Abstract
LipL32 is the major outer membrane protein in pathogenic Leptospira. It is highly conserved throughout pathogenic species and is expressed in vivo during human infection. While these data suggest a role in pathogenesis, a function for LipL32 has not been defined. Outer membrane proteins of gram-negative bacteria are the first line of molecular interaction with the host, and many have been shown to bind host extracellular matrix (ECM). A search for leptospiral ECM-interacting proteins identified the major outer membrane protein, LipL32. To verify this finding, recombinant LipL32 was expressed in Escherichia coli and was found to bind Matrigel ECM and individual components of ECM, including laminin, collagen I, and collagen V. Likewise, an orthologous protein found in the genome of Pseudoalteromonas tunicata strain D2 was expressed and found to be functionally similar and immunologically cross-reactive. Lastly, binding activity was mapped to the C-terminal 72 amino acids. These studies show that LipL32 and an orthologous protein in P. tunicata are immunologically cross-reactive and function as ECM-interacting proteins via a conserved C-terminal region.
Pathogens have evolved a diverse array of mechanisms to survive in the host environment. During the infectious cycle, a pathogen must enter the host, evade the immune response, adhere to tissue, colonize, and finally exit the host to initiate a new infection. Pathogenic species of Leptospira have evolved to complete the infectious cycle while maintaining the capacity to survive in the environment. Leptospira has been found in a wide variety of vertebrate species and in humans causes the disease leptospirosis (18), which is believed to be the most widespread of all zoonoses (1), thus contributing to the high morbidity and mortality rate from Leptospira worldwide. The cycle of human infection starts through direct or indirect contact with the urine of carrier animals whose renal tubules are colonized by leptospires. Leptospira can survive in warm aquatic environments and enter humans via submerged mucosal surfaces or broken skin. Leptospires evade the immune response and then spread to most internal organs, with severe forms of the disease causing major pulmonary damage (28-30).
A paradigm in bacterial pathogenesis is emerging whereby mammalian extracellular matrix (ECM) molecules interact with bacterial cell surface proteins (21). These interactions can be used by the bacterium to enter the host (19), evade the immune response (7), or adhere to tissues (27) as a prelude to tissue colonization. Since the proteins that comprise the ECM are highly conserved throughout the animal kingdom (14, 31) and are available to extracellular zoonotic pathogens, such as Leptospira, it is hypothesized that leptospires have an array of microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) (21). Indeed, the proteins LigA, LigB (3), and Lsa24 (2) have been identified as leptospiral MSCRAMMs. LigA and Lig B bind multiple ECM molecules, and Lsa 24 binds laminin. While these proteins are clearly involved in leptospiral/ECM interactions, we propose that Leptospira has additional proteins that interact with ECM.
This work describes the characterization of the major surface-exposed protein, LipL32 (6, 11), as a leptospiral MSCRAMM. Additional experiments using a closely related protein from the marine bacterium Pseudoalteromonas tunicata (13) showed similar activity. Lastly, the ECM-interacting activity was mapped to the highly conserved C-terminal region. These studies are the first to indicate a conserved function for LipL32 in two phylogenetically distant bacterial species.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Leptospira interrogans serovar Manilae is a clinical isolate (17) that was kindly provided by N. Koizumi, National Health Institute, Tokyo, Japan. L. interrogans and Leptospira biflexa serovar Patoc were maintained in EMJH medium at 30°C as described previously (15). P. tunicata strain D2 was grown in marine broth as described previously (13).
Cell adhesion assay.
Wells of 96-well plates were coated overnight with Matrigel ECM (Becton Dickinson), fibronectin, laminin, or bovine serum albumin (BSA) (Sigma) solution (50 μl of a 100-μg/ml solution in phosphate-buffered saline [PBS]) at 4°C. The next day, the wells were washed four times with 100 μl of PBS. Fifty microliters of leptospires grown to a density of 2 × 108 to 1 × 109 cells/ml was added in triplicate to the experimental wells of the plate in EMJH medium and incubated for 1 to 3 h at 30°C. Nonadherent cells were gently aspirated from the wells The wells were then rinsed gently four to six times with 100 μl of EMJH base (Difco) solution. Twenty-five microliters of 200-μg/ml trypsin in PBS was then added to each well to allow cell detachment for 5 min at 37°C. The detached cells were then counted by dark-field microscopy in a Helber counting chamber, with at least three replicates per condition per day. Biological replicates were performed by repeating the experiment at least three times on different days. Values were compared by Student's two-tailed t test.
Expression and purification of recombinant proteins.
