Abstract
Dendritic cells (DC) are believed to play an important role in the initiation of innate and adaptive immune responses to infection, including respiratory tract infections, where respiratory DC (RDC) perform this role. In this report, we examined the susceptibilities of isolated murine RDC to influenza virus infection in vitro and the effect of the multiplicity of infection (MOI) on costimulatory ligand upregulation and inflammatory cytokine/chemokine production after infection. We found that the efficiency of influenza virus infection of RDC increased with increasing MOIs. Furthermore, distinct subpopulations of RDC differed in their susceptibilities to influenza virus infection and in the magnitude/tempo of costimulatory ligand expression. Additional characterization of the CD11c-positive (CD11c+) RDC revealed that the identifiable subsets of RDC differed in susceptibility to infection, with CD11c+ CD103+ DC exhibiting the greatest susceptibility, CD11c+ CD11bhi DC exhibiting intermediate susceptibility, and CD11c+ B220+ plasmacytoid DC (pDC) exhibiting the least susceptibility to infection. A companion analysis of the in vivo susceptibilities of these RDC subsets to influenza virus revealed a corresponding infection pattern. The three RDC subsets displayed different patterns of cytokine/chemokine production in response to influenza virus infection in vitro: pDC were the predominant producers of most cytokines examined, while CD103+ DC and CD11bhi DC produced elevated levels of the murine chemokine CXCL1 (KC), interleukin 12p40, and RANTES in response to influenza virus infection. Our results indicate that RDC are targets of influenza virus infection and that distinct RDC subsets differ in their susceptibilities and responses to infection.
Dendritic cells (DC) are a distinct lineage of hematopoietic mononuclear cells that are believed to play a critical role in the induction of adaptive immune responses and may also serve a direct role as effector cells in the innate immune responses to infection (4, 15, 30). DC are found both within secondary lymphoid organs (e.g., the spleen and lymph nodes) and at peripheral sites/body surfaces (e.g., the gastrointestinal tract, the genitourinary tract, and the respiratory tract). DC localized to body surfaces act as sentinels, monitoring these sites for the presence of foreign antigen (in particular, infectious microorganisms). These peripheral DC capture antigen (by direct antigen uptake and/or infection of the DC) and, as a result of this encounter, undergo an activation/maturation process. The activated DC then transport the antigen from the body surface to secondary lymphoid organs, where the induction of the adaptive immune responses occurs (3, 16).
DC isolated from secondary lymphoid organs or peripheral sites demonstrate considerable heterogeneity in both phenotype (i.e., cell surface marker expression) and function and can be categorized into distinct subsets. Both the number of distinct DC subsets and the distribution of these DC subsets can differ dramatically depending on the sites or tissues of origin (2, 12, 34).
The respiratory tract (RT) is a major site of antigen encounters with DC. Respiratory DC (RDC) are distributed throughout the upper and lower RT. In the RT, RDC are localized to both large and small airways, as well as to the lung parenchyma (21, 33). Airway-resident RDC are intimately associated with the airway epithelium, are uniformly distributed along the airways, and have a relatively high turnover rate. By contrast, RDC found in alveoli and the lung parenchyma are less uniformly distributed and have a lower turnover rate than airway-associated RDC (32). RDC have been divided into distinct subsets based on a number of criteria, most notably cell surface marker expression (27, 39). Recently, an important link has been made by Beaty et al. and Sung et al. between the anatomical localization of RDC and the expression of cell surface markers (5, 36). These investigators identified three prominent subsets of CD11c-positive (CD11c+) RDC in the normal mouse lung based on the expression of major histocompatibility complex (MHC) class II and the differential expression of CD103, CD11b, and B220. Of particular note is the finding that airway-resident RDC are predominately CD103+ and major producers of interleukin 12p40 (IL-12p40). By contrast, CD103− CD11bhi DC are preferentially localized to the lung parenchyma, and upon activation, these cells are potent producers of a range of inflammatory mediators. B220+ plasmacytoid DC (pDC) represent a third distinct, but less abundant, RDC subset found in the parenchyma of the normal lung (13, 27, 36).
Influenza virus is a major human pathogen, and infection by this virus can produce severe injury to the RT. Influenza virus infection has also been an extremely useful experimental model to examine the induction and development of the adaptive immune responses in the RT. Indeed, RDC that have migrated from the lungs to the lung draining lymph nodes have been implicated as the major antigen-presenting cells (APC) for the induction of adaptive immune CD8+ T-cell response to infection with the virus (6, 17, 23, 38). Furthermore, our laboratory has recently obtained evidence that the size of the inoculum dose of influenza virus used for experimental infection and the tempo of the early virus replication in the RT control the magnitude of the adaptive immune CD8+ T-cell response, possibly through an effect on RDC function (24).
Much of the information on the interaction of influenza virus with DC has come from studies employing splenic DC or DC of bone marrow origin derived from in vitro culture. Such studies have revealed that bone marrow-derived DC can be infected by influenza virus and that influenza virus infection of these DC triggers cytokine (e.g., IL-6 and IL-12) and type 1 interferon production. Furthermore, efficient induction of CD8+ T-cell responses in vitro appears to require direct infection of DC by live virus (7, 8, 26, 28, 29), although presentation of influenza virus antigen from noninfectious sources through cross-presentation by DC has been reported (1).
In spite of the presumed critical role of RDC in the induction of adaptive immune responses to influenza virus infection and the likely role of RDC as innate immune effectors during influenza virus infection of the lungs, minimal information is available on the interaction/infection of RDC with influenza virus and no information is available on the responses of individual RDC subsets to influenza virus infection. In this report, we examine the interaction of purified RDC with infectious influenza virus in vitro. We demonstrate that three major RDC subsets differ in their susceptibilities to influenza virus infection, with the CD103+ RDC subset exhibiting the greatest susceptibility to infection. We also evaluate maturation marker expression by infected RDC and the effect of virus multiplicity of infection (MOI) on RDC maturation and on the production of cytokines/chemokines by activated RDC. We observe that CD103+ DC are the dominant producers of IL-12p40 and that the production of this cytokine is selectively suppressed at increasing MOIs. Related experiments examined the susceptibility of RDC and these RDC subsets to influenza virus infection in vivo. The significance of these findings in defining the role of RDC in respiratory virus infection is discussed.
MATERIALS AND METHODS
Mice.
Ten- to 14-week-old female BALB/cAnNTac mice (H-2d) were purchased from Taconic Farms (Germantown, NY) and maintained in a specific-pathogen-free environment. All experiments were performed in accordance with regulatory standards and guidelines approved by the University of Virginia Animal Care and Use Committee.
Antibodies.
The following monoclonal antibodies (MAbs) were used in the analysis of RDC populations. Anti-MHC class I-fluorescein isothiocyanate (FITC) (SF1-1.1), MHC class II-FITC (AMS-32.1), CD11c-phycoerythrin (PE)-Cy7 (HL3), CD45R/B220-PerCP (RA3-6B2), CD45-PerCP-Cy5.5 (30-F11), SiglecF-PE (E50-2440), CD86-PE (GL-1), CD40-PE (3/23), CD80-PE (16-10A1), CD103-FITC (M/290), and CD11b-PE-Cy7 (M1/70) were purchased from BD Pharmingen (San Diego, CA). Anti-CD86-allophycocyanin (GL1), CD40-allophycocyanin (1C10), CD80-allophycocyanin (30-F11), CD103-PE (2E7), and CD11b-Alexa Fluor 750-allophycocyanin (M1/70) were purchased from eBiosciences (San Diego, CA). Anti-CD103-Alexa Fluor 647 was purchased from Biolegend (San Diego, CA). Goat F(ab′)2 anti-mouse immunoglobulin G-allophycocyanin was purchased from Caltag Laboratories (Carlsbad, CA). Anti-plasmacytoid dendritic cell antigen (PDCA)-PE was purchased from Miltenyi Biotec GmbH (Auburn, CA). The monoclonal anti-NP (H16) was a gift from Walter Gerhard (Wistar Institute, Pennsylvania). The anti-NP (H16) was conjugated to Alexa Fluor 555 dye in accordance with the conjugation kit purchased from Molecular Probes (Eugene, OR).
