Abstract
Surface proteins of tick-borne, intracellular bacterial pathogens mediate functions essential for invasion and colonization. Consequently, the surface proteome of these organisms is specifically relevant from two biological perspectives, induction of protective immunity in the mammalian host and understanding the transition from the mammalian host to the tick vector. In this study, the surface proteome of Anaplasma marginale, a tick-transmitted bacterial pathogen, was targeted by using surface-specific cross-linking to form intermolecular bonds between adjacent proteins. Liquid chromatography and tandem mass spectroscopy were then employed to characterize the specific protein composition of the resulting complexes. The surface complexes of A. marginale isolated from erythrocytes of the mammalian host were composed of multiple membrane proteins, most of which belong to a protein family, pfam01617, which is conserved among bacteria in the genus Anaplasma and the closely related genus Ehrlichia. In contrast, the surface proteome of A. marginale isolated from tick cells was much less complex and contained a novel protein, AM778, not identified within the surface proteome of organisms from the mammalian host. Immunization using the cross-linked surface complex induced protection against high-level bacteremia and anemia upon A. marginale challenge of cattle and effectively recapitulated the protection induced by immunization with whole outer membranes. These results indicate that a surface protein subset of the outer membrane is capable of inducing protective immunity and serves to direct vaccine development. Furthermore, the data support that remodeling of the surface proteome accompanies the transition between mammalian and arthropod hosts and identify novel targets for blocking transmission.
Outer membrane proteins of tick-borne, intracellular bacterial pathogens mediate functions necessary for survival, replication, and transmission. Thus, surface-expressed proteins are logical candidates for vaccine development targeted to either induce protective immunity in the mammalian host or prevent colonization of the tick vector. Comprehensive identification of the surface proteome is a critical step in this vaccine development, a process which has been dramatically accelerated by genome sequencing. Recently, complete genome sequences have been reported for several animal and human pathogens in the family Anaplasmataceae, including Anaplasma marginale, Anaplasma phagocytophilum, Ehrlichia chaffeensis, Ehrlichia canis, Ehrlichia ruminantium, and Neorickettsia sennetsu. Nonetheless, identification of the surface proteome in these bacterial pathogens has been constrained by several factors. First, compared to other gram-negative bacteria, the cell wall of these organisms is unusual, in part, because it lacks lipopolysaccharide (LPS) (10). Consequently, prediction models, such as PSORT, are often inaccurate and have failed to correctly predict the location of several proteins confirmed to be surface exposed through biochemical and immunologic labeling techniques (3). Second, the genomes of these organisms generally encode a large number of hypothetical proteins and proteins that lack homology to proteins with known or predicted function in other, more comprehensively studied bacteria. Third, the abundance and immunodominance of a single family of outer membrane proteins, designated pfam01617, shared by organisms within the family Anaplasmataceae, may mask other less abundant but immunologically or functionally significant surface components. Thus, direct analysis of the surface proteome of pathogens within the genera Anaplasma and Ehrlichia is required to define critically important immunologic and functional features of the surface-exposed molecules (7, 8, 12).
In the context of disease prevention, the surface proteome of these organisms is particularly important from two biological perspectives, induction of protective immunity in the mammalian host and understanding the transition from the mammalian host to the tick vector. In A. marginale, purified outer membranes induce protection against acute bacteremia and severe disease, while many well-described individual native and recombinant proteins are poorly protective (20, 21, 23-25). The protection-inducing outer membrane preparation is complex and contains in excess of 25 proteins, not all of which are surface exposed (12). Consequently, the first objective of this study was to define specific members of surface-expressed protein complexes and test whether these complexes induce protective immunity that recapitulates the immunity induced by outer membrane immunization.
The second objective of this study was to compare the surface proteome of A. marginale isolated from tick cells and erythrocytes. The surface proteome of many tick-borne bacterial pathogens is remodeled during the transition between mammalian and arthropod hosts. The most clearly defined example is Borrelia burgdorferi, the causative agent of Lyme disease, which modulates surface lipoprotein expression in order to survive in different host tissues. For example, OspA is essential for colonization and survival of this spirochete within the tick midgut (19, 34). In contrast, OspC is required to infect the mammalian host and is up-regulated during tick feeding in preparation for transmission to a new host (29, 30). Alterations that occur in the surface-exposed proteome upon the transition between host species are less well defined in Anaplasma and Ehrlichia spp., and investigations have been confined to a single protein family, pfam01617. However, comparison of expression of specific outer membrane proteins within this protein family has revealed host cell-dependent switching and shifts in levels of expression of these closely related proteins (6, 27, 31).
The experiments described here use surface-specific cross-linking to target surface proteins followed by liquid chromatography and tandem mass spectroscopy (LC-MS/MS) coupled with searches of the A. marginale genome to identify the components of surface-expressed protein complexes. First, the surface complexes expressed in A. marginale isolated from erythrocytes were characterized. The complexes were then used as an immunogen to test whether this outer membrane subset would recapitulate the protective immunity induced by whole outer membranes. Second, the A. marginale surface complexes isolated from erythrocytes and tick cells were compared in order to define the global changes in surface expression that accompany the transition between host cells and to identify any molecules uniquely expressed in tick cells. We report the results of these experiments and discuss their significance in terms of next steps in vaccine development and understanding mechanisms of tick-borne transmission.
MATERIALS AND METHODS
A. marginale isolation. (i) Erythrocytes.
Calf C1149 was splenectomized and inoculated with the St. Maries strain of A. marginale. Erythrocytes from this calf were cryopreserved as stabilate in liquid nitrogen when parasitemia reached 18.6%, as previously described (4). Intact A. marginale cells were isolated from this stabilate. Six ml of stabilate was placed in each 50-ml Oakridge tube. Samples were washed five to seven times in phosphate-buffered saline (PBS) to remove hemoglobin and cryopreservative by centrifugation (Avanti J-25; Beckman Coulter) at 35,000 × g for 20 min. The washed erythrocytes from two Oakridge tubes (the equivalent of 12 ml of stabilate) were combined, and the volume was brought up to 15 ml with PBS for sonication. Sonication (Branson digital sonifier 450; 400-W maximum output) was done at 70% of maximum for 4 min total in 30-s intervals. The samples were then brought up to a 50-ml volume with PBS and centrifuged at 35,000 × g for 20 min. The resulting pellet was placed in a 1.5-ml Eppendorf tube and centrifuged at 15,800 × g for 15 min. The top, pale pink, fluffy portion of the pellet (erythrocyte membranes) was discarded, while the dark pink, bottom portion of the pellet (A. marginale) was saved and washed two to three times with PBS by centrifugation at 15,800 × g for 20 min. Isolated A. marginale cells were resuspended in 0.2 to 0.5 ml of PBS, divided into 100-μl aliquots, and stored at −80°C.
