Abstract
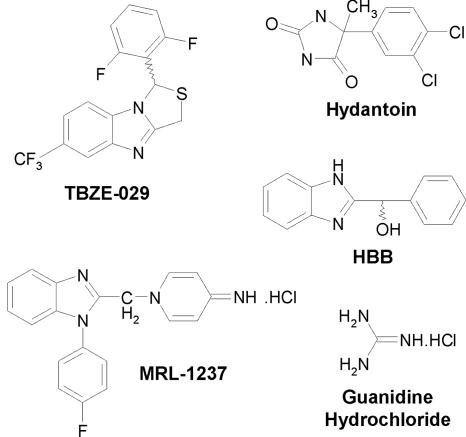
TBZE-029 {1-(2,6-difluorophenyl)-6-trifluoromethyl-1H,3H-thiazolo[3,4-a]benzimidazole} is a novel selective inhibitor of the replication of several enteroviruses. We show that TBZE-029 exerts its antiviral activity through inhibition of viral RNA replication, without affecting polyprotein processing. To identify the viral target of TBZE-029, drug-resistant coxsackievirus B3 (CVB3) was selected. Genotyping of resistant clones led to the identification of three amino acid mutations in nonstructural protein 2C, clustered at amino acid positions 224, 227, and 229, immediately downstream of NTPase/helicase motif C. The mutations were reintroduced, either alone or combined, into an infectious full-length CVB3 clone. In particular the mutations at positions 227 and 229 proved essential for the altered sensitivity of CVB3 to TBZE-029. Resistant virus exhibited cross-resistance to the earlier-reported antienterovirus agents targeting 2C, namely, guanidine hydrochloride, HBB [2-(alpha-hydroxybenzyl)-benzimidazole], and MRL-1237 {1-(4-fluorophenyl)-2-[(4-imino-1,4-dihydropyridin-1-yl)methyl]benzimidazole hydrochloride}. The ATPase activity of 2C, however, remained unaltered in the presence of TBZE-029.
Entero- and rhinoviruses are involved in a wide range of infections in humans and animals. Among the species in the enterovirus genus are the coxsackieviruses, which have been reported to be associated with various clinical manifestations, including myocarditis, pancreatitis, meningitis, and encephalitis. Other clinically relevant enteroviruses are poliovirus, which can cause paralytic poliomyelitis, and echovirus, causing aseptic meningitis or encephalomyelitis (33, 36, 43). Rhinoviruses are the main pathogens associated with the common cold, and, although usually mild and self-limiting, rhinovirus infections have an enormous socioeconomical impact (47, 59).
Enteroviruses belong to the family Picornaviridae, which consists of small, nonenveloped viruses containing a single-stranded positive-strand RNA [(+)RNA] genome of 7.5 kb, which is covalently linked to a small viral protein (VPg) at its 5′ end and polyadenylated at its 3′ end (49). The genomic RNA has a long, highly structured 5′ noncoding region, which contains the internal ribosome entry site, necessary for translation initiation, and a shorter 3′ noncoding region preceding the poly(A) tract, which are both thought to be involved in RNA replication and translation (9). The coding region encodes a single polyprotein that will eventually be cleaved to generate 4 structural and 10 nonstructural proteins (either mature or in their precursor form). The icosahedral capsid of the virus is formed by 60 protomers, each one assembled by the four structural proteins, designated VP1 to -4. The nonstructural region comprises two proteases, the viral RNA-dependent RNA polymerase and seven other proteins that are involved in viral replication (8).
One of the most conserved nonstructural viral proteins among picornaviruses is 2C. Although this protein is an indispensable component of the replication complex, its exact role in viral replication has remained elusive. Protein 2C appears to be multifunctional, and this entails multiple interactions (23). The amino acid sequence of 2C contains three conserved motifs which are typically found in NTP-binding proteins (motifs A and B) or in members of helicase superfamily III (motif C) (28-31, 69). In fact, ATPase activity has been demonstrated for several picornavirus 2C proteins (37, 57, 60) whereas so far, every attempt to demonstrate in vitro RNA helicase activity has failed. Protein 2C contains two regions involved in RNA binding (58). It has been suggested that, in poliovirus, this binding occurs at the 3′ cloverleaf of (−)RNA (3-5); for echovirus, RNA binding was reported to occur in a nonspecific way (38). The amino-terminal region of the protein was predicted to form an amphipathic helix (48), providing the major determinant for its membrane-binding properties, although the C-terminal portion of 2C was also found to interact with membranes (21, 22, 64). The intracellular membranes, on which replication complexes are localized, undergo extensive rearrangements upon viral infection, and it has been shown that these structural changes are also in part ascribed to the presence of 2C/2BC (1, 11-13, 16, 63, 64). Furthermore, protein 2C has been shown to contain a zinc-binding cysteine-rich motif involved in RNA replication (50), to possess properties involved in regulating the activity of the poliovirus 3C protease (6), and to be implicated in the process of uncoating (41). A study using the poliovirus inhibitor hydantoin [5-(3,4-dichlorophenyl)methylhydantoin] suggested a role for 2C during encapsidation (67), although the findings on which this conclusion was based were disputed in a later study (68).