Preparation and administration of Flt3L plasmid.
The pUMVC3-hFlex (Flt3 ligand [Flt3L]) plasmid was obtained from Hardy Kornfeld (University of Massachusetts Medical School, Worcester, MA). Escherichia coli One Shot Top10 competent cells (Invitrogen Life Technologies, Carlsbad, CA) were transformed with the plasmid and grown in large quantity. The Flt3L plasmid was purified from the transformed bacteria using the EndoFree Plasmid Mega Kit (Qiagen, Valencia, CA). Subsequently, the purified plasmids were run through a Detoxigel column (Pierce Chemical Company, Rockford, IL) to remove any residual endotoxin in the plasmid preparation. No residual endotoxin was detected in the plasmid preparation as measured by Limulus amebocyte lysate assay (Charles River Laboratories, Wilmington, MA). Injection of plasmid DNA was performed as described previously. Briefly, Flt3L plasmids were resuspended in sterile 0.9% NaCl at a final concentration of 5 μg/ml, and 2 ml of the solution was rapidly administrated intravenously over 10 seconds into the tail vein of each mouse. This procedure was repeated 6 days after the first plasmid administration. The lungs of plasmid-treated mice were isolated for RDC preparation 6 days after the second plasmid administration.
Preparation of single-lung-cell suspension.
Mice were sacrificed by cervical dislocation. The lungs were perfused via the right ventricle of the heart with 5 ml phosphate-buffered saline (PBS) to remove the intravascular pool of cells from the lung vasculature. The lungs were minced and then digested in 183 U/ml type II collagenase (Worthington, Lakewood, NJ) in Iscove's modified Dulbecco's medium (IMDM) (Gibco BRL, Gaithersburg, MD) at 37°C in 5% CO2 for 30 min. Afterward, the minced lung tissue was passed though cell strainers (BD) and washed three times with Iscove modified Eagle medium. The lung cell pellets were either resuspended in MACS buffer (PBS supplemented with 0.5% bovine serum albumin [BSA] Σ and 2 mM EDTA [Promega Corporation, Madison, WI]) for DC isolation or in fluorescence-activated cell sorter (FACS) lysing solution (BD, San Jose, CA) for antigen detection.
Purification of total RDC and RDC subsets.
All magnetic microbeads used for cell isolation were purchased from Miltenyi Biotec GmbH. For total RDC isolation, lung alveolar macrophages were first removed as described previously (35). Briefly, single-lung-cell suspensions were incubated with PE-conjugated anti-SiglecF antibody, and then the cells were magnetically labeled with anti-PE microbeads. SiglecF+ cells were separated by positive selection using an AutoMACS Separator (Miltenyi Biotec GmbH, Auburn, CA) according to the manufacturer's protocol. The remaining SiglecF− cells were then incubated with anti-CD11c magnetic microbeads (N418), followed by positive selection to yield CD11c+ SiglecF− cells.
For CD11c+ CD103+ RDC isolation, CD103+ lung cells were selected based on the expression of CD103. Total lung cell suspension was incubated with FITC-conjugated anti-CD103 antibody (M290), followed by incubation with anti-FITC microbeads. Subsequently, the microbeads were removed from the CD103+ cell surface using the anti-FITC MultiSort kit (Miltenyi Biotec GmbH) in accordance with the manufacturer's protocol. Then, the recovered CD103+ lung cells were incubated with anti-CD11c microbeads, and CD11c+ CD103+ RDC were enriched after positive microbead selection.
For lung pDC isolation, total lung single-cell suspension was incubated with anti-mPDCA-1 microbeads, and mPDCA-1+ cells were enriched after positive selection.
For CD103− CD11bhi DC isolation, total single-lung-cell suspension was enriched for CD11c+ cells by positive selection with anti-CD11c microbeads. The resulting CD11c+ cells were incubated with anti-CD103-PE, -CD11b-PE Cy-7, and -MHC class II-FITC for 10 min on ice, and then individual cells that were MHC class IIhi CD103− CD11bhi were sorted on a FACSVantage SE flow cytometer (BD) at the Flow Cytometry Core Facility (University of Virginia). The purity of the cells isolated either by magnetic-bead selection or by FACS sorting was determined by flow cytometry to range from 80 to 99%.
Virus and infection.
The mouse-adapted virus type A Japan/305/57 (H2N2) used in this study was grown in 10-day embryonated hen eggs and stored at −80°C. Mice were infected with different doses of virus diluted in serum-free Iscove modified Eagle medium. The mice were lightly anesthetized with halothane prior to receiving influenza virus in a final volume of 50 μl.
For the infection of DC in vitro, purified DC were seeded at a concentration of 1.2 × 106, typically, in a 6- or 24-well flat-bottom plate (Corning Incorporated, Corning, NY) and incubated for 1 h in complete medium (IMDM supplemented with 2 mM l-glutamine, 10% fetal bovine serum [Atlanta Biologicals, Norcross, GA], 100 U/ml penicillin [Gibco BRL], 100 μg/ml streptomycin [Gibco BRL], and 50 mM 2-ME). At the end of incubation, the cells were washed once with prewarmed IMDM to remove residual serum proteins and nonadherent cells in the cultures. Fresh prewarmed serum-free IMDM (1 ml for 6-well plates or 0.3 ml for 24-well plates) was added to the wells. A dose of A/Japan/305/57 virus corresponding to an MOI of 0.1, 1, or 10 was added, and then the cells were incubated on ice for 10 min, followed by 45 min of incubation at 37°C in 5% CO2. At the end of the incubation, the cells were washed once and cultured in complete medium. We routinely found that, after in vitro culture, total uninfected RDC routinely retained 75 to 80% viability over the first 22 h of culture. For virus-infected DC, we typically observed that, by 22 h postinfection (p.i.), DC viability ranged from 65 to 70% at an MOI of 10, with slightly greater viability of DC infected at an MOI of 1 and viability comparable to that of uninfected control cells among the RDC exposed to an MOI of 0.1 in culture for 22 h. When cultured for 44 h in vitro, we found that the viabilities of both uninfected and infected DC had dropped to approximately 50%.
Cytokine/chemokine detection.
Supernatants from cultures of control infected total RDC or RDC subsets were collected at various time points p.i. and frozen at −20°C. The amounts of cytokines and chemokines in the supernatants were quantified using Bio-plex cytokine assay (Bio-Rad Laboratories, Inc., Hercules, CA). Alpha/beta interferon (IFN-α/β) production in the cultured supernatants was measured using an IFN-α/β enzyme-linked immunosorbent assay (ELISA) kit (PBL Biomedical Laboratories, New Brunswick, NJ).
Intracellular and surface staining.
Identification of RDC subsets and detection of costimulatory molecule expression were carried out on total lung cell suspensions from Flt3L-treated or untreated mice by fixation of cells in FACS lysing solution for 10 min at room temperature, followed by resuspension of the fixed cells in FACS buffer (PBS supplemented with 0.5% BSA and 0.02% NaN3). Nonspecific antibody binding was blocked by incubating the cells in FACS buffer containing 5% goat serum (Jackson Immunoresearch, West Grove, PA) for 20 min on ice. Then, the cells were incubated with MAbs against CD45, CD11c, MHC class II, SiglecF, CD103, PDCA, CD40, CD80, and CD86 for 30 min on ice.
To detect intracellular viral protein in total RDC and RDC subsets from either in vitro infection cultures or infected lungs, cells were harvested and fixed in FACS lysing solution for 10 min at room temperature. Nonspecific antibody binding was blocked by incubating the cells for 20 min on ice in permeabilization buffer (PBS supplemented with 0.5% BSA, 0.02% NaN3, and 0.5% saponin [Calbiochem, La Jolla, CA]) containing 5% goat serum. Then, either the unconjugated anti-NP (H16) or Alexa Fluor 555 anti-NP was added to the wells that contained cells from in vitro culture or from infected lungs, respectively, followed by incubation for 30 min on ice. At the end of the incubation, cells from infected lungs were resuspended in FACS buffer, and then the cells were stained for surface marker expression with a cocktail of MAbs against CD11c, MHC class II, CD103, B220, or CD11b for 30 min on ice. For cells from in vitro cultures, the bound unconjugated anti-NP MAbs were then detected by incubating the cells with goat F(ab′)2 anti-mouse immunoglobulin G-allophycocyanin for 30 min on ice. For surface costimulatory molecule detection, cells were resuspended in FACS buffer and stained with a cocktail of MAbs against CD86, CD40, CD80, MHC class I, and MHC class II for 30 min on ice.
