denoted by gray arrows. BCAM gene designations are according to a preliminary annotation of the B. cenocepacia J2315 genome (http://www.sanger.ac.uk/Projects/B_cenocepacia/). (A) Three-gene cluster located on chromosome 2 comprising BCAM0379 (atsR), BCAM0380, and BCAM0381. Black arrows represent genes located at the boundaries of the three-gene cluster containing atsR. Solid black bars indicate the regions analyzed by RT-PCR shown in panel B. (B) RT-PCR analysis of the intergenic regions of BCAM0381-BCAM 0380 and BCAM0380-atsR. Lanes: 1 to 3, RT, no RT, and DNA for BCAM0381-BCAM0380; 4 to 6, RT, no RT, and DNA for BCAM0380-atsR. White arrows indicate bands of expected PCR product size.
Mutation in atsR induces a hyperadhesive phenotype.
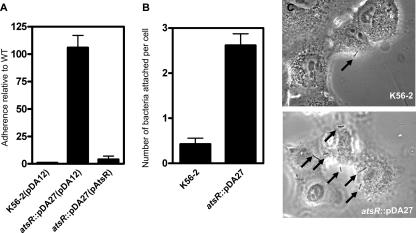
As surface attachment is the first step in biofilm formation, B. cenocepacia DFA21 was tested for adherence on abiotic surfaces by use of an assay that measures the strength of bacterial adhesion after extensive washes with buffer and that also allows a comparison of the adherence levels of different strains within the same six-well plate (see Materials and Methods). For a given strain, the ratio of output to input bacteria can vary from experiment to experiment because of fluctuations in the manual washing procedure. However, the relative adherence of the strains examined in the same plate, compared to that of the wild type in the control well, is conserved between experiments. This adhesion assay revealed that DFA21 is 100-fold more adherent than the wild-type strain, whose adhesion properties allows the adherence of approximately 0.1% of the starting inoculum. As expected, complementation of DFA21 with pAtsR decreased adherence to wild-type levels (Fig. 3A). To determine whether DFA21 displayed similar hyperadhesive properties to human cells, A549 lung epithelial cells were incubated with DFA21 or the wild-type strain. After only 1 h of incubation, DFA21 exhibited a sixfold-increased adherence (P = 0.0002) compared to the parental strain (Fig. 3B and C). Together, these results suggest that the atsR::pDA27 mutant, DFA21, is more adherent than the wild-type strain to both abiotic surfaces and lung epithelial cells.
FIG. 3.
Adhesion assays. (A) Adhesion assay to a polystyrene surface of bacterial strains with the control vector pDA12 and the pAtsR plasmids, as indicated in parentheses. atsR indicates the DFA21 mutant. The adhesion values are shown relative to the value for the control strain, B. cenocepacia K56-2(pDA12), which was set as 1. (B) Adhesion assay on A549 human lung epithelial cells. A549 cells were incubated with B. cenocepacia K56-2 and B. cenocepacia DFA21 (indicated by atsR) for 1 h at 37°C, washed, and stained, and the adherent bacteria were enumerated. Data shown are the means of three independent experiments. Error bars represent the standard deviations. (C) Representative images of A549 cell adherence by B. cenocepacia K56-2 and B. cenocepacia DFA21 (atsR) are shown. Arrows point to adherent bacteria. WT, wild type.
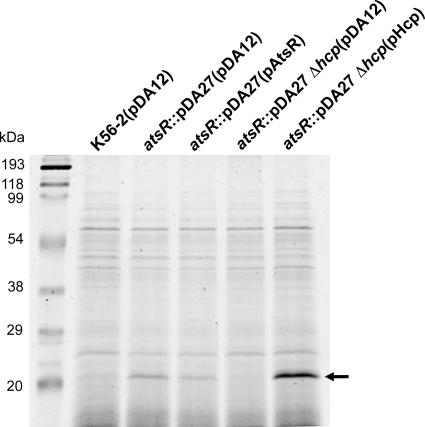
Mutation in atsR increases expression and secretion of an Hcp-like protein.
SDS-PAGE analysis of concentrated culture supernatants revealed a small protein with an apparent mass of ∼21 kDa that was hypersecreted by DFA21 compared to what was seen for the wild type (Fig. 4). DFA21 carrying pAtsR also secreted the 21-kDa protein, albeit at an intermediate level compared to that of wild-type K56-2(pDA12) and DFA21 (pDA12). The apparent mass of the secreted protein, together with the RetS-like properties of AtsR, led us to hypothesize that the 21-kDa polypeptide is an Hcp-like protein. Hcp-like proteins are specifically secreted by the T6SSs of several gram-negative bacteria (34, 40, 44), and Hcp secretion is the hallmark of a functional T6SS activity in many bacterial species (39). In-gel trypsin digestion and mass spectrometry were performed on the 21-kDa protein. Acquired data were submitted for screening via the MASCOT search engine and two peptides were identified, matching with a score of 121 to DUF796, a protein of unknown function corresponding to Hcp.
FIG. 4.
