Abstract
We reinvestigated the clonal diversity and dynamics of Streptococcus mitis and two other abundant members of the commensal microbiota of the upper respiratory tract, Streptococcus oralis and Streptococcus infantis, to obtain information about the origin of frequently emerging clones in this habitat. A culture-independent method was used, based on cloning and sequencing of PCR amplicons of the housekeeping gene gdh, which shows remarkable, yet species-specific, genetic polymorphism. Samples were collected from all potential ecological niches in the oral cavity and pharynx of two adults on two occasions separated by 2 years. Based on analysis of close to 10,000 sequences, significant diversity was observed in populations of all three species. Fluctuations in the relative proportions of individual clones and species were observed over time. While a few clones dominated, the proportions of most clones were very small. The results show that the frequent turnover of S. mitis, S. oralis, and S. infantis clones observed by cultivation can be explained by fluctuations in the relative proportions of clones, most of which are below the level of detection by the traditional culture technique, possibly combined with loss and acquisition from contacts. These findings provide a platform for understanding the mechanisms that govern the balance within the complex microbiota at mucosal sites and between the microbiota and the mucosal immune system of the host.
Emerging insight at the molecular level supports the new paradigm that the human body and its commensal microbiota constitute a highly integrated “superorganism.” The benefits for both the microbiota and the host are substantial and multifaceted and apparently build on a state of mutual tolerance. The commensal microbiota becomes established immediately after birth and develops into complex ecosystems characteristic of the many different niches present on the surfaces of the mucosal membranes and the skin. At the gross level, the composition of this microbiota is very stable during the entire lifetime of an individual, and the compositions for different individuals are similar. However, at the detailed level the interpersonal and interspecies differences are considerable. Thus, each bacterial species is often represented by numerous clones that appear to be specific to the individual or to members of a family, and in contrast to traditional concepts of climax ecosystems, the species niches often appear to be maintained by a succession of clones rather than by stably colonizing clones (6, 15). The nature of the ecological pressure driving these dynamic changes in the microbial population is not known.
The mitis group streptococci, like Streptococcus mitis and Streptococcus oralis, are attractive models for analysis of the intricate relationship between commensals and the host. These organisms are abundant members of the microbiota on all surfaces in the oral cavity and pharynx from birth and throughout life, and they can be easily accessed for sampling (1, 8, 12, 19, 27, 29, 34). Although true commensals, many of the mitis group streptococci are opportunistic pathogens implicated in dental caries (4, 36), in subacute bacterial endocarditis (17, 23), in bacteremia in immunocompromised patients (13, 16, 35, 38), and occasionally in cases of extraoral diseases, such as brain abscesses, meningitis, eye infections, and pneumonia (3, 5, 25, 26, 31). Moreover, S. mitis, in particular, is strikingly closely related to Streptococcus pneumoniae despite distinct differences in its pathogenic potential (M. Kilian, K. Poulsen, T. Blomqvist, L. S. Håvarstein, M. Bek-Thomsen, and U. B. S. Sørensen, submitted for publication). Studies of these streptococci may therefore provide important insight into molecular determinants of the different lifestyles of commensals and pathogens.
Previous cross-sectional and longitudinal studies focusing on the S. mitis population in newborns and infants demonstrated that there are numerous clones assigned to this species in the respiratory tract of each individual. Of the multiple genotypes detected in infants sampled at intervals of weeks to months over a 1-year period, only a few were isolated on more than one occasion (15, 20). Isolates from adults showed the same degree of clonal diversity, but some of the clones colonizing adults were more stably present, although their abundance fluctuated (15, 20). Similar population kinetics was observed for S. oralis in dental plaque (2). Three hypotheses were considered to explain the origin of the multiple new clones that appear to be acquired over time by infants and adults (15). Continuous acquisition over time from close contacts undoubtedly contributes to the process, but the limited sharing of detectable S. mitis genotypes between family members observed in two independent studies suggests that this is not the main factor (14, 20). Frequent emergence of new genotypes in an individual as a result of homologous recombination within the pool of streptococci carried by the individual is an attractive hypothesis for these naturally transformation-competent bacteria, but this hypothesis is not supported by experimental data for S. mitis (15). Rather, the presence of largely different clonal S. mitis populations in different niches in the upper respiratory tract (15, 20) points to transmission between different habitats combined with fluctuations in the abundance of individual clones, the levels of most of which are below the level detectable by the traditional culture technique. In support of this theory a recent study demonstrated that there is intraoral transfer of S. mitis clones in neonates and that the representation of such clones in samples from different habitats varied (20).
Attempts to study populations of S. mitis and S. oralis have been hampered by difficulties in differentiation of these two species. Introduction of the species Streptococcus infantis (18) and Streptococcus pseudopneumoniae (3) further emphasized the problems of differentiation based on traditional phenotypic tests and 16S rRNA sequences, and recent results reported by us demonstrated that neither phenotypic characteristics nor analysis of 16S rRNA sequences allows differentiation of these species (16; Kilian et al., submitted). With this background, it is fair to conclude that previous studies of S. mitis populations may have encompassed several of these mitis group species, which may have affected conclusions concerning the degree of diversity of populations of S. mitis and S. oralis carried by single individuals. We recently demonstrated that S. mitis, S. oralis, and S. infantis indeed do constitute distinct populations of bacteria, although the populations are less coherent than traditionally expected for bacterial species (Kilian et al., submitted). Unambiguous differentiation of these species and, more surprisingly, of the majority of individual clones of these species is feasible based on the sequence of the glucose dehydrogenase gene (gdh) (Kilian et al., submitted).
