Abstract
Salmonella enterica serotype Typhimurium causes an acute inflammatory reaction in the ceca of streptomycin-pretreated mice. We determined global changes in gene expression elicited by serotype Typhimurium in the cecal mucosa. The gene expression profile was dominated by T-cell-derived cytokines and genes whose expression is known to be induced by these cytokines. Markedly increased mRNA levels of genes encoding gamma interferon (IFN-γ), interleukin-22 (IL-22), and IL-17 were detected by quantitative real-time PCR. Furthermore, the mRNA levels of genes whose expression is induced by IFN-γ, IL-22, or IL-17, including genes encoding macrophage inflammatory protein 2 (MIP-2), inducible nitric oxide synthase (Nos2), lipocalin-2 (Lcn2), MIP-1α, MIP-1β, and keratinocyte-derived cytokine (KC), were also markedly increased. To assess the importance of T cells in orchestrating this proinflammatory gene expression profile, we depleted T cells by using a monoclonal antibody prior to investigating cecal inflammation caused by serotype Typhimurium in streptomycin-pretreated mice. Depletion of CD3+ T cells resulted in a dramatic reduction in gross pathology, a significantly reduced recruitment of neutrophils, and a marked reduction in mRNA levels of Ifn-γ, Il-22, Il-17, Nos2, Lcn2, and Kc. Our results suggest that T cells play an important role in amplifying inflammatory responses induced by serotype Typhimurium in the cecal mucosa.
Salmonella enterica serotype Typhimurium elicits intestinal inflammation in the terminal ileum and colon of patients that is characterized by a massive neutrophil influx (3, 17). The generation of intestinal inflammatory responses during serotype Typhimurium infection can be studied using streptomycin-pretreated mice (1). It has recently been proposed that the orchestration of serotype Typhimurium-induced intestinal inflammation can be broken down into mechanisms involved in its induction and mechanisms involved in the amplification of inflammatory responses in tissue (29).
Several factors for the initial induction of inflammatory responses have been proposed using tissue culture models. These include cytokine and hepoxilin A3 production induced by the invasion-associated type III secretion system (T3SS-1) of serotype Typhimurium in intestinal epithelial cells (2, 9, 11, 16, 21), the activation of Toll-like receptors in epithelial cells and macrophages through pathogen-associated molecular patterns (12, 22, 23, 34, 35, 39), and the stimulation of cytosolic nucleotide-binding and oligomerization domain-like receptors by a T3SS-1-dependent translocation of flagellin into the cytosol of macrophages (6, 18, 33).
Compared to factors involved in the initial induction of inflammatory responses, relatively little is known about the mechanisms involved in the subsequent amplification of inflammatory responses in the intestinal mucosa. It has been proposed that mononuclear cells (macrophages and/or dendritic cells) play an important role in initiating the amplification of responses against serotype Typhimurium in tissue by secreting specific cytokines (29). For example, in other models of infection, mononuclear cells produce interleukin-23 (IL-23) and IL-18, two cytokines which stimulate T cells to produce IL-17 and gamma interferon (IFN-γ), respectively (4, 6-8, 18, 30, 37). These data suggest that the production of cytokines by T cells may be a central component of mechanisms that help amplify inflammatory responses in tissue. However, the potential contribution of T-cell-mediated amplification mechanisms to the development of the acute inflammatory reaction developing in the intestinal mucosa during serotype Typhimurium infection has not yet been investigated. Here we determined the gene expression profile elicited by serotype Typhimurium in the intestinal mucosa of streptomycin-pretreated mice and assessed the contribution of T cells to the orchestration of intestinal inflammation.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Serotype Typhimurium strain IR715 is a fully virulent, nalidixic acid-resistant derivative of isolate ATCC 14028 and was used in all experiments (32). Bacteria were cultured aerobically at 37°C in Luria-Bertani (LB) broth.
Purification of MAb.
Hybridoma lines producing monoclonal antibodies (MAb) (rat immunoglobulin G [IgG]) against murine CD3 were purchased from ATCC (CRL-1975). Hybridoma lines were expanded and maintained in Iscove's modified Dulbecco's medium (Gibco catalog no. 12440) containing 10% fetal bovine serum and 5% antibiotic-antimycotic solution (Gibco catalog no. 15240-062). Cells were incubated at 37°C with 5% CO2. Every 2 to 3 days, media were replaced and stored at −20°C until ready for MAb isolation. Medium supernatant containing MAb was diluted 1:1 with 20 mM sodium phosphate in phosphate-buffered saline (PBS), pH 7.0, and pumped through a protein G column (GE HiTrap catalog no. 17-0404-01) using a Bio-Rad peristaltic pump (Biologic LP) set at a rate of 1 ml/min. Columns were then washed using 10 ml of 20 mM sodium phosphate in PBS, pH 7.0, at a rate of 1 ml/min. Columns were eluted using 5 ml of 0.1 M glycine-HCl, pH 2.7. The eluent was aliquoted in 1.5-ml Eppendorf tubes containing 100 μl of 1 M Tris-HCl, pH 9.0, to adjust the pH. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed to qualitatively ascertain the purity of the MAb. Aliquots were then dialyzed against sterile PBS at 4°C for 48 h and subsequently filter sterilized by passage through a 0.2-μm syringe filter (Pall catalog product no. 4612). Protein concentrations were determined using a Bio-Rad DC protein assay (catalog no. 500-0113 through -0115) as described by the manufacturer.
Animal experiments.
To study inflammation in the cecum, 8-week-old streptomycin-pretreated C57BL/6 mice were infected orally with Salmonella serotypes as described previously (1). In brief, groups of four mice were inoculated with streptomycin (0.1 ml of a 200-mg/ml solution in PBS) intragastrically. Mice were inoculated intragastrically 24 h later either with sterile LB broth or with bacteria (0.2 ml containing approximately 1 × 109 CFU/ml). At the indicated time points after infection, mice were euthanized and samples of the cecum were collected for the isolation of mRNA and histopathological analysis. For bacteriologic analysis, cecal contents, mesenteric lymph nodes, and the liver were homogenized and serial 10-fold dilutions were spread on agar plates containing the appropriate antibiotics.
For T-cell depletion experiments, mice were injected intraperitoneally once daily with 0.15 mg/animal of anti-CD3 MAb or an isotype control (purified rat IgG from Sigma) for 3 days. On the last day of antibody treatment, mice were pretreated with streptomycin as described above. Mice were inoculated intragastrically 24 h later either with sterile LB broth or with bacteria (0.2 ml containing approximately 1 × 109 CFU/animal). At 48 h after infection, mice were euthanized and samples of the cecum were collected for isolation of RNA and histopathological analysis. The spleen from each animal was collected to obtain splenocyte populations for analysis.
Gene expression profiling.
For analysis of changes in gene expression after serotype Typhimurium infection, tissue samples of murine cecum were collected, immediately snap-frozen in liquid nitrogen at the site of necropsy, and stored at −80°C until processing. RNA was then extracted from snap-frozen tissue with Tri reagent (Molecular Research Center) according to the instructions of the manufacturer.