Secretion assay. SDS-PAGE analysis of concentrated culture supernatants recovered from strains containing the control plasmid pDA12 or the plasmids pAtsR and pHcp, as indicated in parentheses. atsR indicates DFA21; atsR Δhcp indicates DFA28. Molecular mass markers in kDa are also indicated. The arrow points to the secreted Hcp polypeptide analysis of concentrated culture supernatants recovered from strains containing the control plasmid pDA12 or the plasmids pAtsR and pHcp, as indicated in parentheses. Molecular mass markers in kDa are also indicated. The arrow points to the position of secreted Hcp polypeptide. WT, wild type.
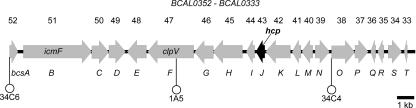
In a previous study, we identified transposon insertions inactivating genes that abolish the infectivity of B. cenocepacia K56-2 in the rat agar bead model of chronic lung infection (22). Further analysis revealed that three of these mutants (1A5, 34C4, and 34C6) had insertions that mapped within genes now referred to as bcsA, bcsF (clpV), and bcsO, which are part of a T6SS gene cluster (Fig. 5). In contrast to other Burkholderia species (44), B. cenocepacia K56-2 contains only one T6SS gene cluster located on chromosome 1, comprising genes organized in three putative transcriptional units (Fig. 5). BCAL0343, located within this cluster, encodes a 167-amino-acid protein orthologous to the Hcp-like proteins secreted by P. aeruginosa (PA0085) and V. cholerae (VCA0017) and has a predicted molecular mass of 18.4 kDa. Similar to other Hcp-like proteins, B. cenocepacia Hcp (BCAL0343) lacks a canonical hydrophobic amino-terminal signal sequence.
FIG. 5.
Genetic map of the T6SS gene cluster. The location of the transposon in the 34C6, 1A5, and 34C4 mutants, which are attenuated for survival in the rat agar bead model (23), is indicated by white circles. This cluster has been designated the bcs cluster for B. cenocepacia survival. The location and direction of transcription of genes are represented by arrows. Locus tags assigned by the Sanger Center are shown above and the bcs annotation of the genes is shown below. The genes icmF (BCAL0351; bcsB), clpV (BCAL0347; bcsF), and hcp (BCAL0343; bcsJ), which are characteristic of T6SSs in other bacteria, are shown.
To confirm that the secreted 21-kDa protein is encoded by hcp, we constructed DFA28 carrying a deletion of hcp in combination with an insertional mutation in atsR. The 21-kDa protein was absent from the concentrated supernatant of the double mutant, DFA28, but its secretion was restored by introducing pHcp, which constitutively expresses Hcp (Fig. 4). We conclude that mutation of atsR leads to hypersecretion of Hcp. To determine whether hcp expression is upregulated in DFA21, quantitative real-time PCR experiments were performed. mRNA expression levels for hcp in the B. cenocepacia K56-2 and DFA21 strains were quantified and normalized to mRNA expression of hisD, which was used as the standard. The expression ratio of DFA21 to the wild type was greater than 5, confirming that the mutation of atsR resulted in an increase of hcp expression compared to what was seen for wild-type K56-2. Since Hcp secretion is indicative of T6SS activity (39), our results suggest that the T6SS is upregulated in DFA21.
Mutation in atsR increases T6SS-dependent growth inhibition of Dictyostelium discoideum by B. cenocepacia.
Virulence of DFA21 was assessed using the social amoeba D. discoideum, a model host that preys on bacteria through its phagocytic feeding behavior. Several pathogenic bacteria resist Dictyostelium predation by producing factors that either actively kill the amoebae (6, 38) or allow intracellular survival and bacterial replication (20, 47). D. discoideum is remarkably similar to mammalian macrophages and infection assays performed with P. aeruginosa, Legionella pneumophila, and Mycobacterium species revealed that the same virulence mechanisms implicated in resistance to Dictyostelium predation also operated against mammalian cells (6, 49).
The susceptibility of B. cenocepacia K56-2 to D. discoideum predation was first tested using a qualitative plaque assay. D. discoideum cells were mixed with either B. cenocepacia K56-2 or K. pneumoniae 18, a strain known to be permissive for D. discoideum growth, and plated on SM agar plates. After a few days, K. pneumoniae was successfully killed by the amoebae, as visualized by clear plaques corresponding to zones where actively feeding and replicating amoebae have phagocytized bacteria. In contrast, no plaques were formed with B. cenocepacia K56-2, demonstrating that wild-type K56-2 is resistant to D. discoideum predation (data not shown). To rule out the possibility that B. cenocepacia K56-2 does not support Dictyostelium growth, the ability of D. discoideum to grow on a thin lawn of B. cenocepacia K56-2 cells was tested by using low-nutrient plates (SM/50, SM/100) to limit bacterial growth. Clear plaques were visualized after a few days, suggesting that low concentrations of B. cenocepacia K56-2 can support D. discoideum growth (data not shown).