Primer pairs (Table 1) were used to amplify the target sequences from genomic DNA obtained from L. interrogans serovar Lai or P. tunicata strain D2. Constructs were produced by ligating the PCR products into the appropriate restriction sites of plasmid pinpoint Xa3 (Promega). Plasmids were transformed into Escherichia coli XL1 blue cells and grown in 100 μg/ml ampicillin to an absorbance of 0.6 at 600 nm. The cells were then induced with 5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 2 h and then centrifuged and stored at −20°C overnight. The cells were thawed and lysed in lysis buffer (50 mM Tris-Cl, 0.2 M NaCl, 2 mM EDTA, 10% glycerol, 0.1% Triton X-100, pH 8.0) with sonication. The lysate was centrifuged at 13,000 × g for 5 min, and the supernatant was applied to a 250-μl column of Soft Link avidin resin (Promega). After binding to the resin, the column was washed with 5 volumes of lysis buffer, and the bound material was then eluted in lysis buffer plus 5 mM biotin. In some instances, the eluted proteins were subjected to a buffer exchange step by gel permeation chromatography in the following buffer: 10 mM HEPES, 0.15 M NaCl, pH 7.4.
TABLE 1.
Primers used in this study
| Construct | Forward primer | Reverse primer |
|---|---|---|
| rLipL32:20-272 | 5′TATAAGCTTTGTGGTGCTTTCGGTGGTCT3′ | 5′TTAACCTAGATCTTTGTTTAAACAG3′ |
| rLipL32:20-106 | 5′TATAAGCTTTGTGGTGCTTTCGGTGGTCT3′ | 5′AAAAGATCTTTACTCACCGATTTCGCCTGTTGG3′ |
| rLipL32: 20-155 | 5′TATAAGCTTTGTGGTGCTTTCGGTGGTCT3′ | 5′AAAAGATCTTTATTTTGCTTTCGCAGCTTTGGCG3′ |
| rLipL32: 20-200 | 5′TATAAGCTTTGTGGTGCTTTCGGTGGTCT3′ | 5′AAAAGATCTTTAGTCGATGTTTTTCAGATCGTC3′ |
| rLipL32:201-272 | 5′TATAAGCTTACTAAAAACTTTTAGTAAGAGG3′ | 5′TTAACCTAGATCTTTGTTTAAACAG3′ |
| r PTD2-05920:19-236 | 5′AAACAGCTGGCAGGTTTTAGCTTAAATAG3′ | 5′AAAAGATCTTTATTTATTGACTGCTTTATG3′ |
Solid-phase binding assays.
Wells of 96-well plates were coated with 5 μg of protein in 50 μl of PBS overnight at 4°C. The wells were then washed four times with 100 μl PBS, and the residual surface was blocked with 100 μl of 4-mg/ml BSA at room temperature for 1 to 2 h and then washed four times with 150 μl PBS. Purified recombinant proteins in a volume of 50 μl were incubated with substrate for 2 h at 37°C. The wells were washed three times with 150 μl of 0.05% Tween 20 in PBS (PBS-T). After the washes, bound protein was stripped from the wells in 25 μl 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. Proteins were then separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane. The membrane was blocked with 5% skim milk buffer in PBS-T and then probed with streptavidin conjugated with horseradish peroxidase (streptavidin-HRP) (Amersham) diluted 1:2,000 in PBS-T. The streptavidin-HRP signal was developed with enhanced chemiluminescence detection reagent (Amersham Biosciences) and exposed to a luminescent-image analyzer (Fujifilm; LAS-3000) and quantified by the bundled software. Values from at least three replicate wells were averaged, and the standard deviation was determined. The values were compared by Student's two-tailed t test.
Sequence analysis.
Protein alignment was performed by the expert protein analysis system (ExPASy) SIM local similarity program alignment tool using the BLOSUM30 comparison matrix with the gap open penalty and gap extension penalty both set at 4. Secondary-structure predictions were performed by the PHDsec structure prediction algorithm (24) found on the Predict protein server (25).
Western blot analysis.
Proteins were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes, which were then blocked with 5% skim milk in PBS-T. Anti-LipL32 antiserum (11) was diluted 1:2,000 in PBS-T and used to probe the blot for 1 h as described previously (4). The antiserum was then removed, and the blot was washed three times for 5 min each time in PBS-T. HRP-conjugated goat anti-rabbit antiserum (Sigma) diluted 1:2,000 was added, and the mixture was incubated for 1 h at 37°C. After the blot was washed as described above, immunoreactive proteins were visualized by chemiluminescence detection.
RESULTS
L. interrogans serovar Manilae binds to the ECM and individual components of the ECM.
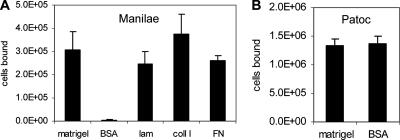
To identify host ECM components that interact with L. interrogans, we developed a leptospiral adhesion assay. Serovar Manilae bound specifically to wells coated with a commercially available preparation of ECM (Matrigel), laminin, collagen I, or fibronectin compared with binding to the BSA control (Fig. 1A). Conversely, the saprophyte L. biflexa bound equally to ECM and BSA (Fig. 1B), indicating that serovar Manilae cells interact specifically with Matrigel ECM.
FIG. 1.
L. interrogans serovar Manilae specifically adheres to surfaces coated with ECM components, while L. biflexa serovar Patoc binds nonspecifically. (A) Wells were coated with Matrigel, BSA, laminin (lam), collagen I (coll I), or fibronectin (FN). The values shown are the averages and standard deviations of the numbers of L. interrogans serovar Manilae cells bound from three replicate wells. Matrigel, laminin, collagen I, and fibronectin binding data were compared to BSA by Student's two-tailed t test with the resulting P values being <0.004. (B) Binding levels of L. biflexa serovar Patoc to Matrigel and BSA were equivalent.