Gene expression profiles in cecal tissues were monitored utilizing a GeneChip mouse expression set 430-2.0 array, representing 45,000 transcripts (Affymetrix, Santa Clara, CA). For each treatment group, total cecal RNA from four mice was pooled and double-stranded cDNA was amplified and biotin labeled with a one-cycle target labeling kit (Affymetrix, Santa Clara, CA) according to the manufacturer's instruction. Biotin-labeled cRNA was hybridized overnight (16 h) to mouse GeneChips and subsequently stained with a streptavidin-phycoerythrin conjugate and scanned with an Affymetrix 3000 scanner at an excitation wavelength of 488 nm. The fluorescence emitted at 570 nm was measured and used to calculate and compare the expression levels of each gene under each experimental condition. To minimize the occurrence of false positives in the array data, a minimum twofold difference in mRNA levels (P value of <0.05; 95% confidence interval) between control and experimental samples was used as the criterion for identifying a “change” in gene expression. Statistical validation was determined through the analysis of at least 11 independent 25-mer oligonucleotide probes for each gene in each sample. Pathway analysis of microarray data was performed using the Ingenuity Pathway Analysis web interface, the DAVID annotation tool, and EASE annotation software (http://david.abcc.ncifcrf.gov/). Statistically overrepresented biological processes (pathways) were determined with better than 95% confidence interval (P value of <0.05).
Real-time PCR.
For quantitative analysis of mRNA levels, 1 μg of RNA from each sample was reverse transcribed in a 50-μl volume (TaqMan reverse transcription reagent; Applied Biosystems), and 4 μl of cDNA was used for each real-time reaction. Real-time PCR was performed using Sybr green (Applied Biosystems) and a 7900HT fast real-time PCR system. The data were analyzed using a comparative cycle threshold method (Applied Biosystems). Increases in cytokine expression in infected mice were calculated relative to the average level of the respective cytokine in four control animals from the corresponding time point after inoculation with sterile LB broth. A list of genes analyzed in this study with the respective primers is provided in Table 1.
TABLE 1.
Primers for real-time PCR
| Gene | Primer pair |
|---|---|
| Gapdha | 5′-TGTAGACCATGTAGTTGAGGTCA-3′ |
| 5′-AGGTCGGTGTGAACGGATTTG-3′ | |
| Il-6 | 5′-GAGGATACCACTCCCAACAGACC-3′ |
| 5′-AAGTGCATCATCGTTGTTCATACA-3′ | |
| Il-12/Il-23p40 | 5′-GGAAGCACGGCAGCAGAATA-3′ |
| 5′-AACTTGAGGGAGAAGTAGGAATGG-3′ | |
| Il-23p19 | 5′-TGTGCCTAGGAGTAGCAGTCCTGA-3′ |
| 5′-TTGGCGGATCCTTTGCAAGCAGAA-3′ | |
| Il-17 | 5′-GCTCCAGAAGGCCCTCAGA-3′ |
| 5′-AGCTTTCCCTCCGCATTGA-3′ | |
| Il-22 | 5′-GGCCAGCCTTGCAGATAACA-3′ |
| 5′-GCTGATGTGACAGGAGCTGA-3′ | |
| Tnf-α | 5′-CATCTTCTCAAAATTCGAGTGACAA-3′ |
| 5′-TGGGAGTAGACAAGGTACAACCC-3′ | |
| Mip-2 | 5′-AGTGAACTGCGCTGTCAATGC-3′ |
| 5′-AGGCAAACTTTTTGACCGCC-3′ | |
| Kc | 5′-TGCACCCAAACCGAAGTCAT-3′ |
| 5′-TTGTCAGAAGCCAGCGTTCAC-3′ | |
| Nos2 | 5′-TTGGGTCTTGTTCACTCCACGG-3′ |
| 5′-CCTCTTTCAGGTCACTTTGGTAGG-3′ | |
| Lcn2 | 5′-ACATTTGTTCCAAGCTCCAGGGC-3′ |
| 5′-CATGGCGAACTGGTTGTAGTCCG-3′ | |
| Mip-1α | 5′-CCATGACACTCTGCAACCAAGT-3′ |
| 5′-TCCGGCTGTAGGAGAAGCA-3′ | |
| Mip-1β | 5′-TGCCCTCTCTCTCCTCTTGCT-3′ |
| 5′-CAGGAAGTGGGAGGGTCAGA-3′ | |
| Ifn-γ | 5′-TCAAGTGGCATAGATGTGGAAGAA-3′ |
| 5′-TGGCTCTGCAGGATTTTCATG-3′ |
Gapdh, gene encoding glyceraldehyde-3-phosphate dehydrogenase.
Flow cytometry.
To prepare splenocytes, single-cell suspensions were prepared by pressing the textured base of a sterile 10-ml syringe on spleens in a circular motion in a 150-mm petri dish. The resulting suspension was passed through a cell strainer (70 μm pore size). Cells were then harvested at 200 × g for 10 min at 4°C. The cell pellet was resuspended in 10 ml ACK buffer (NH4Cl, 8.29 g/liter; KHCO3, 1 g/liter; Na2EDTA·2H2O, 37.2 mg/liter) and incubated at room temperature for 10 min. An additional 10 ml of ACK buffer was added and incubated at room temperature for five more minutes. Cells were centrifuged at 200 × g for 10 min at 4°C. The resulting pellet was adjusted to a concentration of approximately 5 × 107 cells/ml in PBS containing 1% bovine serum albumin (BSA).
A volume of 50 μl of the splenocyte solution was treated with 0.25 μg of anti-mouse CD16/32 (eBioscience clone 93) for 1 h at 4°C. Aliquots were subsequently stained for 1 h at 4°C with phycoerythrin-conjugated CD3 (BD clone 17A2). The amount of each MAb used was determined in a titration experiment. Cells were washed with 300 μl of 1% BSA in PBS and centrifuged at 200 × g. Cells were fixed in 4% Formalin overnight at 4°C. Cells were washed twice as described above and resuspended in 300 μl of 1% BSA in PBS for flow cytometry. Cells were analyzed on a FACScan instrument, and results were analyzed using FlowJo software (TreeStar, Inc., Ashland, OR).
Histopathology.
Tissue samples were fixed in formalin, processed according to standard procedures for paraffin embedding, sectioned at 5 μm, and stained with hematoxylin and eosin. A veterinary pathologist scored inflammatory changes using a blind-sample analysis. Neutrophil counts were determined per high (×40)-magnification microscopy, and numbers were averaged from 10 microscopic fields per animal.
Statistical analysis.
Microarray data were analyzed using model-based algorithms (dChip; http://biosun1.harvard.edu/complab/dchip) and t tests. Genes with significantly (P < 0.05) altered expression were subjected to hierarchical clustering and then to functional and statistical analyses of the genes in each subcluster. Changes in mRNA levels measured by real-time PCR underwent logarithmic transformation, and percent values underwent angular transformation prior to analysis by Student's t test.