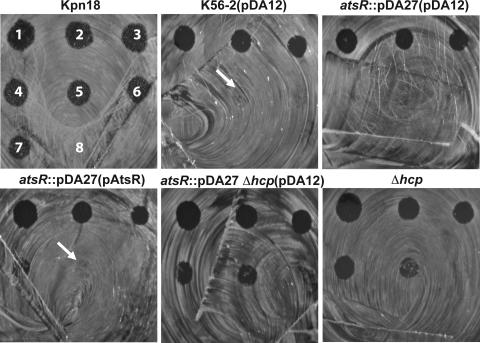
The virulence of DFA21 relative to that of the parental strain was quantitatively characterized by spotting serial dilutions of amoeba cells on bacterial lawns (Fig. 6). This assay measures the lowest number of amoeba cells required for effective bacterial predation. The higher the number of amoebae required for bacterial killing, the higher is the ability of bacteria to resist predation. In the control experiment, D. discoideum formed clear plaques on the K. pneumoniae 18 lawn up to the lowest dilution tested, which contained 781 amoeba cells (Fig. 6). In contrast, D. discoideum at dilutions containing fewer than 3,125 amoebae did not form plaques on B. cenocepacia K56-2 (Fig. 6). On DFA21, 12,500 Dictyostelium cells was the lowest amount deposited on the plate leading to the formation of a visible plaque (Fig. 6), suggesting that atsR inactivation increases the resistance of this strain to predation either by avoiding amoeba killing or by inhibiting Dictyostelium growth. The introduction of pAtsR into DFA21 restored the susceptibility of this mutant to wild-type levels of amoeba predation. DFA27 and DFA28 mutants were even more susceptible to predation than wild-type K56-2, allowing the formation of clear plaques at dilutions containing 3,125 amoebae. Together, these results suggest that there is a correlation between Hcp secretion and resistance to D. discoideum predation.
FIG. 6.
Quantitative plaque assay. D. discoideum cells were applied as droplets onto a lawn of bacterial strains without plasmid or with the control vector pDA12 and pAtsR plasmids, as indicated in parentheses. atsR indicates DFA21; atsR Δhcp indicates DFA28; Δhcp indicates DFA27. The numbers of Dictyostelium cells applied were 50,000 (spot 1, as labeled for K. pneumoniae 18 [upper left]), 25,000 (spot 2), 12,500 (spot 3), 62,500 (spot 4), 3,125 (spot 5), 1,562 (spot 6), and 781 (spot 7); SorC buffer alone is shown for spot 8. After 3 days at 22°C, the ability of Dictyostelium cells to create plaques in the bacterial lawn was recorded. WT, wild type.
To test whether secreted factors were at least in part responsible for the growth inhibition of D. discoideum, amoebae were incubated with HL5 medium or with filtered culture supernatants of Staphylococcus aureus RN6390 or B. cenocepacia K56-2 and DFA21 prior to plating on a K. pneumoniae 18 lawn. S. aureus RN6390 was used as a positive control of cytotoxicity, as this strain secretes an alpha-toxin that leads to rapid cell lysis (17). As expected, no plaques were formed when Dictyostelium cells were preincubated with S. aureus RN6390 culture supernatants. No significant difference in the numbers of plaques formed was observed when amoebae were preincubated with HL5 medium or with culture supernatants of DFA21 or B. cenocepacia K56-2 wild type (data not shown). Under the conditions tested, our results suggest that the growth inhibition of DFA21 and the parental strain toward Dictyostelium is not due to secreted effectors alone but may also require bacterium-amoeba contact.
The T6SS from B. cenocepacia K56-2 mediates actin rearrangements in mammalian macrophages.
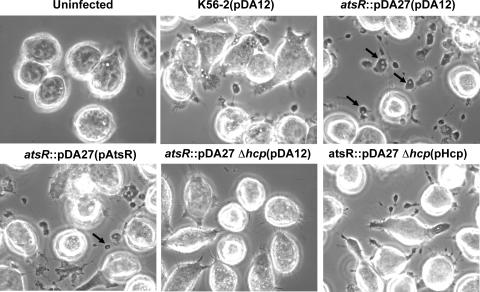
ANA-1 murine macrophages were used as a mammalian model system to investigate interactions between DFA21 and phagocytic cells. Morphological changes of ANA-1 macrophages were observed at 4 h postinfection with live DFA21 compared to K56-2 (Fig. 7). The macrophages displayed globular protrusions that included vacuoles and bacterium-containing vacuoles (Fig. 7; also see Fig. 9 below). The macrophages excluded the membrane-impermeant dye trypan blue, indicating that the cells retain their membrane integrity (data not shown). Heat-inactivated DFA21 did not induce protrusion formation, suggesting that it is an active process that requires metabolically active bacteria (data not shown).
FIG. 7.
Phase-contrast microscopy of infected ANA-1 macrophages. The infections were performed at an MOI of 50:1 for 4 h with parental and mutant strains containing pDA12, pAtsR, or pHcp as indicated in parentheses. atsR indicates DFA21; atsR Δhcp indicates DFA28. Black arrows indicate vacuole-containing protrusions. WT, wild type.