In the present study we reinvestigated the clonal diversity and dynamics of S. mitis in potential habitats in the oral cavity and pharynx by using a culture-independent method previously used to explore microbial diversity in complex habitats, such as soil (24), the bovine rumen (37), the oral cavity (1), and other environmental and human habitats. In addition to S. mitis, this study included S. oralis and S. infantis and was based on cloning and sequencing of close to 10,000 PCR amplicons of the housekeeping gene gdh amplified from pooled samples collected from two adults on two occasions separated by 2 years.
MATERIALS AND METHODS
Sample collection.
Samples were collected at two times separated by 2 years from two females; subject A was Caucasian and was 26 years old, and subject B was Asian and was 32 years old. Each sample was a pool of bacteria collected with cotton swabs from all surfaces of the teeth and mucosal membranes in the oral cavity and the pharynx. After collection, the samples were immediately transferred to and pooled in 1 ml of buffer (1.5 mM sodium citrate, 15 mM sodium chloride). Neither of the two subjects was treated with antibiotics during the 2-year observation period. The subjects were included based on informed consent.
DNA extraction and PCR amplification.
Bacteria in the samples were harvested by centrifugation and resuspended in 300 μl of a lysozyme solution (0.05 M Tris-HCl [pH 8], 20% sucrose, 1 mg/ml lysozyme, 100 U/ml mutanolysin). After incubation at 37°C for 1 h, 250 μl of lysis buffer (10% Sarkosyl, 0.5 M EDTA, MilliQ H2O) and 100 μl of a 10-mg/ml proteinase K solution were added. After incubation at 55°C for 2 h, an additional 100 μl of the 10-mg/ml proteinase K solution was added, and the preparation was incubated at 37°C overnight. DNA was extracted with phenol-chloroform (1:1) and precipitated with ethanol (96%). The quality and amount of DNA were determined by electrophoresis on a 1% agarose gel. An approximately 650-bp internal fragment of the glucose-6-phosphate dehydrogenase gene (gdh) was amplified with primers 5′-ATG GAC AAA CCA GCN AGY TT-3′ (forward primer) and 5′-GCT TGA GGT CCC ATR CTN CC-3′ (reverse primer). Previous use of this primer set with a comprehensive collection of mitis, anginosus, salivarius, and mutans group streptococci revealed that it amplifies the gdh target sequence in all strains of S. oralis, S. mitis, S. infantis, S. pneumoniae, and S. pseudopneumoniae and in some strains of the taxon “Streptococcus mitis biovar 2,” which according to recent phylogenetic analyses is a distinct species more closely related to S. oralis than to S. mitis. No other Streptococcus species gave amplicons with this primer set (Kilian et al., submitted). Duplicate PCR amplification reactions were carried out for each sample using 50-μl (total volume) mixtures containing approximately 3 ng of whole-cell DNA as the template, 10 μM of each primer, and Platinum PCR SuperMix (Invitrogen Corporation, California). The amplification conditions comprised 2 min of incubation at 94°C, followed by 35 cycles of 30 s at 94°C, 30 s at 55°C, and 1 min at 72°C and then a final extension for 7 min at 72°C. Amplification products were visualized by electrophoresis on a 1% agarose gel supplemented with ethidium bromide.
Genomic libraries and sequencing.
Libraries of the gdh amplicons were prepared for each sample using pools of the two independent amplification products. PCR amplification products were ligated into the pCR 4-TOPO cloning vector (Invitrogen) and cloned into Escherichia coli One Shot electrocompetent cells (Invitrogen) by electroporation performed with a Bio-Rad electroporator (Bio-Rad Laboratories, Hercules, CA). Cloning and transformation reactions were performed as described by the supplier. Transformants were plated onto YET/2 agar (Invitrogen) supplemented with 100 μg/ml ampicillin and incubated overnight at 37°C. From the libraries constructed using baseline samples from subjects A and B, approximately 1,800 and 3,700 colonies, respectively, were picked for sequencing. From the two libraries constructed using the 2-year follow-up samples, approximately 2,500 clones from each subject were sequenced. Plasmid purification and sequencing of the inserts were performed at the J. Craig Venter Science Foundation Joint Technology Center (Rockville, MD).
Sequence analysis.
Raw sequences were processed using Lucy, version 1.20p (http://www.tigr.org/software/sequencing.shtml), a program that performs automated sequence quality assessment, confidence reassurance, vector trimming, and vector removal. The initial alignment of amplified sequences from each of the individuals was performed with the Clustal software in MEGA3.1 (21). The sequences were manually edited, and sequences shorter than 400 bp were omitted. Corrected sequences were grouped, and representative sequences were identified with the program FastgroupII available at http://phage.sdsu.edu/research/tools/fastgroup/. Representatives of the identified sequence variants were resequenced in the opposite direction for quality control. Potential chimeric molecules artificially assembled during the PCR were identified by comparing phylogenetic trees based on the 5′ end and the 3′ end of representative sequences. Phylogenetic associations of identified gdh alleles were determined by including gdh sequences from the type strains of S. mitis (NCTC12261), S. oralis (NCTC11427), S. infantis (GTC849), S. pneumoniae (NCTC7465), and S. pseudopneumoniae (CCUG49455), as well as two reference strains of the taxon known as “S. mitis biovar 2,” in the phylogenetic analysis. The analysis was performed with MEGA3.1 by using the Minimum Evolution algorithm and the Kimura two-parameter model (21). The robustness of the clusters obtained was evaluated by bootstrap analysis using 500 resamplings.
