Abstract
Flavivirus nonstructural (NS) proteins are involved in RNA replication and modulation of the host antiviral response; however, evidence is mounting that some NS proteins also have essential roles in virus assembly. Kunjin virus (KUN) NS2A is a small, hydrophobic, transmembrane protein that is part of the replication complex and inhibits interferon induction. Previously, we have shown that an isoleucine (I)-to-asparagine (N) substitution at position 59 of the NS2A protein blocked the production of secreted virus particles in cells electroporated with viral RNA carrying this mutation. We now show that prolonged incubation of mutant KUN NS2A-I59N replicon RNA, in an inducible BHK-derived packaging cell line (expressing KUN structural proteins C, prM, and E), generated escape mutants that rescued the secretion of infectious virus-like particles. Sequencing identified three groups of revertants that included (i) reversions to wild-type, hydrophobic Ile, (ii) pseudorevertants to more hydrophobic residues (Ser, Thr, and Tyr) at codon 59, and (iii) pseudorevertants retaining Asn at NS2A codon 59 but containing a compensatory mutation (Thr-to-Pro) at NS2A codon 149. Engineering hydrophobic residues at NS2A position 59 or the compensatory T149P mutation into NS2A-I59N replicon RNA restored the assembly of secreted virus-like particles in packaging cells. T149P mutation also rescued virus production when introduced into the full-length KUN RNA containing an NS2A-I59N mutation. Immunofluorescence and electron microscopy analyses of NS2A-I59N replicon-expressing cells showed a distinct lack of virus-induced membranes normally present in cells expressing wild-type replicon RNA. The compensatory mutation NS2A-T149P restored the induction of membrane structures to a level similar to those observed during wild-type replication. The results further confirm the role of NS2A in virus assembly, demonstrate the importance of hydrophobic residues at codon 59 in this process, implicate the involvement of NS2A in the biogenesis of virus-induced membranes, and suggest a vital role for the virus-induced membranes in virus assembly.
West Nile virus (WNV), dengue virus, yellow fever virus (YFV), and Japanese encephalitis virus are members of the arthropod-borne flaviviruses known to cause serious disease in humans (9). The 1999 New York strain of WNV (NY99) has been shown to cause fever and flu-like symptoms, extending to meningitis, encephalitis, and polio-like myelitis (26). Since its introduction into the United States, approximately 27,000 cases (with over 1,000 fatalities) have been reported for the NY99 strain of WNV (CDC November 2007; www.cdc.gov/ncidod/dvbid/westnile/surv&control.htm). Kunjin virus (KUN) is a nonpathogenic subtype of WNV endemic to Australia (7). The KUN genome is a positive-strand RNA molecule of 11,022 bp (11). KUN RNA is translated into one long open reading frame encoding a polyprotein that is cleaved posttranslationally into three structural (C, prM, and E) and seven nonstructural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (4). The structural proteins constitute the virus particle, while the NS proteins are primarily involved in RNA replication and modulation of the host response (12, 13, 18). While much work has focused on the involvement of the structural proteins in flavivirus assembly, evidence suggests that NS proteins are also involved (16, 17, 28).
Flavivirus NS2A is a small (231 amino acids), hydrophobic, multifunctional, membrane-associated protein involved in RNA replication (2, 21). NS2A binds with high specificity to the 3′ untranslated region (UTR) of viral RNA and to other components of the replication complex (21). In addition, NS2A has roles in modulating the host-antiviral interferon response (18, 19, 20, 25) and assembly/secretion of virus particles (16, 17). A KUN full-length infectious clone (pAKUN) containing an amino acid mutation at position 59 in NS2A (from isoleucine to asparagine, I59N) proved extremely inefficient at forming plaques in BHK cells compared to the plaque-forming ability of wild-type (WT) virus (11, 17). In other studies with YFV, a Lys-to-Ser mutation at position 190 in NS2A blocked the production of virus particles, although the secretion of empty prM-E particles remained unimpaired (16). Radiolabeling of the structural proteins revealed proper processing of C-prM-E precursor, suggesting that cleavage events were not responsible for the packaging defect. Compensatory mutations (Asp to Val, Ala, or Gly) were found in the N-terminal helicase domain at NS3 codon 343; the mutations restored the secretion of virus particles, thus implicating a role for NS3 in virus assembly and a possible interaction between NS2A and NS3 during this process (16). These findings for YFV and our previous results with KUN illustrate that WT NS2A protein is required for the production of flavivirus particles.
In this study, we analyze (pseudo)revertants generated from prolonged incubation of KUN replicon RNA containing an NS2A-I59N mutation in packaging cells producing KUN structural proteins. Through reverse transcription-PCR (RT-PCR), sequencing, and site-directed mutagenesis of replicon RNAs, we have examined the effect of these (pseudo)reversions on RNA replication, assembly, and secretion of virus-like particles (VLPs) and established the requirement for hydrophobic residues at codon 59 in this process. We have also isolated a compensatory Thr-to-Pro mutation at NS2A codon 149; the mutation rescues an assembly/secretion defect caused by the NS2A-I59N mutation. Immunofluorescence and electron microscopy analysis showed a role for hydrophobic residues at codon 59 in the formation of specific virus-induced membranes and indicated that these membranes may contribute to virus assembly.
MATERIALS AND METHODS
Cell culture.
tetKUNCprME-packaging BHK21 (8), BHK21, and VERO cells were maintained in Dulbecco's modified minimal essential medium (DMEM; Invitrogen) supplemented with 5% fetal bovine serum (FBS; Invitrogen) at 37°C in a 5% CO2 incubator. tetKUNCprME-packaging BHK21 cells were additionally supplemented with G418 (100 mg/ml) and puromycin (3 mg/ml) to maintain, and with doxycycline (5 mg/ml) to suppress, the expression of KUN structural proteins.
Plasmid construction. (i) repPACβgal constructs.
repPACβgal mutant constructs were generated by QuikChange PCR using primer pairs (Table 1) and an intermediate plasmid template containing a 3.8-kb HindIII fragment encoding partial NS1, NS2A, and NS2B proteins and also containing almost the entire NS3 gene (27). PCR was performed with Pfu DNA polymerase (Promega), followed by DpnI (New England Biolabs) digestion and transformation into Escherichia coli DH5α cells. Mutated AvrII fragments were then cloned back into the replicon repPACβgal and sequence verified. AvrII fragments encoding NS2A-I59N and the double-mutation NS2A-I59N-T149P were subsequently cloned into repZeo and the full-length infectious clone FLSDXpro_HDVr (17).
TABLE 1.