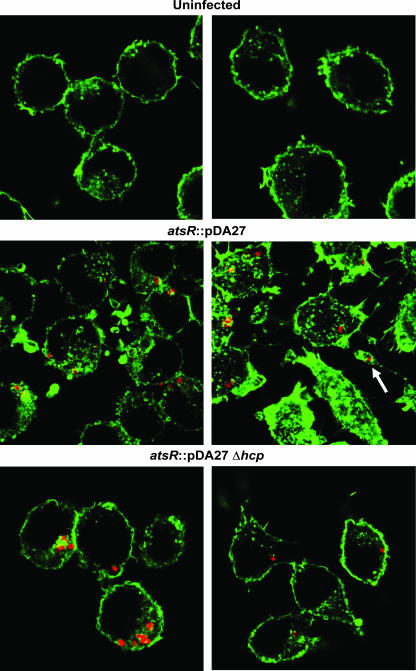
FIG. 9.
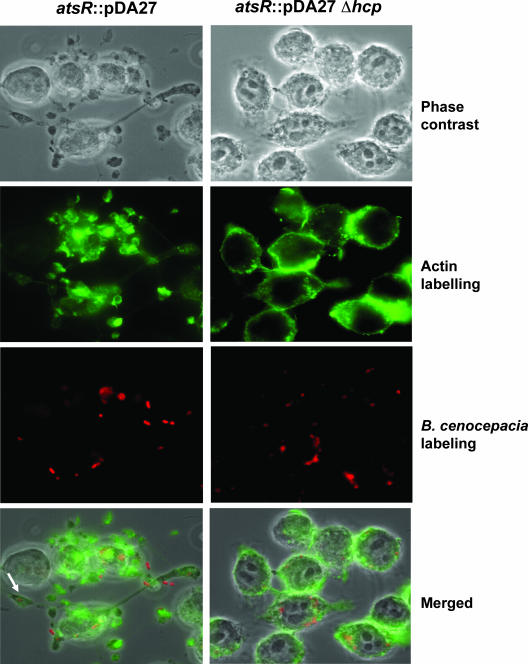
Actin distribution of infected ANA-1 macrophages. Representative confocal images of uninfected ANA-1 macrophages or ANA-1 macrophages infected with DFA21 (indicated as atsR) or DFA28 (indicated as atsR Δhcp) at an MOI of 50:1 for 4 h. Green fluorescence denotes filamentous actin. Red fluorescence denotes individual B. cenocepacia bacteria. The white arrow indicates a protrusion with a bacterium-containing vacuole.
To distinguish whether hyperadherence or Hcp hypersecretion was associated with the DFA21-mediated protrusions, ANA-1 cells were infected with DFA28, which has a hyperadherent phenotype (data not shown) but does not secrete Hcp (and potentially other T6SS effectors). DFA28 did not cause any detectable morphological changes in ANA-1 cells, whereas introduction of pHcp into DFA28 restored the ability of these bacteria to cause cellular protrusions. In addition, protrusions were still formed when ANA-1 cells were infected with DFA28(pAtsR), which secretes Hcp but does not have a hyperadherent phenotype, thus confirming that the formation of protrusions is mediated by the T6SS. Moreover, tissue culture medium supernatants from macrophages infected with DFA21 did not induce protrusions when added to wells containing uninfected ANA-1 cells, suggesting that the morphological changes require cell-to-cell contact between bacteria and macrophages (data not shown).
The morphological changes of the ANA-1 macrophages suggested an involvement of the cellular cytoskeleton. To elucidate in part the mechanism by which these protrusions were formed, ANA-1 cells were infected as previously described with DFA21 and filamentous actin was detected using phalloidin. In noninfected controls, filamentous actin was localized in the cytoplasm in a peripheral ring (data not shown; also see Fig. 9), while filamentous actin was significantly reorganized when the cells were infected with DFA21 (Fig. 8 and 9). Actin clumps could be found alone and in association with bacterium-containing vacuoles and also in the cytoplasmic protrusions (Fig. 8). In contrast, ANA-1 cells infected with DFA28 had a peripheral actin distribution very similar to that of uninfected cells (Fig. 8 and 9). Thus, DFA21 through the secretion of Hcp can induce actin rearrangements within macrophages, leading to the formation of cellular protrusions.
FIG. 8.
Actin distribution of infected ANA-1 macrophages. Representative fluorescence micrographs of ANA-1 macrophages infected with strains DFA21 (shown as atsR) and DFA28 (shown as atsR Δhcp) at an MOI of 50:1 for 4 h are shown. Green fluorescence denotes filamentous actin. Red fluorescence denotes individual B. cenocepacia bacteria. The white arrow indicates a protrusion with a bacterium-containing vacuole. WT, wild type.
DISCUSSION
In this study, we have identified a putative sensor kinase-response regulator hybrid (AtsR) involved in the regulation of B. cenocepacia K56-2 virulence genes. AtsR contains one response regulator domain but lacks any DNA binding or Hpt (histidine phosphotransfer) domain, suggesting that other proteins are involved in the phosphorelay. The atsR gene is part of a three-gene operon together with BCAM0380 and BCAM0381, whose functions are unknown. We have preliminary evidence suggesting that BCAM0381 (which contains a response regulator domain as well as a helix-turn-helix binding motif) is part of the AtsR signaling pathway, but its position in the regulatory cascade is not yet elucidated (D. Aubert and M. Valvano, unpublished data).