Estimation of diversity.
Rarefaction curves and richness estimates were calculated with the program EstimateS, version 8.0, available at http://viceroy.eeb.uconn.edu/EstimateS. The sample-based Mao Tau method was used to compute the rarefaction curves. The bias-corrected Chao1 estimator of species richness was calculated after 100 randomizations of sampling without replacement.
Sequence divergence within species.
The range of divergence within the targeted gdh sequence in a collection of epidemiologically independent isolates of S. mitis, S. oralis, and S. infantis was determined on the basis of pairwise distances calculated using MEGA3.1. Only isolates that were distinct at other gene loci were included. The gdh sequences included 62 sequences from S. mitis, 47 sequences from S. oralis, and 12 sequences from S. infantis. The origin of these strains has been described previously (16; Kilian et al., submitted).
Nucleotide sequence accession numbers.
Representative gdh nucleotide sequences have been deposited in the GenBank database (http://www.ncbi.nlm.nih.gov/) under accession numbers EU486460 to EU486523 and EU486526 to EU486538.
RESULTS
Sequence divergence within species.
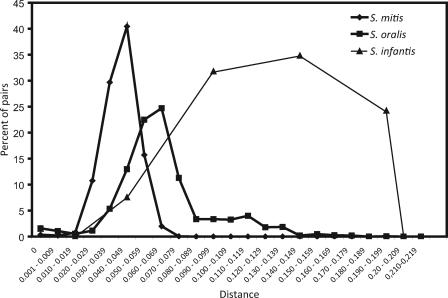
The distribution of gdh pairwise sequence distances among the 121 independent isolates of S. mitis (n = 62), S. oralis (n = 47), and S. infantis (n = 12) is shown in Fig. 1. In agreement with our previous observation based on analysis of several housekeeping genes (Kilian et al., submitted), the range of interstrain gdh sequence distances was significantly larger than that found in traditional pathogenic bacteria, such as S. pneumoniae (0.010 ± 0.001 [mean ± standard error of the mean]). Based on these observations, we selected an allele cutoff of 4 nucleotide differences corresponding to a distance of approximately 0.01 (1.0%). Although this cutoff may have resulted in a marginal underestimate of the total number of clones with no relevance for the aims of this study, it eliminated biases due to Taq polymerase-induced errors, which have been estimated to be within 0.2% of amplified nucleotides in a sequence (32). Although based on a limited collection of strains, the significant sequence divergence calculated for strains assigned to S. infantis (Fig. 1) raises doubts about the coherence of this species.
FIG. 1.
Distribution of pairwise sequence distances for a fragment of the gdh gene in collections of independent isolates of S. mitis (n = 62), S. oralis (n = 47), and S. infantis (n = 12). By comparison, the range of genetic distances among alleles of housekeeping genes of S. pneumoniae is 0 to 0.012. The lower cutoff distance used to define distinct alleles in this study was 0.01.
Identification of genotypes.
Sequencing of the gdh gene insert libraries constructed on the basis of PCR amplicons from S. mitis, S. oralis, and S. infantis clones present in the baseline and follow-up samples resulted in a total of 9,846 sequences. All sequences from each individual were aligned and corrected manually. Sequences that spanned less than 400 bp of the expected 619 bp were omitted. Of 4,628 sequences from baseline samples, 4,317 were used in subsequent analyses. Chimeric molecules were identified by comparing trees based on the first 100 nucleotides of the 5′ end and the last 100 nucleotides of the 3′ end of all observed gdh alleles. In 4,317 baseline sequences, a total of 90 potential chimeras (2.0%) were identified. For the majority of chimeric sequences we were able to identify the component molecules, which were derived from other alleles observed in our samples. Six sequences for which discordant tree positions were obtained for the two ends but for which we failed to identify both of the component sequences were considered natural recombinant forms and were included in the subsequent analysis. This is in agreement with the concept that homologous recombination resulting in mosaic gene structures has been an important factor in the long-term evolution of streptococci belonging to the species studied (Kilian et al., submitted).