Oligonucleotides used for construction of plasmids, RT-PCR, and sequencinga
| Primer name | Sequence (5′ to 3′)a |
|---|---|
| JNS1SacII-F | GGGCACCGCGGACCTGCCACCCGCACCb |
| J2A-R | CATGTCGGTCGATTTTCCAG |
| NS2A-F | TACAACGCTGACATGATTG |
| NS2A59N-F | CTATGTCAATCTGGTGGGGGCGGCCTTTG |
| NS2A59N-R | CACCAGATTGACATAGCGTAACACATCAG |
| NS2A59R-F | CTATGTCAGGCTGGTGGGGGCGGCCTTTG |
| NS2A59R-R | CACCAGCCTGACATAGCGTAACACATCAG |
| NS2A59S-F | CTATGTCAGTCTGGTGGGGGCGGCCTTTG |
| NS2A59S-R | CACCAGACTGACATAGCGTAACACATCAG |
| NS2A59T-F | CTATGTCACTCTGGTGGGGGCGGCCTTTG |
| NS2A59T-R | CACCAGAGTGACATAGCGTAACACATCAG |
| NS2A59V-F | CTATGTCGTTCTGGTGGGGGCGGCCTTTG |
| NS2A59V-R | CACCAGAACGACATAGCGTAACACATCAG |
| NS2A59Y-F | CTATGTCTATCTGGTGGGGGCGGCCTTTG |
| NS2A59Y-R | CACCAGATAGACATAGCGTAACACATCAG |
| NS2A149P-F | ACGTTCCCCACAACATCTAATGTGGTCGTC |
| NS2A149P-R | TGTTGTGGGGAACGTTATCGCTCTCAATA |
| NS2A198S-F | CAAAAAAGTCAGGAGCCAGTCTGCTATGTT |
| NS2A198S-R | CTCCTGACTTTTTTGCAGCTGCACTTC |
| PstIZeoframe-F | CAACCTGCAGAAGCCAAGTTGACCAGTGCCGc |
| NsiIZeo-R | GCGCATGCATCGTCCTGCTCCTCGGCCACd |
Introduced nucleotide mutations are shown in boldface, and inserted nucleotides are shown in boldface italics.
The SacII site is underlined.
The PstI site is underlined.
The NsiI site is underlined.
(ii) repZeo constructs.
repZeo constructs were generated by cloning the Zeocin resistance sequence into the NsiI site of SP6KUNrep2 (modified from pKUNrep2, with the cytomegalovirus promoter replaced with an SP6 promoter) (30). The zeocin fragment was PCR amplified by using high-fidelity Pfu polymerase (Promega) from the plasmid pIZ/V5-His (Invitrogen) using a forward primer incorporating a PstI restriction site and a reverse primer incorporating an NsiI restriction site (Table 1). Mutant repZeo constructs were generated via QuikChange PCR as described above.
In vitro transcription and electroporation.
All replicon and full-length infectious clone cDNA templates were linearized with XhoI (New England Biolabs) and purified using phenol-chloroform extraction and ethanol precipitation. In vitro RNAs were transcribed using 1 μg linearized DNA template, 10× SP6 transcription buffer (Roche), 1 mM rNTP/CAP mix (Promega; New England Biolabs), 40 U SP6 RNA polymerase (Roche), and 20 U RNasin (Promega) in a 20-μl reaction mixture at 37°C for 2 h. Template DNA was removed by the addition of 1 U of DNase and incubation at 37°C for 15 min. Cells were washed three times in ice-cold diethyl pyrocarbonate-phosphate buffered saline (PBS) and prepared at a final concentration of 5 × 106/ml. Four hundred microliters of cells was mixed with ∼20 μg RNA in a 0.2-cm cuvette (Bio-Rad) and pulsed twice with a 10-s interval by using a Bio-Rad Gene Pulser II (∞ Ω, 25 μF, 1.5 KV). Cells were left on ice for 5 min postelectroporation, resuspended in cell culture medium, seeded, and incubated for various periods for examination of RNA replication and VLP production. VLP titers in culture fluids from electroporated cells were determined by infecting new cells and staining the cells in situ with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) to detect β-galactosidase (β-Gal) expression or by immunofluorescent analysis with either anti-NS1 or anti-NS3 antibodies.
Isolation of (pseudo)revertants generated from tetKUNCprME-packaging BHK21 cells electroporated with repPACβgal NS2A-I59N mutant replicon RNA.
Replicon RNA was transcribed and electroporated into tetKUNCprME-packaging BHK21 cells, and cells were seeded in either a 60-mm2 dish (500 μl) or a 96-well plate (200 μl per well). After prolonged incubation for 9 days, 60 mm2 dishes were fixed with 4% formaldehyde and stained in situ with X-Gal. Culture fluids from individual wells of the 96-well plate were collected at 9 days after transfection, clarified by low-speed (700 × g) centrifugation and used to infect new tetKUNCprME-packaging cells in 24-well plates. Total cellular RNA from infected cells was then isolated at 2 days postinfection by using the TRIzol reagent (Invitrogen) according to the manufacturer's recommendations. SuperScript III one-step RT-PCR with Platinum Taq DNA polymerase (Invitrogen) was used (as recommended by the manufacturer) to amplify the NS2A gene using the oligonucleotides JNS1SacII-F and J2A-R, followed by sequencing with the oligonucleotide NS2A-F (Table 1).
Northern blot hybridization.
Total cellular RNA was purified from RNA-electroporated cells by using TRIzol reagent according to manufacturer's recommendations. Six micrograms of total RNA was run on a 1.5% agarose-2% formaldehyde denaturing gel and transferred to a HybondN membrane (Amersham) overnight. Membrane-bound RNA was cross-linked by UV exposure for 5 min and hybridized in ExpressHyb solution (Clontech) with 32P-labeled probes (Megaprime DNA labeling kit; Amersham) specific for the KUN 3′ UTR and β-actin sequences.
Immunofluorescence and X-Gal staining. (i) Immunofluorescence analyses.
Immunofluorescence analyses were performed on cells grown on glass coverslips and fixed with 4% paraformaldehyde for 10 min. VLP titrations were performed using the mouse monoclonal KUN anti-NS1 antibody 4G4 (supplied by Roy Hall, University of Queensland), while localization studies utilized the rabbit KUN anti-NS3 antibody. The secondary antibodies AF488 anti-mouse, AF488 anti-rabbit, and AF594 anti-mouse were purchased from Molecular Probes/Invitrogen.
(ii) repPACβgal analysis.
An analysis of the replication and spread of repPACβgal mutant replicon RNA was performed on 4% formaldehyde-fixed cells by staining with X-Gal at 37°C overnight.
Virus infection, plaque assay, and VLP titration.
BHK21 and VERO cells were seeded in six-well plates and infected with KUN strain FLSDX and mutant NS2A-I59N-T149P virus at a multiplicity of infection of 0.1 for 2 h. Cells were washed three times with cell culture medium before being replaced with DMEM supplemented with 2% FBS. Fifty-microliter aliquots of culture fluids were harvested at 0, 12, 24, 36, 48, 72, 96, and 120 h postadsorption and stored at −80°C. Plaque assays were performed in six-well plates by infecting 80% confluent monolayers of BHK21 cells with dilutions of virus-containing culture fluids for 2 h. Cells were overlaid with 2 ml of medium containing 70% DMEM, 2.5% FBS, 15 mM sodium hydrogen carbonate, 5 U penicillin-streptomycin, 25 mM HEPES, 2 mM GlutaMAX, and 0.35% low-melting-point agarose and incubated for 4 days at 37°C with 5% CO2. Cells were fixed by adding 1 ml of 20% formaldehyde directly onto the overlay and incubating for 30 min at room temperature. Cells were rinsed with water and stained with 0.2% crystal violet for 20 min, and plaques were counted. Twenty-four-well plates seeded with VERO cell monolayers (∼80% confluent) were infected with 200-μl dilutions of VLP-containing culture fluids for 2 h and allowed to incubate in 300 μl cell culture medium at 37°C with 5% CO2. At 48 h postinfection, cells were fixed and stained in situ with X-Gal (repPACβgal constructs) or anti-NS1 antibodies (repZeo constructs) and the numbers of positive cells were counted.
Sucrose density gradient separation and ELISA.