(ii) ISE6 tick cells.
ISE6 cells infected with the St. Maries strain of A. marginale were grown to confluence (15, 16). Infection of the cells was monitored by examining Giemsa-stained cytospins. When approximately 80% of the ISE6 cells were infected, the cells were flushed from the bottom of the flask, and lysed by repeated aspiration (five times) through a bent, 0.437-mm diameter needle. A. marginale cells were separated from the lysate by filtration through a 2-μm GMF-150, 25-mm-diameter filter (Whatman, Florham Park, NJ) into 1.5-ml Eppendorf tubes (28). The filtrate was washed three times in PBS by centrifugation at 11,000 × g for 11 min. After the final wash, the initial bodies were resuspended in 0.2 to 0.5 ml PBS, divided into 100-μl aliquots, and frozen at −80°C.
Cross-linking outer membrane proteins.
A. marginale proteins isolated from erythrocytes were cross-linked with disulfosuccinimidyltartrate (sulfo-DST), bis(sulfosuccinimidyl)suberate (BS3), 3,3′-dithiobis[sulfosuccinimidylpropionate] (DTSSP), ethylene glycolbis(sulfosuccinimidylsuccinate) (sulfo-EGS), and sulfosuccinimidyl 2-[m-azido-o-nitrobenzamido]ethyl-1,3′-dithiopropionate (sulfo-SAND). The same cross-linkers, with the exception of sulfo-SAND, were used to cross-link the A. marginale isolated from ISE6 tick cells. All cross-linkers were made by Pierce (Rockford, IL), except sulfo-DST, which was made by Soltec Ventures (Beverly, MA). Sulfo-DST (6.4-Å spacer arm), BS3 (11.4-Å spacer arm), DTSSP (12-Å spacer arm), and sulfo-EGS (16.1-Å spacer arm) are membrane impermeable, homobifunctional, and have N-hydroxysuccinimide (NHS)-ester groups which react with primary amines. Sulfo-SAND (18.5-Å spacer arm) is also membrane impermeable but is heterobifunctional with an amine-reactive N-hydroxysuccinimide-ester and a photoactivatible nitrophenyl azide (Table 1). Optimization of the cross-linking reaction was done in PBS using from 0.1 to 5.0 mM of linking agent and up to 20 mM of DTSSP and 10 mM of sulfo-DST. A 3.0 mM concentration of linking agent was determined to be optimal. A. marginale isolated from erythrocytes was treated with all cross-linkers, separated by gel electrophoresis, and submitted for LC-MS/MS. LC-MS/MS was done on A. marginale isolated from ISE6 cells and treated with DTSSP and BS3. For LC-MS/MS, 200 μg of isolated A. marginale was cross-linked with 3 mM cross-linker in PBS and a final volume of 82 μl for 30 min with intermittent gentle mixing. Cross-linking was halted with 20 mM Tris pH 7 for 15 min. A. marginale materials were then pelleted by centrifugation at 15,800 × g for 15 min. The A. marginale was resuspended in 50 μl of lysis buffer (50 mM Tris, 1% Nonidet P-40, pH 8), and 30 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. In the case of DTSSP and sulfo-SAND, which each have a cleavable disulfide bond within the spacer arm, the SDS-PAGE buffer lacked a reducing agent. To prepare immunogen, a final concentration of 3 mM DTSSP was used to cross-link A. marginale isolated from erythrocytes. Cross-linking was done with aliquots of 500 μg of protein and with PBS in a final volume of 208 μl.
TABLE 1.
Properties of the selected cross-linking agents
| Linking agent | Spacer arm (Å)a | Reactive group | Functional groupb | Cleaving reagent(s) |
|---|---|---|---|---|
| Sulfo-DST | 6.4 | NHS-ester | Amine | None |
| BS3 | 11.4 | NHS-ester | Amine | None |
| DTSSP | 12.0 | NHS-ester | Amine | Thiols |
| Sulfo-EGS | 16.1 | NHS-ester | Amine | Hydroxylamine |
| Sulfo-SAND | 18.5 | NHS-ester/aryl azide | Amine, nonselective | Thiols |
Length of the spacer arm in angstroms.
Target of reactive group.
Gel electrophoresis.
Cross-linking was confirmed by SDS-PAGE using 4 to 20% prepoured polyacrylamide gels (Bio-Rad, Hercules, CA). Gels were then stained with Sypro ruby (Bio-Rad) overnight, destained for 2 h with 70% acetic acid and 10% methanol, and rinsed with water, and proteins bands were visualized with UV light. For LC-MS/MS and immunization, the complexed proteins were separated from other cellular components by gel electrophoresis using a 1.5-mm-thick 1 to 10% polyacrylamide separating gel (35.5:1 acrylamine:bis-acrylamide; Fisher, Rockford, IL) with 0.8% agarose (SeaKem HGT) and an 0.8% agarose stacking gel in a vertical system (1, 5, 9). Samples were boiled for 3 min and loaded into the appropriate well. Electrophoresis occurred in running buffer with 0.05 M Tris, 0.384 M glycine, and 0.1% SDS at pH 8.3 at 12 to 20 V for 24 to 36 h (1).
LC-MS/MS and A. marginale database search.
After electrophoresis, gels were stained overnight with Sypro ruby, as described above. Large-molecular-size bands representing complexes and protein bands between 15 and 40 kDa were excised and destained overnight in 50% methanol and 5% acetic acid in water. In-gel trypsin digests were done as previously described (12). The trypsin-digested samples were analyzed by LC-MS/MS as previously described using an Esquire HCT electrospray ion trap (Bruker Daltonics, Billerica, MA) and LC Packings Ultimate Nano high-performance liquid chromatography system with the following modifications of the LC procedure (13). An LC Packings monolithic column, PS-DVB, was used to separate the trypsin fragments using 0.1% formic acid buffers. A flow rate of 800 nl/min with five steps of linear gradient was used as follows: 5% buffer B at 3 min, 15% buffer B at 15 min, 30% buffer B at 60 min, 65% buffer B at 95 min, and then 100% buffer B at 95.1 min and held at 100% until 115 min. Buffer A was composed of 0.1% formic acid with 3% acetonitrile. Buffer B was composed of 0.1% formic acid with 95% acetonitrile. MS/MS fragment ion lists were compared, using a local MASCOT server (www.matrixscience.com), to the genome of the St. Maries strain of A. marginale (3). Identification of complexed proteins was based on peptides with ion scores of >47 with a high likelihood that the peptide match was not a random event (P < 0.0001), while identification of noncomplexed proteins was based on peptides with ion scores of ≥18 and P values of <0.05.