The labeled cells were detected by flow cytometry using FACSCaliber (BD, Mountain View, CA) or FACS Canto (BD, Mountain View, CA) and were analyzed using Flowjo software (Tree Star). For these analyses of intracellular viral protein accumulation and cell surface costimulatory molecule upregulation, we analyzed only those cells within the population that fell within the live-cell fraction (gate) based on the light scatter properties of the cultured infected or uninfected RDC as further defined by fluorescent-dye exclusion.
Statistics analysis.
The means ± standard deviations (SD) of multiple experiments were calculated by Excel (Microsoft). Statistical significance was determined using two-tailed Student t tests.
RESULTS
Identification of CD11c+ cells from the lungs of healthy and Flt3L-treated mice.
We wanted to characterize the responses of isolated RDC to infection with type A influenza virus at different MOIs in vitro. Since RDC are included among the CD45+ CD11c+ cells found in the murine lungs (20), we identified CD45+ CD11c+ cells as the initial step in characterizing their response to influenza virus infection. Total lung cell suspensions were prepared from the lungs of naïve (uninfected) BALB/c mice and analyzed for the expression of the hematopoietic cell marker CD45 by flow cytometry. Figure 1A shows the light scatter properties (Fig. 1A1) and CD45 staining (Fig. 1A2) of total lung cells from naïve mice. As Fig. 1A2 shows, approximately 30% of cells in the normal lung were of hematopoietic origin. In companion experiments, we simultaneously examined total lung cell suspensions for cell surface expression of CD45, CD11c, MHC class II, and SiglecF using a cocktail of antibodies directed to these markers.
FIG. 1.
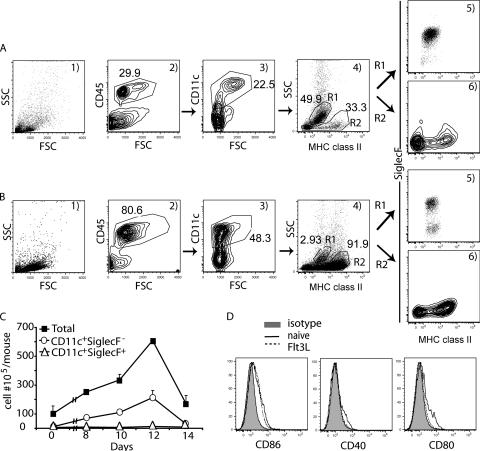
Characterization of the lung CD11c+ population from untreated and Flt3L-treated mice. Single-cell suspensions from the lungs of naïve mice (A) and day 12 Flt3L-treated mice (B) were labeled with MAbs against CD11c, MHC class II, SiglecF, and CD45 and analyzed by flow cytometry. (C) Kinetics of the cell accumulation in the lung at the indicated time after the first Flt3L administration. Single-lung-cell suspensions from untreated (day 0) and day 8, 10, 12, and 14 Flt3L-treated mice were stained with the same MAbs as for panels A and B. The numbers of CD11c+ SiglecF+ and CD11c+ SiglecF− cells were calculated based on the total leukocyte count and the percentage of these two populations as described for panels A and B (mean ± SD; n ≥ 3). (D) Comparison of costimulatory ligand expression on RDC from naïve and Flt3L-treated mice. Lung cell suspensions from naïve or Flt3L-treated mice were labeled with MAbs against CD11c, MHC class II, CD86, CD40, and CD80. The cells were gated on CD11c+ SiglecF−. The data in panels A, B, and D are representative of >3 independent experiments. FSC, forward scatter; SSC, side scatter.
As Fig. 1A3 illustrates, CD11c+ cells accounted for 20% to 25% of the normal lung CD45+ cells. When the gated CD11c+ cells were analyzed for MHC class II expression (Fig. 1A4, MHC class II versus side scatter), two distinct populations of CD11c+ cells were identified. One of these populations (Fig. 1A4, R1) was comprised of cells with little or no MHC class II expression and high side scatter. These cells represented approximately 50% of the total CD11c+ cells and had the light scatter properties attributed to alveolar macrophages. Alveolar macrophages have recently been reported to express not only CD11c, but also the lectin SiglecF (35, 36). The population of CD11c+ cells within the R1 gate was found to be SiglecF+ (Fig. 1A5). By contrast, the remaining CD11c+ cells (Fig. 1A4, R2) consisted of cells that displayed a range of MHC class II expression levels and a low side scatter profile. Unlike alveolar macrophages the CD11c+ cells in R2 were SiglecF− (Fig. 1A6). These CD11c+ SiglecF− lung cells, which would be expected to include both MHC class II high- and low-expressing RDC, represented the initial lung cell population for our analysis of virus infection. The cells typically represented approximately 1% to 2% of the total cells in lung suspensions, and usually <2 × 105 of these CD11c+ SiglecF− cells could be liberated from an individual mouse lung set.
Because of the limited number of CD11c+ SiglecF− cells available from normal lungs for in vitro infection with influenza virus, we employed a strategy to increase the total number of CD11c+ SiglecF− lung cells available for analysis using a plasmid (Flt3L)-based administration method (25). When cell suspensions were prepared from the lungs of mice exposed to this Flt3L induction regimen, the light scatter properties of the isolated cells (Fig. 1B1) were similar to those of cells from untreated lungs (Fig. 1A1). However, in contrast to the untreated lungs, there was a marked increase in the fraction of CD45+ cells, which now represented up to 80% of the total lung cells (Fig. 1B2). Furthermore, as expected, the fraction of CD45+ cells that were CD11c+ doubled in the lungs of Flt3L-treated mice to approximately 50% of the total hematopoietic cells isolated from the lungs (Fig. 1B3). As with normal lungs, two distinct populations of CD11c+ cells were identified based on MHC class II expression and light scatter properties (Fig. 1 B4). However, the alveolar macrophages (Fig. 1B4, R1, and B5) represented a small fraction (2%) of the total CD11c+ cells, while the RDC containing a CD11c+ SiglecF− cell fraction (Fig. 1B4, R2, and B6) now represented 90% of the CD11c+ cells. The recovery of CD11c+ SiglecF− cells from lung cell suspensions was increased at least 45-fold compared to those from untreated mice when cells were harvested at day 12 after Flt3L treatment (Fig. 1C).
This strategy of Flt3L induction increased the number of CD11c+ SiglecF− RDC available for analysis. Although the light scatter properties and marker expression of the CD11c+ SiglecF− cells isolated from the lungs of Flt3L-treated mice were comparable to those of cells from normal lungs, exposure to the Flt3L plasmid could have resulted in the RDC activation. To evaluate this possibility, we compared the expression of the DC-associated maturation markers CD40, CD80, and CD86 on CD11c+ SiglecF− cells isolated from the lungs of healthy and Flt3L-treated mice. As Fig. 1D shows, RDC from the treated mice exhibited comparable levels of expression of these activation markers, suggesting that RDC isolated from the lungs of Flt3L-treated mice had undergone minimal, if any, activation.
Responses of RDC to influenza virus infection at different MOIs.
We used CD11c+ SiglecF− cells isolated from the lungs of Flt3L-treated mice as the source of RDC for in vitro infection with influenza virus A/JAPAN/305/57. Isolated RDC were infected at the indicated MOIs and analyzed, along with uninfected RDC, for intracellular influenza virus nucleocapsid protein and costimulatory ligand expression at 4, 10, and 22 h p.i. by flow cytometry.