Expression of KUN structural proteins was induced by the removal of doxycycline from tetKUNCprMErepZeo-packaging BHK21 cells. Culture fluids were collected at 24-h intervals for 5 days, and VLP titers were determined. Samples from 72 h were concentrated by using an Amicon Ultra-15 centrifugal filter unit with an Ultracel-100 membrane before being loaded onto a 10 to 40% continuous sucrose gradient (prepared in DMEM) and subjected to centrifugation at 38,000 rpm for 2 h using a Beckman SW41 rotor. Twenty-four fractions were collected from the top of the gradient and analyzed for the presence of prM-E particles (by E capture enzyme-linked immunosorbent assay [ELISA]) and infectious VLPs (by infectivity assay). E capture ELISA was performed as described previously (10). Briefly, mouse monoclonal ascites anti-E antibody 3.91D (provided by Roy Hall) was coated onto the bottom of flat-bottom 96-well plates in 100 μl/well coating buffer (25 mM NaHCO3, 25 mM Na2CO3, pH 9.6) at 1 μg/ml. Plates were washed with PBS-T (PBS, 0.05% Tween 20), and nonspecific binding sites were blocked by the addition of 300 μl/well TENTC buffer (50 mM Tris, 1 mM EDTA, 150 mM NaCl, 0.2% casein, 0.05% Tween 20, pH 8.0) for 1 h at 37°C. Sucrose gradient fractions (100 μl/well) were added and incubated at room temperature for 1 h. Plates were washed five times with PBS-T and incubated for 1 h at room temperature with 100 μl/well of 1/5,000 dilution of horseradish peroxidase (HRP)-conjugated anti-E monoclonal antibody 6B6C-1-HRP (provided by John Roehrig, Centers for Disease Control and Prevention, Fort Collins, CO). Plates were washed 10 times with PBS-T, followed by the addition of 100 μl/well ABTS [2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid)] substrate (50 mM Na2HPO4, 25 mM citric acid, 0.0075% H2O2, 1 mM ABTS). Plates were incubated in the dark for 15 min before absorbance was read at 405 nm by using an Anthos Lucy 2 plate reader (Anthos Labtec Instruments).
Resin-embedded electron microscopy.
Transfected cells were fixed with 3% glutaraldehyde in PBS for 2 h. The cell pellets were postfixed in 1% osmium tetroxide (in 0.1 M cacodylate buffer) and incubated overnight at 4°C in 2% uranyl acetate-80% acetone. Pellets were then dehydrated with an acetone series and embedded in Epon. Ultrathin sections (∼60 nm) were cut with a Diatome diamond knife and stained and contrasted with uranyl acetate and lead citrate before being viewed with a JOEL 1010 electron microscope at 80 kV. Images were captured on a MegaView III side-mounted charge-coupled device camera (Soft Imaging Systems) and processed in Adobe Photoshop.
RESULTS
Hydrophobic residues at codon 59 are essential for NS2A function in assembly and secretion of virus-like particles.
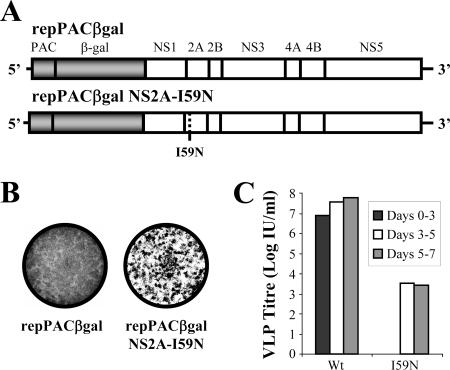
Our previous studies identified an Ile-to-Asn mutation in NS2A which blocked the production of secreted virus from transfected full-length RNA and that of secreted virus-like particles from replicon RNA cotransfected with Semliki Forest virus replicon vector expressing KUN structural genes (17). However, in our earlier studies, full-length pAKUN RNA containing the NS2A-I59N mutation did eventually produce secreted infectious virus in transfected cells after a prolonged incubation (11). Viral RNA from the recovered virus contained a reversion to the WT Ile at position 59 of NS2A (17). In order to further analyze the role of residue 59 in NS2A in the assembly and/or secretion of VLPs and to identify additional mutations that were able to rescue the defect caused by the I59N mutation, replicon RNA containing the NS2A-I59N mutation (repPACβgal NS2A-I59N [Fig. 1A]) was electroporated into tetKUNCprME-packaging BHK21 cells and cells were incubated for up to 7 days. In situ X-Gal staining after 7 days of incubation showed distinct foci of β-Gal-expressing cells (Fig. 1B), suggesting a rescue of the defect in the assembly/secretion of VLPs caused by the NS2A-I59N mutation. This rescue was subsequently confirmed by positive in situ staining of VERO cells infected with culture fluids from repPACβgal NS2A-I59N RNA-transfected packaging cells harvested 3 to 7 days posttransfection, but not from samples harvested earlier (0 to 3 days) (Fig. 1C). However, the titers of VLPs secreted from mutant replicon RNA-transfected cells were approximately 10,000-fold lower than corresponding VLP titers secreted from WT replicon RNA-transfected cells (Fig. 1C).
FIG. 1.
Production of virus-like particles in tetKUNCprME-packaging BHK21 cells transfected with repPACβgal replicon RNA containing NS2A-I59N mutation after extended incubation. (A) Schematic representation of puromycin (PAC)-selectable and β-Gal-expressing KUN replicon constructs used for electroporation into tetKUNCprME-packaging BHK-21 cells. Heterologous gene inserts are gray, and the KUN nonstructural genes (as indicated) are colored white. NS2A-I59N mutation is shown by a broken line. (B) Detection of β-Gal expression in tetKUNCprME-packaging BHK-21 cells electroporated with KUN replicon RNAs (A) at 7 days posttransfection by using X-Gal staining after fixation with 4% formaldehyde. (C) Infectious VLP titers in culture fluids of tetKUNCprME-packaging BHK-21 cells electroporated with WT and NS2A-I59N mutant RNAs 3, 5, and 7 days postelectroporation. The titers were determined by infectivity assay on fresh VERO cells as described in Materials and Methods.