LipL32 is an ECM-interacting protein.
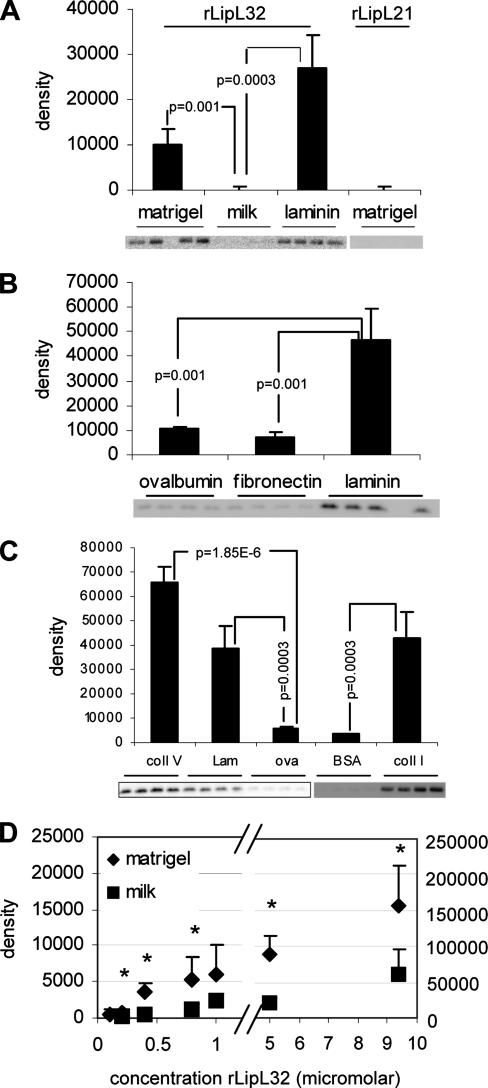
To identify potential leptospiral ECM-interacting proteins, serovar Manilae total membranes separated by two-dimensional gel electrophoresis were probed with biotinylated Matrigel. Several protein spots that bound to biotin-Matrigel were observed (data not shown). The most intense spot from each molecular mass was identified by mass spectrometry as LipL32. No other Matrigel-binding proteins, such as the Lig proteins, were identified, presumably due to their low abundance (3), especially compared to LipL32. We then examined whether a recombinant form of mature LipL32 bound ECM components in a solid-phase binding assay. Recombinant LipL32 (rLipL32) and another abundant leptospiral outer membrane protein, LipL21 (rLipL21), were expressed as fusion proteins containing an N-terminal biotin acceptor peptide tag that became biotinylated upon expression in E. coli (23). Purified biotinylated proteins were added singly or in combination to wells coated with Matrigel, laminin, fibronectin, collagen I, or collagen V. Ovalbumin, skim milk, or BSA served as a control. rLipL32 bound specifically to Matrigel, laminin, collagen I, and collagen V, but not to fibronectin or to the control proteins (Fig. 2A, B, and C). In contrast, rLipL21 did not bind Matrigel (Fig. 2A). When rLipL21 and rLipL32 were coincubated with Matrigel- and BSA-coated wells, only rLipL32 was recovered from Matrigel-coated wells (data not shown). A dose-response curve for the binding of rLipL32 to Matrigel (Fig. 2D) showed statistically significant binding to Matrigel over BSA at rLipL32 concentrations as low as 200 nM. The binding kinetics exhibited a biphasic profile. The first 0.1 to 1 μM phase followed a standard saturation curve, while the points in the 5 to 10 μM range exhibited 10 to 30 times more rLipL32 bound and higher BSA binding values.
FIG. 2.
rLipL32 specifically binds Matrigel, laminin, collagen I, and collagen V. (A to C) Wells of a 96-well plate were coated in quadruplicate with Matrigel, milk, or laminin (A); ovalbumin, fibronectin, or laminin (B); or collagen V (coll V), laminin (lam), ovalbumin (ova), bovine serum albumin (BSA), or collagen I (coll I) (C). The blots below the graphs show rLipL32 or rLipL21 recovered from each binding substrate. The bands were analyzed by densitometry, with the average and standard deviation of each binding condition shown in the bar graphs as indicated. Conditions were compared by Student's two-tailed t test, with the resulting P values indicated. (D) Concentrations of rLipL32 were added to Matrigel- or BSA-control-treated wells, and the amounts of bound protein were determined as described for panels A to C in triplicate. The asterisks above the data points indicate P values of <0.04 for the amount of rLipL32 bound to Matrigel compared to BSA (control).
A LipL32 homolog is present in the marine bacterium P. tunicata strain D2.