The percentage of influenza virus-infected (NP-expressing) RDC increased with increasing MOIs and time p.i. (Fig. 2A and B), with up to 60% to 70% of RDC infected at an MOI of 10 by 22 h p.i. (Fig. 2C). The detection of viral NP in the RDC exposed to influenza virus required de novo viral gene expression and was not attributable to NP from the input virions in the infectious inoculum. When RDC were exposed to infectious virus and examined for viral NP expression after <1 h of culture at 4°C or at 37°C for up to 18 h of culture in the presence of a protein synthesis inhibitor, no viral NP was detected in the flow cytometry-based assay (data not shown). Comparable absence of NP detection was observed when RDC were exposed to a comparable inoculum of inactivated influenza virus (K. Hargadon and T. J. Braciale, unpublished observations). Also, in NP+ cells, this viral protein was distributed within the exposed RDC in the diffuse nuclear and granular cytoplasmic pattern characteristic of influenza virus-infected cells (X. Hao, unpublished observations).
FIG. 2.
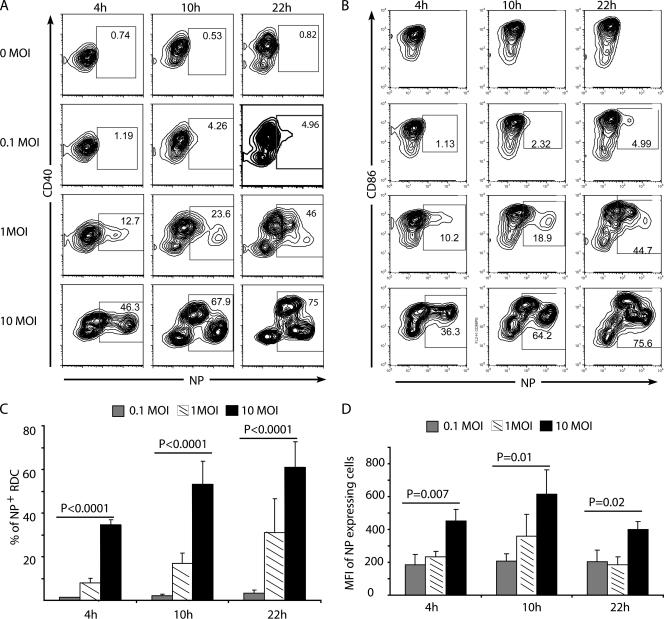
Response of CD11c+ SiglecF− cells to influenza virus infection. Isolated CD11c+ SiglecF− cells were infected in vitro with influenza virus at MOIs of 0.1, 1, and 10 or left untreated (0 MOI). At 4, 10, and 22 h p.i., cells were harvested, fixed, and stained for intracellular NP and surface CD40 and CD86. (A) Kinetics of surface CD40 and intracellular NP expression in CD11c+ SiglecF− cells. (B) Kinetics of surface CD86 and intracellular NP expression in CD11c+ SiglecF− cells. (C and D) Cumulative data on the percentages of NP+ cells and the mean fluorescence intensities (MFI) of NP+ cells at different MOIs (mean + SD; n ≥ 3). The data in panels A and B are representative of three independent experiments.
At the highest MOI employed, a significant fraction of the infected RDC (30% to 40%) expressed high levels of NP as early as 4 h p.i. (Fig. 2C). It is noteworthy that with increasing MOIs, not only was there the expected increase in the percentage of NP+ cells (Fig. 2C), but the levels of NP expression (mean fluorescence intensity per cell) also increased as a function of the MOI (Fig. 2D), a result previously reported with influenza virus infection of bone marrow-derived DC (29). This finding suggests that in infected DC, viral gene expression was at least partly regulated by the number of input (infecting) viral genomes.
Analysis of the kinetics of NP expression in relation to the tempo of upregulation of the costimulatory ligands CD40 (Fig. 2A) and CD86 (Fig. 2B) was also revealing. In uninfected RDC, there was a progressive upregulation of these two costimulatory ligands over time in culture. The subpopulation of high-NP-expressing RDC detected in the culture at an MOI of 10 at 4 h p.i. had likewise upregulated both CD40 and CD86 expression. In the culture infected at an MOI of 1 analyzed at the same time point, as expected, a much smaller fraction of the cells were NP+ and at a somewhat lower intensity of NP expression. Importantly, at 10 and 22 h p.i. in the cultures at an MOI of 10, the subpopulation of high-NP-expressing RDC demonstrated early in infection (i.e., at 4 h p.i.) did not upregulate these costimulatory ligands (Fig. 2A and B). At these two later time points after infection, a second subpopulation of infected cells that expressed lower levels of NP but retained CD40 and CD86 expression over time in culture was now discernible. This second subpopulation of CD40+/CD86+ infected RDC, which expressed NP more slowly and at lower levels, was also demonstrated in the RDC infected at an MOI of 1. We also noted a similar correlation between the tempo and intensity of NP expression and the level of expression of CD80 and MHC class I, while MHC class II was relatively uniformly upregulated on uninfected and infected RDC and retained at similar high levels throughout the course of infection (unpublished data).
Proinflammatory mediator production by RDC following influenza virus infection.
RDC likely play a dual role in the host responses to respiratory infection with pathogens like influenza virus; that is, they serve as a vehicle for antigen delivery to the lymph nodes from the RT and may also serve as innate immune effectors through the release of proinflammatory cytokines/chemokines (17, 22, 31). We examined the impacts of influenza virus infection of RDC at different MOIs on the kinetics of the production of a panel of inflammatory mediators using the Bio-plex bead assay.
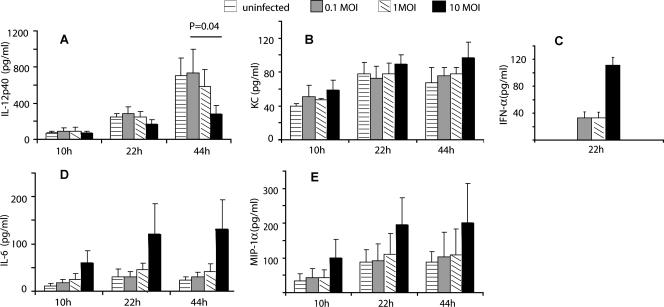
As Fig. 3 shows, there was considerable variability in the production of the five mediators. The production of IFN-α (Fig. 3C), which for technical reasons could be evaluated at only one time point (i.e., 22 h), was greatest at the highest MOI examined. This was a trend that was also observed for two of the mediators, IL-6 (Fig. 3D) and MIP-1α (Fig. 3E), but it did not achieve statistical significance. The notable exception to this trend was IL-12p40 (Fig. 3A). The production of IL-12p40 decreased with increasing MOI, and this decreased cytokine synthesis reached statistical significance relative to uninfected RDC by 44 h in culture. It is noteworthy that, overall, influenza virus infection of the RDC had only a modest effect on the production of the five mediators examined compared to the response observed in uninfected RDC triggered to activate by in vitro culture.
FIG. 3.
Cytokine production by CD11c+ SiglecF− cells after influenza virus infection. Isolated CD11c+ SiglecF− cells were infected in vitro with influenza virus at MOIs of 0.1, 1, and 10 or untreated (0 MOI). Supernatants were collected at 10, 22, and 44 h p.i., and the amounts of cytokines and chemokines in the supernatants were measured using the Bio-plex cytokine assay and IFN-α ELISA kits (mean + SD; n ≥ 3).
Identification and characterization of three distinct subsets of RDC.
The analysis shown in Fig. 2 indicated that different subpopulations of RDC were differentially susceptible to influenza virus infection (as a function of the MOI) and exhibited different patterns of costimulatory ligand expression. These findings raised the possibility that distinct subsets of RDC found in the normal murine RT respond differentially to influenza virus infection. Several recent studies have indicated that there are at least three prominent subsets of CD11c+ RDC in the RT of naïve mice: CD103+ DC, CD103− CD11bhi (CD11bhi) DC, and pDC (36). To determine whether our findings on differential susceptibilities of RDC to influenza virus infection reflected the responses of specific RDC subsets, we first determined if these RDC subsets were represented among the CD11c+ SiglecF− cells isolated from the lungs of Flt3L-treated mice.