Immunization and challenge. (i) Preparation of protein complexes for immunization.
Approximately 2.5 mg of cross-linked, solubilized A. marginale was loaded in each gel. The top approximately 3 mm of each stacking gel containing the complexes was excised and placed in 12.5 ml of 1× PBS. A portion of the gel was stained with Sypro ruby to confirm cross-linking. The complexes were released from the agarose by boiling for 3 min. The mixture of complexes, PBS, and melted agarose was centrifuged at 35,000 × g for 20 min to form a soft agarose pellet. The agarose pellet was saved. The protein complexes within the supernatant were then concentrated with a 15-ml, 50,000 molecular weight cutoff Centriprep centrifugal filter device (Millipore, Billerica, MA) and combined with the agarose pellet. The immunogen was homogenized by repeated aspiration with a 0.467-mm-diameter needle and 6-ml syringe. The presence of the protein complex in the immunogen was confirmed with gel electrophoresis and Sypro ruby staining.
(ii) Preparation of outer membranes for immunization.
Outer membranes of A. marginale from calf C1149 were isolated using sucrose density gradient centrifugation. A. marginale was purified as described above, resuspended in 20% sucrose in 10 mM HEPES, and sonicated on ice for 4 min at the maximum setting to disrupt the membranes. The supernatant was layered on a sucrose gradient and centrifuged at 82,000 × g for 20 h, as previously described (12, 32).
(iii) Animals.
The bovine lymphocyte antigen-DRB3 alleles of 15 Holstein calves were determined by the PCR-restriction fragment length polymorphism method and sequencing exon 2 of the DRB3 gene (18, 26, 33). The animals were divided into three groups of five animals per group. One animal with the following DRB3 alleles was in each group: DRB3*1101/*0101, DRB3*1501/*0101, and DRB3*1501/*1501. One animal in the outer membrane-immunized group and complex-immunized group had DRB3*0101/*0101, while the corresponding animal in the adjuvant-only group was half-matched, with DRB3*1101/*0101. The remaining animal in the complex-immunized group had DRB3*2002/*0101, and the remaining animals in the outer membrane and adjuvant-only groups had DRB3*1501/*1201 and DRB3*0101/*1201, respectively. The calves were immunized five times at 3-week intervals with approximately 35 μg of either outer membranes or complexes suspended in 1 mg of saponin in a total volume of 1 ml. The third group of five calves was similarly immunized on the same schedule using 1 mg of saponin only.
(iv) Challenge.
Four months after the last immunization, calves were challenged intravenously with approximately 1 × 104 of A. marginale (St. Maries strain) in 1 ml of Hanks’ balanced salt solution. The A. marginale was acquired from calf 31919, which had 3.1% infected erythrocytes and a packed cell volume (PCV) of 33%. Starting 10 days postchallenge, all calves were bled daily and the PCV and percent erythrocytes containing bacteria were determined. Data analysis for the percent infected erythrocytes began the first day that 1% of erythrocytes were infected (day 26 postchallenge) in any animal. Data analysis for PCV started the first day a group of animals had a mean 30% decrease in PCV, which corresponded to 36 days postchallenge. Repeated-measures analysis of variance on data ranks (Friedman's analysis of variance) was used to analyze the data, as they were not normally distributed.
SDS-PAGE and immunoblotting.
Sonicated pellets of A. marginale (St. Maries strain)-infected erythrocytes from calf C1149 were stored in proteinase inhibitor buffer at −80°C, as previously described (17). The equivalent of 1 × 108 infected erythrocytes was loaded in each well and electrophoresed at 70 to 80 V. After transfer to nitrocellulose, proteins were detected using monoclonal antibodies or bovine immune serum. To determine immunoglobulin G (IgG) titers, serum from the immunized animals was serially diluted from 1:300 to 1:30,000, and antibody binding was detected with horseradish peroxidase-labeled goat anti-bovine IgG (Kirkegaard & Perry Laboratories, Gaithersburg, MD) diluted to 1:4,000 and developed with an ECL Western blotting detection system (Pierce, Rockford, IL). Monoclonal antibody AnaF16C1, which detects Msp5, was diluted to 2 μg/ml, and anti-Omp9 monoclonal antibody 121/1055 was diluted to 4 μg/ml. Antibody binding was detected with goat anti-mouse antibody diluted to 1:10,000 using the Western Star chemiluminescence system (Applied Biosystems, Foster City, CA).
RESULTS
Cross-linking the A. marginale outer membrane.
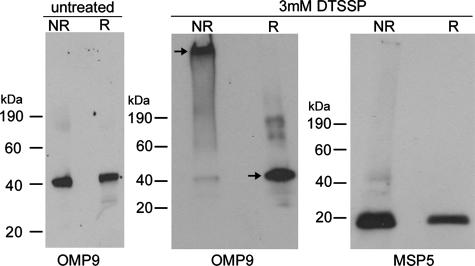
Cross-linking of A. marginale isolated from erythrocytes and tick cells resulted in the formation of a single, large-molecular-size band, consistent with protein complex formation. To ensure that the large-molecular-size band was a specific result of the cross-linking, DTSSP cross-linked A. marginale proteins were analyzed with Western blotting under both reducing and nonreducing conditions. DTSSP has a disulfide bond in the spacer arm, which is cleavable with reducing agents. In cross-linked samples, Omp9, used as a marker protein, was present in the large-molecular-size band, indicating complex formation (Fig. 1). However, upon treatment of the cross-linked complexes with a reducing agent (β-mercaptoethanol), Omp9 was released from the complexes and migrated to its expected size of 40 kDa. Omp9 from non-cross-linked A. marginale migrated to 40 kDa under both reducing and nonreducing conditions, indicating the presence of Omp9 within the large complexes was due to cross-linking rather than the electrophoresis conditions. As a control, Msp5, which was not identified in the intermolecular bound complexes (as reported below), was identified in both the cross-linked and reduced samples at the expected molecular size (Fig. 1).
FIG. 1.
Effective and specific A. marginale surface protein cross-linking. Intact bacteria isolated from erythrocytes were untreated or treated with 3 mM DTSSP and electrophoresed under nonreducing (NR) and reducing (R) conditions. Monoclonal antibodies 121/1055 and AnaF16C1 were used to detect Omp9 and Msp5, respectively.