Microarray data accession numbers.
Microarray data were deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database under the microarray data accession number GSE10594.
RESULTS
Kinetics of cytokine production in the cecum during serotype Typhimurium infection.
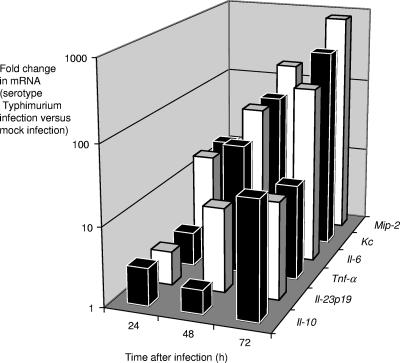
We performed a pilot experiment to determine the kinetics of cytokine production in the cecal mucosa of mice. Six groups of four streptomycin-pretreated mice were inoculated with sterile LB broth (mock infection) or with serotype Typhimurium. One group of mock-infected mice and one group of serotype Typhimurium-infected mice were euthanized at 24, 48, and 72 h after infection. RNA from the cecum was extracted, and the expression levels of neutrophil chemoattractants (macrophage inflammatory protein 2 [MIP-2] and keratinocyte-derived cytokine [KC]), proinflammatory cytokines (tumor necrosis factor alpha [TNF-α], IL-6, and IL-23p19), and anti-inflammatory cytokines (IL-10) for each animal were determined by quantifying the respective mRNA levels using real-time PCR (Fig. 1). This analysis revealed that proinflammatory cytokines were strongly induced in the cecal mucosa by 48 h after serotype Typhimurium infection, and this time point was chosen for subsequent studies.
FIG. 1.
Time course of cytokine responses in the ceca of streptomycin-pretreated mice during serotype Typhimurium infection. For each time point, changes in gene expression observed for serotype Typhimurium-infected mice (n = 4) compared to that observed for mock-infected mice (n = 4) were determined. The graph shows geometric means of increases (n-fold) in Mip-2, Kc, Il-6, Tnf-α, Il23p19, and Il-10 expression over time.
Characterization of serotype Typhimurium infection by gene expression profiling of the cecal mucosa.
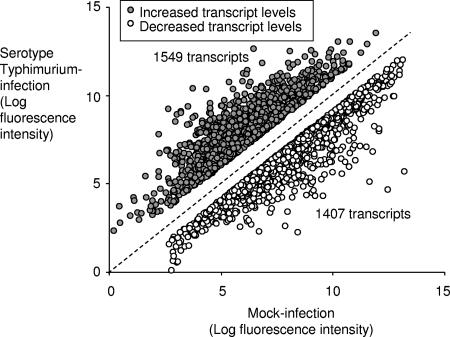
We reasoned that if an amplification of inflammatory responses occurs during serotype Typhimurium infection, then genes whose expression is induced by these amplification mechanisms would be expected to dominate the list of genes whose transcript levels are increased in infected intestinal tissue. Groups of four streptomycin-pretreated mice were inoculated with sterile LB broth (mock infection) or with serotype Typhimurium, and RNA from the cecum was extracted 48 h after infection. Compared to levels for mock infection, levels of 1,549 transcripts were elevated during serotype Typhimurium infection (≥2-fold change, P < 0.05), while levels of 1,407 transcripts were decreased (Fig. 2).
FIG. 2.
Serotype Typhimurium-induced increases and decreases in mRNA levels in the cecal mucosa. The graph shows the changes in expression for genes whose transcript levels were significantly increased (n = 1,549) or decreased (n = 1,407) 48 h after serotype Typhimurium infection compared to levels after mock infection.
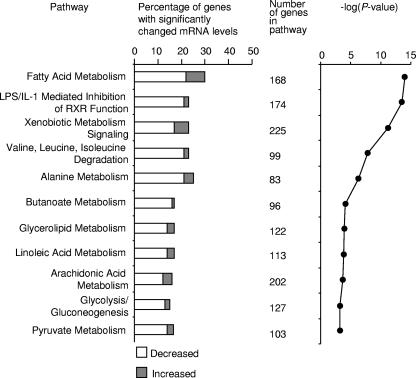
A large majority of the genes whose transcript levels were reduced in the cecum during serotype Typhimurium infection were associated with the metabolic functions of the intestinal epithelium (Fig. 3). Multiple genes involved in metabolism of lipids, xenobiotics, carboxylic acids, carbohydrates, and amino acids showed substantially decreased mRNA levels compared to levels for healthy uninfected controls. Additionally, genes that control retinoid X receptor-based signaling in response to lipopolysaccharide and/or IL-1 were found at reduced mRNA levels.
FIG. 3.
Gene expression profile of transcripts that were significantly decreased in the cecal mucosa during serotype Typhimurium infection. Biological analysis of microarray data was performed using the Affymetrix NetAFFX web interface and the DAVID (http://david.abcc.ncifcrf.gov/) annotation tool. Statistically overrepresented (P < 0.05) biological processes within subclusters were identified using EASE (http://david.abcc.ncifcrf.gov/). Biological processes that were significantly overrepresented among decreased transcripts are indicated. LPS, lipopolysaccharide; RXR, retinoid X receptor.
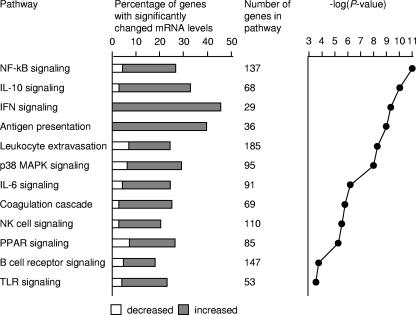
The profile of genes whose transcript levels increased during serotype Typhimurium infection in the cecum was dominated by responses related to innate and adaptive immunity (Fig. 4). Genes whose mRNA levels were increased most dramatically in serotype Typhimurium-infected tissue compared to mock-infected tissue included Il-22 and Ifn-γ (Table 2), which encode two cytokines that can be produced by T cells (4, 15, 30). IL-22 and IFN-γ synergize to increase the expression of nitric oxide synthase (iNOS) (40), and mRNA levels of these genes were also markedly increased after serotype Typhimurium infection (Table 2). Other IFN-γ-inducible genes whose mRNA levels were markedly increased during serotype Typhimurium infection included genes encoding IFN-induced protein 44 (Ifi44), an IFN-inducible GTPase (Gvin1), and Cxcl10 (Ip-10) (Table 2). IL-22 is produced by a subset of T cells that also secrete IL-17 (15). We have shown recently that serotype Typhimurium infection of streptomycin-pretreated mice results in a marked increase in mRNA levels of Il-17 in the cecal mucosa by 48 h after infection (24). Although changes in the level of Il-17 mRNA could not be detected by microarray analysis, serotype Typhimurium infection resulted in markedly increased mRNA levels of genes whose expression is induced by IL-17, including Mip-2 (38), Lcn2 (lipocalin-2) (28), Mip-1α (38), Kc (5), and Mip-1β (38) (Table 2). Overall, gene expression profiling suggested that many of the genes whose mRNA levels were increased most dramatically during serotype Typhimurium infection either encoded cytokines that were potentially derived from T cells or encoded products whose expression is regulated by these cytokines.