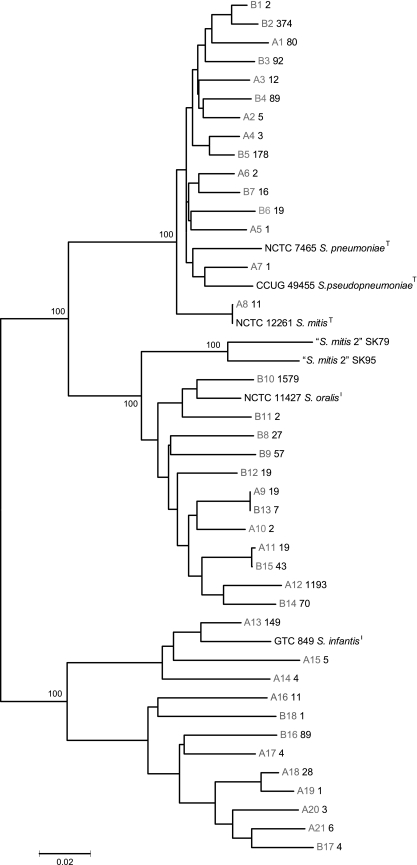
Sorting of all sequences, after exclusion of assumed chimeras, resulted in identification of a total of 39 gdh alleles. Phylogenetic analysis of these sequences together with reference gdh sequences of the type strains of S. mitis, S. oralis, S. infantis, S. pneumoniae, and S. pseudopneumoniae by the Minimum Evolution algorithm in MEGA3.1 identified three distinct clusters supported by significant bootstrap values, each of which contained one type strain (Fig. 2). The only exception was the mitis cluster, which, in agreement with our previous observations (Kilian et al., submitted), included the S. pneumoniae and S. pseudopneumoniae sequences. We excluded the possibility that sequences in the libraries originated from S. pneumoniae, which is a rare organism in the oral cavity and only occasionally colonizes the pharynx of healthy adults. Furthermore, according to our previous observations, gdh sequences of S. pneumoniae are monophyletic. We previously observed that S. pseudopneumoniae has gdh alleles that cluster with S. mitis alleles (Kilian et al., submitted). As no information on the occurrence of this recently described species is available, we cannot exclude the possibility that some of the clones ascribed to S. mitis may represent S. pseudopneumoniae. None of the gdh sequences amplified from the samples clustered with the sequences from two reference strains of “S. mitis biovar 2.”
FIG. 2.
Consensus Minimum Evolution tree showing phylogenetic associations and genetic diversity of gdh alleles amplified from samples from two individuals. Bootstrap values, based on 500 replicates, are shown for the major branches. Alleles from subjects A and B are designated A and B, respectively. The boldface values that follow the allele designations are the numbers of identical sequences in the sequenced samples of the gene libraries.
The numbers of sequences assigned to each ofthe three species based on the cluster analysis and the numbers of clones of each species detected in the two individuals are shown in Table 1. The majority of sequences from subjects A and B (1,233 sequences [79%] and 1,804 sequences [68%], respectively) clustered with the S. oralis reference sequence and represented a total of four clones in subject A and eight clones in subject B. The relative proportions of sequences identified as S. mitis and S. infantis were different in the two subjects. In subject A, S. mitis and S. infantis accounted for 7% (115) and 14% (211) of all sequences and comprised eight and nine genotypes, respectively. In contrast, S. mitis was the dominant species of these two species in subject B and accounted for 29% (770) of all sequences, representing seven distinct genotypes. Only 4% (94) of all sequences in subject B were identified as S. infantis sequences; these sequences represented three clones of the species.
TABLE 1.
Numbers of sequences allocated to S. mitis, S. oralis, and S. infantis by cluster analysis and inferred numbers of genotypes detected for each species in two individuals at two observation times separated by 2 years
| Subject | Observation time |
S. oralis
|
S. mitis
|
S. infantis
|
Total no. of genotypes | |||
|---|---|---|---|---|---|---|---|---|
| No. of sequences | No. of genotypes | No. of sequences | No. of genotypes | No. of sequences | No. of genotypes | |||
| A | 1 | 1,233 | 4 | 115 | 8 | 211 | 9 | 21 |
| 2 | 40 | 6 | 2,007 | 15 | 37 | 10 | 31 | |
| B | 1 | 1,804 | 8 | 770 | 7 | 94 | 3 | 18 |
| 2 | 72 | 9 | 1,791 | 13 | 216 | 3 | 25 | |
| Total | 1 | 3,037 | 12 | 885 | 15 | 305 | 12 | 39 |
| 2 | 112 | 15 | 3,798 | 28 | 253 | 13 | 56 | |
As demonstrated by the number of sequences assigned to each allele (Fig. 2), significant differences in the proportions of individual sequences were observed. In both of the individuals a single genotype accounted for 70% or more of the identified sequences.
Coverage of alleles represented in gene libraries.
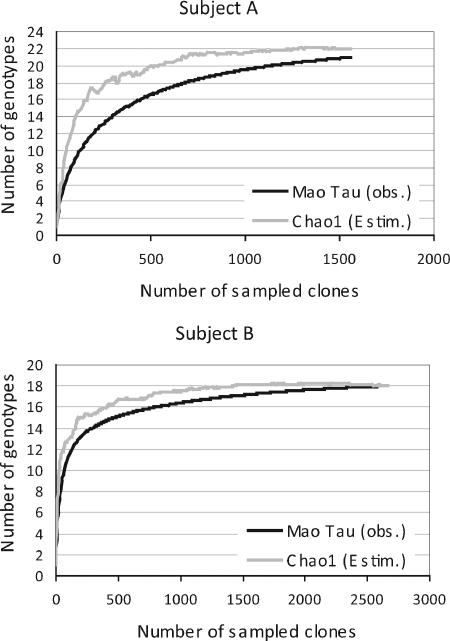
The total number of genotypes present in each of the two individuals was estimated by the nonparametric estimator of total richness Chao1. Based on these calculations, the number of alleles present in subject A was ∼22 (95% confidence interval [CI], 21 to 31). Thus, the 21 alleles observed (95% CI, 18 to 24) represented 95.5% (95% CI, 77.4 to 85.7%) of the estimated alleles. The estimated total richness in subject B was 18, and 18 alleles (range, 17 to 19 alleles) were observed, indicating that the coverage was 100% (95% CI, 94.7 to 105.9%) (Fig. 3).