The pattern of cross-linking in A. marginale isolated from erythrocytes and tick cells was similar with all cross-linkers used, with the exception of sulfo-DST, as visualized in 4 to 20% polyacrylamide gels. However, the banding patterns of non-cross-linked A. marginale proteins and those treated with sulfo-DST were identical, even when using up to 10 mM of cross-linker (data not shown). Sulfo-DST has the shortest spacer arm, 6.4 Å, which likely forms intra- rather than intermolecular bonds. Additionally, in the samples treated with sulfo-DST, peptides from many of the known and predicted outer membrane proteins, including Msp2, Msp3, Msp4, Omp4, Omp7, Omp8, Omp9, Omp10, and OpAG2, were identified by LC-MS/MS at their expected molecular sizes, thus confirming that minimal to no cross-linking occurred with sulfo-DST (data not shown).
Composition of the surface protein complexes.
All proteins detected in the cross-linked complexes of A. marginale isolated from erythrocytes, except four, were known or predicted to be surface expressed, consistent with the surface specificity of cross-linking. This indicated that gel electrophoresis was an effective means of separating the complexes from other, non-cross-linked proteins. In A. marginale cells isolated from erythrocytes, the BS3- and DTSSP-linked complexes were of identical composition except for four proteins. The common members of these complexes included Msp1a, Msp2, Msp3, Msp4, Omp1, Omp7, Omp8, Omp9, OpAG2, Am779, and Am854, while Omp11, Am780, Am1011, and VirB10 were identified only in the BS3-linked complex (Table 2). The concordance of results obtained with BS3 and DTSSP likely reflects the similarity in these two cross-linkers. BS3 has an 11.6-Å spacer arm, while DTSSP has a 12-Å spacer arm; all other features of these two cross-linkers are identical. The protein complexes that resulted from treatment with sulfo-EGS (16.1 Å) and sulfo-SAND (18.5 Å) contained seven and six proteins, respectively. The seven proteins identified in the sulfo-EGS-linked complex were all also identified in the BS3-linked complex and included Msp2, Msp3, Omp7, Am854, Omp8, OpAG2, and VirB10. Additional outer membrane proteins Msp4, Omp1, Omp9, and Omp11, which were identified in the BS3-linked complex, were also all identified in appropriately sized bands between 25 and 37 kDa in the sulfo-EGS-treated sample, indicating these proteins were detectable in the sulfo-EGS samples but were not associated with the complexes. The sulfo-SAND-linked complexes had three proteins in common with all the other linked complexes, Msp2, Msp3, and Omp7, and an additional three proteins unique to the sulfo-SAND-linked complex, Am1051, Am366, and Am712. In the sulfo-SAND-treated samples, Am854, Omp8, OpAG2, Msp4, Omp1, Omp9, and Omp11, which were present in complexes formed by other cross-linkers, were detected from bands between 25 and 37 kDa, indicating these proteins were detectable but not associated with the complexes. Sulfo-SAND is the only cross-linker used that has a photoactivated reactive group, which in concert with the long spacer arm (18.5 Å) likely accounts for the lesser diversity of intermolecular cross-linked proteins in the complexes compared to the other cross-linkers. Msp5, OpAG3, Ana29, and Omp13 were consistently identified at the appropriate molecular sizes in samples treated with BS3, DTSSP, sulfo-EGS, and sulfo-SAND but were unassociated with the complexes.
TABLE 2.
Composition of surface protein complexes in A. marginale cells isolated from erythrocytes
| Protein | Surface location basis (reference)a | PSORT prediction (score) | MASCOT ion scoreb
|
|||
|---|---|---|---|---|---|---|
| BS3 (11.4 Å) | DTSSP (12 Å) | Sulfo-EGS (16.1 Å) | Sulfo-SAND (18.5 Å) | |||
| Msp2 | Exptl (12, 24) | 1,127 | 1,951 | 858 | 116 | |
| Msp3 | Exptl (12, 24) | 1,087 | 1,204 | 858 | 206 | |
| Omp7 | Exptl (12)c | 166 | 338 | 66 | 99 | |
| PSORT | Outer membrane (0.309) | |||||
| Am854 | Exptl (12)c | 102 | 78 | 53 | ||
| PSORT | Outer membrane (0.790) | |||||
| Omp8 | PSORT | Outer membrane (0.414) | 100 | 103 | 58 | |
| OpAG2 | Exptl (11) | 166 | 113 | 51 | ||
| Am779 | PSORT | Outer membrane (0.936) | 53 | 87 | ||
| Msp1a | Exptl (24) | 100 | 101 | |||
| Msp4 | Exptl (24) | 98 | 78 | |||
| Omp1 | PSORT | Outer membrane (0.166) | 60 | 70 | ||
| Omp9 | PSORT | Outer membrane (0.530) | 257 | 135 | ||
| Am1011 | Not predicted | 55 | ||||
| Am780 | Not predicted | 63 | ||||
| Omp11 | PSORT | Outer membrane (0.795) | 64 | |||
| VirB10 | Exptl (12)c | 51 | 58 | |||
| Am1051 | Not predicted | 52 | ||||
| Am366 | Exptl (12)c | 51 | ||||
| Am712 | Not predicted | 50 | ||||
Surface exposure was determined based on experimental evidence (reference in parentheses) or a PSORT prediction for gram-negative bacteria (http://psort.nibb.ac.jp/form.html).
MASCOT ion scores that were greater than 47 for the linking agent (spacer arm lengths indicated in parentheses) and were significant (P < 0.0001). Included is the highest ion score from multiple samples.
Proteins were previously identified in whole outer membranes but with unconfirmed surface expression.
Immunization and challenge.
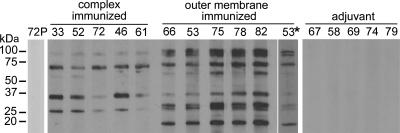
Groups of five calves each were immunized with DTSSP-linked complexes, whole outer membranes, or, as an adjuvant control, saponin. The IgG titer for each vaccinate in the complex-immunized and whole outer membrane-immunized groups was 10,000. In Western blot assays using A. marginale isolated from erythrocytes as antigen, sera from the complex-immunized animals bound a subset of the proteins recognized by sera from animals immunized with whole outer membranes (Fig. 2). Sera from outer membrane-immunized animals recognized six predominant bands between approximately 19 kDa and 100 kDa, with doublets at 100 kDa and 25 kDa (Fig. 2). In contrast, sera from the surface complex-immunized animals recognized a subset of these proteins, with the notable absence of antibody to the 19-kDa protein, representing Msp5. This was consistent with the detection of Msp5 in whole outer membranes, but not the surface complexes. Additionally, single bands rather than doublets were present at 100 kDa and 25 kDa (Fig. 2). This difference in reactivity between the surface complex-immunized and whole outer membrane-immunized groups was observed at both greater (1:10,000) and lesser (1:300) serum dilutions (data not shown). No proteins were recognized by preimmune sera or sera from the animals immunized with adjuvant only (Fig. 2).