There is no approved antiviral therapy for the treatment of picornavirus infections (59). Several molecules have previously been reported to inhibit picornavirus replication, some of which have entered clinical trials (20). As protein 2C is highly conserved among picornaviruses and as its presence is indispensable for efficient viral replication, this protein is an attractive target for viral inhibition. Presumably the best-studied picornavirus inhibitor targeting the 2C protein so far is guanidine hydrochloride (GuaHCl) (14) (Fig. 1). Early studies with this compound revealed that resistance to and/or dependence on guanidine maps to the 2C region of poliovirus (2, 52-54). Guanidine was shown to inhibit a 2C function that is required for the initiation of (−)RNA but not (+)RNA synthesis or RNA elongation (7). Bienz and coworkers showed that guanidine prevents the association of 2BC with host membrane structures during viral replication (13). Moreover, it was shown that guanidine at millimolar concentrations inhibits ATP hydrolysis (51), although other studies were not able to confirm this observation (39, 57). Another compound that has been known for decades and that selectively blocks enterovirus replication is 2-(alpha-hydroxybenzyl)-benzimidazole (HBB) (24-27) (Fig. 1). Resistance to (or dependence on) HBB of echovirus is caused by single amino acid mutations in protein 2C (32, 39). Also, compound 1-(4-fluorophenyl)-2-[(4-imino-1,4-dihydropyridin-1-yl)methyl]benzimidazole hydrochloride (MRL-1237) (Fig. 1) was shown to inhibit the replication of several enteroviruses, and MRL-1237-resistant poliovirus mutants were shown to carry a mutation in the 2C gene (61). Finally, poliovirus mutants resistant to hydantoin (Fig. 1), which blocks postsynthetic cleavages and assembly of poliovirus, were shown to carry mutations scattered throughout the 2C sequence (67, 68), and hence this compound also appears to target enterovirus protein 2C.
FIG. 1.
Structural formulae of TBZE-029 and earlier reported enterovirus inhibitors targeting protein 2C.
In the present study, we characterized the antiviral activity of TBZE-029 {1-(2,6-difluorophenyl)-6-trifluoromethyl-1H,3H-thiazolo[3,4-a]benzimidazole} (Fig. 1), a compound that we recently described as an inhibitor of the replication of several enteroviruses (19), and identified protein 2C as its molecular target.
MATERIALS AND METHODS
Cells and viruses.
Vero cells (ATTC CCL-81) or Buffalo green monkey (BGM) cells were grown in minimal essential medium (Gibco, Merelbeke, Belgium) supplemented with 10% heat-inactivated fetal bovine serum (Integro, Leuvenheim, The Netherlands), 5% bicarbonate (Gibco, Merelbeke, Belgium), and 5% l-glutamine (Gibco, Merelbeke, Belgium). Cells were grown at 37°C in a 5% CO2 incubator. Coxsackievirus B3 was derived from plasmid p53CB3/T7, which contains a full-length cDNA of CVB3 strain Nancy behind a T7 RNA promoter (70). For assays involving virus growth, 2% fetal bovine serum was used instead of 10%.
Compounds.
TBZE-029 was synthesized as previously described (15). Enviroxime and MRL-1237 were synthesized by G. Pürstinger (Institut für Pharmazie, Universität Innsbruck, Austria). GuaHCl was purchased from Sigma, and HBB was from ChemPacific (Baltimore, MD). Rupintrivir was a kind gift from Amy Patick (Pfizer, La Jolla, CA). All compounds were solubilized in dimethyl sulfoxide (DMSO) and stored at 4°C. For working solutions, the DMSO stocks were diluted in minimal essential medium to the desired concentration.
In vitro RNA transcription and transfection.