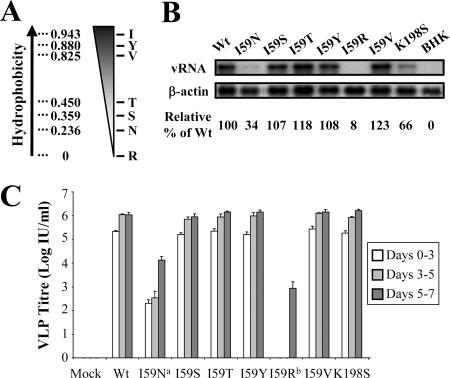
To identify the mutations in replicon RNA that allowed the assembly and secretion of VLPs in packaging cells, repPACβgal NS2A-I59N RNA-transfected cells were seeded into the wells of a 96-well plate and allowed to incubate for 9 days. Recovered, secreted VLPs from each well were used to infect fresh nonpackaging cells in a 96-well plate, and replicon RNA in infected cells was then analyzed by RT-PCR and sequencing analysis of the entire genome. Three types of mutations were identified (Table 2), including a reversion to WT Ile (groups 1 to 3), pseudoreversions to hydrophobic residues Ser, Thr, and Tyr at codon 59 (groups 4 to 8) (Fig. 2A), and a compensatory mutation, Thr-to-Pro at NS2A position 149 (group 9). To confirm the role of hydrophobic residues at NS2A codon 59, pseudoreversions were introduced into the plasmid repPACβgal. A number of additional mutations were also introduced for the following reasons: (i) NS2A-I59R was designed to determine whether the assembly and secretion of VLPs would be impaired by a hydrophilic residue at position 59; (ii) NS2A-I59V was introduced to assess the possible effect of Val residue at this position, found to be conserved in other flaviviruses (e.g., Japanese encephalitis, yellow fever, tick-borne encephalitis, and St. Louis encephalitis); and (iii) NS2A-K198S was used to examine the potential role in KUN virus assembly/secretion of this residue which is likely to correspond to the residue K190 in YFV NS2A, which has previously been shown to block YFV assembly/secretion (16). Mutant replicon RNAs were transfected into normal BHK21 cells and packaging BHK21 cells for an analysis of RNA replication and production of infectious particles, respectively. Northern blot hybridization of total RNA purified from transfected BHK21 cell lysates showed comparable levels of RNA replication between WT, I59S, I59T, I59Y, and I59V RNAs (100%, 107%, 118%, 108%, and 123%, respectively [Fig. 2B]), while the replication efficiencies of I59N and K198S mutant RNAs were less efficient (34% and 66%, respectively [Fig. 2B]). I59R RNA appeared to be severely impaired in replication (8% [Fig. 2B]). VLP production efficiencies were similar between WT, I59S, I59T, I59V, and I59Y RNAs (Fig. 2C), verifying the essential role of a hydrophobic amino acid residue at codon 59 of KUN NS2A in the assembly/secretion of virus particles. RT-PCR and sequencing analysis of replicon RNAs isolated from VLPs recovered in day-7 culture fluid confirmed the retention of introduced mutations. Delayed recovery of low amounts of VLPs from I59N- and I59R-transfected packaging cells (Fig. 2C) indicates the appearance of reversions able to rescue assembly late in replication/packaging. Indeed, RT-PCR and sequencing analysis of replicon RNAs isolated from VLPs in day-7 culture fluid identified reversions to Ile or Tyr in the I59N mutant and to Ile or Val in the I59R mutant (data not shown).
TABLE 2.
Amino acid substitutions in NS2A found in replicon RNAs from rescued secreted VLPs
| Group | No. of clones | Amino acid replacing indicated amino acid in indicated codon
|
|||
|---|---|---|---|---|---|
| Ala in NS2A-30 | Ser in NS2A-33 | Asn in NS2A-59 | Thr in NS2A-149 | ||
| 1 | 32 | -a | - | Ile | - |
| 2 | 1 | Pro | - | Ile | - |
| 3 | 1 | - | Thr | Ile | - |
| 4 | 3 | - | - | Ser | - |
| 5 | 3 | - | - | Thr | - |
| 6 | 15 | - | - | Tyr | - |
| 7 | 1 | - | Thr | Tyr | - |
| 8 | 2 | - | - | - | Pro |
Represents the same residues as in input RNA.
FIG. 2.
Replication and packaging of NS2A 59 pseudorevertants. (A) Hydrophobicity plot of amino acid residues found in pseudorevertants and cloned into NS2A codon 59 of repPACβgal cDNA. (B) Northern blot analysis of total cellular RNA from BHK-21 cells 2 days postelectroporation with various mutant KUN replicon RNAs. The probes were 32P-labeled cDNA fragments specific for the KUN 3′ UTR and for β-actin. Efficiencies of replication are expressed as percentages of accumulated viral RNAs relative to WT and normalized to β-actin (shown under the blots). (C) Infectious titers of VLPs in culture fluids of tetKUNCprME-packaging BHK-21 cells electroporated with mutant repPACβgal RNAs and harvested at 3, 5, and 7 days postelectroporation. The titers were determined by infectivity assay on VERO cells as described in Materials and Methods. Error bars indicate standard deviations. a, RT-PCR and sequencing on culture fluids revealed (pseudo)reversions to Ile and Tyr at NS2A codon 59; b, RT-PCR and sequencing on culture fluids revealed (pseudo)reversions to Ile and Val at NS2A codon 59.
These results suggest that hydrophobic residues are required at position 59 in the assembly/secretion of virus particles and that the packaging defect observed in cells transfected with I59N and I59R mutant RNA is rescued by (pseudo)reversions to hydrophobic residues at position 59 after prolonged replication. Interestingly, the K198S mutation did not have any noticeable effect on the efficiency of VLP production/secretion (Fig. 2C), demonstrating substantial differences between KUN and YFV in the location of amino acids/regions in corresponding NS2A proteins involved in virus assembly/secretion.
Compensatory mutation at NS2A codon 149 rescues the defect in virus assembly/secretion caused by the NS2A-I59N mutation.
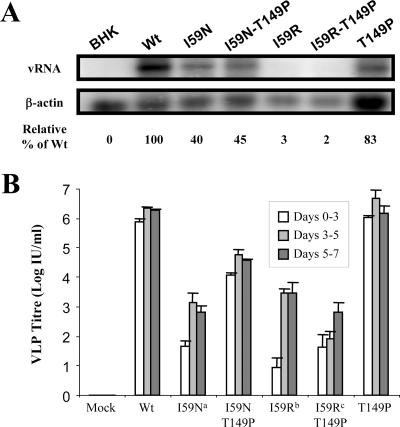
In addition to (pseudo)revertants with hydrophobic residues at NS2A codon 59, pseudorevertants retaining NS2A-I59N but containing a compensatory mutation at NS2A codon 149 (Thr-to-Pro) were also isolated during prolonged replication of replicon RNA in packaging cells. When these two mutations were introduced simultaneously into replicon RNA and Northern blot hybridization was performed, NS2A-I59N-T149P mutant RNA exhibited a decreased replication efficiency compared to that of WT RNA (45% [Fig. 3A]). However, this replication efficiency was similar to the reduced replication efficiency of NS2A-I59N mutant RNA (40% [Fig. 3A]). An examination of packaging efficiency showed decreased VLP production for I59N-T149P compared to that for WT from days 0 to 3 posttransfection, most likely attributed to the decreased efficiency in RNA replication (Fig. 3B). RT-PCR and sequencing analyses of culture fluid at day 9 confirmed the retention of NS2A-I59N-T149P mutations (data not shown).
FIG. 3.
Ability of Thr-to-Pro mutation at NS2A codon 149 to rescue packaging defects of two different I59 mutants. (A) Replication efficiencies of mutant RNAs in transfected BHK-21 cells. Total cellular RNA was purified 48 h postelectroporation, and accumulated viral RNAs were detected by Northern blotting using 32P-labeled probes specific for the KUN 3′ UTR and β-actin. The efficiencies of RNA replication are expressed as percentages relative to the WT and normalized to β-actin. (B) Infectious titers of VLPs in culture fluids of tetKUNCprME-packaging BHK-21 cells electroporated with mutant repPACβgal RNAs and harvested at 3, 5, and 7 days postelectroporation. The titers were determined by infectivity assay on fresh VERO cells as described in Materials and Methods. Error bars indicate standard deviations. a, RT-PCR and sequencing on culture fluids revealed (pseudo)reversions to Tyr at NS2A codon 59; b, RT-PCR and sequencing on culture fluids revealed (pseudo)reversions to Ile NS2A codon 59; c, RT-PCR and sequencing on culture fluids revealed (pseudo)reversions to Trp NS2A codon 59.