FIG. 2.
Immunization using isolated surface complexes induced a subset of antigen-specific antibodies compared to immunization using whole outer membranes. Antigens in A. marginale isolated from bovine erythrocytes were detected with serum from immunized animals that was diluted 1:3,000. A different exposure is shown for outer membrane-immunized animal 53* to more clearly indicate the doublet at 100 kDa. No specific antibodies were detected in animals prior to immunization (shown for animal 72 [72P]) or in animals inoculated with adjuvant alone.
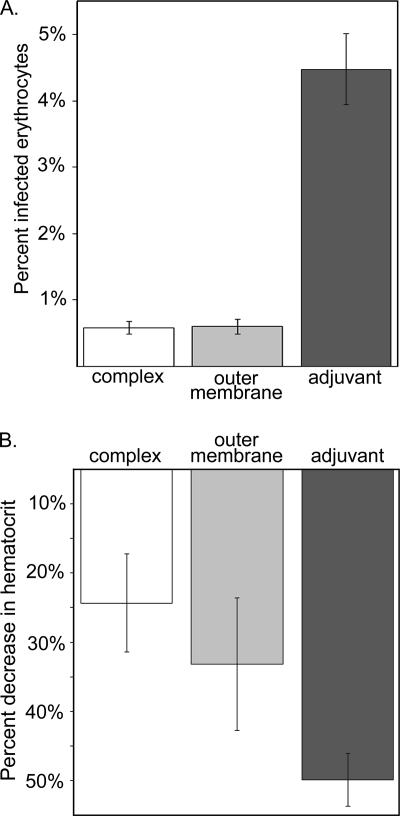
Upon challenge with A. marginale, calves immunized with either outer membranes or DTSSP-linked complexes were similarly, and significantly, protected from bacteremia compared to animals that received only adjuvant (P < 0.01). There was no difference in protection between outer membrane- and complex-immunized animals. This similar level of protection is consistent with the hypothesis that the surface complexes are the portion of the whole outer membrane immunogen responsible for inducing protective immunity against acute, high-level bacteremia. The mean percentage of erythrocytes infected with A. marginale was 0.60% in the groups immunized with the surface complexes and whole outer membranes. The group that received only adjuvant had a mean of 4.48% infected erythrocytes (Fig. 3A). In addition, the degree of protection against disease, measured by the severity of A. marginale-induced anemia, was similar, with no significant difference between surface complex-immunized animals and outer membrane-immunized animals. The animals receiving adjuvant only had a significantly lower PCV than the animals receiving either outer membranes or complexes (P < 0.05). The mean percentage decrease in hematocrit was 19% for the complex-immunized group and 28% for the outer membrane-immunized group. In contrast, the mean percentage decrease in the adjuvant control was 45% (Fig. 3B). Differences among groups were not attributable to major histocompatibility complex haplotype, as these were balanced between the groups, and there was no association between major histocompatibility complex haplotype and protection against either bacteremia or disease severity.
FIG. 3.
Surface complex immunization recapitulates the protection induced by whole A. marginale outer membrane immunization. A. Protection against high-level bacteremia B. Protection against severe anemia.
Surface proteins of A. marginale isolated from tick cells.
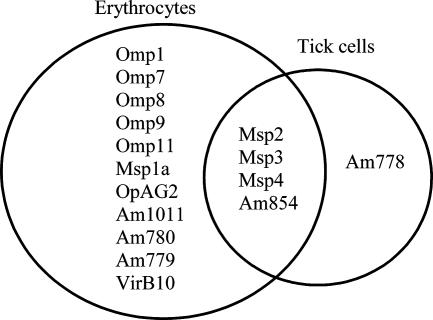
Because DTSSP and BS3 resulted in maximum cross-linking in A. marginale isolated from erythrocytes, these two cross-linkers were used to target the outer membrane proteins of A. marginale isolated from ISE6 cells. Similar to A. marginale isolated from erythrocytes, cross-linking of A. marginale from tick cells resulted in large-molecular-size protein complexes. As previously described, using BS3 as a cross-linker, at least 15 proteins were present in surface complexes from A. marginale isolated from erythrocytes (Table 2). In contrast, only five proteins, Msp2, Msp3, Msp4, Am778, and Am854, were identified in complexes from A. marginale isolated from tick cells and cross-linked with BS3 (Fig. 4). Am778, Msp4, and HtpG were the only proteins identified in the DTSSP-linked complexes. However, peptides representing Msp5, Omp1, Omp4, Omp7, Omp8, and Omp9 were identified in non-cross-linked A. marginale from tick cells, indicating that a variety of proteins could be detected although they were not localized to the surface.
FIG. 4.
Comparison of members of the protein complexes from A. marginale isolated from erythrocytes and ISE6 tick cells. Samples were cross-linked with 3 mM BS3.
Identification of hypothetical proteins.
Multiple proteins that had not been previously detected, had been annotated as hypothetical, and had no homology to proteins with known function were identified in these samples (Table 3). Several of these proteins, Am778, Am779, Am780, Am1051, and Am712, were present in surface-exposed complexes. The majority of these proteins are conserved among at least two other members of the Anaplasmataceae, and 4 (Am649, Am347, Am1248, and Am630) have homologs in only one other species within this family, while 13 have no significant homology with other known proteins (Table 3).
TABLE 3.