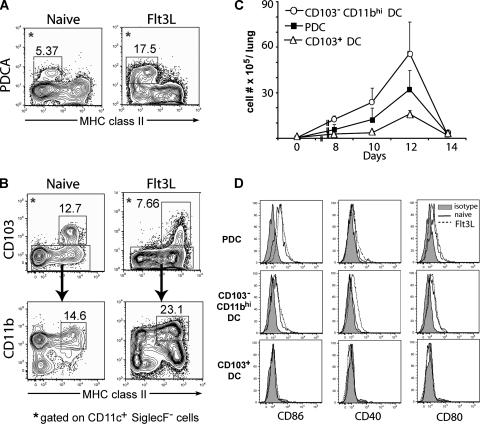
For this analysis, total lung cell suspensions were prepared from untreated and Flt3L-treated mice, and the CD45+ CD11c+ Siglec F− cell population was identified by flow cytometry and further analyzed for the expression of MHC class II and for markers defining these distinct RDC subsets. As Fig. 4A shows, cells that displayed low levels of MHC class II and expressed the bone marrow stromal marker Ag-1/mPDCA-1, characteristic of pDC, were detected in both lungs, with pDC representing a more prominent component of the CD11c+ SiglecF− cell population from Flt3L-treated mice (17% versus 5%). The CD103+ DC and CD11bhi DC accounted for 13% and 12%, respectively, of CD11c+ SiglecF− cells from untreated mice (Fig. 4B). After Flt3L treatment, the fraction of CD103+ DC subset was slightly reduced to approximately 7% of the CD11c+ SiglecF− cells, while that of CD11bhi DC increased to approximately 21% of the CD11c+ SiglecF− cells (Fig. 4B). However, in response to the ligand exposure, the absolute number of each of these RDC subsets in the RT did increase substantially, with pDC, CD103+ DC, and CD103− CD11bhi DC increasing 180-fold, 40-fold, and 200-fold, respectively, compared to their counterparts in the RT from untreated mice, with a peak number of these RDC subsets detectable at day 12 after Flt3L treatment (Fig. 4C).
FIG. 4.
Identification of three RDC subsets in the lungs of untreated and Flt3L-treated mice. Lung cell suspensions from untreated day 12 Flt3L-treated mice were labeled with MAbs against CD11c, MHC class II, CD11b, CD103, and mPDCA-1 and analyzed by flow cytometry. The cells were gated on CD11c+ SiglecF− cells. (A) Identification and enumeration of pDC in both naïve and Flt3L-treated mice based on the expression mPDCA-1 and MHC class II. (B) Identification of CD103+ DC and CD11bhi DC based on the expression of MHC class II, CD103, and CD11b. (C) Kinetics of the accumulation of the number of RDC subsets in the lungs after the first Flt3L administration. Lung cell suspensions from untreated and day 8, 10, 12, and 14 Flt3L-treated mice were stained with the same MAbs as in panels A and B. The number of each subset was calculated based on the total leukocyte count and the percentage of these three subsets as defined for panels A and B (mean + SD; n ≥ 3). (D) Costimulatory ligand expression on RDC subsets from untreated and Flt3L-treated mice. RDC subsets were defined as for panels A and B. The data in panels A, B, and D are representative of three independent experiments.
We also examined the expression of the costimulatory ligands CD86, CD80, and CD40 on these three subsets of RDC freshly isolated from Flt3L-treated and control mice. As Fig. 4D and E show, the expression levels of these markers on the surfaces of RDC from Flt3L-treated mice were comparable to that of control RDC.
We also noted (Fig. 4B) an increase in the proportion of CD11blo MHC class IIhi cells detected within the CD11c+ SiglecF− cell population from treated mice. The precise nature and role of these cells are not clear; but they may represent precursors to more differentiated RDC subsets and are recruited to the lungs in increased numbers after the stimulus of Flt3L.
Differential sensitivities of RDC subsets to influenza virus infection in vitro.
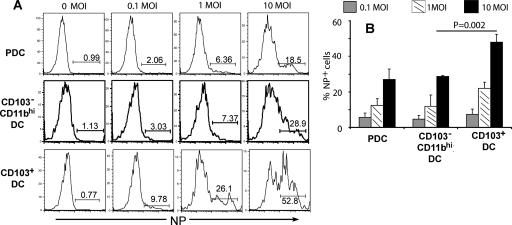
In the murine model, there is evidence that the different subsets of splenic DC may differ in their susceptibilities to viral and/or bacterial infection (11, 18, 40). In view of the findings shown in Fig. 2 demonstrating differential responsiveness of the total CD11c+ Siglec F− cell population to influenza virus infection, we examined the susceptibility of each of these isolated RDC subsets to influenza virus infection.
Figure 5 shows the results of the analysis of viral NP expression at 22 h p.i. for each of these purified RDC subsets isolated from lungs of Flt3L-treated mice and infected at the indicated MOI. Each of the three RDC subsets showed the expected increase in the percentage of influenza virus NP-expressing cells with increasing MOIs, with the CD103+ RDC exhibiting slightly higher NP expression per cell (Fig. 5A). The CD103+ DC also exhibited the greatest susceptibility to influenza virus infection, with approximately 50% of the cells infected at an MOI of 10 (Fig. 5B). The infected CD103+ DC also demonstrated reduced expression of CD86 compared to uninfected control cells at 22 h p.i. (unpublished data). The CD11bhi RDC subset was less susceptible to influenza virus infection than its CD103+ counterpart, with only approximately 30% of the cells demonstrating NP expression at the 22-h time point (Fig. 5B). pDC displayed a degree of susceptibility to infection similar to that of CD11bhi DC. Both CD11bhi DC and pDC upregulated expression of CD86/CD40 in response to infection and retained high levels of these costimulatory ligands comparable to the level displayed by cultured uninfected control cells (unpublished data).
FIG. 5.
In vitro infections of three different RDC subsets with influenza virus. The three RDC subsets (i.e., CD103+, CD11bhi, and PDC) were isolated and infected as described in Materials and Methods. (A) Intracellular NP expression in RDC subsets at 22 h p.i. The data are representative of three independent experiments. (B) Cumulative data on the percentage of NP+ cells (mean + SD; n ≥ 3).
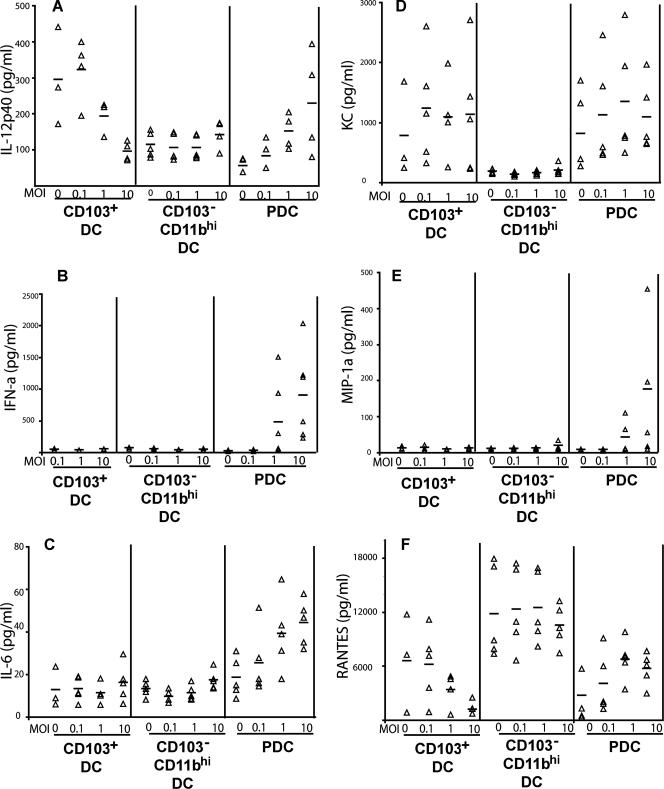
Differential cytokine/chemokine production by RDC subsets after influenza virus infection.