To analyze whether the 149 Pro residue alone could affect virus assembly and secretion and rescue assembly and secretion of mutant replicon RNA with Arg at codon 59, repPACβgal mutants NS2A-T149P and NS2A-I59R-T149P were generated. Mutant RNAs were transfected into normal and packaging BHK21 cells for an analysis of RNA replication and production of infectious particles, respectively. Northern blot hybridization of total RNA purified from BHK21 cell lysates showed a negligible effect on viral RNA replication of (i) the individual NS2A-T149P mutation compared to that of the WT RNA (83% and 100%, respectively [Fig. 3A]) and (ii) NS2A-I59R-T149P mutations compared to that of the NS2A-I59R mutant RNA (2% and 3%, respectively [Fig. 3A]). Efficiencies of VLP production in packaging cells were similar between WT and NS2A-T149P RNAs (∼106 VLPs/ml), clearly demonstrating that this mutation alone does not have any effect on virus assembly/secretion. Interestingly, NS2A-T149P mutation did not appear to rescue the defect in VLP assembly/secretion caused by I59R mutation (Fig. 3B) early (0 to 3 days) in transfection, possibly due to extremely inefficient RNA replication. Some production of VLPs occurred later in transfection (3 to 7 days) for both I59R and I59R-T149P RNAs, suggesting the occurrence of revertants in I59 position; however, no difference in VLP titers between these two RNAs was detected (Fig. 3B).
To determine whether the I59N-T149P double mutation shown to restore assembly of VLPs was also able to rescue virus production from the full-length RNA, these mutations were introduced into the FLSDXpro_HDVr infectious cDNA clone (17). Indeed, the transfection of mutant RNA resulted in the recovery of infectious virus by 24 h of postelectroporation, with a titer of ∼107 IU/ml (data not shown). The retention of the NS2A-I59N-T149P mutation in the recovered virus was confirmed by RT-PCR and sequencing analyses of virus-infected cell lysates. Plaque assays of recovered virus in BHK21 and VERO cells showed that mutant NS2A-I59N-T149P virus produced smaller plaques than did the WT KUN virus (Fig. 4A). Virus growth kinetic studies in BHK21 and VERO cells revealed that mutant virus replicated with an efficiency ∼2- to 15-fold lower than that of WT virus (Fig. 4B and C), with the difference being most profound in VERO cells (Fig. 4C).
FIG. 4.
Characterization of KUN virus containing a Thr-to-Pro compensatory mutation at codon 149 that rescues the packaging defect caused by NS2A-I59N mutation. (A) Plaque morphology of WT and mutant viruses. BHK21 and VERO cells were infected with the viruses at a multiplicity of infection of 0.1 PFU/cell and stained with crystal violet at 4 and 5 days postinfection, respectively. (B and C) Growth kinetics of the WT and mutant viruses on BHK21 and VERO cells, respectively. Culture fluids from infected cells were collected every 12 h for 3 days, and viral titers were determined by plaque assay as described in Materials and Methods.
NS2A-I59N mutation in replicon RNA does not affect processing of structural proteins and secretion of prM-E particles in packaging cells.
Characterization of NS2A-I59N mutant RNA has proven difficult in systems involving replicons or full-length clones due to variable transfection efficiencies, impaired replication efficiency, and the rapid appearance of reversions/compensatory mutations. The generation of a cell line which would stably produce replicating replicon RNA and at the same time allow inducible expression of structural proteins could (i) eliminate the problems associated with the variability of RNA transfection and replication efficiencies and (ii) provide a system in which the induction of structural protein expression can be regulated. Therefore, we have generated such a cell line, tetKUNCprMErepZeo, by transfecting replicon RNAs (WT and NS2A-I59N) expressing the zeocin (Zeo) resistance gene into tetKUNCprME packaging cells and selecting stable cells in the presence of Zeo (Fig. 5A). RT-PCR and sequencing analyses confirmed the retention of the I59 or I59N mutation in corresponding established cell lines. Northern blot hybridization of total RNA purified from cell lysates showed that levels of RNA replication between WT and I59N mutant RNAs in these cell lines were similar (Fig. 5B). An infectious assay performed with culture fluids showed that when the expression of structural proteins was induced by the removal of doxycycline, high titers of secreted infectious VLPs were produced from cells carrying WT replicon RNA; this high-titer secretion started from day 2 postinduction, reaching up to ∼107 VLPs/ml by day 5 after induction (Fig. 5C). In contrast, the induction of structural protein expression in cells carrying NS2A-I59N mutant replicon RNA resulted in very inefficient production of secreted infectious VLPs, with titers not exceeding 103 VLPs/ml by day 5 (Fig. 5C). However, the detection of any VLPs at all suggests the appearance of reversions able to partially rescue VLP assembly/secretion. Indeed, RT-PCR and sequencing analysis of RNA isolated from recovered VLPs detected reversion to the WT Ile at position 59 (data not shown).
FIG. 5.
Secretion of prM-E particles is not affected by NS2A-I59N mutation in a cell line stably expressing both a structural gene cassette and replicon RNA. (A) Schematic representation of production of secreted prM-E particles and replicon RNA-containing VLPs in tetKUNCprMErepZeo cells. Stably expressing cells were generated as described in Materials and Methods. (B) Northern blot analysis of RNA replication in tetKUNCprME-packaging BHK-21 cells stably expressing WT and NS2A-I59N-mutated repZeo replicon RNAs using 32P-labeled probes specific for the KUN 3′ UTR and β-actin. CMV, cytomegalovirus; DOX, doxycycline; PAC, puromycin; SV40, simian virus 40; TRE, tetracycline-responsive element. (C) Production of VLPs containing repZeo RNAs in culture fluids harvested every 24 h for 5 days after induction of structural protein expression. VLP titers were determined by infectivity assay on VERO cells using immunofluorescence staining with anti-NS1 antibodies, as described in Materials and Methods. Error bars indicate standard deviations. (D) Sucrose gradient separation of prM-E particles and VLPs. Culture fluids were collected from tetKUNCprME-packaging BHK-21 cells stably expressing WT or NS2A-I59N-mutated repZeo RNAs 3 days postinduction of structural proteins, concentrated, and placed on a 10 to 40% continuous sucrose gradient. Fractions were taken from the top to the bottom of the gradient and analyzed by E capture ELISA. The detection of infectious VLPs in gradient fractions was performed by an infectivity assay of VERO cells using immunofluorescence staining with anti-NS1 antibodies.
While VLP production was impaired in packaging cells stably transfected with I59N replicon RNA early in replication prior to the appearance of reversions, it is not clear whether this mutation has an effect on the assembly or secretion of VLPs. To analyze the effect of NS2A-I59N mutation in replicon RNA on the secretion of prM-E particles in tetKUNCprMErepZeo cells, culture fluids at 72 h postinduction of structural protein expression were harvested, concentrated, and loaded onto a 10 to 40% continuous sucrose gradient for separation. Fractions were collected for the detection of prM-E particles (by E capture ELISA) and infectious VLPs (by infectivity titration assay). E capture ELISA analysis of gradient fractions showed a distinct peak of activity between fractions 6 and 14 for both WT and NS2A-I59N mutant samples, which correlates with the sedimentation density characteristic of empty prM-E particles (5, 13) (Fig. 5D). However, a peak in ELISA readings between fractions 21 and 23 corresponding to RNA-containing VLPs was detected only for the WT, not for I59N samples. VLP titration using immunofluorescence analysis of VLP-infected cells with anti-NS1 antibodies confirmed the presence of an RNA-expressing NS1 gene in WT VLP fractions 21 to 23 as well as in adjacent fractions 17 to 20 and 24, but not in corresponding fractions from NS2A-I59N samples (Fig. 5D). The efficient secretion of prM-E particles from packaging cells stably expressing mutant NS2A-I59N replicon RNA demonstrates that this mutation does not affect the secretion of virus particles and is therefore likely to be responsible for the block in their assembly. The results are in agreement with YFV studies in which a mutation in NS2A that blocked virus production did not have any effect on the secretion of prM-E particles (16).