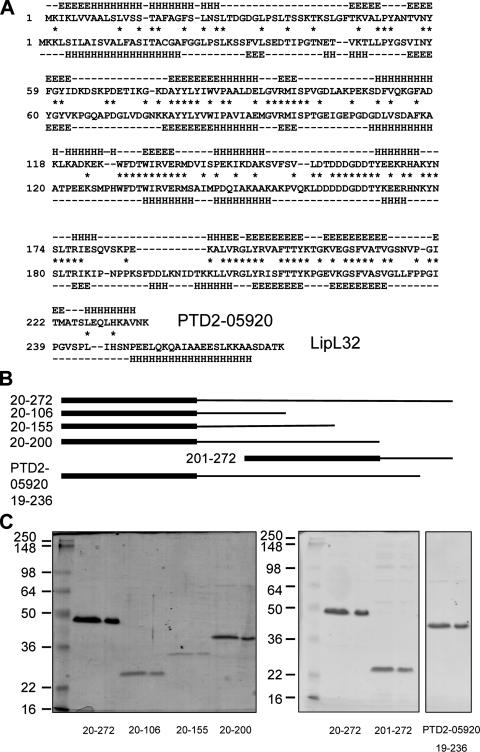
Bioinformatics analysis revealed the presence of a gene, PTD2-05920, similar to lipL32, in the marine bacterium P. tunicata strain D2, with the encoded proteins sharing significant sequence identity in several regions throughout their sequences (Fig. 3A). Secondary-structure predictions of the two proteins likewise suggested similarity at the structural level, even in regions with little sequence similarity (Fig. 3A). Interestingly, the protein encoded by PTD2-05920 contains a signal peptidase I cleavage site, in contrast to LipL32, which has a signal peptidase II site and has been shown experimentally to be lipidated (11).
FIG. 3.
Sequence analysis of LipL32 and an orthologous protein, PTD2-05920, from P. tunicata. (A) Alignment of PTD2-05920 and LipL32 sequences. Identical amino acids are indicated by asterisks. Predicted β-sheet regions are designated by E, while α-helices are denoted by H. A predicted random coil is denoted by a dash. (B) rLipL32 deletion derivatives and PTD2-05920 proteins used in this study. The N-terminal biotin acceptor peptide tag is denoted by the thick lines at the left, while LipL32 or PTD2-05920 sequence is shown as a thin line to the right (drawn to scale). The amino acid numbers comprising each construct are shown on the left. (C) Purification of LipL32 deletion derivatives and PTD2-05920. The positions of standard molecular mass markers (kDa) are shown on the left. Purified proteins were resolved in duplicate lanes as indicated: rLipL32, 20 to 272; rLipL32 deletion, 20 to 106; rLipL32 deletion, 20 to 155; rLipL32 deletion, 20 to 200; rLipL32 deletion, 201 to 272; and recombinant PTD2-05920, 19 to 236.
rPTD2-05920 is recognized by anti-LipL32 serum.
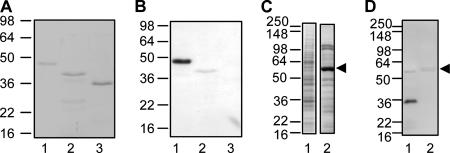
PTD2-05920 (amino acids 19 to 236) was expressed with an N-terminal biotin acceptor peptide tag, purified (Fig. 3B and C), and tested by Coomassie blue and Western blot analyses with rLipL21 and rLipL32 as control proteins. While all proteins were detected by Coomassie blue staining (Fig. 4A), only rPTD2-05920 and rLipL32 were recognized by anti-LipL32 antiserum (Fig. 4B). The cellular expression of PTD2-05920 was also examined in P. tunicata by Coomassie blue staining (Fig. 4C) and anti-LipL32 Western blotting (Fig. 4D). Immunoreactivity was confined to the 32-kDa protein corresponding to LipL32 in the L. interrogans sample (Fig. 4D, lane 1). There was no reaction with PTD2-05920 in the P. tunicata whole-cell lysate. We observed some nonspecific binding to the highly abundant P. tunicata protein, which was identified by mass spectrometry as the unrelated protein PTD2-07619 (data not shown).
FIG. 4.
rPTD2-05920 is recognized by anti-LipL32 serum. (A) rLipL32 (lane 1), rPTD2-05920 (lane 2), and rLipL21 (lane 3) were analyzed by Coomassie blue staining of SDS-PAGE-separated proteins. The positions of standard molecular mass markers (kDa) are shown on the left. rPTD2-05920 migrates at a lower apparent molecular mass than rLipL32 due to the fact that it is 36 amino acids smaller. (B) The same proteins as in panel A were analyzed with anti-LipL32 antiserum by Western blotting. (C) The lysate from 108 L. interrogans cells (lane 1) and the lysate from P. tunicata strain D2 (14 μg) (lane 2) were analyzed by Coomassie blue staining of SDS-PAGE-separated proteins. A highly abundant protein in P. tunicata is indicated by the arrowhead. (D) The same proteins as in panel C were probed with anti-LipL32 antiserum, showing reactivity with LipL32 (lane 1) but not with PTD2-05920. Nonspecific binding to the prominent P. tunicata band (identified as PTD2-07619) (see Results) is indicated by the arrowhead.
rPTD2-05920 binds ECM.