There is a substantial body of evidence indicating that distinct DC subsets differ in their profiles of cytokine/chemokine production in response to inflammatory stimuli, such as virus or bacterial infections (11, 19, 40). We therefore investigated proinflammatory mediators released by these three RDC subsets at 22 h. p.i. in response to influenza virus infection at increasing MOIs (Fig. 6). pDC, although not efficiently infected by influenza virus, were prominent producers of most of these mediators. As expected, these DC were the predominant producers of IFN-α (Fig. 6B) but also showed elevated production of MIP-1α (Fig. 6E), as well as IL-6 (Fig. 6C) and IL-12p40 (Fig. 6A), with infection at increasing MOIs. The CD103+ DC produced variable levels of the murine cytokine CXCL1 (KC) (Fig. 6D) and low levels of IL-6 (Fig. 6C) independently of the MOI. Importantly, the CD103+ DC demonstrated progressive decrease in the production of IL-12p40 (Fig. 6A), as well as RANTES (Fig. 6F). The drop in IL-12p40 production in total RDC at high MOIs (Fig. 3A) was accounted for, in part at least, by the reduced production of IL-12p40 by the CD103+ RDC infected at high MOIs. The CD11bhi DC were weak producers of most of the mediators, with the exception of RANTES, where the cells appeared to be the predominant producers of the chemokine (Fig. 6F). Overall, these results support the view that different subsets of isolated RDC may differ both quantitatively and qualitatively in proinflammatory mediator production in response to influenza virus infection.
FIG. 6.
Cytokine and chemokine production by RDC subsets after influenza virus infection. The three RDC subsets in Fig. 4A and B were isolated and infected as indicated in Materials and Methods. At 22 h p.i., supernatants were harvested and the amounts of cytokines and chemokines were measured using the Bio-plex cytokine assay. IFN-α production was measured with an IFN-α ELISA kit. The triangles represent the values of individual experiments for the indicated cytokines and chemokines from RDC, and the mean values are indicated by the bars (n = 3 to 5).
Influenza virus infection of RDC subsets within the RT.
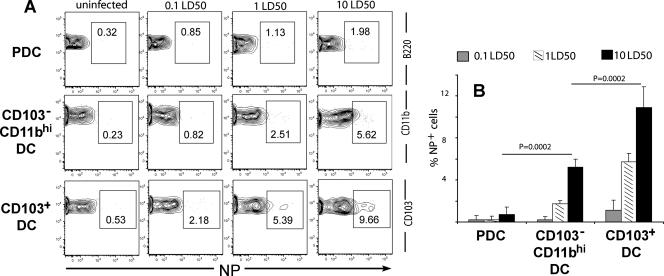
The in vitro analysis described above suggested that the different subsets of RDC present in the lungs were differentially susceptible to influenza virus infection, with the CD103+ DC subsets exhibiting the greatest susceptibility. We therefore wished to determine the extent of infection (percentage of viral NP+ cells) of each of these RDC subsets isolated from influenza virus-infected lungs and analyzed directly ex vivo. Previous analyses by us (23) and others (6) indicated that RDC begin to undergo accelerated migration out of the RT to the draining lymph nodes within 18 to 24 h after experimental influenza virus infection. Although this is a time when the virus titers have not reached maximum levels in the infected lungs, direct ex vivo analysis of RDC infection at 18 h p.i. would minimize the depletion of infected RDC due to accelerated migration of the RDC in response to infection.
To carry out this analysis, we infected mice at three different intranasal-inoculum doses and harvested lungs 18 h later for direct ex vivo analysis of viral NP expression. Not surprisingly, at this early time point after intranasal virus inoculation, only a small percentage of infected RDC was detected (Fig. 7). However, the CD103+ RDC subset was most susceptible to infection, examined by NP expression, over the range of intranasal-inoculum doses, with the CD11bhi subset exhibiting intermediate sensitivity to infection (Fig. 7A), as was likewise observed in vitro (Fig. 5). It is noteworthy that over a range of inocula in repeat experiments, a distinct hierarchy of susceptibility to infection was observed: CD103+ DC > CD11bhi DC > pDC (Fig. 7B).
FIG. 7.
In vivo detection of infected RDC subsets. Mice were infected with 0.1, 1, and 10 50% lethal doses of influenza virus intranasally. Mice that received the diluent Isocove's medium served as negative controls. (A) At 18 h after intranasal infection, lung cell suspensions from the control and infected mice were labeled with MAbs against surface CD11c, MHC class II, CD11b, CD103, B220, and intracellular NP. RDC subsets were gated as in Fig. 4A and B. The data in Fig. 7A represent three independent experiments. (B) Cumulative data on the percentage NP+ in each RDC subset (mean + SD; n ≥ 3).
DISCUSSION
In this report, we examined the responses of total lung CD11c+ cells enriched for RDC and specific RDC subsets to influenza virus infection. We demonstrated that the pattern of infection of total unseparated RDC (CD11c+ SiglecF− cells) by influenza virus was not uniform. A fraction of this total cell population was readily infected by influenza virus and expressed high levels of influenza virus nucleocapsid protein as early as 4 h p.i., particularly at the highest MOI employed (i.e., 10). A second fraction of cells appeared more resistant to infection and expressed nucleocapsid protein over a more prolonged time interval. This second subpopulation also upregulated cell surface costimulatory ligands and sustained high-level expression of these ligands throughout the course of in vitro infection. Based on these findings, we went on to examine whether the apparent heterogeneity in the response to influenza virus infection, and in particular the apparent differences in sensitivity to infection by the virus, reflected the responses of distinct RDC subsets present in the culture to infection. We demonstrated that different RDC subsets showed differential susceptibilities to influenza virus infection in vitro: CD103+ DC were the most susceptible to infection, with CD11bhi DC demonstrating intermediate susceptibility to infection and pDC the least. These in vitro observations were corroborated by in vivo studies on the susceptibilities of these three RDC subsets to infection. Finally, the differences in the responses of these RDC subsets to infection were also reflected in the MOI-dependent production of at least one cytokine, IL-12, by the different RDC subsets in response to virus infection.
Several different DC subsets have been identified in the lungs, each with distinct properties indicating functional specialization (12, 22, 39). In the mouse, RDC express CD11c, which serves as a conventional marker for the identification and isolation of this lung cell type. RDC can be further subdivided based on the level of MHC class II expression; the expression of various cell surface markers, most notably the myeloid cell marker CD11b; and more recently the differential expression of the CD103 integrin alpha chain (36). The expression of the last two markers is of considerable interest, as they distinguish the anatomical localization of these two RDC subsets. Our initial analysis of the susceptibilities of isolated CD11c+ lung cells to infection with influenza virus in vitro (Fig. 2) suggested that different subsets of RDC may be differentially susceptible to infection with type A influenza virus. Two sets of observations supported this hypothesis. First, in our analysis of the susceptibilities of purified CD103+ RDC, CD11bhi RDC, and pDC to influenza virus infection, a clear difference in susceptibility was noted (Fig. 5), with CD103+ RDC representing the most susceptible RDC subset and pDC the least susceptible subset. Second, when isolated from the lungs of influenza virus-infected mice and analyzed directly ex vivo, the CD103+ RDC subset also demonstrated the greatest percentage of influenza virus-infected (NP-expressing) cells (Fig. 7). While in vitro infection of RDC at an MOI as high as 10 may appear to be artificially high, in vivo exposure of RDC (particularly the airway-localized CD103+ RDC to an MOI as high as 10) is possible, if not likely, because of the very high local lung virus titers achieved early in the infected RT of mice (24). Nonetheless, it is noteworthy that, in spite of infection of RDC in vitro with virus at an MOI as high as 10 or after high-dose infection in vivo, only a fraction of the RDC were susceptible to infection. The mechanistic basis for this resistance to infection remains to be defined.
The finding that the CD103+ cells are the RDC subset most susceptible to infection among those examined is particularly intriguing, as these cells have been demonstrated to localize to the epithelial mucosa of the large (conducting) and small airways, forming a dense intraepithelial/subepithelial web (36). The close anatomical proximity of this RDC subset to the respiratory epithelium, the major target of productive influenza virus infection in the RT, make this RDC subset a likely candidate for efficient infection by influenza virus. Indeed, among the CD11c+ cells, which migrate from the lungs to the draining mediastinal lymph nodes in response to influenza virus infection (23), only the CD103+ RDC subset is infected by the virus and is a potent stimulator of naïve virus-specific CD8+ T cells (T. S. Kim and T. J. Braciale, unpublished data). While these findings might argue for a prominent/dominant role of CD103+ RDC in the orchestration of the adaptive immune response to influenza virus because of their susceptibility to infection and proximity in vivo to epithelial cells, such an interpretation should be considered with caution. As this report demonstrates, the three RDC subsets analyzed are each susceptible to infection with influenza virus in vitro and in vivo. Furthermore, both the CD103+ DC and the CD11bhi RDC subsets can trigger the induction of response by antigen-specific CD8+ and CD4+ T cells after in vitro infection of the RDC and coculture with naïve T cells (X. Hao and T. J. Braciale, unpublished data). At present, the precise role of each of these RDC subsets in the induction of adaptive immune responses to influenza virus is uncertain and remains to be elucidated, although recent studies suggest that the CD103+ DC and the CD11bhi RDC may have distinct and complementary roles in the induction of T-cell responses to soluble antigen introduced into the RT (5, 14).