FIG. 4.
Gene expression profile of transcripts that were significantly increased in the cecal mucosa during serotype Typhimurium infection. Biological analysis of microarray data was performed using the Affymetrix NetAFFX web interface and the DAVID (http://david.abcc.ncifcrf.gov/) annotation tool. Statistically overrepresented (P < 0.05) biological processes within subclusters were identified using EASE (http://david.abcc.ncifcrf.gov/). Biological processes that were significantly overrepresented among increased transcripts are indicated. MAPK, mitogen-activated protein kinase; PPAR, peroxisome proliferator-activated receptor; TLR, Toll-like receptor.
TABLE 2.
List of transcripts related to immunity and inflammation whose levels were elevated more than 10-fold (P < 0.05) in serotype Typhimurium-infected cecal tissue
| Gene | Gene expression profiling of mRNA from ceca of serotype Typhimurium-infected mice compared to those of mock-infected mice
|
Avg fold change determined by real-time PCR confirmation | |
|---|---|---|---|
| Fold change | P value | ||
| Cxcl2 (Mip-2) | 97.9 | 0.001 | 168 |
| Cxcl5 | 62.3 | 0.001 | NDa |
| Il-1α | 57.2 | 0.002 | ND |
| Cccl3 (Mip-1a) | 53.9 | 0.002 | 17.3 |
| Ifn-γ | 53.9 | 0.002 | 554 |
| Irg1 | 40.3 | 0.001 | ND |
| S100a8 (calgranulin A) | 37.0 | 0.001 | ND |
| S100a9 (calgranulin B) | 32.5 | 0.001 | ND |
| Nos2 (iNOS) | 30.6 | 0.008 | 56.9b |
| Il-22 | 27.9 | 0.002 | 579 |
| Il-6 | 26.7 | 0.002 | 44.0 |
| Lcn2 (lipocalin 2) | 26.6 | 0.003 | 58.3 |
| Fpr-rs2 | 25.5 | 0.003 | ND |
| Il-1β | 21.0 | 0.004 | ND |
| Cxcl1 (Kc) | 20.6 | 0.005 | 83.7 |
| Mmp3 | 20.5 | 0.003 | ND |
| Ccl4 (Mip-1β) | 20.4 | 0.003 | 22.7 |
| Il-33 | 18.8 | 0.001 | ND |
| Ccl2 | 17.4 | 0.003 | ND |
| Il-1rn (IL-1 receptor antagonist) | 15.9 | 0.006 | ND |
| Il-18bp (IL-18 binding protein) | 15.8 | 0.002 | ND |
| Ccl7 | 15.6 | 0.007 | ND |
| Sell (lymphocyte selectin) | 15.2 | 0.005 | ND |
| Mmp8 | 14.0 | 0.007 | ND |
| Ltf (lactoferrin) | 13.6 | 0.017 | ND |
| Ifi44 (IFN-induced protein 44) | 13.6 | 0.004 | ND |
| Mmp10 | 13.4 | 0.007 | ND |
| Ccr5 | 12.4 | 0.004 | ND |
| Cxcl10 (Ip-10) | 11.9 | 0.002 | ND |
| Gvin1 (IFN-inducible GTPase) | 11.6 | 0.006 | ND |
| Cxcl11 | 11.1 | 0.006 | ND |
| Cxcl9 | 10.7 | 0.005 | ND |
| Itk (IL-2-inducible T-cell kinase) | 10.7 | 0.037 | ND |
| Tnf-α | 10.2 | 0.011 | 85.8 |
ND, not determined.
Determined for mock-depleted C57BL/6 mice.
Real-time PCR analysis demonstrates a dramatic increase in mRNA levels for Il-22, Ifn-γ, and Il-17 during serotype Typhimurium infection.
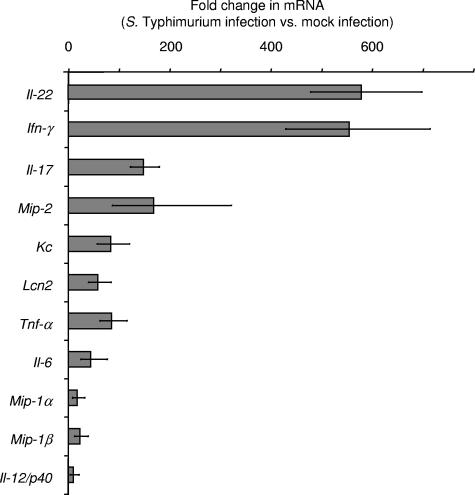
To confirm results from gene expression profiling, mRNA levels of cytokines for selected genes were monitored by quantitative real-time PCR at 48 h after infection by using the ceca from groups of four serotype Typhimurium-infected and four mock-infected mice. Quantitative measurements by real-time PCR showed that Il-22 (579-fold), Ifn-γ (554-fold), and Il-17 (149-fold) were the cytokine genes whose mRNA levels were elevated most dramatically after serotype Typhimurium infection (Fig. 5). Furthermore, the mRNA levels of genes regulated by IL-22, IL-17, and/or IFN-γ were also markedly increased, including Mip-2 (168-fold), Kc (84-fold), Lcn2 (58-fold), Mip-1α (17-fold), and Mip-1β (23-fold). In summary, real-time PCR analysis suggested that marked increases in the mRNA levels of Il-22, Il-17, and Ifn-γ were among the changes in the gene expression profile that exhibited the greatest magnitudes. Furthermore, our data confirmed the marked increase in mRNA levels of genes induced by IL-22, IL-17, and IFN-γ, which had been suggested by gene expression profiling (Table 2).
FIG. 5.
Cytokine expression elicited by serotype Typhimurium in streptomycin-pretreated mice 48 h after infection, as measured by quantitative real-time PCR. Changes in gene expression observed for serotype Typhimurium-infected mice (n = 4) compared to that observed for mock-infected mice (n = 4) were determined. Data are shown as geometric means of changes (n-fold) ± standard errors, determined for RNA from individual mice.
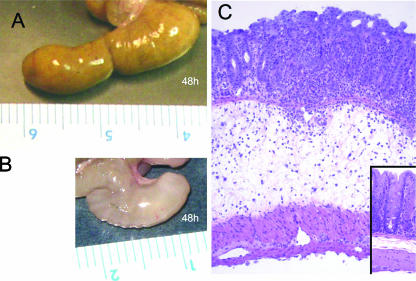
The detection of increased mRNA levels of genes associated with immunity and inflammation (Fig. 4 and 5) was accompanied by the development of pathological changes in the cecal mucosa (Fig. 6). At 48 h after inoculation, the ceca from serotype Typhimurium-infected mice contained little contents and had severe gross pathological changes, characterized by reduced size and thickening of the cecal wall (Fig. 6B). At 48 h after infection, the histopathology was characterized by epithelial erosion, neutrophil infiltration in the mucosa, and marked edema in the submucosa (Fig. 6C), which was consistent with previous reports describing cecal inflammation in this model (1, 24).