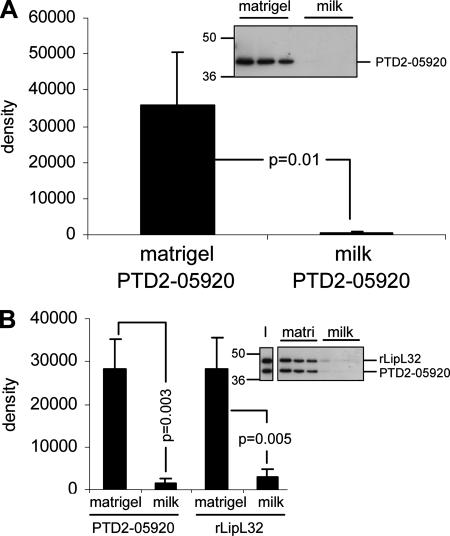
rPTD2-05920 was tested for ECM-binding activity in the solid-phase ECM-binding assay. When tested singly, rPTD2-05920 bound specifically to Matrigel (Fig. 5A). When equivalent amounts of rPTD2-05920 and rLipL32 were coincubated, the two proteins were recovered in equivalent amounts specifically from Matrigel-coated wells (Fig. 5B).
FIG. 5.
rPTD2-05920 binds Matrigel. (A) rPTD2-05920 was added to Matrigel- or milk-treated wells in triplicate. The blot shows rPTD2-05920 recovered from each binding substrate. The positions of standard molecular mass markers (kDa) are shown on the left. The bands were analyzed by densitometry, with the averages and standard deviations shown in the bar graph. Conditions were compared by Student's two-tailed t test, with the resulting P values indicated. (B) Equivalent amounts of rLipL32 and rPTD2-05920 were incubated with Matrigel (matri) or milk wells and analyzed as described for panel A. The input ratio of rLipL32 and rPTD2-05920 is shown in lane I.
ECM-binding activities of truncated LipL32 derivatives.
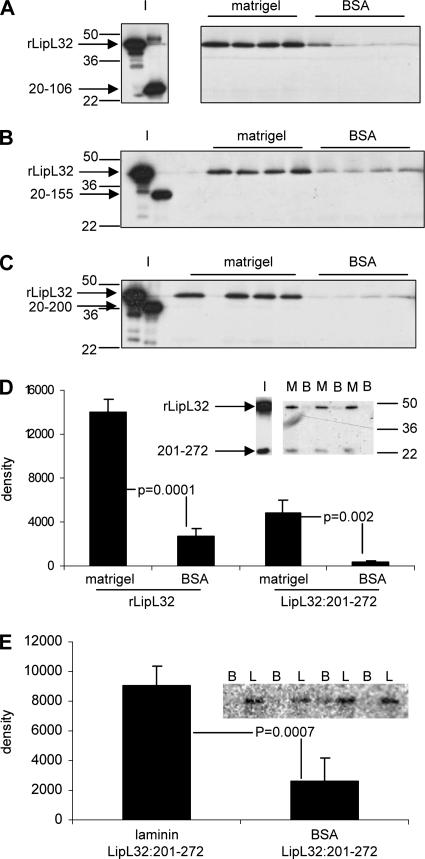
A series of clones expressing fragments of LipL32 were constructed in order to define the region(s) of LipL32 involved in binding to Matrigel ECM. The secondary-structure predictions of LipL32 and PTD2-05920 were used to design truncation constructs for the expression of protein fragments that were not disrupted within predicted alpha-helical or beta-sheet domains. These proteins were expressed as fusion proteins with an N-terminal biotin acceptor peptide tag and purified (Fig. 3B and C). Solid-phase binding assays were performed with rLipL32:20-106, rLipL32:20-155, rLipL32:20-200, and rLipL32:201-272 alone or mixed with full-length, mature rLipL32:20-272. None of the C-terminally truncated proteins bound to Matrigel alone (data not shown) or when mixed with full-length rLipL32:20-272 (Fig. 6A, B, and C). However, a construct comprising the 72 C-terminal amino acids (rLipL32:201-272) bound Matrigel (data not shown) and laminin (Fig. 6E) over control levels when incubated in isolation. When rLipL32:201-272 was coincubated with full-length rLipL32:20-272 at a 1:3 input ratio, both proteins bound Matrigel over control levels at similar ratios (Fig. 6D).
FIG. 6.