Prior to in vitro RNA transcription, plasmid p53CB3/T7 was linearized with SalI (Promega, Leiden, The Netherlands). The digest was purified (gel and PCR purification kit; Promega, Leiden, The Netherlands), and 2.5 μg of DNA was used for in vitro RNA transcription (Ribomax large-scale RNA production system; Promega, Leiden, The Netherlands). The transcription reaction was carried out at 37°C for 4 h, after which the reaction mixture was DNase treated and the RNA was purified (RNA cleanup system; Qiagen, Venlo, The Netherlands). Transfections were carried out in 25-cm2 culture flasks on Vero cells, grown to ∼75% confluence. Reaction mixtures, containing 2 ml OptiMEM (Gibco, Merelbeke, Belgium), 2.5 μg of purified RNA, and 10 μl of DMRIE-C transfection reagent (Invitrogen, Merelbeke, Belgium) were incubated for 4 h at 37°C. Subsequently, the medium was replaced with fresh growth medium and incubation was continued until the cultures exhibited extensive cytopathic effect (CPE). At this point, flasks were subjected to three rounds of freezing and thawing, and the collected supernatant was titrated for infectious-virus content as described earlier (19).
Multicycle CPE reduction assays.
The antiviral activity of the selected compounds was initially determined using an MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium]-based CPE reduction assay. Briefly, cells grown to confluence in 96-well plates were infected with 100 50% cell culture infective doses (CCID50) of virus. After an adsorption period of 2 hours at 37°C, virus was removed and serial dilutions of the compounds were added. The cultures were further incubated at 37°C for 3 days, until complete CPE was observed in the infected and untreated virus control (VC). After removal of the medium, 90 μl culture medium and 10 μl MTS-phenazine methosulfate (Promega, Leiden, The Netherlands) were added to each well. After an incubation period of 2 h at 37°C the optical density (OD) of each well was read at 498 nm in a microplate reader. CPE values were calculated as follows: %CPE = 100·(ODCC − ODCVB3+Compound)/(ODCC − ODVC). In this formula, ODCC corresponds to the OD of the uninfected and untreated cell cultures, ODVC represents the OD of the infected and untreated cell cultures, and ODCVB3+Compound represents the OD of the CVB3-infected cell cultures treated with a given concentration of compound. The 50% effective concentration (EC50) was defined as the concentration of compound that offered 50% protection against virus-induced CPE and was calculated using logarithmic interpolation.
Effect on viral RNA and infectious-virus yield in multicycle growth assays.
Vero cells, grown to confluence in 48-well plates, were infected with 100 CCID50 of virus. After an adsorption period of 2 hours at 37°C, virus was removed and serial dilutions of the compounds were added. The cultures were further incubated at 37°C for 3 days, until complete CPE was observed in the VC. At this point, viral RNA was isolated from the supernatant using a viral RNA isolation kit (Macherey-Nagel, Düren, Germany) and quantified using real-time reverse transcription-quantitative PCR (RT-qPCR). Quantification of infectious-virus yields was done by end point titration. Serial 10-fold dilutions were tested in six replicative wells of 96-well plates. Virus titers were calculated and expressed as CCID50 values (55).
Analysis of viral RNA accumulation with subgenomic replicon pCB3/T7-Luc.
Accumulation of viral (+)RNA was monitored by transfecting cells (in the presence or absence of TBZE-029) with RNA derived from the SalI-linearized plasmid p53CB3/T7-Luc, which contains a subgenomic CVB3 replicon carrying a luciferase gene in place of the capsid-coding P1 region. At 2 h, 4 h, 6 h, and 8 h posttransfection, the cells were washed three times with phosphate-buffered saline (PBS) and lysed in 100 μl of lysis buffer, and the luciferase activity was measured in a liquid scintillation counter with the luciferase assay system according to the recommendations of the manufacturer (Promega, Leiden, The Netherlands). Luciferase activity was expressed in (relative) light units.
Quantitative analysis of CVB3 RNA by real-time RT-qPCR assays.