FIG. 6.
Gross pathological (A and B) and histopathological (C) appearance of the murine cecum. (A) Cecum of a streptomycin-pretreated mouse 48 h after inoculation with sterile LB broth, with a normal appearance. (B) Cecum of a streptomycin-pretreated mouse 48 h after inoculation with serotype Typhimurium. Note the reduced size of the cecum, which was devoid of contents and had a whitish discoloration and gelatinous appearance, indicating severe edema. (C) Murine cecum 48 h after infection with serotype Typhimurium, shown at low magnification (×10) to illustrate edema in the submucosa. The inset at the bottom right shows a section of the cecum from an animal 48 h after inoculation with sterile LB broth at the same magnification (×10).
T cells are major contributors to intestinal inflammation elicited by serotype Typhimurium.
Gene expression profiling and real-time PCR analysis suggested that the host response to serotype Typhimurium infection is dominated by a dramatic increase in the mRNA levels of Il-22, Il-17, and Ifn-γ and of genes whose expression can be induced by the cytokines encoded by these three genes (Table 2; Fig. 5). Since T cells are a potential source of IL-22, IL-17, and IFN-γ (4, 15, 30), these data raised the possibility that genes whose mRNA levels were increased most dramatically during serotype Typhimurium infection may participate in a T-cell-mediated amplification of responses in tissue. The intestine is the largest reservoir of T cells in the human body, and this cell type is present abundantly throughout the lamina propria, thus making this cell type well suited for amplifying inflammatory responses. To assess the importance of T cells in orchestrating inflammation, two groups of four mice were treated with rat anti-mouse CD3 MAb to deplete T cells. As a control, two groups of four mice were treated with nonspecific rat IgG control antibody (mock depletion). All four groups of mice were streptomycin pretreated. Four CD3-depleted and four mock-depleted mice were inoculated with sterile LB broth (mock infection), and the remaining two groups were infected with serotype Typhimurium. At 48 h after inoculation, organs were collected for analysis.
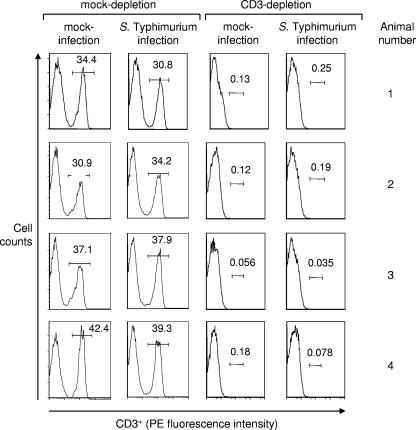
Spleens from all animals were processed for flow cytometry to monitor T-cell depletion. Splenocytes from mock-depleted mice contained on average 36.2% CD3+ T cells. This percentage remained unchanged when mock-depleted mice were infected with serotype Typhimurium (35.5% CD3+ T cells) (Fig. 7). In contrast, CD3+ T cells composed on average 0.12% of the splenocyte population in mice treated with the anti-mouse CD3 MAb. Similarly, splenocytes from CD3-depleted mice infected with serotype Typhimurium contained on average 0.12% of CD3+ T cells. These data suggested that T cells had been depleted effectively by treatment with rat anti-mouse CD3 MAb.
FIG. 7.
Fractions of T cells present in splenocyte populations of mice (n = 8) treated with anti-mouse CD3 MAb (CD3 depletion) or mice (n = 8) treated with nonspecific IgG antibody (mock depletion). Four mock-depleted and four CD3-depleted mice were infected with serotype Typhimurium, while the remaining animals were mock infected. Splenocytes were isolated 48 h after infection, and cells were analyzed by flow cytometry. The number in each box indicates the fraction of cells within the lymphocyte population that was scored CD3+. PE, phycoerythrin.
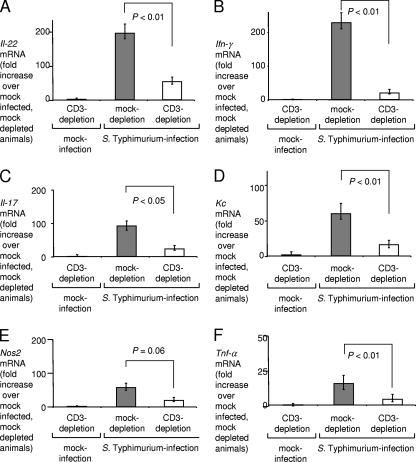
RNA was extracted from the cecum to investigate the development of proinflammatory changes in mRNA levels in each group. Mock infection of CD3-depleted mice resulted in mRNA levels of proinflammatory cytokines similar to those found after mock infection of mock-depleted mice. However, compared to mock infection of mock-depleted mice, serotype Typhimurium infection of mock-depleted mice resulted in markedly increased mRNA levels of genes encoding inflammatory cytokines, including Il-22 (197-fold) (Fig. 8A), Ifn-γ (228-fold) (Fig. 8B), Il-17 (92-fold) (Fig. 8C), Kc (60-fold) (Fig. 8D), Nos2 (57-fold) (Fig. 8E), and Tnf-α (16-fold) (Fig. 8F). In contrast, this marked induction of proinflammatory cytokine expression was not observed to occur during serotype Typhimurium infection of CD3-depleted mice (Fig. 8). CD3 depletion dramatically reduced the ability of serotype Typhimurium to elicit increased mRNA levels of Il-22, Il-17, and Ifn-γ and of genes regulated by the encoded cytokines. These data suggested that T cells were an important source of IL-22, IL-17, and IFN-γ production in the intestinal mucosa 48 h after serotype Typhimurium infection.
FIG. 8.
Changes in gene expression elicited 48 h after serotype Typhimurium infection in the cecal mucosa of mock-depleted mice (filled bars) or CD3-depleted mice (open bars), as measured by quantitative real-time PCR. Data are expressed as changes of mRNA levels over mRNA levels detected in mock-infected, mock-depleted mice. Data represent geometric means ± standard errors. Statistical significance of differences is indicated by P values.
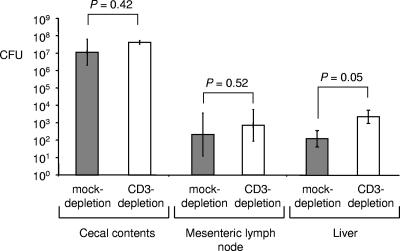
To determine whether attenuated inflammatory responses in CD3-depleted mice were due to a large decrease in the bacterial load, bacteria were recovered from cecal contents, the mesenteric lymph node, and the liver. CD3 depletion resulted in increased bacterial loads in the liver (Fig. 9) (P = 0.05). More bacteria were also recovered from cecal contents and mesenteric lymph nodes of CD3-depleted mice than from those of mock-depleted mice, but these differences were not significant. In conclusion, CD3-depleted mice exhibited similar (i.e., in the cecum and mesenteric lymph node) or increased (i.e., in the liver) bacterial burdens, while expression of inflammatory cytokines was markedly reduced. These data suggested that reduced cytokine responses in CD3-depleted mice were not caused by a reduction in bacterial numbers.