The C-terminal 72 amino acids of LipL32 are necessary and sufficient for binding to Matrigel and laminin. (A) rLipL32 was coincubated with rLipL32 deletion 20 to 106 at the input ratio shown in blot I. This mixture was incubated with Matrigel- or BSA-treated wells in quadruplicate. The blot shows that only rLipL32 was recovered from the Matrigel-treated wells. The positions of standard molecular mass markers (kDa) are shown on the left. (B) rLipL32 deletion 20 to 155 was coincubated with rLipL32 at the input ratio shown in lanes I. Proteins were added to Matrigel- or BSA-treated wells as for panel A. Only rLipL32 was recovered from Matrigel-treated wells. (C) rLipL32 deletion 20 to 200 was coincubated with rLipL32 and analyzed for binding as for panel A. Only rLipL32 was recovered in the Matrigel-treated wells. (D) rLipL32 deletion 201 to 272 was coincubated with rLipL32 at the input ratio indicated by lane I. Both proteins were recovered from Matrigel-treated wells (denoted by M) at similar ratios. Neither protein was recovered from BSA-treated wells (denoted by B). The positions of standard molecular mass markers are shown on the right. The blot was quantified by densitometry, with the averages and standard deviations from triplicate wells presented graphically. Conditions were compared by Student's two-tailed t test, with the resulting P values indicated. (E) rLipL32 deletion 201 to 272 was incubated with laminin (L) or BSA control (B) in quadruplicate. The blot shows the amount of rLipL32 deletion 201 to 272 recovered. The bands were analyzed by densitometry as in panel D.
DISCUSSION
LipL32 is one of the most highly studied leptospiral proteins, because it is thought to be important during human pathogenesis. Genetic data show that the LipL32 gene is highly conserved in pathogenic species and is absent in closely related saprophytic species (11, 22). Proteomic studies have shown LipL32 to be the major surface-exposed outer membrane protein from laboratory-cultured strains (4-6). Evidence for the direct involvement of LipL32 in mammalian pathogenesis is also strong. First, proteomics studies using leptospires isolated from an individual with the severe pulmonary form of leptospirosis that were applied to a guinea pig model of leptospirosis likewise showed that LipL32 is expressed during infection (20). Second, immunohistochemistry studies showed that LipL32 is expressed in leptospires from kidney tissue of infected mammals (11). Finally, anti-LipL32 antibodies are made during human infection (10). These studies indicate selective pressure to retain this protein that dominates the cell-surface architecture of Leptospira during laboratory culture and infection. However the function of LipL32 has proven elusive, since it has no similarity to any other protein with a known function and reports suggesting hemolytic activity (12) have yet to be substantiated.
LipL32 was identified as an ECM-binding protein in this study, following the observation that L. interrogans serovar Manilae cells adhered specifically to Matrigel, laminin, collagen I, and fibronectin. Previous studies showed that there was some strain-specific variation in LipL32 expression levels (11). Therefore, it is speculated that there may be a correlation between expression and cellular interaction with ECM. Since LipL32 expression is high under laboratory culture conditions that are osmotically similar to environmental reservoirs, this may reflect the biological role of LipL32 as a protein that initiates the interaction with ECM during the transition from environment to host. However, experiments using anti-LipL32 antiserum as a potential adhesion-blocking reagent failed to block adhesion, as tested by an in vitro adhesion assay (D. E. Hoke and B. Adler, unpublished results). This is probably due to redundancy of adhesion molecules in Leptospira (e.g., LigA, LigB, and LSA24).
rLipL32 was used to model the binding properties of native LipL32. These studies showed selective binding to Matrigel ECM and the individual components laminin and collagens I and V, but not to fibronectin. In the multiplicity of ECM ligands that it binds, LipL32 is similar to the LigA and LigB proteins, which bind collagens I and IV, laminin, and especially fibronectin and fibrinogen (3). Work with other bacterial ECM-binding proteins that bind disparate ECM molecules showed that separate domains are responsible for these multiple interactions (26). Future work will be aimed at determining the domains of LipL32 required for molecular interaction with different components of the ECM.
The binding curve for the interaction of rLipL32 with Matrigel ECM appears complex, with the detection of a small amount of rLipL32 bound at the 0.2 to 1 μM concentration range and 10 to 30 times more bound at the 5 to 10 μM concentration range. This is hypothesized to be due to the heterogeneous nature of this ECM preparation, in which LipL32 can bind to the major constituents of Matrigel, which are laminin and collagen IV (16), with different affinities. While this theory requires future confirmation, including formally testing the binding activity for collagen IV, it is apparent that LipL32 displays complex binding activity consistent with its potential to bind multiple ligands within Matrigel ECM.
The first wave of bacterial genomic information was largely skewed to human pathogens. Analysis of these pathogen genomes indicated that LipL32 is restricted to the genus Leptospira. However, recent genome projects have included environmental bacteria, allowing the identification of a highly similar gene, PTD2-05920, in the marine surface-associated bacterium P. tunicata (8, 9, 13). Our work shows that rPTD2-05920 is immunologically and functionally similar to LipL32. Interestingly, native PTD2-05920 was not detected in broth-cultured P. tunicata, indicating that the protein is not expressed under in vitro growth conditions. While almost nothing is known about gene or protein expression in P. tunicata, it is logical to suggest that PTD2-05920 may be differentially expressed when required for adhesion.
P. tunicata was originally isolated from the marine tunicate Ciona intestinalis, in which it is believed to protect the host surface from the colonization of a variety of fouling organisms. While little is known about this interaction, it is interesting to note that C. intestinalis is a primitive chordate that has many of the genes necessary for ECM synthesis (14). Therefore, it is possible that the common function of LipL32 and PTD2-05920 as ECM-binding proteins is exploited by these divergent bacteria for the common biological goal of interacting with their host. C. intestinalis is an invertebrate that diverged most recently before the point of vertebrates, making it a model for vertebrate evolution (14). While ECM genes have been shown to be associated with the evolution of multicellular organisms and higher organismal complexity, LipL32 may be a component of the molecular biology of bacterial evolution allowing expansion from primitive chordates to mammalian hosts.