Real-time qPCR was performed on the ABI Prism 7000 sequence detection system (Applied Biosystems, Foster City, CA). Primers and probes were developed with Primer Express software (Applied Biosystems) as described elsewhere (46). The following primers and probe were used: a forward primer specific for nucleotides 2937 to 2957 (5′-ACG AAT CCC AGT GTG TTT TGG-3′), a reverse primer specific for nucleotides 3003 to 2982 (5′-TGC TCA AAA ACG GTA TGG ACA T-3′), and a TaqMan probe specific for nucleotides 2960 to 2977 (5′-6-carboxyfluorescein-CGA GGG AAA CGC CCC GCC-6-carboxytetramethylrhodamine-3′). Each reaction was performed in 25 μl of a PCR reagent mixture (One-Step RT-qPCR mixture; Eurogentec, Seraing, Belgium) containing 900 nM of each primer and 200 nM of the specific TaqMan probe. The PCR consisted of a reverse transcription step (30 min at 48°C), a Taq activation step (10 min at 95°C), and 50 cycles of denaturation (15 s at 94°C) and annealing (1 min at 60°C). The RNA copy number in each sample was determined by a standard curve generated by increasing the copy number of a synthetic transcript corresponding to 67 nucleotides of the CVB3 genome.
Time of drug addition studies.
Vero cells, grown to confluence in 24-well culture plates, were infected with 1 × 104 CCID50 of coxsackievirus B3. After an adsorption period of 1 h at 37°C, virus was removed and replaced with 500 μl growth medium. At 1-hour intervals, 500 μl of medium containing a 2× compound solution was added (final concentration, 80 μM). At 8 hours postinfection (p.i.), the supernatant of the infected cultures was collected and viral RNA was quantified using real-time RT-qPCR.
Analysis of viral polyprotein processing in vivo.
BGM cells, grown to confluence in 24-well plates, were infected with 2 × 107 CCID50 of coxsackievirus B3. At 5 h 15 min p.i., the medium was replaced with 300 μl methionine-free medium. Thirty minutes later, the cultures were pulse-labeled in methionine-free medium containing 1 μl/well of [35S]Met in the absence or presence of TBZE-029 (final concentration, 80 μM) for 15 min. At 6 h p.i. (5 h 15 min plus 30 min plus 15 min), cells were washed twice with PBS and lysed in cold lysis buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, and 0.05% sodium dodecyl sulfate (SDS). Translation products were analyzed on a 10% polyacrylamide gel containing SDS. The gels were fixed in 30% methanol-10% acetic acid, rinsed in DMSO, fluorographed with 20% 2,5-diphenyloxazole in DMSO, dried, and exposed to Kodak XAR film.
Single-cycle growth curves.
Confluent Vero cell monolayers in 24-well plates were infected with virus at a multiplicity of infection (MOI) of 1 for 30 min at 37°C. The cells were washed three times with PBS, supplied with the medium (in the presence or absence of TBZE-029), and further incubated. At 2 h, 4 h, 6 h, and 8 h p.i., cells were disrupted by three cycles of freezing and thawing. Virus titers were determined by end point titration.
Generation of TBZE-029-resistant virus.
Drug-resistant virus was generated by growing virus in the presence of increasing concentrations of TBZE-029 on confluent Vero cultures in 48-well culture plates. After 4 to 5 days, culture supernatant was collected from those cultures that exhibited full CPE in the presence of the highest concentration of compound. This virus was used for a successive round of infection, a procedure that was repeated until full CPE was noticed at concentrations of TBZE-029 that did not allow replication of wild-type virus. Subsequently, the resistant-virus pool was subjected to two rounds of plaque purification (in the presence of compound), and individual clones were used for sequencing.
Site-directed mutagenesis.
Seven mutant CVB3 clones, containing either single or multiple amino acid replacements at positions 224, 227, and/or 229 in protein 2C, were constructed. The seven clones were designated CVB3[A224V], CVB3[I227V], CVB3[A229V], CVB3[A224V-I227V], CVB3[A224V-A229V], CVB3[I227V-A229V], and CVB3[A224V-I227V-A229V]. The corresponding synthetic oligonucleotides (and their complementary reverse oligonucleotides) were used for site-directed mutagenesis: 5′-GTC TTG GCA TCG ACC AAT GTA GGA TCT ATT AAT GCT CCA ACC G-3′, 5′-GTC TTG GCA TCG ACC AAT GCA GGA TCT GTT AAT GCT CCA ACC G-3′, 5′-GTC TTG GCA TCG ACC AAT GCA GGA TCT ATT AAT GTT CCA ACC G-3′, 5′-GTC TTG GCA TCG ACC AAT GTA GGA TCT GTT AAT GCT CCA ACC G-3′, 5′-GTC TTG GCA TCG ACC AAT GTA GGA TCT ATT AAT GTT CCA ACC G-3′, 5′-GTC TTG GCA TCG ACC AAT GCA GGA TCT GTT AAT GTT CCA ACC G-3′, and 5′-GTC TTG GCA TCG ACC AAT GTA GGA TCT GTT AAT GTT CCA ACC G-3′. The mutated sequences are underlined. Site-directed mutagenesis was performed with plasmid p53CB3/T7 using the XL Blue large site-directed mutagenesis kit (Stratagene, Amsterdam, The Netherlands), according to the manufacturer's instructions. After mutagenesis, the individual clones were verified by sequencing. Next, a 711-bp fragment containing the desired mutations was isolated using the enzymes BssHII and XbaI and reintroduced into an original, nonmutagenized clone of the same plasmid, p53CB3/T7. From these mutants, RNA transcripts and infectious viruses were generated as described above.