FIG. 9.
Recovery of serotype Typhimurium from the organs of mock-depleted mice (filled bars) or CD3-depleted mice (open bars). Bars represent geometric means ± standard errors of CFU/organ (cecal contents) or CFU/g tissue (mesenteric lymph node and liver). Statistical significance of differences is indicated by P values.
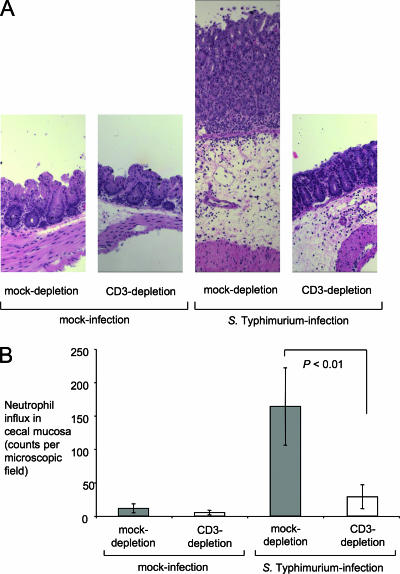
We next investigated the biological consequences of the blunted cytokine responses observed in T-cell-depleted mice by investigating the development of cecal inflammation. At 48 h after inoculation, ceca from mock-depleted mice infected with serotype Typhimurium contained little contents and had severe gross pathological changes, characterized by reduced size and thickening of the cecal wall. This appearance was typical of the gross pathological changes elicited by serotype Typhimurium at 48 h after infection in this model (Fig. 6B). In contrast, ceca from serotype Typhimurium-infected CD3-depleted mice did not show gross pathological changes and appeared similar to ceca from mock-infected mice (Fig. 6A). Histopathological evaluation revealed epithelial erosion, neutrophil infiltration in the mucosa, and edema in the submucosa of mock-depleted mice infected with serotype Typhimurium. These pathological changes were reduced drastically in serotype Typhimurium-infected CD3-depleted mice (Fig. 10A). Compared to serotype Typhimurium-infected mock-depleted mice, serotype Typhimurium-infected CD3-depleted mice recruited markedly reduced numbers of neutrophils into to the cecal mucosa (Fig. 10B), which correlated well with the reduced levels detected in this group of mRNA levels for the neutrophil chemoattractant gene Kc (Fig. 8D) and the reduced transcript levels for Il-17 (Fig. 7C), encoding a cytokine controlling expression of neutrophil chemoattractants (5, 38).
FIG. 10.
Effect of CD3 depletion on the histopathology of the murine cecum 48 h after infection with serotype Typhimurium. (A) All panels are shown at the same magnification (×10) to illustrate the magnitude of edema in serotype Typhimurium-infected animals. The two panels on the left show the normal appearance of the cecal mucosa in mock-infected animals. The second panel from the right shows the cecum of a mock-depleted animal 48 h after serotype Typhimurium infection. Note the marked thickening of the intestinal wall due to edema in the submucosa. The right panel shows the appearance of the cecal mucosa in a CD3-depleted animal 48 h after serotype Typhimurium infection. (B) Neutrophil recruitment into the cecal mucosa. The numbers of neutrophils per microscopic field were determined by a veterinary pathologist during a blind examination of slides from the cecal mucosa. Data represent means ± standard errors. Statistical significance of differences is indicated by P values.
DISCUSSION
Serotype Typhimurium causes a localized gastroenteritis in humans that is characterized by a massive neutrophil influx in the terminal ileum and colon. Upon translocation through the intestinal epithelium, bacteria are found mainly within mononuclear cells (i.e., macrophages or dendritic cells) or neutrophils in the lamina propria (25, 27). Some of the cytokines released by mononuclear cells after their stimulation with bacteria may help to amplify inflammatory responses in tissue (29). The gene expression profiles elicited in macrophages or epithelial cells infected with serotype Typhimurium have been described previously (23, 26, 39). However, since only a very small fraction of cells in infected tissue contain bacteria, cytokine production by these cells may be limited in scope. In contrast, responses that are amplified effectively in tissue would be expected to be responsible for the most prominent changes in gene expression observed in the cecal mucosa during serotype Typhimurium infection. Consistent with this idea, we found that mRNA levels upregulated most dramatically during serotype Typhimurium infection of the cecal mucosa included genes encoding T-cell-derived cytokines (i.e., IL-22, IL-17, and IFN-γ) and genes whose expression is known to be regulated by these cytokines. The predominance of these responses in vivo was not apparent from previous studies of gene expression profiles determined in tissue culture (23, 26, 39). Results from in vivo gene expression profiling were consistent with a central role of T cells in amplifying responses in tissue. Importantly, CD3+ T-cell depletion resulted in a dramatic attenuation of intestinal inflammatory responses during serotype Typhimurium infection. These data provided compelling support for the idea that T cells play an important role in amplifying local inflammatory responses during infection of the intestinal mucosa by serotype Typhimurium.
It is currently not clear by which mechanisms T cells in the cecal mucosa are stimulated to produce IL-22, IL-17, and IFN-γ during serotype Typhimurium infection. Previous studies have implicated mononuclear cells as possible sources for cytokines that would stimulate the release of IL-22, IL-17, and IFN-γ by T cells. For example, serotype Typhimurium flagellin is translocated by the T3SS-1 into the cytosol of macrophages (33), where it is sensed by Ipaf, a cytosolic nucleotide-binding and oligomerization domain-like receptor (6, 18). Recognition of flagellin by Ipaf leads to activation of caspase 1, which in turn cleaves precursors of IL-18 and IL-1β into their active forms (10, 20). Examination of splenic tissue from mice shows that IL-18 can stimulate antigen-experienced T cells to rapidly secrete IFN-γ during bacterial infection by an antigen-independent mechanism, thereby significantly amplifying early effector responses in vivo (30). In a mouse model of Klebsiella pneumoniae lung infection, bacterial stimulation of Toll-like receptors on dendritic cells results in IL-23 production, which in turn triggers the rapid production of IL-17 by T cells. This paracrine IL-23-mediated mechanism also triggers the release of IL-17 from T cells in a mouse model of Mycobacterium bovis infection (36). However, additional work is required to determine whether these pathways also participate in the amplification of inflammatory responses in the intestinal mucosa during serotype Typhimurium infection.