FIG. 3.
Rarefaction analysis based on the gdh clone libraries generated from samples collected from subjects A and B. The curves reflecting the observed [Mao Tau (obs.)] and estimated [Chao1 (Estim.)] richness values plotted as a function of the number of sampled clones demonstrate that virtually all genotypes were detected.
Follow-up examination.
To evaluate the stability and dynamics of clones of the three species in question, samples were obtained from the two individuals 2 years after the baseline observation, which provided a valid test of stability. A total of 5,121 valid gdh sequences were generated from these samples. Among these sequences 56 distinct alleles were identified after sorting and exclusion of chimeras. As described above, the phylogenetic affiliation of the sequences was determined by cluster analysis (data not shown). In agreement with the results of the baseline examination, three distinct clusters corresponding to S. mitis, S. oralis, and S. infantis were revealed by the phylogenetic analysis of the 56 alleles. In contrast to the baseline observations, S. mitis was the dominant species in both individuals, accounting for 96% (2,007) and 86% (1,791) of all the sequences, and was represented by increased numbers of genotypes (15 and 13 genotypes, respectively). Accordingly, the relative proportions of sequences identified as S. oralis sequences were reduced to 2 and 3% in the two subjects, but these sequences comprised more genotypes than the numbers determined for the baseline observation (six and nine genotypes in subjects A and B, respectively, versus four and eight genotypes for the baseline observation). In subject A, S. infantis was the least abundant species, accounting for only 2% of all sequences, but it was represented by 10 distinct genotypes, compared with values of 14% and nine genotypes for the baseline observation. In subject B, S. infantis accounted for 10% of all sequences, compared with 4% for the baseline observation, and was still represented by three genotypes (Table 1).
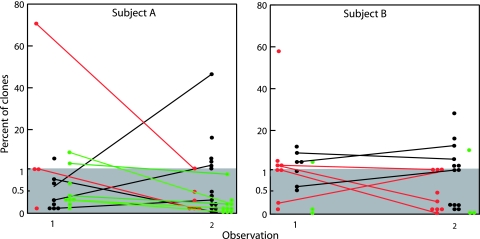
Figure 4 shows the longitudinal aspects deduced from the results of the two observations separated by 2 years. The data showed that 18 of the 39 genotypes present at the baseline observation time were redetected 2 years later (Fig. 4). In subject A, the 11 redetected genotypes included four S. mitis clones, two S. oralis clones, and five S. infantis clones. The seven clones that were stably present in subject B included three S. mitis clones and four S. oralis clones, whereas none of the three initially identified S. infantis clones were redetected after 2 years. Of the 56 clones detected in the two subjects at the time of the second observation, 38 were not detected at the initial sampling time. The abundance of persistent genotypes was observed to fluctuate; some genotypes showed significant fluctuations (e.g., clone A12, which represented 75% of all sequences in the initial sample but only 1% of the sequences in the second sample), while others showed minor variations (Fig. 4).
FIG. 4.
Dynamics of clones of S. mitis (black circles and lines), S. oralis (red circles and lines), and S. infantis (green circles and lines) detected on two occasions separated by 2 years in samples from all niches within the oral cavity and pharynx of two adult subjects. Circles connected by a line represent a clone detected on both occasions. Other clones were detected on only one occasion.
DISCUSSION
In this study we reexamined the population diversity and dynamics of S. mitis and two other important members of the commensal microbiota of the human oral cavity and pharynx using a highly sensitive nonculture detection method. The study design made it possible to evaluate the hypothesis that these anatomical sites and their many distinct ecological niches house a large number of clones, the levels of most of which are below the level detectable by traditional cultivation (15). We have proposed that the apparent acquisition and loss of clones observed in cultivation studies (11, 15, 20) to some extent may be a result of fluctuations in the relative proportions of clones in the microbiota (15). This study confirmed this hypothesis.
To fulfill the aim of the study, we needed a method with sensitivity and discriminatory power that exceeded the sensitivity and discriminatory power of traditional culture techniques. The PCR-based strategy used has been successfully employed in studies of the biodiversity of complex microbial ecosystems (1, 24, 28, 37). Instead of 16S rRNA sequences, which reveal species diversity, our target marker was the gdh housekeeping gene. Due to the unusual sequence divergence in housekeeping genes of S. mitis, S. oralis, and S. infantis, which exceeds the sequence divergence of corresponding genes in pathogens like S. pneumoniae by a factor 10 (Kilian et al., submitted) (Fig. 1), this method is capable of detecting individual clones of the three commensal species with a high degree of accuracy (Kilian et al., submitted). Sequencing of close to 10,000 clones from the gdh libraries generated from the samples collected from the two subjects on two occasions separated by 2 years ensured detection of clones that constituted minor proportions (<0.1%) of the populations of the three species.