Sequencing.
PCR fragments that cover the entire CVB3 genome were generated and analyzed using the cycle sequencing method (ABI Prism Big Dye Terminator cycle sequencing ready reaction kit). Both DNA strands were sequenced. Sequencing data were obtained using an ABI 373 automated sequence analyzer (Applied Biosystems, Lennik, Belgium), and sequences were analyzed using the Vector NTI software (Invitrogen, Merelbeke, Belgium).
Cloning and expression of wild-type and an active-site mutant coxsackievirus B3 protein 2C.
The cDNA encoding the 2C domain of CVB3 was amplified by PCR with the following primers: forward primer, GGGGACAAGTTTGTACAAAAAAGCAGGCTTAGAAAACCTGTACTTCCAGGGTaacaatagctggcttaagaaatttac (the recombination site is in uppercase letters; the tobacco etch virus protease cleavage site is underlined and uppercase and is followed by the gene-specific sequence [lowercase]); reverse primer, GGGGACCACTTTGTACAAGAAAGCTGGGTCTTATTActggaacagtgcctcaagcg (the recombination site is in uppercase letters; stop codons [underlined, uppercase] are followed by the gene-specific sequence [lowercase]). The resulting PCR product was cloned via the Gateway system (Invitrogen, Merelbeke, Belgium) into the compatible pDEST17 expression vector in order to fuse the gene of interest with N-terminal ATGs (His6, His6-maltose binding protein, His6-glutathione S-transferase [GST], His6-Trx). A soluble-expression screening was performed for each fusion in order to define the best fusion expressed in its best condition (10). The His6-GST-2C was chosen and expressed solubly in Rosetta pLysS, at 20°C in 16 g/liter tryptone-10 g/liter yeast extract-5 g/liter NaCl containing 100 μg/ml ampicillin (2 YT)-ampicillin-chloramphenicol medium. Similarly, an active-site mutant of 2C which carried the K135A mutation in NTPase motif A was cloned and expressed. This mutation was introduced using site-directed mutagenesis on the CVB3 full-length clone pCB53/T7 as described earlier using the following synthetic oligonucleotide (and its complementary reverse oligonucleotide): 5′-AGC CCT GGT GCC GGC GCG TCG GTG GCA ACA AAC TT-3′, where the mutated sequence is underlined.
Production and purification of protein 2C of coxsackievirus B3.
A volume of 60 ml of an overnight preculture was used to inoculate 2 liters of 2YT medium supplemented with ampicillin and chloramphenicol. The culture was grown at 37°C until the OD at 600 nm reached 0.5. Induction was then performed overnight with 500 μM IPTG (isopropyl-β-d-thiogalactopyranoside) at 20°C. The culture was stopped by centrifuging for 10 min at 6,000 × g. The pellet was resuspended in lysis buffer (50 mM Tris, 300 mM NaCl, 10 mM imidazole [pH 8], 0.1% Triton X-100, 5% glycerol, 0.25 mg/ml lysozyme, EDTA-free antiprotease cocktail [Roche, Brussels, Belgium]) before being stored at −80°C. Cells were lysed by sonication. The lysate was clarified by centrifugation at 16,000 × g for 30 min. The purification consisted of two steps. First immobilized-metal affinity chromatography was performed on a 5-ml Hisprep column using the Akta Xpress (GE Healthcare, Diegem, Belgium) system. After a washing step at 50 mM imidazole, the protein was eluted in 50 mM Tris, 300 mM NaCl, and 250 mM imidazole, pH 8. Both GST alone and the GST-2C fusion were copurified. The two proteins were separated by gel filtration using a HiLoad 16/60 Superdex200 equilibrated in 10 mM Tris, 300 mM NaCl, and 2 mM dithiothreitol, pH 7.5. Purity was checked on an acrylamide SDS-polyacrylamide gel electrophoresis gel colored with Coomassie blue. The pool corresponding to the GST-2C fusion (first peak) was concentrated using an Amicon Ultra centrifugal filter unit (30,000-Da cutoff; Millipore, Brussels, Belgium) over 1 mg/ml and diluted in 50% glycerol for storage at −20°C. For the K135A mutant, an additional affinity purification was performed on glutathione-Sepharose 4B (GE Healthcare) following the gel filtration step. The purification was carried out in the gel filtration buffer, and elution was in the buffer supplemented with 10 mM reduced glutathione. The remaining reduced glutathione was then removed by subsequent washes during the protein concentration.