NS2A-I59N mutation blocks induction of virus-specific membrane structures.
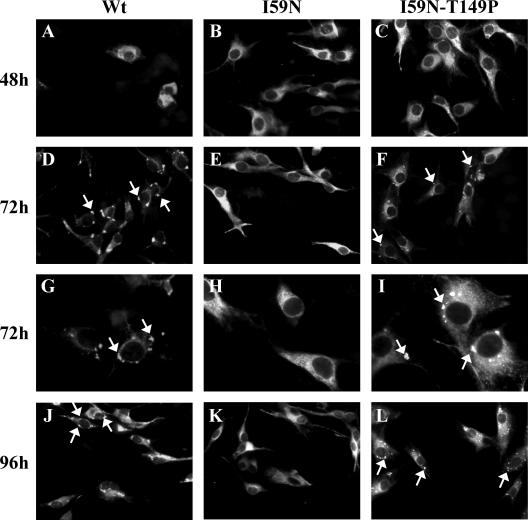
KUN virus assembly was previously suggested to occur in endoplasmic reticulum (ER) membranes continuous with characteristic virus-induced membrane structures, termed convoluted membranes or paracrystalline arrays, and vesicle packets (23). Convoluted membranes/paracrystalline arrays and vesicle packets have long been identified as the intracellular sites of flavivirus polyprotein translation/processing and replication, respectively (21, 31, 32). To determine whether the block in virus assembly exhibited by the NS2A-I59N mutation may be caused by alterations in the formation of virus-induced membranes, immunofluorescence analysis with anti-NS3 antibodies was performed. We have used this antibody extensively to identify the membrane sites of flavivirus replication (31). At 48 h postelectroporation, cells expressing all RNAs (mutants and WT) showed perinuclear and diffuse cytoplasmic staining (Fig. 6A to C). However, the labeling pattern changed dramatically in cells transfected with the WT replicon RNA at 72 h and 96 h postelectroporation, showing characteristic distinct foci within cytoplasmic-ER compartments (Fig. 6D, G, and J), as observed previously (22). In contrast, the staining of cells transfected with mutant NS2A-I59N RNA remained diffuse (Fig. 6E, H, and K), indicating the lack of profound membrane alterations characteristic of WT RNA replication. As NS2A-I59N mutant RNA replicated less efficiently than did WT RNA, it is possible that the changes in membrane induction could be due to the differences in RNA replication efficiency. To test for this possibility, membrane induction by compensatory mutant RNA NS2A-I59N-T149P, which exhibited a replication efficiency similar to that of the NS2A-I59N mutant RNA (Fig. 2B), was analyzed at 72 h and 96 h posttransfection. Staining with anti-NS3 antibodies showed labeling patterns similar to that seen in cells transfected with WT RNA (Fig. 6F, I, and L), thus confirming that the lack of foci seen in NS2A-I59N-transfected cells was not due to the less efficient RNA replication. To confirm the changes in membrane structures caused by the NS2A-I59N mutation observed by immunofluorescence analysis, electron microscopy analysis of resin-embedded sections prepared from cells at 72 h posttransfection was performed. Cells transfected with WT RNA showed a very distinct pattern of virus-induced membrane structures, while NS2A-I59N RNA-transfected cells did not, thus confirming immunofluorescence results (Fig. 7A, B, C, and D, respectively). These results provide direct demonstration of the effect of the NS2A-I59N mutation on changes in the formation of virus-induced membrane structures and therefore link these membranes with flavivirus assembly. However, the compensatory Thr-to-Pro mutation at NS2A position 149 was able to induce membrane structures that were visually similar to those induced by WT KUN replicon RNA (Fig. 7E and F). There was some variation observed with the NS2A-I59N-T149P mutant that may reflect the lower level of replication compared to WT replicon RNA. This additionally correlates with our previous observations comparing different replicating KUN replicon constructs (22).
FIG. 6.
Lack of characteristic virus-induced membranes in BHK21 cells transfected with mutant NS2A-I59N replicon RNA. (A) BHK-21 cells were electroporated with WT and mutant repPACβgal RNAs, fixed with 4% paraformaldehyde at 48 h, 72 h, and 96 h posttransfection, and stained with anti-NS3 antibodies, as described Materials and Methods. Virus-induced foci are highlighted by arrows. All images are taken at ×300 magnification except those for panels G to I, which were taken at ×680 magnification.
FIG. 7.
Ultrastructure of BHK21 cells containing NS2A-I59N mutant replicon RNA. Cells were resin embedded at 72 h postelectroporation as described in Materials and Methods. Arrows indicate vesicle packets. Abbreviations: CM, convoluted membranes; PC, paracrystalline arrays; Nuc, nucleus. Bars, 500 nm.
These results indicate that the T149P compensatory mutation rescues the assembly defect of the NS2A-I59N mutation and restores the ability to induce characteristic membrane structures, thus providing a convincing argument for the role of these membranes in virus assembly.
DISCUSSION
We have previously identified an amino acid substitution at codon 59 of NS2A (Ile-to-Asn) in the full-length infectious clone pAKUN which blocked production of virus particles (17). In this study, we used replicon RNA repPACβgal (17) and a trans-packaging system (packaging cells) (8) to further analyze the role of this mutation in the assembly and/or secretion of viral particles. We showed that after a prolonged incubation (more than 3 days) of repPACβgal NS2A-I59N mutant RNA in tetKUNCprME-packaging cells, the production and spread of VLPs was partially restored. RT-PCR and sequencing of RNA isolated from recovered VLPs identified reversions of Asn 59 to the WT Ile or to other hydrophobic residues, Ser, Thr, and Tyr, as well as a compensatory Thr-to-Pro mutation at position 149. The identification of revertants, pseudorevertants, and compensatory mutation in NS2A allowing packaging of mutant RNAs into secreted VLPs confirms the role of NS2A in the assembly and/or secretion of virus particles.
NS2A is a small, hydrophobic protein containing a number of predicted transmembrane domains, although the exact membrane topology of NS2A is yet to be determined. Taking into account the fact that Ile59 is located within a stretch of 10 hydrophobic residues (Fig. 8), it is possible that it may be embedded within the membrane and interacts with other transmembrane domains of NS2A or other membrane-associated viral proteins involved in virus assembly. The possibility that the I59N mutation may affect the properties of NS2A protein via altering its processing and/or stability can be ruled out, as we have not detected any differences in NS2A processing or stability between WT and I59N mutants using Western blot analysis of NS2A expressed from NS1-NS2A cassette (see Fig. S1 in the supplemental material). YFV studies with a virus assembly-deficient NS2A mutant identified compensatory mutations (Asp to Val, Ala, or Gly, noticeably all changes to hydrophobic residues) in NS3 at codon 343 which rescued a block in virus assembly caused by a Lys-to-Ser mutation at codon 190 (corresponding to codon 198 in KUN) in NS2A (16), thus suggesting NS2A-NS3 interactions during virus assembly. Our studies with I59N mutation in KUN, however, failed to identify any compensatory mutations in NS3 or anywhere else in the genome except in NS2A. Indeed, the Thr-to-Pro mutation at position 149 in KUN NS2A was able to compensate the virus assembly defect caused by I59N mutation. The identification of a compensatory T149P mutation in KUN NS2A suggests a potential interaction between codons 59 and 149, which may be required to maintain NS2A function in KUN virus assembly. The reason for the difference in location of the mutations blocking and rescuing virus assembly between our studies and YFV studies is not clear. To determine whether the region in YFV NS2A implicated in the production of virus particles is also essential to KUN assembly, a mutant was constructed in which the single amino acid substitution shown to block production of virus particles in YFV (K190S) was introduced into the amino acid position 198 in NS2A of KUN replicon; this position likely corresponds to amino acid 190 in YFV NS2A, as identified by a sequence alignment (K198S mutant [Fig. 8]). However, this mutation had no effect on the assembly and secretion of replicon RNA-containing KUN VLPs. This result and the results with compensatory mutations suggest that YFV and KUN may differ with regards to which specific regions/residues in NS2A may play a role in virus assembly.