By nature, our method allows comparison of only one gene locus, which theoretically limits its discriminatory power. However, comprehensive phylogenetic analysis of a large collection of isolates supported by polyphasic taxonomic analysis demonstrated that gdh sequences provided unambiguous and reliable information about species affiliation and that sharing of the same allele by more strains is rare (Kilian et al., submitted), as confirmed by the data shown in Fig. 1. In contrast, recent studies demonstrated that no known phenotypic trait can be used to differentiate the species in question (16). As a consequence, previous conclusions concerning the diversity of S. mitis and S. oralis populations (2, 14, 15, 20) conceivably were based on mixtures of species.
Artifacts in the form of chimeric sequences are a potential risk in PCR-based analyses of complex microbiota. In this context it may be important that the streptococci in question are naturally competent for transformation and that the mosaic structures of genes reflect the fact that homologous recombination contributed to their evolution (9, 10, 16, 22, 30). Therefore, our attempt to eliminate artificial chimeric sequences by discarding the sequences showing discordant phylogenetic affiliations at the two ends may, theoretically, have resulted in an underestimate of the natural diversity among the sequences.
S. oralis, S. mitis, and S. infantis are all members of the mitis group, which currently includes 12 recognized species. We have previously demonstrated that the primer set used in this study amplifies the target gdh sequence exclusively in all strains of S. mitis, S. oralis, S. infantis, S. pneumoniae, and S. pseudopneumoniae and in some strains previously assigned to “S. mitis biovar 2” (Kilian et al., submitted). Phylogenetic analysis of gdh sequences revealed distinct clusters of S. oralis and S. infantis. However, S. pneumoniae and S. pseudopneumoniae each constitute single lineages within a distinct cluster of multiple lineages, the remainder of which currently belong to S. mitis (Kilian et al., submitted). These phylogenetic relationships are also reflected in the tree shown in Fig. 2. Although some gdh alleles obtained in the present study were closely related to the reference sequences for S. pneumoniae, we excluded the possibility that they originated from this species based on our previous observation that gdh sequences of S. pneumoniae form a monophyletic subcluster (Kilian et al., submitted). However, we cannot exclude the possibility that some of the gdh sequences represent the recently described organism S. pseudopneumoniae, for which no information on occurrence is available. As indicated by the separate clustering of the two reference strains of “S. mitis biovar 2,” none of the sequences amplified from the samples represented this taxon. In agreement with our previous observation based on comprehensive phylogenetic analysis, “S. mitis biovar 2” represents a distinct species that is more closely related to S. oralis than to S. mitis.
At the time of the first sampling, a total of 39 distinct gdh alleles (21 alleles in subject A and 18 alleles in subject B) were detected in the two individuals. According to the total richness estimate, these numbers represent approximately 95.5 and 100% of the total number of distinct alleles in the libraries established for the two subjects. We equated these figures with the number of clones while recognizing that this may have caused a marginal underestimate of the actual number of clones due to potential sharing of gdh alleles by distinct clones (Table 1). Of the 39 clones present at the time of the baseline observation, 18 were redetected at the time of the second observation. Combined with the fact that 38 of 56 clones identified at the second sampling time were not detected at the initial sampling time, these observations suggest that acquisition and loss contribute to the strikingly unstable population dynamics of each of the three species, but we cannot exclude the possibility that some of these clones were present on both occasions but below the detection level. The suggestion that acquisition contributes to this phenomenon is in agreement with the previously observed transfers of S. oralis clones among cohabiting couples (2) and occasional sharing of S. mitis clones by infants and their parents (11, 14).
Possible biases introduced by the PCR-based technique prevent the relative abundance of a genotype from being precisely determined from its proportional representation among amplified sequences. Yet previous studies demonstrated good agreement between the proportions of bacterial species detected by in situ hybridization and the relative proportions in clone libraries (7, 33). On this basis, Fig. 4 clearly demonstrates that the majority of the clones carried by the two individuals studied accounted for minor proportions of the populations of the respective species. It is conceivable that such clones would have been missed by cultivation even if multiple isolates were examined. Furthermore, Fig. 4 shows examples of fluctuations from minor proportions to predominance and vice versa. This finding is in agreement with our hypothesis that part of the apparent acquisition and loss of clones observed in cultivation studies may be explained by such fluctuations (11, 15, 20). Furthermore, our longitudinal data demonstrate that such fluctuations also affect the balance between individual species. While S. oralis was the predominant species among the three species harbored by the two individuals at the baseline sampling time, both subjects exhibited a shift toward predominance of S. mitis 2 years later (Table 1). Due to the fact that this study was based on pooled samples from multiple surfaces in the oral cavity and pharynx, we could not discern if such fluctuations are confined to particular habitats.
In conclusion, this study revealed significant clonal diversity within populations of S. mitis, S. oralis, and S. infantis carried by individuals in the upper respiratory tract. The total number of clones, the demonstration of numerous clones in proportions that are not detectable by traditional culture methods, and the observed significant changes in proportions over time demonstrate that the populations of commensal streptococci in the oral cavity and pharynx are constantly changing due to significant fluctuations in the relative proportions of existing clones and species possibly combined with loss and acquisition from close contacts. One important biological implication of these findings is that, in contrast to individual clones of potential pathogens, like the closely related organism S. pneumoniae, commensal bacteria are not subject to rapid elimination by the adaptive mucosal immune system. Rather, the observed fluctuations in abundance of individual clones of commensal streptococci are due to interactions within the complex microbiota, the nature of which is largely unknown.