The unseparated CD11c+ RDC (Fig. 3) produced several different inflammatory mediators. In the cases of several mediators (e.g., the murine cytokine CXCL1 [KC] and MIP-1α), the time courses of chemokine secretion by uninfected RDC undergoing maturation in vitro and RDC infected at different MOIs were compatible (Fig. 3B) or at least did not reach statistical significance (Fig. 3E). It appears, therefore, that direct ex vivo infection of these mature lung tissue DC by influenza virus provides only a modest stimulus for the production of some proinflammatory mediators beyond that observed with uninfected RDC. This could reflect suppression of the innate host response in the infected cells by influenza virus or, perhaps more provocatively, mucosally localized DC like RDC may only weakly respond in situ to inflammatory stimuli, like virus infection, in order to prevent the development of excessive local inflammation in the RT. In keeping with this concept, we found that elevated IL-6 production (Fig. 3D) was only observed after infection at the highest MOI (also observed for IFN-α). By contrast, the secretion of IL-12p40 was suppressed after infection at the highest MOI employed (Fig. 3A), suggesting an immunosuppressive effect of influenza virus infection on the production of certain critical mediators.
Analysis of the cytokine responses of individual RDC subsets (Fig. 6) revealed the cellular sources of these mediators. As expected, lung pDC were the major producers of IFN-α among the RDC, and production of this cytokine was dependent upon the MOI. pDC were also prominent producers of most of the other mediators examined, in keeping with their likely role as early innate immune effectors critical in the early host response to virus infection (9). Although the CD11bhi RDC subset has been implicated as an important producer of a variety of proinflammatory mediators in the RT (5), the chemokine RANTES was the predominant inflammatory mediator produced by this RDC subset (Fig. 6F).
IL-12 is believed to play a critical role in the induction of adaptive immune responses through its ability to drive the differentiation of activated T cells, and in particular CD8+ T cells, into fully functional effector cells (10). As noted above, there was a progressive decrease in the secretion of the IL-12p40 molecule with increasing MOIs in both the unseparated RDC (Fig. 3A) and the CD103+ RDC subset (Fig. 6A). The MOI-dependent decrease in IL-12p40 production by CD103+ RDC is most easily explained by influenza virus-induced inhibition of host protein synthesis, as this RDC subset is most susceptible to infection. If CD103+ RDC serve as the dominant APC for the induction of a CD8+ T-cell response to influenza virus, then infection of this cell type at a high MOI in the RT could impair the cells' capacity to effectively present antigen and activate antiviral CD8+ T cells after their migration into the lymph nodes from the infected lungs through dysregulation of IL-12 production, as previously suggested by us (24). Exposure of RDC to the rapid and high-level lung virus replication after a high-inoculum-dose infection in vivo could mimic the impact of in vitro infection at a high MOI on the function/antigen-presenting capability of CD103+ RDC and result in the defective CD8+ T-cell response to high-dose influenza virus infection. A test of this possibility awaits isolation and detailed characterization of this and other RDC subsets isolated from mice infected with different virus inocula.
As demonstrated here (Fig. 1) and elsewhere (36), the alveolar macrophages are the most highly represented CD11c+ cell subset in the normal murine lung, with total RDC and individual RDC subsets representing smaller fractions of the total CD11c+ cells. Isolation of sufficient numbers of individual RDC subsets from normal mouse lungs to carry out the analyses described here was not feasible. We therefore developed a strategy to increase the numbers of RDC isolated from the lung by relying on the DC growth factor Flt3L to generate expanded numbers of RDC in the lungs for subsequent analysis and in vitro characterization. Although treatment with Flt3L has been reported to increase the total number of RDC in the lungs (27, 37), exposure to Flt3L may also increase the maturation/activation state of the RDC. This could be particularly problematic using the plasmid-based approach employed here to express this growth factor. However, we did not note any significant difference in the level of maturation/costimulatory ligand expression among total RDC or individual RDC subsets isolated from the lungs of healthy and Flt3L-treated mice (Fig. 1 and 4). Thus, for the three RDC subsets analyzed in vitro in this report, we believe that the use of RDC isolated from the lungs after Flt3L-induced expansion closely mimicked the corresponding RDC subsets isolated from the normal lungs. We did, however, identify a population of CD11blow/intermediate-expressing cells with variable levels of MHC class II expression that were increased in the lungs of Flt3L-treated mice (Fig. 4). We do not know the nature of the cells, but they likely represent precursors to the more mature RDC subsets. These lung CD11c+ cells likely were recently mobilized from the bone marrow to the lungs as a result of growth factor treatment.
In summary, in this report, we have shown that RDC can be efficiently infected by influenza virus. Importantly, for the first time, we provided evidence that different RDC subsets display differential susceptibilities to influenza virus infection and differential cytokine production. Our results suggest a complex interaction among RDC subsets and other innate immune cells responding to influenza virus infection.
Acknowledgments
This work was supported by AI-15608, AI-37293, AI-57168, and HL-33391.
Footnotes
Published ahead of print on 19 March 2008.
REFERENCES
- 1.Albert, M. L., B. Sauter, and N. Bhardwaj. 1998. Dendritic cells acquire antigen from apoptotic cells and induce class I-restricted CTLs. Nature 39286-89. [DOI] [PubMed] [Google Scholar]
- 2.Asselin-Paturel, C., A. Boonstra, M. Dalod, I. Durand, N. Yessaad, C. Dezutter-Dambuyant, A. Vicari, A. O'Garra, C. Biron, F. Briere, and G. Trinchieri. 2001. Mouse type I IFN-producing cells are immature APCs with plasmacytoid morphology. Nat. Immunol. 21144-1150. [DOI] [PubMed] [Google Scholar]
- 3.Banchereau, J., F. Briere, C. Caux, J. Davoust, S. Lebecque, Y.-J. Liu, B. Pulendran, and K. Palucka. 2000. Immunobiology of dendritic cells. Annu. Rev. Immunol. 18767-811. [DOI] [PubMed] [Google Scholar]
- 4.Banchereau, J., and R. M. Steinman. 1998. Dendritic cells and the control of immunity. Nature 392245-252. [DOI] [PubMed] [Google Scholar]
- 5.Beaty, S. R., C. E. Rose, Jr., and S.-S. J. Sung. 2007. Diverse and potent chemokine production by lung CD11bhigh dendritic cells in homeostasis and in allergic lung inflammation. J. Immunol. 1781882-1895. [DOI] [PubMed] [Google Scholar]
- 6.Belz, G. T., C. M. Smith, L. Kleinert, P. Reading, A. Brooks, K. Shortman, F. R. Carbone, and W. R. Heath. 2004. Distinct migrating and nonmigrating dendritic cell populations are involved in MHC class I-restricted antigen presentation after lung infection with virus. Proc. Natl. Acad. Sci. USA 1018670-8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bender, A., M. Albert, A. Reddy, M. Feldman, B. Sauter, G. Kaplan, W. Hellman, and N. Bhardwaj. 1998. The distinctive features of influenza virus infection of dendritic cells. Immunobiology 198552-567. [DOI] [PubMed] [Google Scholar]
- 8.Cella, M., M. Salio, Y. Sakakibara, H. Langen, I. Julkunen, and A. Lanzavecchia. 1999. Maturation, activation, and protection of dendritic cells induced by double-stranded RNA. J. Exp. Med. 189821-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colonna, M., G. Trinchieri, and Y.-J. Liu. 2004. Plasmacytoid dendritic cells in immunity. Nat. Immunol. 51219-1226. [DOI] [PubMed] [Google Scholar]