Identification of previously hypothetical A. marginale proteins
| Gene identity | MASCOT ion scorea | % Sequence coverageb | PSORT predictionc | Host celld | BLAST homologye |
|---|---|---|---|---|---|
| Am778f | 940 | 42 | Outer membrane, 0.925 | ISE6 | A. phagocytophilum, Ehrlichia spp., Rickettsia spp., Wolbachia spp. |
| Am779 | 87 | 17 | Outer membrane, 0.936 | RBC | A. phagocytophilum, Ehrlichia spp., Rickettsia spp., Wolbachia spp. |
| Am1097 | 49 | 20 | Periplasmic space, 0.944; outer membrane, 0.383 | ISE6 | A. phagocytophilum, Ehrlichia spp., Rickettsia spp., Wolbachia spp. |
| Am202 | 43 | 6 | Inner membrane, 0.808; outer membrane, 0.790 | RBC | A. phagocytophilum, Ehrlichia spp., Neorickettsia sennetsu, Wolbachia spp. |
| Am862 | 120 | 20 | Cytoplasm, 0.097 | RBC | A. phagocytophilum, Ehrlichia spp., N. sennetsu, Wolbachia spp. |
| Am780 | 63 | 21 | Inner membrane, 0.145 | RBC | A. phagocytophilum, Ehrlichia spp., Wolbachia spp. |
| Am419 | 27 | 17 | Inner membrane, 0.274 | RBC | A. phagocytophilum, Ehrlichia spp., Wolbachia spp. |
| Am391 | 85 | 39 | Inner membrane, 0.164 | RBC | A. phagocytophilum, Ehrlichia spp., Wolbachia spp. |
| Am742 | 57 | 15 | Inner membrane, 0.217 | RBC | A. phagocytophilum, Ehrlichia spp., Wolbachia spp. |
| Am660 | 153 | 28 | Cytoplasm, 0.134 | RBC | A. phagocytophilum, N. sennetsu, Wolbachia spp. |
| Am1037 | 22 | 2 | Periplasmic space, 0.938 | ISE6 | A. phagocytophilum, Ehrlichia spp. |
| Am1051 | 62 | 8 | Inner membrane, 0.172 | RBC | A. phagocytophilum, Ehrlichia spp. |
| Am823 | 22 | 4 | Inner membrane, 0.584 | RBC | A. phagocytophilum, Ehrlichia spp. |
| Am1048 | 21 | 1 | Inner membrane, 0.423 | ISE6 | A. phagocytophilum, Ehrlichia spp. |
| Am375 | 85 | 21 | Periplasmic space, 0.939 | RBC, ISE6 | A. phagocytophilum, Ehrlichia spp. |
| Am649 | 180 | 32 | Inner membrane, 0.323 | RBC, ISE6 | A. phagocytophilum |
| Am347 | 28 | 9 | Inner membrane, 0.179 | RBC | A. phagocytophilum |
| Am1248 | 24 | 19 | Inner membrane, 0.542 | RBC | Ehrlichia spp. |
| Am630 | 40 | 5 | Outside, 0.671 | RBC | Wolbachia spp. |
| Am540 | 41 | 5 | Inner membrane, 0.688 | RBC | Nonconserved |
| Am368 | 18 | 1 | Inner membrane, 0.391 | RBC | Nonconserved |
| Am550 | 21 | 3 | Periplasmic space, 0.842 | RBC | Nonconserved |
| Am1359 | 21 | 10 | Inner membrane, 0.501 | ISE6 | Nonconserved |
| Am346 | 1159 | 64 | Plasma membrane, 0.600 | RBC | Nonconserved |
| Am1041 | 19 | 23 | Inner membrane, 0.480 | RBC | Nonconserved |
| Am1225 | 77 | 11 | Cytoplasm, 0.081 | RBC | Nonconserved |
| Am1226 | 69 | 41 | Cytoplasm, 0.353 | RBC, ISE6 | Nonconserved |
| Am185 | 21 | 3 | Cytoplasm, 0.179 | RBC | Nonconserved |
| Am265 | 35 | 10 | Cytoplasm, 0.102 | RBC, ISE6 | Nonconserved |
| Am712 | 50 | 8 | Inner membrane, 0.172 | RBC | Nonconserved |
| Am354 | 27 | 14 | Inner membrane, 0.457 | RBC | Nonconserved |
| Am359 | 32 | 9 | Inner membrane, 0.361 | RBC | Nonconserved |
MASCOT ion scores of ≥18 and significant at P < 0.05 are reported. Data reported are for the highest ion score when a protein was identified in multiple samples.
Percentage of coverage by the identified peptide(s).
Highest score for subcellular location as predicted by PSORT prediction for gram-negative bacteria (http://psort.nibb.ac.jp/form.html).
Host cell from which A. marginale was isolated. RBC, red blood cells.
Included are organisms with proteins having an E value of less than 1e-5 when aligned with the A. marginale protein using NCBI BLAST (2.2.17).
Proteins shown in bold are those identified in surface-expressed complexes. The others were identified only in bacterial lysates.
DISCUSSION
The hypothesis that A. marginale surface protein complexes recapitulate the protective immunity induced by whole outer membrane protein immunization was accepted based on the significant protection against both high-level bacteremia and anemia. The cross-linked surface proteins represent a subset of the whole outer membrane immunogen, as demonstrated by both the reactivity of the postimmunization sera and by the LC-MS/MS analysis. Many of the identified proteins in the cross-linked immunogen had been previously described and localized to the surface, including Msp1a, Msp2, Msp3, Msp4, and OpAG2 (11, 24). The remaining identified proteins had been predicted to be surface expressed or were known only as hypothetical proteins; these included Omp1, Omp7, Omp8, Omp9, Am779, and Am854. Omp1, Omp7, Omp8, and Omp9 are invariant proteins known to be expressed at high levels in bovine erythrocytes (17). Am854, a component of the outer membrane immunogen, is a peptidoglycan-associated lipoprotein, and as such is likely a structural component of the outer membrane (12). Little is known about Am779, which was a hypothetical protein, now confirmed to be surface expressed.
To date, a single A. marginale surface protein capable of reproducibly inducing immunity has not been identified, while immunization with either the outer membrane or the complexes has now been shown to induce protection against both acute bacteremia and anemia. This difference between the efficacy of the complex immunogens versus single proteins may reflect the need for broad epitope diversity of antibody targeting or, alternatively, may reflect the importance of linked recognition induced by proteins in a membrane-associated complex. For example, linkage of a T-cell epitope-rich membrane protein to a membrane protein bearing B-cell epitopes results in enhanced antibody production to the B-cell antigen (14). The role that linked recognition of antigen plays in the induction of protective immunity will be tested by immunization with DTSSP-linked complexes with an intact spacer arm or with complexes that have been subjected to chemical reduction to break the cross-linker spacer arms.