Acknowledgments
This study was supported by grants from the Faculty of Health Sciences, Aarhus University, the Velux Foundation, and the Karen Elise Jensen Foundation.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 3 March 2008.
REFERENCES
- 1.Aas, J. A., B. J. Paster, L. N. Stokes, I. Olsen, and F. E. Dewhirst. 2005. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 435721-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alam, S., S. R. Brailsford, S. Adams, C. Allison, E. Sheehy, L. Zoitopoulos, E. A. Kidd, and D. Beighton. 2000. Genotypic heterogeneity of Streptococcus oralis and distinct aciduric subpopulations in human dental plaque. Appl. Environ. Microbiol. 663330-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arbique, J. C., C. Poyart, P. Trieu-Cuot, G. Quesne, M. G. Carvalho, A. G. Steigerwalt, R. E. Morey, D. Jackson, R. J. Davidson, and R. R. Facklam. 2004. Accuracy of phenotypic and genotypic testing for identification of Streptococcus pneumoniae and description of Streptococcus pseudopneumoniae sp. nov. J. Clin. Microbiol. 424686-4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowden, G. H., J. Ekstrand, B. McNaughton, and S. J. Challacombe. 1990. Association of selected bacteria with the lesions of root surface caries. Oral Microbiol. Immunol. 5346-351. [DOI] [PubMed] [Google Scholar]
- 5.Carratala, J., B. Roson, A. Fernandez-Sevilla, F. Alcaide, and F. Gudiol. 1998. Bacteremic pneumonia in neutropenic patients with cancer: causes, empirical antibiotic therapy, and outcome. Arch. Intern. Med. 158868-872. [DOI] [PubMed] [Google Scholar]
- 6.Caugant, D. A., B. R. Levin, and R. K. Selander. 1981. Genetic diversity and temporal variation in the E. coli population of a human host. Genetics 98467-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dedysh, S. N., T. A. Pankratov, S. E. Belova, I. S. Kulichevskaya, and W. Liesack. 2006. Phylogenetic analysis and in situ identification of Bacteria community composition in an acidic Sphagnum peat bog. Appl. Environ. Microbiol. 722110-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diaz, P. I., N. I. Chalmers, A. H. Rickard, C. Kong, C. L. Milburn, R. J. Palmer, Jr., and P. E. Kolenbrander. 2006. Molecular characterization of subject-specific oral microflora during initial colonization of enamel. Appl. Environ. Microbiol. 722837-2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dowson, C. G., A. Hutchison, N. Woodford, A. P. Johnson, R. C. George, and B. G. Spratt. 1990. Penicillin-resistant viridans streptococci have obtained altered penicillin-binding protein genes from penicillin-resistant strains of Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 875858-5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feil, E. J., J. M. Smith, M. C. Enright, and B. G. Spratt. 2000. Estimating recombinational parameters in Streptococcus pneumoniae from multilocus sequence typing data. Genetics 1541439-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fitzsimmons, S., M. Evans, C. Pearce, M. J. Sheridan, R. Wientzen, G. Bowden, and M. F. Cole. 1996. Clonal diversity of Streptococcus mitis biovar 1 isolates from the oral cavity of human neonates. Clin. Diagn. Lab. Immunol. 3517-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frandsen, E. V., V. Pedrazzoli, and M. Kilian. 1991. Ecology of viridans streptococci in the oral cavity and pharynx. Oral Microbiol. Immunol. 6129-133. [DOI] [PubMed] [Google Scholar]
- 13.Han, X. Y., M. Kamana, and K. V. Rolston. 2006. Viridans streptococci isolated by culture from blood of cancer patients: clinical and microbiologic analysis of 50 cases. J. Clin. Microbiol. 44160-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hohwy, J., and M. Kilian. 1995. Clonal diversity of the Streptococcus mitis biovar 1 population in the human oral cavity and pharynx. Oral Microbiol. Immunol. 1019-25. [DOI] [PubMed] [Google Scholar]
- 15.Hohwy, J., J. Reinholdt, and M. Kilian. 2001. Population dynamics of Streptococcus mitis in its natural habitat. Infect. Immun. 696055-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoshino, T., T. Fujiwara, and M. Kilian. 2005. Use of phylogenetic and phenotypic analyses to identify nonhemolytic streptococci isolated from bacteremic patients. J. Clin. Microbiol. 436073-6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu, R. B., and F. Y. Lin. 2006. Effect of penicillin resistance on presentation and outcome of nonenterococcal streptococcal infective endocarditis. Cardiology 105234-239. [DOI] [PubMed] [Google Scholar]
- 18.Kawamura, Y., X. G. Hou, Y. Todome, F. Sultana, K. Hirose, S. E. Shu, T. Ezaki, and H. Ohkuni. 1998. Streptococcus peroris sp. nov. and Streptococcus infantis sp. nov., new members of the Streptococcus mitis group, isolated from human clinical specimens. Int. J. Syst. Bacteriol. 48921-927. [DOI] [PubMed] [Google Scholar]