FIG. 8.
Sequence alignment of the flavivirus NS2A protein. The same residues are indicated as dots within the alignment, while different residues are shown. Dashes represent deletions. The KUN amino acid residues 59, 149, and 198 and corresponding amino acids in other flaviviruses at this position are indicated by boxes.
Using the stable cell line teKUNCprMErepZeo independently expressing (i) structural proteins from integrated plasmid DNA and (ii) actively replicating mutant replicon RNA, we have shown that I59N mutation in NS2A had no effect on the processing of structural proteins and secretion of prM-E particles. The results agree with YFV studies which also showed no effect of the mutation in NS2A blocking virus assembly on processing of structural proteins and secretion of prM-E particles. In addition, our electron microscopy studies of the tetKUNCprMErepZeo cells after the induction of structural protein expression failed to detect any virus-like particles in the cell cytoplasm/ER (data not shown). Taken together, these findings suggest that NS2A is likely to be involved in the assembly of virus particles rather than in their secretion.
Our immunofluorescence analysis using KUN NS3-specific antibodies as markers of intracellular RNA replication sites showed that cells expressing NS2A-I59N mutant replicon RNA exhibited diffuse cytoplasmic staining, suggesting the absence of characteristic virus-induced membrane structures. Electron microscopy analysis of resin-embedded sections confirmed the immunofluorescence results. In contrast, cells expressing I59N-T149P-mutated NS2A showed distinct stained membrane foci by immunofluorescence and membrane structures similar to those detected in WT replicon-transfected cells by electron microscopy analysis. These results implicate the involvement of NS2A in the formation of virus-induced membranes. Our recent studies with KUN and the studies of others with dengue virus demonstrated a role for NS4A in the induction of virus-specific membranes (24, 29). However, the potential role of NS2A protein in the induction of virus-specific membranes was not examined in these studies. The results presented here indicate that NS2A may also be involved in the induction of virus-specific membranes and thus complement NS4A for this function.
We previously proposed that NS2A may facilitate virus assembly by transporting RNA or partially assembled nucleocapsid to subcellular compartments where assembly occurs (17). Binding assays involving purified KUN NS2A protein reveal strong interaction of NS2A with KUN RNA and other components of the replication complex (NS3 and NS5) (21). Based on these and our other results on the characterization of intracellular sites of KUN RNA replication, we hypothesized that NS2A transports the partially assembled replication complex to the sites of RNA replication via hydrophobic interactions with NS4A in the presence of RNA (14, 17). Together, the close proximity of sites of RNA replication to the sites of virus assembly (23), the association of NS2A with RNA (21), and the demonstrated coupling between RNA replication and virus assembly (15) strongly favor a model in which the NS2A is directly involved in transporting RNA from sites of RNA replication across virus-induced membranes to adjacent sites of virus assembly (17). Alternatively, the transportation of nascent RNA to the sites of virus assembly by NS2A may be facilitated indirectly, with NS2A acting as a viroporin. Viroporins are typically small (60 to 120 amino acids), hydrophobic viral proteins that are able to form an amphipathic α-helix (6). The insertion of these proteins into membranes, followed by their oligomerization, creates a hydrophilic pore, allowing low-molecular-weight molecules to cross the membrane. Additional hydrophobic regions, stretches of basic amino acids, and domains rich in aromatic amino acids interact with membranes, disturbing the organization of the lipid bilayer, further enhancing membrane permeability and causing membrane instability and rearrangement (6).
Interestingly, poliovirus small hydrophobic nonstructural protein 2B was shown to cause membrane permeability (1). Fluorescence resonance energy transfer and Western blot analysis under nonreducing conditions revealed that poliovirus 2B protein forms tetramers associated with membranes (common among membrane-disrupting proteins) (1). Intriguingly, polar amino acid substitutions within stretches of hydrophobic amino acids in poliovirus 2B protein greatly reduced membrane permeability (1). Significantly, studies involving the flavivirus Japanese encephalitis virus revealed that membrane permeability is caused by the small hydrophobic nonstructural proteins, including NS2A (3). As viroporins are often associated with permeabilization of the plasma membrane, it is plausible to speculate that flavivirus NS2A may act by forming pores within intracellular membranes, thus allowing the transport of newly synthesized RNA or nucleocapsid to the sites of virion assembly. Mutation of the hydrophobic Ile residue to the small, polar Asn residue may prevent membrane pore formation, leading to the discrepancy in membrane rearrangements observed between WT and mutant NS2A-I59N RNAs, thus preventing the transport of RNA or nucleocapsid across membranes for virion assembly. Within the membranes, hydrophobic interactions between codons 59 and 149 (Fig. 8) may be involved in forming proper tertiary protein structure/conformation. The compensatory mutation to Pro at position 149 may provide more flexibility to the amino acid backbone and restore interaction with mutated codon 59 within the membrane, resulting in the restoration of tertiary structure/conformation of NS2A protein and the reformation of membrane pores.
In summary, our results confirm the role for the flavivirus NS2A protein in virus assembly, demonstrate the requirement for hydrophobic residues at codon 59 in production of virus particles, and identify a compensatory mutation within NS2A at codon 149 that rescues this defect. Immunofluorescence and electron microscopy analyses implicate the involvement of NS2A in the formation of virus-induced membranes, and suggest a role for these membranes in virus assembly. Further studies are needed to elucidate the exact mechanisms by which the flavivirus nonstructural protein NS2A is able to participate in the assembly of virus particles.
Supplementary Material
Acknowledgments
We thank Roy Hall (University of Queensland) for providing mouse monoclonal anti-NS1 antibodies (4G4) and mouse monoclonal anti-E antibodies (3.91D) and John Roehrig (Centers for Disease Control and Prevention, Fort Collins, CO) for providing HRP-conjugated anti-E antibodies (6B6C-1-HRP).
This research was supported by the grants to A.A.K. from the National Health and Medical Research Council of Australia.