While some bacterial MSCRAMMs have been shown to utilize similar binding domains (26), there is nothing in the LipL32 sequence that would indicate a particular binding domain a priori. Therefore, a series of deletion constructs was made, identifying the C-terminal 72 amino acids as necessary and sufficient for binding. This region thus constitutes a novel ECM-binding sequence. A comparison of LipL32 and PTD2-05920 C termini revealed a highly conserved region of 29 amino acids with 75% identity, which is therefore a likely candidate for the minimal binding region of the two proteins.
The interaction of a bacterium with its host is the result of a response to environmental stimuli acting through multiple adhesion molecules. While the adhesive contribution of the leptospiral Lig proteins is proposed to be controlled by changes in osmolarity (3), LipL32 function may be affected by posttranslational events, such as proteolytic cleavage. Studies have suggested that the C terminus of LipL32 is shed, while the N terminus remains attached to the cell surface (4, 20). Since this work implicates the C terminus of LipL32 as the region that binds ECM, cleavage may modulate the LipL32/ECM interaction. Thus, LipL32 is proposed to be a component of an adhesive program for Leptospira and possibly other host-associated bacteria.
Acknowledgments
The anti-LipL32 antiserum was provided by David A. Haake, as were helpful comments and discussion. Thanks are due to Staffan Kjelleberg for collaborative assistance in this project.
This work was supported by a grant from the Australian National Health and Medical Research Council, Canberra, Australia.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 19 February 2008.
REFERENCES
- 1.Anonymous. 1999. Leptospirosis worldwide, 1999. Wkly. Epidemiol. Rec. 74217-223. [PubMed] [Google Scholar]
- 2.Barbosa, A. S., P. A. Abreu, F. O. Neves, M. V. Atzingen, M. M. Watanabe, M. L. Vieira, Z. M. Morais, S. A. Vasconcellos, and A. L. Nascimento. 2006. A newly identified leptospiral adhesin mediates attachment to laminin. Infect. Immun. 746356-6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choy, H. A., M. M. Kelley, T. L. Chen, A. K. Moller, J. Matsunaga, and D. A. Haake. 2007. Physiological osmotic induction of Leptospira interrogans adhesion: LigA and LigB bind extracellular matrix proteins and fibrinogen. Infect. Immun. 752441-2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cullen, P. A., S. J. Cordwell, D. M. Bulach, D. A. Haake, and B. Adler. 2002. Global analysis of outer membrane proteins from Leptospira interrogans serovar Lai. Infect. Immun. 702311-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cullen, P. A., D. A. Haake, and B. Adler. 2004. Outer membrane proteins of pathogenic spirochetes. FEMS Microbiol. Rev. 28291-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cullen, P. A., X. Xu, J. Matsunaga, Y. Sanchez, A. I. Ko, D. A. Haake, and B. Adler. 2005. Surfaceome of Leptospira spp. Infect. Immun. 734853-4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dinkla, K., M. Rohde, W. T. Jansen, J. R. Carapetis, G. S. Chhatwal, and S. R. Talay. 2003. Streptococcus pyogenes recruits collagen via surface-bound fibronectin: a novel colonization and immune evasion mechanism. Mol. Microbiol. 47861-869. [DOI] [PubMed] [Google Scholar]
- 8.Egan, S., S. James, C. Holmstrom, and S. Kjelleberg. 2001. Inhibition of algal spore germination by the marine bacterium Pseudoalteromonas tunicata. FEMS Microbiol. Ecol. 3567-73. [DOI] [PubMed] [Google Scholar]
- 9.Egan, S., T. Thomas, C. Holmstrom, and S. Kjelleberg. 2000. Phylogenetic relationship and antifouling activity of bacterial epiphytes from the marine alga Ulva lactuca. Environ. Microbiol. 2343-347. [DOI] [PubMed] [Google Scholar]
- 10.Guerreiro, H., J. Croda, B. Flannery, M. Mazel, J. Matsunaga, M. Galvao Reis, P. N. Levett, A. I. Ko, and D. A. Haake. 2001. Leptospiral proteins recognized during the humoral immune response to leptospirosis in humans. Infect. Immun. 694958-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haake, D. A., G. Chao, R. L. Zuerner, J. K. Barnett, D. Barnett, M. Mazel, J. Matsunaga, P. N. Levett, and C. A. Bolin. 2000. The leptospiral major outer membrane protein LipL32 is a lipoprotein expressed during mammalian infection. Infect. Immun. 682276-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hauk, P., N. S. Romero, S. A. Vasconcellos, M. E. Genovez, R. J. Ward, M. Schattner, R. M. Goméz, and P. L. Ho. 2005. Expression and characterization of HlyX hemolysin from Leptospira interrogans serovar Copenhageni: potentiation of hemolytic activity by LipL32. Biochem. Biophys. Res. Commun. 3331341-1347. [DOI] [PubMed] [Google Scholar]