The T-cell products IL-22, IL-17, and IFN-γ likely play an important role in the amplification of intestinal inflammatory responses. IL-17 is important for efficient neutrophil recruitment, as shown by its ability to induce the production of neutrophil chemoattractants (CXC chemokines) in human bronchial epithelial cells in vitro (13) and its ability to stimulate neutrophil recruitment in rat airways in vivo upon intratracheal instillation (14). Consistent with these findings, CD3+ T-cell depletion blunted Il-17 expression during serotype Typhimurium infection and resulted in a dramatic attenuation of neutrophil recruitment in the cecal mucosa. Furthermore, IL-22, IL-17, and/or IFN-γ can activate various effector mechanisms of the innate immune system, including expression of iNOS (19, 40), lipocalin-2 (28), and human β-defensin (13). The induction of these antimicrobial responses may contribute to a reduction in the numbers of competing microorganisms, which has been shown to be beneficial for serotype Typhimurium during intestinal colonization (31). A beneficial effect of inflammation on the recovery of serotype Typhimurium from cecal contents is observed at 5 days after streptomycin treatment, when normal composition of the intestinal microbiota is restored. However, at the time point chosen for our study (i.e., at 3 days after streptomycin treatment), normal composition of the intestinal microbiota had not yet been restored, which may explain why we did not observe that intestinal inflammation increased the numbers of serotype Typhimurium bacteria in the cecal contents. Instead, we observed increased translocation of serotype Typhimurium to the liver in CD3-depleted mice, which may be due to a defect in inducing neutrophil influx and antimicrobial responses in the intestinal mucosa of these animals. In conclusion, our data suggest that one function of intestinal T cells is to amplify innate immune responses by producing cytokines that regulate innate immune functions.
Acknowledgments
This investigation was conducted in a facility constructed with support from Research Facilities Improvement Program grant number C06 RR12088-01 from the National Center for Research Resources, National Institutes of Health. Work in A.J.B.'s laboratory was supported by Public Health Service grants AI040124, AI044170, and AI065534. T.A.P. and R.L.S. are recipient of fellowships from CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brasília, Brazil). I.G. was supported by Public Health Service grant AI06055.
Editor: A. Camilli
Footnotes
Published ahead of print on 17 March 2008.
REFERENCES
- 1.Barthel, M., S. Hapfelmeier, L. Quintanilla-Martínez, M. Kremer, M. Rohde, M. Hogardt, K. Pfeffer, H. Russmann, and W.-D. Hardt. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 712839-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, L. M., S. Hobbie, and J. E. Galán. 1996. Requirement of cdc42 for salmonella-induced cytoskeletal and nuclear responses. Science 2742115-2118. [DOI] [PubMed] [Google Scholar]
- 3.Day, D. W., B. K. Mandal, and B. C. Morson. 1978. The rectal biopsy appearances in Salmonella colitis. Histopathology 2117-131. [DOI] [PubMed] [Google Scholar]
- 4.D'Orazio, S. E., M. J. Troese, and M. N. Starnbach. 2006. Cytosolic localization of Listeria monocytogenes triggers an early IFN-gamma response by CD8+ T cells that correlates with innate resistance to infection. J. Immunol. 1777146-7154. [DOI] [PubMed] [Google Scholar]
- 5.Dubin, P. J., and J. K. Kolls. 2007. IL-23 mediates inflammatory responses to mucoid Pseudomonas aeruginosa lung infection in mice. Am. J. Physiol. Lung Cell Mol. Physiol. 292L519-L528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franchi, L., A. Amer, M. Body-Malapel, T. D. Kanneganti, N. Ozoren, R. Jagirdar, N. Inohara, P. Vandenabeele, J. Bertin, A. Coyle, E. P. Grant, and G. Nunez. 2006. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat. Immunol. 7576-582. [DOI] [PubMed] [Google Scholar]
- 7.Happel, K. I., P. J. Dubin, M. Zheng, N. Ghilardi, C. Lockhart, L. J. Quinton, A. R. Odden, J. E. Shellito, G. J. Bagby, S. Nelson, and J. K. Kolls. 2005. Divergent roles of IL-23 and IL-12 in host defense against Klebsiella pneumoniae. J. Exp. Med. 202761-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Happel, K. I., M. Zheng, E. Young, L. J. Quinton, E. Lockhart, A. J. Ramsay, J. E. Shellito, J. R. Schurr, G. J. Bagby, S. Nelson, and J. K. Kolls. 2003. Cutting edge: roles of Toll-like receptor 4 and IL-23 in IL-17 expression in response to Klebsiella pneumoniae infection. J. Immunol. 1704432-4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hardt, W. D., L. M. Chen, K. E. Schuebel, X. R. Bustelo, and J. E. Galan. 1998. S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell 93815-826. [DOI] [PubMed] [Google Scholar]
- 10.Hersh, D., D. M. Monack, M. R. Smith, N. Ghori, S. Falkow, and A. Zychlinsky. 1999. The salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc. Natl. Acad. Sci. USA 962396-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hobbie, S., L. M. Chen, R. J. Davis, and J. E. Galan. 1997. Involvement of mitogen-activated protein kinase pathways in the nuclear responses and cytokine production induced by Salmonella typhimurium in cultured intestinal epithelial cells. J. Immunol. 1595550-5559. [PubMed] [Google Scholar]
- 12.Huang, F. C., A. Werne, Q. Li, E. E. Galyov, W. A. Walker, and B. J. Cherayil. 2004. Cooperative interactions between flagellin and SopE2 in the epithelial interleukin-8 response to Salmonella enterica serovar Typhimurium infection. Infect. Immun. 725052-5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kao, C. Y., Y. Chen, P. Thai, S. Wachi, F. Huang, C. Kim, R. W. Harper, and R. Wu. 2004. IL-17 markedly up-regulates beta-defensin-2 expression in human airway epithelium via JAK and NF-kappaB signaling pathways. J. Immunol. 1733482-3491. [DOI] [PubMed] [Google Scholar]
- 14.Laan, M., Z. H. Cui, H. Hoshino, J. Lotvall, M. Sjostrand, D. C. Gruenert, B. E. Skoogh, and A. Linden. 1999. Neutrophil recruitment by human IL-17 via C-X-C chemokine release in the airways. J. Immunol. 1622347-2352. [PubMed] [Google Scholar]
- 15.Liang, S. C., X. Y. Tan, D. P. Luxenberg, R. Karim, K. Dunussi-Joannopoulos, M. Collins, and L. A. Fouser. 2006. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J. Exp. Med. 2032271-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCormick, B. A., C. A. Parkos, S. P. Colgan, D. K. Carnes, and J. L. Madara. 1998. Apical secretion of a pathogen-elicited epithelial chemoattractant activity in response to surface colonization of intestinal epithelia by Salmonella typhimurium. J. Immunol. 160455-466. [PubMed] [Google Scholar]