We accept the hypothesis that transition of the bacterium from the erythrocyte to the tick cell is accompanied by remodeling of the surface of A. marginale. These differences are not attributable to differences in the quantity of A. marginale isolated from erythrocytes compared to tick cells, as we controlled for this by using approximately 10-fold more tick cell-derived organisms in the analyses. There are several possible explanations for the decreased number of proteins in surface-expressed complexes of A. marginale isolated from tick cells, including down-regulation of protein expression, altered export of proteins such that the proteins are expressed but not exported to the surface of the bacterium, or altered protein display such that the proteins are surface expressed but are masked by other surface molecules and are consequently inaccessible to the cross-linking reagents. The degree to which ISE6 cells replicate in the environment of the tick midgut is unknown. Thus, additional experiments are required to confirm that similar membrane remodeling occurs during infection of the tick midgut. However, previous studies comparing the amount of Omp1, Omp4, Omp7, Omp8, and Omp9 expressed in erythrocytes and IDE8 tick cells indicated that markedly lower amounts of these proteins are present in tick cells compared to erythrocytes (17). These decreased amounts of protein reflected quantitative decreases in levels of transcript in both IDE8 cells and ex vivo tick midguts compared to erythrocytes, indicating decreased protein expression of these members of pfam01617 is transcriptionally regulated similarly in both in vitro tick cells and the tick midgut (17).
Proteins identified both on the surface of A. marginale from erythrocytes and from tick cells include Msp2, Msp3, Msp4, and Am854. It was previously shown that Msp2, Msp3, and Msp4 are also expressed in midguts and salivary glands of infected ticks ex vivo (2, 22). In these experiments, Am778 was the only protein identified that was unique to A. marginale residing in tick cells and, furthermore, had higher ion scores (492) than any of the other peptides identified in complexes from A. marginale cells isolated from tick cells, suggesting large quantities of this protein. Quantitation of Am778 transcript and protein from A. marginale grown in different host cells will be required to determine if the mass spectroscopy findings reported here truly reflect differential gene expression. Am778 was originally annotated as a hypothetical protein and is encoded by a locus that also encodes Am779 and Am780. Am779 and Am780 were also identified in complexes of surface proteins from A. marginale isolated from erythrocytes (Table 2). Thus, all three of these proteins are surface expressed. Additionally, all three genes are highly conserved among the genera of the family Anaplasmataceae, including A. phagocytophilum, E. canis, E. chaffeensis, E. ruminantium, and Wolbachia spp. (7). The conspicuous exception is the absence of these genes in Neorickettsia sennetsu, the only sequenced member of the genus Neorickettsia. N. sennetsu does not reside within the tick during any portion of its life cycle, suggesting that differential expression or display of the proteins from this locus may play a role in colonization of the tick by Anaplasma spp. and Ehrlichia spp. Further experimentation will be required to determine the role, if any, expression of these proteins has in colonization of either tick or mammalian host tissues.
These experiments expand our knowledge of the A. marginale proteome in three ways. First, the characterized surface-expressed protein complexes of A. marginale isolated from erythrocytes induce protective immunity similar to that induced by purified outer membranes. Thus, the outer membrane components which induce protective immunity are now more narrowly defined and allow further definition of the composition of an effective vaccine. Second, the differences in the surface proteome of A. marginale colonizing tick cells and mammalian cells are more globally defined. The regulatory mechanisms and role that specific molecules, such as Am778, play in the colonization of the tick cell remain to be determined. Third, the overall knowledge of the A. marginale proteome is expanded by confirmation of expression of many proteins previously identified as hypothetical.
Acknowledgments
We are grateful for the excellent technical assistance of Beverly Hunter, Ralph Horn, Anna George, Shawn Sanders, Sam Nielsen, and Gordon Armstrong and the statistical assistance of Marc Evans and Scott Williams.
This research was supported by NIH R01 AI44005 and R01 AI053692, USDA ARSCRIS 5348-32000-027-00D and USDA-ARS cooperative agreement 58-5348-3-0212, and by NIH-NCRR grant no. 1 S10 RR017805-01. Susan Noh was partially supported by K08 AI052412.
Editor: R. P. Morrison
Footnotes
Published ahead of print on 3 March 2008.
REFERENCES
- 1.Affinity Biologicals. 2007. Immunoblotting of von Willebrand factor polymers in plasma and platelets using peroxidase conjugated antibody and chemiluminescence detection: protocol. Affinity Biologicals, Ancaster, Ontario, Canada.
- 2.Barbet, A. F., R. Blentlinger, J. Yi, A. M. Lundgren, E. F. Blouin, and K. M. Kocan. 1999. Comparison of surface proteins of Anaplasma marginale grown in tick cell culture, tick salivary glands, and cattle. Infect. Immun. 67102-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brayton, K. A., L. S. Kappmeyer, D. R. Herndon, M. J. Dark, D. L. Tibbals, G. H. Palmer, T. C. McGuire, and D. P. Knowles, Jr. 2005. Complete genome sequencing of Anaplasma marginale reveals that the surface is skewed to two superfamilies of outer membrane proteins. Proc. Natl. Acad. Sci. USA 102844-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eriks, I. S., D. Stiller, W. L. Goff, M. Panton, S. M. Parish, T. F. McElwain, and G. H. Palmer. 1994. Molecular and biological characterization of a newly isolated Anaplasma marginale strain. J. Vet. Diagn. Investig. 6435-441. [DOI] [PubMed] [Google Scholar]
- 5.Gabelli, C., D. G. Stark, R. E. Gregg, and H. B. Brewer. 1986. Separation of apolipoprotein B species by agarose-acrylamide gel electrophoresis. J. Lipid Res. 27457-460. [PubMed] [Google Scholar]
- 6.Ganta, R. R., C. Cheng, E. C. Miller, B. L. McGuire, L. Peddireddi, K. R. Sirigireddy, and S. K. Chapes. 2007. Differential clearance and immune responses to tick cell-derived versus macrophage culture-derived Ehrlichia chaffeensis in mice. Infect. Immun. 75135-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ge, Y., and Y. Rikihisa. 2007. Identification of novel surface proteins of Anaplasma phagocytophilum by affinity purification and proteomics. J. Bacteriol. 1897819-7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ge, Y., and Y. Rikihisa. 2007. Surface-exposed proteins of Ehrlichia chaffeensis. Infect. Immun. 753833-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henderson, N. S., L. G. Nijtmans, J. G. Lindsay, E. Lamantea, M. Zeviani, and I. J. Holt. 2000. Separation of intact pyruvate dehydrogenase complex using blue native agarose gel electrophoresis. Electrophoresis 212925-2931. [DOI] [PubMed] [Google Scholar]