- 10.Curtsinger, J. M., D. C. Lins, and M. F. Mescher. 2003. Signal 3 determines tolerance versus full activation of naive CD8 T cells: dissociating proliferation and development of effector function. J. Exp. Med. 1971141-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalod, M., T. Hamilton, R. Salomon, T. P. Salazar-Mather, S. C. Henry, J. D. Hamilton, and C. A. Biron. 2003. Dendritic cell responses to early murine cytomegalovirus infection: subset functional specialization and differential regulation by interferon alpha/beta. J. Exp. Med. 197885-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Heer, H. J., H. Hammad, M. Kool, and B. N. Lambrecht. 2005. Dendritic cell subsets and immune regulation in the lung. Semin. Immunol. 17295-303. [DOI] [PubMed] [Google Scholar]
- 13.De Heer, H. J., H. Hammad, T. Soullie, D. Hijdra, N. Vos, M. A. M. Willart, H. C. Hoogsteden, and B. N. Lambrecht. 2004. Essential role of lung plasmacytoid dendritic cells in preventing asthmatic reactions to harmless inhaled antigen. J. Exp. Med. 20089-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Del Rio, M.-L., J.-I. Rodriguez-Barbosa, E. Kremmer, and R. Forster. 2007. CD103− and CD103+ bronchial lymph node dendritic cells are specialized in presenting and cross-presenting innocuous antigen to CD4+ and CD8+ T cells. J. Immunol. 1786861-6866. [DOI] [PubMed] [Google Scholar]
- 15.Grayson, M. H. 2006. Lung dendritic cells and the inflammatory response. Ann. Allergy Asthma Immunol. 96643-651. [DOI] [PubMed] [Google Scholar]
- 16.Guermonprez, P., J. Valladeau, L. Zitvogel, C. Thery, and S. Amigorena. 2002. Antigen presentation and T cell stimulation by dendritic cells. Annu. Rev. Immunol. 20621-667. [DOI] [PubMed] [Google Scholar]
- 17.Hamilton-Easton, A., and M. Eichelberger. 1995. Virus-specific antigen presentation by different subsets of cells from lung and mediastinal lymph node tissues of influenza virus-infected mice. J. Virol. 696359-6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henri, S., J. Curtis, H. Hochrein, D. Vremec, K. Shortman, and E. Handman. 2002. Hierarchy of susceptibility of dendritic cell subsets to infection by Leishmania major: inverse relationship to interleukin-12 production. Infect. Immun. 703874-3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hochrein, H., K. Shortman, D. Vremec, B. Scott, P. Hertzog, and M. O'Keeffe. 2001. Differential production of IL-12, IFN-alpha, and IFN-gamma by mouse dendritic cell subsets. J. Immunol. 1665448-5455. [DOI] [PubMed] [Google Scholar]
- 20.Holt, P. G., S. Haining, D. J. Nelson, and J. D. Sedgwick. 1994. Origin and steady-state turnover of class II MHC-bearing dendritic cells in the epithelium of the conducting airways. J. Immunol. 153256-261. [PubMed] [Google Scholar]
- 21.Holt, P. G., M. A. Schon-Hegrad, M. J. Phillips, and P. G. McMenamin. 1989. Ia-positive dendritic cells form a tightly meshed network within the human airway epithelium. Clin. Exp. Allergy 19597-601. [DOI] [PubMed] [Google Scholar]
- 22.Lambrecht, B. N., J. B. Prins, and H. C. Hoogsteden. 2001. Lung dendritic cells and host immunity to infection. Eur. Respir. J. 18692-704. [PubMed] [Google Scholar]
- 23.Legge, K. L., and T. J. Braciale. 2003. Accelerated migration of respiratory dendritic cells to the regional lymph nodes is limited to the early phase of pulmonary infection. Immunity 18265-277. [DOI] [PubMed] [Google Scholar]
- 24.Legge, K. L., and T. J. Braciale. 2005. Lymph node dendritic cells control CD8+ T cell responses through regulated FasL expression. Immunity 23649-659. [DOI] [PubMed] [Google Scholar]
- 25.Liu, F., Y. K. Song, and D. Liu. 1999. Hydrodynamics-based transfection in animals by systemic administrataion of plasmid DNA. Gene Ther. 61258-1266. [DOI] [PubMed] [Google Scholar]
- 26.Macatonia, S. E., P. M. Taylor, S. C. Knight, and B. A. Askonas. 1989. Primary stimulation by dendritic cells induces antiviral proliferative and cytotoxic T cell responses in vitro. J. Exp. Med. 1691255-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masten, B. J., G. K. Olson, D. F. Kusewitt, and M. F. Lipscomb. 2004. Flt3 ligand preferentially increases the number of functionally active myeloid dendritic cells in the lungs of mice. J. Immunol. 1724077-4083. [DOI] [PubMed] [Google Scholar]
- 28.Nonacs, R., C. Humborg, J. P. Tam, and R. M. Steinman. 1992. Mechanisms of mouse spleen dendritic cell function in the generation of influenza-specific, cytolytic T lymphocytes. J. Exp. Med. 176519-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oh, S., J. M. McCaffery, and M. C. Eichelberger. 2000. Dose-dependent changes in influenza virus-infected dendritic cells result in increased allogeneic T-cell proliferation at low, but not high, doses of virus. J. Virol. 745460-5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peebles, R. S., Jr., and B. S. Graham. 2001. Viruses, dendritic cells and the lung. Respir. Res. 2245-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piqueras, B., J. Connolly, H. Freitas, A. K. Palucka, and J. Banchereau. 2006. Upon viral exposure, myeloid and plasmacytoid dendritic cells produce 3 waves of distinct chemokines to recruit immune effectors. Blood 1072613-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schon-Hegrad, M. A., O. J. McMenamin, and P. G. Holt. 1991. Abstract studies on the density, distribution, and surface phenotype of intraepithelial class II major histocompatibility complex antigen (Ia)-bearing dendritic cells (DC) in the conducting airways. J. Exp. Med. 1731345-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sertl, K., T. Takemura, E. Tschachler, V. J. Ferrans, M. A. Kaliner, and E. M. Shevach. 1986. Dendritic cells with antigen-presenting capability reside in airway epithelium, lung parenchyma, and visceral pleura. J. Exp. Med. 163436-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shortman, K., and Y.-J. Liu. 2002. Mouse and human dendritic cell subtypes. Nat. Rev. Immunol. 2151-161. [DOI] [PubMed] [Google Scholar]
- 35.Stevens, W. W., T. S. Kim, L. M. Pujanauski, X. Hao, and T. J. Braciale. 2007. Detection and quantitation of eosinophils in the murine respiratory tract by flow cytometry. J. Immunol. Methods 32763-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sung, S. S., S. M. Fu, C. E. Rose, Jr., F. Gaskin, S. T. Ju, and S. R. Beaty. 2006. A major lung CD103 αE-β7 integrin-positive epithelial dendritic cell population expressing Langerin and tight junction proteins. J. Immunol. 1762161-2172. [DOI] [PubMed] [Google Scholar]
- 37.Swanson, K. A., Y. Zheng, K. M. Heidler, Z. D. Zhang, T. J. Webb, and D. S. Wilkes. 2004. Flt3-ligand, IL-4, GM-CSF, and adherence-mediated isolation of murine lung dendritic cells: assessment of isolation technique on phenotype and function. J. Immunol. 1734875-4881. [DOI] [PubMed] [Google Scholar]
- 38.Vermaelen, K. Y., I. Carro-Muino, B. N. Lambrecht, and R. A. Pauwels. 2000. Specific migratory dendritic cells rapidly transport antigen from the airways to the thoracic lymph nodes. J. Exp. Med. 19351-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Von Garnier, C., L. Filgueira, M. Wikstrom, M. Smith, J. A. Thomas, D. H. Strickland, P. G. Holt, and P. A. Stumbles. 2005. Anatomical location determines the distribution and function of dendritic cells and other APCs in the respiratory tract. J. Immunol. 1751609-1618. [DOI] [PubMed] [Google Scholar]
- 40.Yrlid, U., and M. J. Wick. 2002. Antigen presentation capacity and cytokine production by murine splenic dendritic cell subsets upon salmonella encounter. J. Immunol. 169108-116. [DOI] [PubMed] [Google Scholar]