We have determined that the inactivation of atsR in B. cenocepacia K56-2 leads to increased biofilm formation and adherence to polystyrene and lung epithelial cells. The genes that are required for biofilm maturation have been identified by transposon mutagenesis in B. cepacia H111 and can be divided into three main classes: genes encoding surface proteins (cable pili and adhesins), genes involved in the biogenesis and maintenance of outer membrane integrity, and genes that encode regulatory factors that affect N-acyl homoserine lactone production (21). Biofilm formation could be advantageous for B. cenocepacia in the CF lung environment in various ways. For example, bacteria could be protected from antibiotics and/or mediators of host defense (10). Also, increased adherence may facilitate epithelial cell invasion as well as the injection of bacterial molecules that are essential for virulence into the cell's cytoplasm. Consequently, biofilm formation plays an important role in the development of persistence leading to chronic inflammation and lung deterioration, which is the primary cause of mortality in CF patients. In the abiotic adhesion assay, a centrifugation step was performed to enhance bacterial contact to the polystyrene surface. Since no centrifugation was performed in the adhesion experiments with A549 cells, this assay might reflect better the natural binding properties of B. cenocepacia. The molecular basis of the increased adherence in DFA21 is unknown. Studies of the P. aeruginosa ΔretS mutant suggest that hyperadherence is related to increased expression of two exopolysaccharide biosynthetic gene clusters (pel and psl). Microarray experiments, currently under way in our laboratory, will provide insight into the genes that are up- or downregulated in an atsR mutant background, as well as the genes involved in the hyperadherence phenotype of DFA21.
We determined that DFA21 overexpresses and hypersecretes an Hcp-like protein. In contrast to what was seen for P. aeruginosa, analysis of the sequenced genome of B. cenocepacia J2315 reveals that it encodes only one Hcp-like protein (34). The B. cenocepacia hcp is likely part of an operon encoding other components of the T6SS, such as a ClpV-like chaperone (BCAL0347) shown in P. aeruginosa to be required for Hcp secretion (34). Thus, hypersecretion of Hcp in DFA21 is likely due to the overexpression of the entire T6SS rather than an overproduction of Hcp alone. Besides Hcp, other proteins are probably specifically secreted by the T6SS and remain to be identified. Orthologs of the genes encoding VgrG proteins from V. cholerae and P. aeruginosa are also found in the B. cenocepacia J2315 genome outside the T6SS locus identified in this study.
Interestingly, the majority of T6SSs studied to date have been discovered in mutant strains that overexpress the system (34, 40, 44), suggesting that T6SS is generally repressed in bacteria grown under laboratory conditions. Expression of the T6SS is tightly regulated and the mechanism of this regulation involves two-component systems, transcriptional activators, and posttranslational regulation through threonine phosphorylation (34, 35, 40, 44). Upregulation of the T6SS by overexpression of the two-component system virAG in Burkholderia mallei (44) or by inactivation of atsR in B. cenocepacia is similar to the results observed with the global regulators RetS and LadS in P. aeruginosa (34, 54). RetS and LadS have reciprocal regulatory activities; therefore, RetS inactivation is equivalent to LadS overexpression and leads to the same phenotypes, including overexpression of the T6SS and increased biofilm formation. This reciprocal regulation controlling the expression of virulence factors is likely to occur in Burkholderia spp. and might be performed by the virAG and atsR homologs.
The D. discoideum plaque assays and macrophage infections performed in this study provide strong evidence that the T6SS plays an important role in B. cenocepacia K56-2 interactions with eukaryotic cells. From a previous screen in our laboratory for B. cenocepacia virulence genes, we identified three transposon insertion mutants (1A5, 34C4, 34C6) that were attenuated in the rat agar bead model of chronic lung infection (22). Further analysis indicated that these mutants had insertions within T6SS genes (Fig. 5 and data not shown), demonstrating that the T6SS is required for the virulence of B. cenocepacia in vivo.
D. discoideum and macrophage infection assays are relevant models for the study of B. cenocepacia. An extensive study performed in our laboratory showed that many clinical and environmental isolates of the BCC associated with CF persist within Acanthamoeba cells (32), suggesting that amoebae might be environmental reservoirs for BCC species. Electron micrographs have also shown that infected amoebae can release vacuoles containing bacteria that are sufficiently small to enter the human lower airway, potentially facilitating BCC colonization of the respiratory tract (32). Acanthamoeba and D. discoideum are remarkably similar to mammalian macrophages and our laboratory showed that BCC bacteria utilize a similar intracellular strategy to persist within amoebae and macrophages by delaying the phagosome-lysosome fusion (26, 27). The exact role of the T6SS in the resistance of B. cenocepacia K56-2 to D. discoideum predation is unknown. We noticed in our plaque assays that the size of the plaques formed by any of the B. cenocepacia strains tested (wild type or hcp mutant) remained unchanged even after 5 days (or more) of incubation, whereas plaques formed on a K. pneumoniae lawn continued to expand. In addition, the plaques formed on any B. cenocepacia lawn were characterized by the absence of fruiting bodies (a multicellular stage from D. discoideum), in contrast to lawns of K. pneumoniae, where fruiting bodies were abundant. These results suggest that B. cenocepacia resists Dictyostelium predation by actively killing the amoebae or inhibiting their growth and that virulence factors other than the T6SS may be involved.