- 10.Lin, M., and Y. Rikihisa. 2003. Ehrlichia chaffeensis and Anaplasma phagocytophilum lack genes for lipid A biosynthesis and incorporate cholesterol for their survival. Infect. Immun. 715324-5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lohr, C. V., K. A. Brayton, V. Shkap, T. Molad, A. F. Barbet, W. C. Brown, and G. H. Palmer. 2002. Expression of Anaplasma marginale major surface protein 2 operon-associated proteins during mammalian and arthropod infection. Infect. Immun. 706005-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopez, J. E., W. F. Siems, G. H. Palmer, K. A. Brayton, T. C. McGuire, J. Norimine, and W. C. Brown. 2005. Identification of novel antigenic proteins in a complex Anaplasma marginale outer membrane immunogen by mass spectrometry and genomic mapping. Infect. Immun. 738109-8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macmillan, H., K. A. Brayton, G. H. Palmer, T. C. McGuire, G. Munske, W. F. Siems, and W. C. Brown. 2006. Analysis of the Anaplasma marginale major surface protein 1 complex protein composition by tandem mass spectrometry. J. Bacteriol. 1884983-4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macmillan, H., J. Norimine, K. A. Brayton, G. H. Palmer, and W. C. Brown. 2008. Physical linkage of naturally complexed bacterial outer membrane proteins enhances immunogenicity. Infect. Immun. 761223-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munderloh, U. G., E. F. Blouin, K. M. Kocan, N. L. Ge, W. L. Edwards, and T. J. Kurtii. 1996. Establishment of the tick (Acari: Ixodidae)-borne cattle pathogen Anaplasma marginale (Rickettsiales: Anaplasmataceae) in tick cell culture. J. Med. Entomol. 33656-664. [DOI] [PubMed] [Google Scholar]
- 16.Munderloh, U. G., Y. Lui, M. Wang, C. Chen, and T. J. Kurtii. 1994. Establishment, maintenance and description of cell lines from the tick Ixodes scapularis. J. Parasitol. 80533-543. [PubMed] [Google Scholar]
- 17.Noh, S. M., K. A. Brayton, D. P. Knowles, J. T. Agnes, M. J. Dark, W. C. Brown, T. V. Baszler, and G. H. Palmer. 2006. Differential expression and sequence conservation of the Anaplasma marginale msp2 gene superfamily outer membrane proteins. Infect. Immun. 743471-3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Norimine, J., and W. C. Brown. 2005. Intrahaplotype and interhaplotype pairing of bovine leukocyte antigen DQA and DQB molecules generate functional DQ molecules important for priming CD4+ T-lymphocyte responses. Immunogenetics 57750-762. [DOI] [PubMed] [Google Scholar]
- 19.Pal, U., and E. Fikrig. 2003. Adaptation of Borrelia burgdorferi in the vector and vertebrate host. Microbes Infect. 5659-666. [DOI] [PubMed] [Google Scholar]
- 20.Palmer, G. H., A. F. Barbet, G. H. Cantor, and T. C. McGuire. 1989. Immunization of cattle with the MSP-1 surface protein complex induces protection against a structurally variant Anaplasma marginale isolate. Infect. Immun. 573666-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palmer, G. H., A. F. Barbet, W. C. Davis, and T. C. McGuire. 1986. Immunization with an isolate-common surface protein protects cattle against anaplasmosis. Science 2311299-1302. [DOI] [PubMed] [Google Scholar]
- 22.Palmer, G. H., K. M. Kocan, S. J. Barron, J. A. Hair, A. F. Barbet, W. C. Davis, and T. C. McGuire. 1985. Presence of common antigens, including major surface protein epitopes, between the cattle (intraerythrocytic) and tick stages of Anaplasma marginale. Infect. Immun. 50881-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palmer, G. H., and T. F. McElwain. 1995. Molecular basis for vaccine development against anaplasmosis and babesiosis. Vet. Parasitol. 57233-253. [DOI] [PubMed] [Google Scholar]
- 24.Palmer, G. H., and T. C. McGuire. 1984. Immune serum against Anaplasma marginale initial bodies neutralizes infectivity for cattle. J. Immunol. 1331010-1015. [PubMed] [Google Scholar]
- 25.Palmer, G. H., S. M. Oberle, A. F. Barbet, W. L. Goff, W. C. Davis, and T. C. McGuire. 1988. Immunization of cattle with a 36-kilodalton surface protein induces protection against homologous and heterologous Anaplasma marginale challenge. Infect. Immun. 561526-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park, Y. H., Y. S. Joo, J. Y. Park, J. S. Moon, S. H. Kim, N. H. Kwon, J. S. Ahn, W. C. Davis, and C. J. Davies. 2004. Characterization of lymphocyte subpopulations and major histocompatibility complex haplotypes of mastitis-resistant and susceptible cows. J. Vet. Sci. 529-39. [PubMed] [Google Scholar]
- 27.Postigo, M., A. Taoufik, L. Bell-Sakyi, E. de Vries, W. I. Morrison, and F. Jongejan. 2007. Differential transcription of the major antigenic protein 1 multigene family of Ehrlichia ruminantium in Amblyomma variegatum ticks. Vet. Microbiol. 122298-305. [DOI] [PubMed] [Google Scholar]
- 28.Rikihisa, Y., N. Zhi, G. P. Wormser, B. Wen, H. W. Horowitz, and K. E. Hechemy. 1997. Ultrastructural and antigenic characterization of a granulocytic ehrlichiosis agent directly isolated and stably cultivated from a patient in New York state. J. Infect. Dis. 175210-213. [DOI] [PubMed] [Google Scholar]
- 29.Schwan, T. G., and J. Piesman. 2000. Temporal changes in outer surface proteins A and C of the Lyme disease-associated spirochete, Borrelia burgdorferi, during the chain of infection in ticks and mice. J. Clin. Microbiol. 38382-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwan, T. G., J. Piesman, W. T. Golde, M. C. Dolan, and P. A. Rosa. 1995. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc. Natl. Acad. Sci. USA 922909-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singu, V., H. Liu, C. Cheng, and R. R. Ganta. 2005. Ehrlichia chaffeensis expresses macrophage- and tick cell-specific 28-kilodalton outer membrane proteins. Infect. Immun. 7379-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tebele, N., T. C. McGuire, and G. H. Palmer. 1991. Induction of protective immunity by using Anaplasma marginale initial body membranes. Infect. Immun. 593199-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Eijk, M. J., J. A. Stewart-Haynes, and H. A. Lewin. 1992. Extensive polymorphism of the BoLA-DRB3 gene distinguished by PCR-RFLP. Anim. Genet. 23483-496. [DOI] [PubMed] [Google Scholar]
- 34.Yang, X. F., U. Pal, S. M. Alani, E. Fikrig, and M. V. Norgard. 2004. Essential role for OspA/B in the life cycle of the Lyme disease spirochete. J. Exp. Med. 199641-648. [DOI] [PMC free article] [PubMed] [Google Scholar]