- 19.Kazor, C. E., P. M. Mitchell, A. M. Lee, L. N. Stokes, W. J. Loesche, F. E. Dewhirst, and B. J. Paster. 2003. Diversity of bacterial populations on the tongue dorsa of patients with halitosis and healthy patients. J. Clin. Microbiol. 41558-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirchherr, J. L., G. H. Bowden, D. A. Richmond, M. J. Sheridan, K. A. Wirth, and M. F. Cole. 2005. Clonal diversity and turnover of Streptococcus mitis bv. 1 on shedding and nonshedding oral surfaces of human infants during the first year of life. Clin. Diagn. Lab. Immunol. 121184-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar, K., K. Tamura, and M. Nei. 2004. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinformatics 5150-163. [DOI] [PubMed] [Google Scholar]
- 22.Kuramitsu, H. K., and V. Trapa. 1984. Genetic exchange between oral streptococci during mixed growth. J. Gen. Microbiol. 1302497-2500. [DOI] [PubMed] [Google Scholar]
- 23.Lalani, T., Z. A. Kanafani, V. H. Chu, L. Moore, G. R. Corey, P. Pappas, C. W. Woods, C. H. Cabell, B. Hoen, C. Selton-Suty, T. Doco-Lecompte, C. Chirouze, D. Raoult, J. M. Miro, C. A. Mestres, L. Olaison, S. Eykyn, E. Abrutyn, and V. G. Fowler, Jr. 2006. Prosthetic valve endocarditis due to coagulase-negative staphylococci: findings from the International Collaboration on Endocarditis Merged Database. Eur. J. Clin. Microbiol. Infect. Dis. 25365-368. [DOI] [PubMed] [Google Scholar]
- 24.Liesack, W., and E. Stackebrandt. 1992. Occurrence of novel groups of the domain Bacteria as revealed by analysis of genetic material isolated from an Australian terrestrial environment. J. Bacteriol. 1745072-5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu, C. H., W. N. Chang, Y. C. Lin, N. W. Tsai, P. C. Liliang, T. M. Su, C. S. Rau, Y. D. Tsai, C. L. Liang, C. J. Chang, P. Y. Lee, H. W. Chang, and J. J. Wu. 2002. Bacterial brain abscess: microbiological features, epidemiological trends and therapeutic outcomes. QJM 95501-509. [DOI] [PubMed] [Google Scholar]
- 26.Møller, K., E. H. Frederiksen, J. H. Wandall, and P. Skinhøj. 1999. Meningitis caused by streptococci other than Streptococcus pneumoniae: a retrospective clinical study. Scand. J. Infect. Dis. 31375-381. [DOI] [PubMed] [Google Scholar]
- 27.Nyvad, B., and M. Kilian. 1990. Comparison of the initial streptococcal microflora on dental enamel in caries-active and in caries-inactive individuals. Caries Res. 24267-272. [DOI] [PubMed] [Google Scholar]
- 28.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 1833770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pearce, C., G. H. Bowden, M. Evans, S. P. Fitzsimmons, J. Johnson, M. J. Sheridan, R. Wientzen, and M. F. Cole. 1995. Identification of pioneer viridans streptococci in the oral cavity of human neonates. J. Med. Microbiol. 4267-72. [DOI] [PubMed] [Google Scholar]
- 30.Poulsen, K., J. Reinholdt, C. Jespersgaard, K. Boye, T. A. Brown, M. Hauge, and M. Kilian. 1998. A comprehensive genetic study of streptococcal immunoglobulin A1 proteases: evidence for recombination within and between species. Infect. Immun. 66181-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prasad, K. N., A. M. Mishra, D. Gupta, N. Husain, M. Husain, and R. K. Gupta. 2006. Analysis of microbial etiology and mortality in patients with brain abscess. J. Infect. 53221-227. [DOI] [PubMed] [Google Scholar]
- 32.Saiki, R. K., D. H. Gelfand, S. Stoffel, S. J. Scharf, R. Higuchi, G. T. Horn, K. B. Mullis, and H. A. Erlich. 1988. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239487-491. [DOI] [PubMed] [Google Scholar]
- 33.Schmitt-Wagner, D., M. W. Friedrich, B. Wagner, and A. Brune. 2003. Phylogenetic diversity, abundance, and axial distribution of bacteria in the intestinal tract of two soil-feeding termites (Cubitermes spp.). Appl. Environ. Microbiol. 696007-6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith, D. J., J. M. Anderson, W. F. King, H. J. van, and M. A. Taubman. 1993. Oral streptococcal colonization of infants. Oral Microbiol. Immunol. 81-4. [DOI] [PubMed] [Google Scholar]
- 35.Tunkel, A. R., and K. A. Sepkowitz. 2002. Infections caused by viridans streptococci in patients with neutropenia. Clin. Infect. Dis. 341524-1529. [DOI] [PubMed] [Google Scholar]
- 36.van Houte, J., J. Lopman, and R. Kent. 1994. The predominant cultivable flora of sound and carious human root surfaces. J. Dent. Res. 731727-1734. [DOI] [PubMed] [Google Scholar]
- 37.Whitford, M. F., R. J. Forster, C. E. Beard, J. Gong, and R. M. Teather. 1998. Phylogentic analysis of rumen bacteria by comparative sequence analysis of cloned 16S rRNA genes. Anaerobe 4153-163. [DOI] [PubMed] [Google Scholar]
- 38.Wisplinghoff, H., R. R. Reinert, O. Cornely, and H. Seifert. 1999. Molecular relationships and antimicrobial susceptibilities of viridans group streptococci isolated from blood of neutropenic cancer patients. J. Clin. Microbiol. 371876-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]