- 17.McGovern, V. J., and L. J. Slavutin. 1979. Pathology of salmonella colitis. Am. J. Surg. Pathol. 3483-490. [DOI] [PubMed] [Google Scholar]
- 18.Miao, E. A., C. M. Alpuche-Aranda, M. Dors, A. E. Clark, M. W. Bader, S. I. Miller, and A. Aderem. 2006. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat. Immunol. 7569-575. [DOI] [PubMed] [Google Scholar]
- 19.Miljkovic, D., and V. Trajkovic. 2004. Inducible nitric oxide synthase activation by interleukin-17. Cytokine Growth Factor Rev. 1521-32. [DOI] [PubMed] [Google Scholar]
- 20.Monack, D. M., W. W. Navarre, and S. Falkow. 2001. Salmonella-induced macrophage death: the role of caspase-1 in death and inflammation. Microbes Infect. 31201-1212. [DOI] [PubMed] [Google Scholar]
- 21.Mrsny, R. J., A. T. Gewirtz, D. Siccardi, T. Savidge, B. P. Hurley, J. L. Madara, and B. A. McCormick. 2004. Identification of hepoxilin A3 in inflammatory events: a required role in neutrophil migration across intestinal epithelia. Proc. Natl. Acad. Sci. USA 1017421-7426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nau, G. J., J. F. Richmond, A. Schlesinger, E. G. Jennings, E. S. Lander, and R. A. Young. 2002. Human macrophage activation programs induced by bacterial pathogens. Proc. Natl. Acad. Sci. USA 991503-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nau, G. J., A. Schlesinger, J. F. Richmond, and R. A. Young. 2003. Cumulative Toll-like receptor activation in human macrophages treated with whole bacteria. J. Immunol. 1705203-5209. [DOI] [PubMed] [Google Scholar]
- 24.Raffatellu, M., R. L. Santos, D. Chessa, R. P. Wilson, S. E. Winter, C. A. Rossetti, S. D. Lawhon, H. Chu, T. Lau, C. L. Bevins, L. G. Adams, and A. J. Bäumler. 2007. The capsule encoding the viaB locus reduces interleukin-17 expression and mucosal innate responses in the bovine intestinal mucosa during infection with Salmonella enterica serotype Typhi. Infect. Immun. 754342-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reis, B. P., S. Zhang, R. M. Tsolis, A. J. Bäumler, L. G. Adams, and R. L. Santos. 2003. The attenuated sopB mutant of Salmonella enterica serovar Typhimurium has the same tissue distribution and host chemokine response as the wild type in bovine Peyer's patches. Vet. Microbiol. 97269-277. [DOI] [PubMed] [Google Scholar]
- 26.Rosenberger, C. M., M. G. Scott, M. R. Gold, R. E. Hancock, and B. B. Finlay. 2000. Salmonella typhimurium infection and lipopolysaccharide stimulation induce similar changes in macrophage gene expression. J. Immunol. 1645894-5904. [DOI] [PubMed] [Google Scholar]
- 27.Santos, R. L., S. Zhang, R. M. Tsolis, A. J. Bäumler, and L. G. Adams. 2002. Morphologic and molecular characterization of Salmonella typhimurium infection in neonatal calves. Vet. Pathol. 39200-215. [DOI] [PubMed] [Google Scholar]
- 28.Shen, F., M. J. Ruddy, P. Plamondon, and S. L. Gaffen. 2005. Cytokines link osteoblasts and inflammation: microarray analysis of interleukin-17- and TNF-alpha-induced genes in bone cells. J. Leukoc. Biol. 77388-399. [DOI] [PubMed] [Google Scholar]
- 29.Srikanth, C. V., and B. J. Cherayil. 2007. Intestinal innate immunity and the pathogenesis of Salmonella enteritis. Immunol. Res. 3761-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Srinivasan, A., R. M. Salazar-Gonzalez, M. Jarcho, M. M. Sandau, L. Lefrancois, and S. J. McSorley. 2007. Innate immune activation of CD4 T cells in salmonella-infected mice is dependent on IL-18. J. Immunol. 1786342-6349. [DOI] [PubMed] [Google Scholar]
- 31.Stecher, B., R. Robbiani, A. W. Walker, A. M. Westendorf, M. Barthel, M. Kremer, S. Chaffron, A. J. Macpherson, J. Buer, J. Parkhill, G. Dougan, C. von Mering, and W. D. Hardt. 2007. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 52177-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stojiljkovic, I., A. J. Bäumler, and F. Heffron. 1995. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J. Bacteriol. 1771357-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun, Y. H., H. G. Rolan, and R. M. Tsolis. 2007. Injection of flagellin into the host cell cytosol by Salmonella enterica serotype Typhimurium. J. Biol. Chem. 28233897-33901. [DOI] [PubMed] [Google Scholar]
- 34.Tallant, T., A. Deb, N. Kar, J. Lupica, M. J. de Veer, and J. A. DiDonato. 2004. Flagellin acting via TLR5 is the major activator of key signaling pathways leading to NF-kappa B and proinflammatory gene program activation in intestinal epithelial cells. BMC Microbiol. 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tükel, C., M. Raffatellu, A. D. Humphries, R. P. Wilson, H. L. Andrews-Polymenis, T. Gull, J. F. Figueiredo, M. Wong, K. S. Michelsen, M. Akcelik, L. G. Adams, and A. J. Bäumler. 2005. CsgA is a pathogen-associated molecular pattern of Salmonella enterica serotype Typhimurium that is recognized by Toll-like receptor 2. Mol. Microbiol. 58289-304. [DOI] [PubMed] [Google Scholar]
- 36.Umemura, M., A. Yahagi, S. Hamada, M. D. Begum, H. Watanabe, K. Kawakami, T. Suda, K. Sudo, S. Nakae, Y. Iwakura, and G. Matsuzaki. 2007. IL-17-mediated regulation of innate and acquired immune response against pulmonary Mycobacterium bovis bacille Calmette-Guerin infection. J. Immunol. 1783786-3796. [DOI] [PubMed] [Google Scholar]
- 37.Ye, P., P. B. Garvey, P. Zhang, S. Nelson, G. Bagby, W. R. Summer, P. Schwarzenberger, J. E. Shellito, and J. K. Kolls. 2001. Interleukin-17 and lung host defense against Klebsiella pneumoniae infection. Am. J. Respir. Cell Mol. Biol. 25335-340. [DOI] [PubMed] [Google Scholar]
- 38.Ye, P., F. H. Rodriguez, S. Kanaly, K. L. Stocking, J. Schurr, P. Schwarzenberger, P. Oliver, W. Huang, P. Zhang, J. Zhang, J. E. Shellito, G. J. Bagby, S. Nelson, K. Charrier, J. J. Peschon, and J. K. Kolls. 2001. Requirement of interleukin 17 receptor signaling for lung CXC chemokine and granulocyte colony-stimulating factor expression, neutrophil recruitment, and host defense. J. Exp. Med. 194519-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeng, H., A. Q. Carlson, Y. Guo, Y. Yu, L. S. Collier-Hyams, J. L. Madara, A. T. Gewirtz, and A. S. Neish. 2003. Flagellin is the major proinflammatory determinant of enteropathogenic Salmonella. J. Immunol. 1713668-3674. [DOI] [PubMed] [Google Scholar]
- 40.Ziesche, E., M. Bachmann, H. Kleinert, J. Pfeilschifter, and H. Muhl. 2007. The interleukin-22/STAT3 pathway potentiates expression of inducible nitric-oxide synthase in human colon carcinoma cells. J. Biol. Chem. 28216006-16015. [DOI] [PubMed] [Google Scholar]