Microarray analyses performed by others (16, 34) indicate that genes associated with chronic infections, such as biofilm-promoting genes, are upregulated in the P. aeruginosa ΔretS mutant, whereas genes associated with acute infections, such as the TTSS-encoding genes, are downregulated. Upregulation of the T6SS in the P. aeruginosa retS mutant suggests that this secretion system could be required for the development of chronic infection. Culture supernatants of the atsR::pDA27 mutant did not cause growth inhibition of D. discoideum or the morphological defects found upon bacterial infection of ANA-1 macrophages, suggesting that the T6SS requires bacterium-cell contact to exert its function. Our data clearly show the formation of cellular protrusions rich in actin that may include vacuoles or bacterium-containing vacuoles. It is currently unknown if formation of these protrusions involving actin rearrangements aid B. cenocepacia to escape from macrophages or delay the phagosome-lysosome fusion, an intracellular survival strategy known to be employed by B. cenocepacia (26). B. cenocepacia possesses other virulence factors, including a TTSS required for virulence in a murine infection model (53), two type IV secretion systems (12), extracellular proteases (5), hemolysin inducing degranulation and cell death in human phagocytes (23), and phospholipases, among others. Further investigations of the B. cenocepacia T6SS, including the molecular mechanism of secretion, are required for a better understanding of the role of this system in the persistence and intracellular survival of this bacterium and other related bacteria.
Acknowledgments
We thank the members of our laboratory for helpful discussions, J. Parkhill for allowing us access to the draft annotation of B. cenocepacia J2315, D. Radzioch (McGill University, Department of Human Genetics, Montreal General Hospital Research Institute, Montreal, Quebec, Canada) for the gift of the ANA-1 cell line, P. Devreotes and the Dicty Stock Center for the gift of the D. discoideum AX3 strain, C. Y. Kang for the gift of the A549 cell line, D. E. Heinrichs for the gift of the S. aureus RN6390 strain, and K. E. Keith for help with confocal microscopy.
This work was supported in part by grants from the Canadian Cystic Fibrosis Foundation and the Canadian Institutes of Health Research (to M.A.V.). D.F.A. was supported by a Postdoctoral Fellowship from the Canadian Institutes of Health Research in partnership with the Association of Medical Microbiology and Infectious Disease Canada. R.S.F. was supported by a Graduate Student Fellowship from the Canadian Cystic Fibrosis Foundation. M.A.V. holds a Canada Research Chair in Infectious Diseases and Microbial Pathogenesis.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 3 March 2008.
REFERENCES
- 1.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang, C., and R. C. Stewart. 1998. The two-component system. Regulation of diverse signaling pathways in prokaryotes and eukaryotes. Plant Physiol. 117723-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coenye, T., and P. Vandamme. 2003. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 5719-729. [DOI] [PubMed] [Google Scholar]
- 4.Cohen, S. N., A. C. Chang, and L. Hsu. 1972. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc. Natl. Acad. Sci. USA 692110-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corbett, C. R., M. N. Burtnick, C. Kooi, D. E. Woods, and P. A. Sokol. 2003. An extracellular zinc metalloprotease gene of Burkholderia cepacia. Microbiology 1492263-2271. [DOI] [PubMed] [Google Scholar]
- 6.Cosson, P., L. Zulianello, O. Join-Lambert, F. Faurisson, L. Gebbie, M. Benghezal, C. Van Delden, L. K. Curty, and T. Kohler. 2002. Pseudomonas aeruginosa virulence analyzed in a Dictyostelium discoideum host system. J. Bacteriol. 1843027-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox, G. W., B. J. Mathieson, L. Gandino, E. Blasi, D. Radzioch, and L. Varesio. 1989. Heterogeneity of hematopoietic cells immortalized by v-myc/v-raf recombinant retrovirus infection of bone marrow or fetal liver. J. Natl. Cancer Inst. 811492-1496. [DOI] [PubMed] [Google Scholar]
- 8.Das, S., and K. Chaudhuri. 2003. Identification of a unique IAHP (IcmF associated homologous proteins) cluster in Vibrio cholerae and other proteobacteria through in silico analysis. In Silico Biol. 3287-300. [PubMed] [Google Scholar]
- 9.de Bruin, O. M., J. S. Ludu, and F. E. Nano. 2007. The Francisella pathogenicity island protein IglA localizes to the bacterial cytoplasm and is needed for intracellular growth. BMC Microbiol. 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desai, M., T. Buhler, P. H. Weller, and M. R. Brown. 1998. Increasing resistance of planktonic and biofilm cultures of Burkholderia cepacia to ciprofloxacin and ceftazidime during exponential growth. J. Antimicrob. Chemother. 42153-160. [DOI] [PubMed] [Google Scholar]