Footnotes
Published ahead of print on 12 March 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Agirre, A., A. Barco, L. Carrasco, and J. L. Nieva. 2002. Viroporin-mediated membrane permeabilization. Pore formation by nonstructural poliovirus 2B protein. J. Biol. Chem. 27740434-40441. [DOI] [PubMed] [Google Scholar]
- 2.Chambers, T. J., D. W. McCourt, and C. M. Rice. 1989. Yellow fever virus proteins NS2A, NS2B and NS4B: identification and partial N-terminal amino acid sequence analysis. Virology 169100-109. [DOI] [PubMed] [Google Scholar]
- 3.Chang, Y. S., C. L. Liao, C. H. Tsao, M. C. Chen, C. I. Liu, L. K. Chen, and Y. L. Lin. 1999. Membrane permeabilization by small hydrophobic nonstructural proteins of Japanese encephalitis virus. J. Virol. 736257-6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coia, G., M. D. Parker, G. Speight, M. E. Byrne, and E. G. Westaway. 1988. Nucleotide and complete amino acid sequences of Kunjin virus: definitive gene order and characteristics of the virus-specified proteins. J. Gen. Virol. 691-21. [DOI] [PubMed] [Google Scholar]
- 5.Gehrke, R., M. Ecker, S. W. Aberle, S. L. Allison, F. X. Heinz, and C. W. Mandl. 2003. Incorporation of tick-borne encephalitis virus replicons into virus-like particles by a packaging cell line. J. Virol. 778924-8933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gonzalez, M. E., and L. Carrasco. 2003. Viroporins. FEBS Lett. 55228-34. [DOI] [PubMed] [Google Scholar]
- 7.Hall, R. A., A. K. Broom, D. W. Smith, and J. S. Mackenzie. 2002. The ecology and epidemiology of Kunjin virus. Curr. Top. Microbiol. Immunol. 267253-269. [DOI] [PubMed] [Google Scholar]
- 8.Harvey, T. J., W. J. Liu, X. J. Wang, R. Linedale, M. Jacobs, A. Davidson, T. T. Le, I. Anraku, A. Suhrbier, P. Y. Shi, and A. A. Khromykh. 2004. Tetracycline-inducible packaging cell line for production of flavivirus replicon particles. J. Virol. 78531-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heinz, F. X., M. S. Collett, R. H. Purcell, E. A. Gould, C. R. Howard, M. Houghton, R. J. M. Moormann, C. M. Rice, and H.-J. Theil. 2000. Family Flaviviridae, p. 859-878. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Seventh report of the International Committee for the Taxonomy of Viruses. Academic Press, San Diego, CA.
- 10.Hunt, A. R., R. A. Hall, A. J. Kerst, R. S. Nasci, H. M. Savage, N. A. Panella, K. L. Gottfried, K. L. Burkhalter, and J. T. Roehrig. 2002. Detection of West Nile virus antigen in mosquitoes and avian tissues by a monoclonal antibody-based capture enzyme immunoassay. J. Clin. Microbiol. 402023-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khromykh, A. A., and E. G. Westaway. 1994. Completion of Kunjin virus RNA sequence and recovery of an infectious RNA transcribed from stably cloned full-length cDNA. J. Virol. 684580-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khromykh, A. A., and E. G. Westaway. 1997. Subgenomic replicons of the flavivirus Kunjin: construction and applications. J. Virol. 711497-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khromykh, A. A., A. N. Varnavski, and E. G. Westaway. 1998. Encapsidation of the flavivirus Kunjin replicon RNA by using a complementation system providing Kunjin virus structural proteins in trans. J. Virol. 725967-5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khromykh, A. A., P. L. Sedlak, K. J. Guyatt, R. A. Hall, and E. G. Westaway. 1999. Efficient trans-complementation of the flavivirus Kunjin NS5 protein but not of the NS1 protein requires its coexpression with other components of the viral replicase. J. Virol. 7310272-10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khromykh, A. A., A. N. Varnavski, P. L. Sedlak, and E. G. Westaway. 2001. Coupling between replication and packaging of flavivirus RNA: evidence derived from the use of DNA-based full-length cDNA clones of Kunjin virus. J. Virol. 754633-4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kummerer, B. M., and C. M. Rice. 2002. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J. Virol. 764773-4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, W. J., H. B. Chen, and A. A. Khromykh. 2003. Molecular and functional analyses of Kunjin virus infectious cDNA clones demonstrate the essential roles for NS2A in virus assembly and for a nonconservative residue in NS3 in RNA replication. J. Virol. 777804-7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, W. J., H. B. Chen, X. J. Wang, H. Huang, and A. A. Khromykh. 2004. Analysis of adaptive mutations in Kunjin virus replicon RNA reveals a novel role for the flavivirus nonstructural protein NS2A in inhibition of beta interferon promoter-driven transcription. J. Virol. 7812225-12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, W. J., X. J. Wang, V. V. Mokhonov, P. Y. Shi, R. Randall, and A. A. Khromykh. 2005. Inhibition of interferon signalling by the New York 99 strain and Kunjin subtype of West Nile virus involves blockage of STAT1 and STAT2 activation by nonstructural proteins. J. Virol. 791934-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, W. J., X. J. Wang, D. C. Clark, M. Lobigs, R. A. Hall, and A. A. Khromykh. 2006. A single amino acid substitution in the West Nile virus nonstructural protein NS2A disables its ability to inhibit alpha/beta interferon induction and attenuates virus virulence in mice. J. Virol. 802396-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mackenzie, J. M., A. A. Khromykh, M. K. Jones, and E. G. Westaway. 1998. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology 245203-215. [DOI] [PubMed] [Google Scholar]
- 22.Mackenzie, J. M., A. A. Khromykh, and E. G. Westaway. 2001. Stable expression of noncytopathic Kunjin replicons simulates both ultrastructural and biochemical characteristics observed during replication of Kunjin virus. Virology 279161-172. [DOI] [PubMed] [Google Scholar]
- 23.Mackenzie, J. M., and E. G. Westaway. 2001. Assembly and maturation of the flavivirus Kunjin virus appear to occur in the rough endoplasmic reticulum and along the secretory pathway, respectively. J. Virol. 7510787-10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller, S., S. Kastner, J. Krijnse-Locker, S. Buhler, and R. Bartenschlager. 2007. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J. Biol. Chem. 2828873-8882. [DOI] [PubMed] [Google Scholar]
- 25.Munoz-Jordan, J. L., G. G. Sanchez-Burgos, M. Laurent-Rolle, and A. Garcia-Sastre. 2003. Inhibition of interferon signalling by dengue virus. Proc. Natl. Acad. Sci. USA 10014333-14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petersen, L. R., and A. A. Marfin. 2002. West Nile virus: a primer for the clinician. Ann. Intern. Med. 137173-179. [DOI] [PubMed] [Google Scholar]
- 27.Pijlman, G. P., N. Kondratieva, and A. A. Khromykh. 2006. Translation of the flavivirus Kunjin NS3 gene in cis but not its RNA sequence or secondary structure is essential for efficient RNA packaging. J. Virol. 8011255-11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pugachev, K. V., F. Guirakhoo, S. W. Ocran, F. Mitchell, M. Parsons, C. Penal, S. Girakhoo, S. O. Pougatcheva, J. Arroyo, D. W. Trent, and T. P. Monath. 2004. High fidelity of yellow fever virus RNA polymerase. J. Virol. 781032-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roosendaal, J., E. G. Westaway, A. A. Khromykh, and J. M. Mackenzie. 2006. Regulated cleavages at the West Nile virus NS4A-2K-NS4B junctions play a major role in rearranging cytoplasmic membranes and Golgi trafficking of the NS4A protein. J. Virol. 804623-4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varnavski, A. N., P. R. Young, and A. A. Khromykh. 2000. Stable high-level expression of heterologous genes in vitro and in vivo by noncytopathic DNA-based Kunjin virus replicon vectors. J. Virol. 744394-4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Westaway, E. G., J. M. Mackenzie, M. T. Kenney, M. K. Jones, and A. A. Khromykh. 1997. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 716650-6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Westaway, E. G., J. M. Mackenzie, and A. A. Khromykh. 2003. Kunjin RNA replication and applications of Kunjin replicons. Adv. Virus Res. 5999-140. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.