- 13.Holmstrom, C., S. James, B. A. Neilan, D. C. White, and S. Kjelleberg. 1998. Pseudoalteromonas tunicata sp. nov., a bacterium that produces antifouling agents. Int. J. Syst. Bacteriol. 481205-1212. [DOI] [PubMed] [Google Scholar]
- 14.Huxley-Jones, J., D. L. Robertson, and R. P. Boot-Handford. 2007. On the origins of the extracellular matrix in vertebrates. Matrix Biol. 262-11. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, R. C., J. Walby, R. A. Henry, and N. E. Auran. 1973. Cultivation of parasitic leptospires: effect of pyruvate. Appl. Microbiol. 26118-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kleinman, H. K., and G. R. Martin. 2005. Matrigel: basement membrane matrix with biological activity. Semin. Cancer Biol. 15378-386. [DOI] [PubMed] [Google Scholar]
- 17.Koizumi, N., and H. Watanabe. 2003. Identification of a novel antigen of pathogenic Leptospira spp. that reacted with convalescent mice sera. J. Med. Microbiol. 52585-589. [DOI] [PubMed] [Google Scholar]
- 18.Levett, P. N. 2001. Leptospirosis. Clin. Microbiol. Rev. 14296-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mulvey, M. A. 2002. Adhesion and entry of uropathogenic Escherichia coli. Cell Microbiol. 4257-271. [DOI] [PubMed] [Google Scholar]
- 20.Nally, J. E., J. P. Whitelegge, S. Bassilian, D. R. Blanco, and M. A. Lovett. 2007. Characterization of the outer membrane proteome of Leptospira interrogans expressed during acute lethal infection. Infect. Immun. 75766-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patti, J. M., B. L. Allen, M. J. McGavin, and M. Hook. 1994. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 48585-617. [DOI] [PubMed] [Google Scholar]
- 22.Picardeau, M., D. M. Bulach, C. Bouchier, R. L. Zuerner, N. Zidane, P. J. Wilson, S. Creno, E. S. Kuczek, S. Bommezzadri, J. C. Davis, A. McGrath, M. J. Johnson, C. Boursaux-Eude, T. Seemann, Z. Rouy, R. L. Coppel, J. I. Rood, A. Lajus, J. K. Davies, C. Médigue, and B. Adler. 2008. Genome sequence of the saprophyte Leptospira biflexa provides insights into the evolution of Leptospira and the pathogenesis of leptospirosis. PLoS One 3e1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Promega. 2007. Abstract for PinPoint Xa protein purification system. www.promega.com/tbs/tm028/tm028.html.
- 24.Rost, B., and C. Sander. 1993. Prediction of protein secondary structure at better than 70% accuracy. J. Mol. Biol. 232584-599. [DOI] [PubMed] [Google Scholar]
- 25.Rost, B., G. Yachdav, and J. Liu. 2004. The PredictProtein server. Nucleic Acids Res. 32W321-W326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwarz-Linek, U., M. Hook, and J. R. Potts. 2006. Fibronectin-binding proteins of gram-positive cocci. Microbes Infect. 82291-2298. [DOI] [PubMed] [Google Scholar]
- 27.Schwarz-Linek, U., M. Hook, and J. R. Potts. 2004. The molecular basis of fibronectin-mediated bacterial adherence to host cells. Mol. Microbiol. 52631-641. [DOI] [PubMed] [Google Scholar]
- 28.Silva, J. J., M. O. Dalston, J. E. Carvalho, S. Setubal, J. M. Oliveira, and M. M. Pereira. 2002. Clinicopathological and immunohistochemical features of the severe pulmonary form of leptospirosis. Rev. Soc. Bras. Med. Trop. 35395-399. [DOI] [PubMed] [Google Scholar]
- 29.Simpson, F. G., K. A. Green, G. J. Haug, and D. L. Brookes. 1998. Leptospirosis associated with severe pulmonary haemorrhage in Far North Queensland. Med. J. Aust. 169151-153. [DOI] [PubMed] [Google Scholar]
- 30.Trevejo, R. T., J. G. Rigau-Perez, D. A. Ashford, E. M. McClure, C. Jarquin-Gonzalez, J. J. Amador, J. O. de los Reyes, A. Gonzalez, S. R. Zaki, W. J. Shieh, R. G. McLean, R. S. Nasci, R. S. Weyant, C. A. Bolin, S. L. Bragg, B. A. Perkins, and R. A. Spiegel. 1998. Epidemic leptospirosis associated with pulmonary hemorrhage—Nicaragua, 1995. J. Infect. Dis. 1781457-1463. [DOI] [PubMed] [Google Scholar]
- 31.Wada, H., M. Okuyama, N. Satoh, and S. Zhang. 2006. Molecular evolution of fibrillar collagen in chordates, with implications for the evolution of vertebrate skeletons and chordate phylogeny. Evol. Dev. 8370-377. [DOI] [PubMed] [Google Scholar]