- 11.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 166127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engledow, A. S., E. G. Medrano, E. Mahenthiralingam, J. J. LiPuma, and C. F. Gonzalez. 2004. Involvement of a plasmid-encoded type IV secretion system in the plant tissue watersoaking phenotype of Burkholderia cenocepacia. J. Bacteriol. 1866015-6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 761648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flannagan, R. S., D. Aubert, C. Kooi, P. A. Sokol, and M. A. Valvano. 2007. Burkholderia cenocepacia requires a periplasmic HtrA protease for growth under thermal and osmotic stress and for survival in vivo. Infect. Immun. 751679-1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flannagan, R. S., T. Linn, and M. A. Valvano. A system for the construction of targeted unmarked gene deletions in the genus Burkholderia. Environ. Microbiol., in press. [DOI] [PubMed]
- 16.Goodman, A. L., B. Kulasekara, A. Rietsch, D. Boyd, R. S. Smith, and S. Lory. 2004. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell 7745-754. [DOI] [PubMed] [Google Scholar]
- 17.Gouaux, E. 1998. Alpha-hemolysin from Staphylococcus aureus: an archetype of beta-barrel, channel-forming toxins. J. Struct. Biol. 121110-122. [DOI] [PubMed] [Google Scholar]
- 18.Govan, J. R., P. H. Brown, J. Maddison, C. J. Doherty, J. W. Nelson, M. Dodd, A. P. Greening, and A. K. Webb. 1993. Evidence for transmission of Pseudomonas cepacia by social contact in cystic fibrosis. Lancet 34215-19. [DOI] [PubMed] [Google Scholar]
- 19.Govan, J. R., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hagele, S., R. Kohler, H. Merkert, M. Schleicher, J. Hacker, and M. Steinert. 2000. Dictyostelium discoideum: a new host model system for intracellular pathogens of the genus Legionella. Cell. Microbiol. 2165-171. [DOI] [PubMed] [Google Scholar]
- 21.Huber, B., K. Riedel, M. Kothe, M. Givskov, S. Molin, and L. Eberl. 2002. Genetic analysis of functions involved in the late stages of biofilm development in Burkholderia cepacia H111. Mol. Microbiol. 46411-426. [DOI] [PubMed] [Google Scholar]
- 22.Hunt, T. A., C. Kooi, P. A. Sokol, and M. A. Valvano. 2004. Identification of Burkholderia cenocepacia genes required for bacterial survival in vivo. Infect. Immun. 724010-4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hutchison, M. L., I. R. Poxton, and J. R. Govan. 1998. Burkholderia cepacia produces a hemolysin that is capable of inducing apoptosis and degranulation of mammalian phagocytes. Infect. Immun. 662033-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Isles, A., I. Maclusky, M. Corey, R. Gold, C. Prober, P. Fleming, and H. Levison. 1984. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J. Pediatr. 104206-210. [DOI] [PubMed] [Google Scholar]
- 25.Kolter, R., M. Inuzuka, and D. R. Helinski. 1978. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell 151199-1208. [DOI] [PubMed] [Google Scholar]
- 26.Lamothe, J., K. K. Huynh, S. Grinstein, and M. A. Valvano. 2007. Intracellular survival of Burkholderia cenocepacia in macrophages is associated with a delay in the maturation of bacteria-containing vacuoles. Cell. Microbiol. 940-53. [DOI] [PubMed] [Google Scholar]
- 27.Lamothe, J., S. Thyssen, and M. A. Valvano. 2004. Burkholderia cepacia complex isolates survive intracellularly without replication within acidic vacuoles of Acanthamoeba polyphaga. Cell. Microbiol. 61127-1138. [DOI] [PubMed] [Google Scholar]
- 28.Lefebre, M. D., and M. A. Valvano. 2002. Construction and evaluation of plasmid vectors optimized for constitutive and regulated gene expression in Burkholderia cepacia complex isolates. Appl. Environ. Microbiol. 685956-5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loutet, S. A., R. S. Flannagan, C. Kooi, P. A. Sokol, and M. A. Valvano. 2006. A complete lipopolysaccharide inner core oligosaccharide is required for resistance of Burkholderia cenocepacia to antimicrobial peptides and bacterial survival in vivo. J. Bacteriol. 1882073-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahenthiralingam, E., T. Coenye, J. W. Chung, D. P. Speert, J. R. Govan, P. Taylor, and P. Vandamme. 2000. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J. Clin. Microbiol. 38910-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahenthiralingam, E., P. Vandamme, M. E. Campbell, D. A. Henry, A. M. Gravelle, L. T. Wong, A. G. Davidson, P. G. Wilcox, B. Nakielna, and D. P. Speert. 2001. Infection with Burkholderia cepacia complex genomovars in patients with cystic fibrosis: virulent transmissible strains of genomovar III can replace Burkholderia multivorans. Clin. Infect. Dis. 331469-1475. [DOI] [PubMed] [Google Scholar]
- 32.Marolda, C. L., B. Hauroder, M. A. John, R. Michel, and M. A. Valvano. 1999. Intracellular survival and saprophytic growth of isolates from the Burkholderia cepacia complex in free-living amoebae. Microbiology 1451509-1517. [DOI] [PubMed] [Google Scholar]
- 33.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 1702575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mougous, J. D., M. E. Cuff, S. Raunser, A. Shen, M. Zhou, C. A. Gifford, A. L. Goodman, G. Joachimiak, C. L. Ordonez, S. Lory, T. Walz, A. Joachimiak, and J. J. Mekalanos. 2006. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 3121526-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mougous, J. D., C. A. Gifford, T. L. Ramsdell, and J. J. Mekalanos. 2007. Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat. Cell Biol. 9797-803. [DOI] [PubMed] [Google Scholar]
- 36.Ortega, X., T. A. Hunt, S. Loutet, A. D. Vinion-Dubiel, A. Datta, B. Choudhury, J. B. Goldberg, R. Carlson, and M. A. Valvano. 2005. Reconstitution of O-specific lipopolysaccharide expression in Burkholderia cenocepacia strain J2315, which is associated with transmissible infections in patients with cystic fibrosis. J. Bacteriol. 1871324-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30295-304. [DOI] [PubMed] [Google Scholar]
- 38.Pukatzki, S., R. H. Kessin, and J. J. Mekalanos. 2002. The human pathogen Pseudomonas aeruginosa utilizes conserved virulence pathways to infect the social amoeba Dictyostelium discoideum. Proc. Natl. Acad. Sci. USA 993159-3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pukatzki, S., A. T. Ma, A. T. Revel, D. Sturtevant, and J. J. Mekalanos. 2007. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. USA 10415508-15513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pukatzki, S., A. T. Ma, D. Sturtevant, B. Krastins, D. Sarracino, W. C. Nelson, J. F. Heidelberg, and J. J. Mekalanos. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. USA 1031528-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rao, P. S., Y. Yamada, Y. P. Tan, and K. Y. Leung. 2004. Use of proteomics to identify novel virulence determinants that are required for Edwardsiella tarda pathogenesis. Mol. Microbiol. 53573-586. [DOI] [PubMed] [Google Scholar]
- 42.Reik, R., T. Spilker, and J. J. Lipuma. 2005. Distribution of Burkholderia cepacia complex species among isolates recovered from persons with or without cystic fibrosis. J. Clin. Microbiol. 432926-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1990. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 44.Schell, M. A., R. L. Ulrich, W. J. Ribot, E. E. Brueggemann, H. B. Hines, D. Chen, L. Lipscomb, H. S. Kim, J. Mrazek, W. C. Nierman, and D. Deshazer. 2007. Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol. Microbiol. 641466-1485. [DOI] [PubMed] [Google Scholar]
- 45.Sheahan, K. L., C. L. Cordero, and K. J. Satchell. 2004. Identification of a domain within the multifunctional Vibrio cholerae RTX toxin that covalently cross-links actin. Proc. Natl. Acad. Sci. USA 1019798-9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith, D. L., L. B. Gumery, E. G. Smith, D. E. Stableforth, M. E. Kaufmann, and T. L. Pitt. 1993. Epidemic of Pseudomonas cepacia in an adult cystic fibrosis unit: evidence of person-to-person transmission. J. Clin. Microbiol. 313017-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Solomon, J. M., A. Rupper, J. A. Cardelli, and R. R. Isberg. 2000. Intracellular growth of Legionella pneumophila in Dictyostelium discoideum, a system for genetic analysis of host-pathogen interactions. Infect. Immun. 682939-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Speert, D. P., D. Henry, P. Vandamme, M. Corey, and E. Mahenthiralingam. 2002. Epidemiology of Burkholderia cepacia complex in patients with cystic fibrosis, Canada. Emerg. Infect. Dis. 8181-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steinert, M., and K. Heuner. 2005. Dictyostelium as host model for pathogenesis. Cell. Microbiol. 7307-314. [DOI] [PubMed] [Google Scholar]
- 50.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69183-215. [DOI] [PubMed] [Google Scholar]
- 51.Suarez, G., J. C. Sierra, J. Sha, S. Wang, T. E. Erova, A. A. Fadl, S. M. Foltz, A. J. Horneman, and A. K. Chopra. 24 October 2007. Molecular characterization of a functional type VI secretion system from a clinical isolate of Aeromonas hydrophila. Microb. Pathog. doi: 10.1016/j.micpath.2007.10.005. [DOI] [PMC free article] [PubMed]
- 52.Sussman, M. 1987. Cultivation and synchronous morphogenesis of Dictyostelium under controlled experimental conditions. Methods Cell Biol. 28 9-29. [DOI] [PubMed] [Google Scholar]
- 53.Tomich, M., A. Griffith, C. A. Herfst, J. L. Burns, and C. D. Mohr. 2003. Attenuated virulence of a Burkholderia cepacia type III secretion mutant in a murine model of infection. Infect. Immun. 711405-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ventre, I., A. L. Goodman, I. Vallet-Gely, P. Vasseur, C. Soscia, S. Molin, S. Bleves, A. Lazdunski, S. Lory, and A. Filloux. 2006. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc. Natl. Acad. Sci. USA 103171-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.West, A. H., and A. M. Stock. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26369-376. [DOI] [PubMed] [Google Scholar]
- 56.Williams, S. G., L. T. Varcoe, S. R. Attridge, and P. A. Manning. 1996. Vibrio cholerae Hcp, a secreted protein coregulated with HlyA. Infect. Immun. 64283-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng, J., and K. Y. Leung. 2007. Dissection of a type VI secretion system in Edwardsiella tarda. Mol. Microbiol. 661192-1206. [DOI] [PubMed] [Google Scholar]