ATPase assay and detection.
ATPase reactions with 20-μl mixtures were performed in buffer O+ (Fermentas, St Leon-Rot, Germany), containing purified CVB3 protein 2CATPase-GST and 10 μM [α-32P]ATP (10 μCi/μl; Hartmann Analytic, Braunschweig, Germany), either in the presence or absence of antiviral compounds. Reactions were performed at 30°C or 37°C and stopped by addition of 80 μl 0.1 M EDTA. One microliter of the reaction mixture was applied to polyethyleneimine cellulose-coated thin-layer chromatography plates (Merck, Overijse, Belgium) and run for 2 h in thin-layer chromatography buffer (0.15 M LiCl, 0.15 M formic acid). The chromatogram was exposed to Biomax MR X-ray film (Kodak, Brussels, Belgium) and analyzed.
RESULTS
TBZE-029 inhibits viral replication in a dose-dependent manner.
From a series of 2,6-dihalophenyl-substituted 1H,3H-thiazolo[3,4-a]benzimidazoles with antienterovirus activity, TBZE-029 (designated compound 4a in reference 19) was selected for mechanistic studies. As recently reported, TBZE-029 inhibits virus-induced CPE formation of several enteroviruses in multicycle-growth assays (19). To verify that the protection against CPE formation was indeed due to inhibition of viral replication, CVB3 RNA from culture supernatant was quantified by means of real-time RT-qPCR. No viral RNA was detectable in the supernatant of infected cultures that had been treated with TBZE-029 at concentrations of 33 μM or higher (Fig. 2A); at a concentration of 3.3 μM, still a 40% reduction in viral RNA was observed compared to the untreated control. These data were corroborated by determining the effect of the compound on infectious-virus yield. The decrease in viral RNA yield was paralleled by a decrease in infectious-virus yield (Fig. 2B).
FIG. 2.
Dose-dependent inhibition of viral replication by TBZE-029. Vero cells grown to confluence in 48-well plates were infected with CVB3, treated with different concentrations of TBZE-029, and incubated for 3 days at 37°C until extensive CPE was noticed. At that point, supernatant was collected and assayed for the presence of viral RNA (by means of real-time RT-qPCR) (A) or infectious-virus yield (by end point titration) (B).
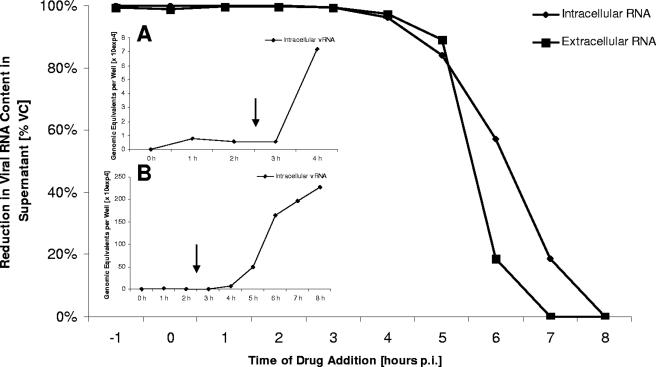
TBZE-029 exerts its antiviral activity at a stage that coincides with viral RNA synthesis and polyprotein processing/synthesis.
To define the stage in the viral replication cycle at which TBZE-029 exerts its antiviral activity, time of drug addition studies were performed. A fixed concentration of TBZE-029 (80 μM) was added at 1-hour intervals, starting at 1 hour preinfection. As shown in Fig. 3, a complete inhibition of viral replication was observed at the end of a single replication cycle (8 h) when TBZE-029 was added within the first 3 hours after infection. When the compound was added at a time point later than 3 h p.i., a gradual loss in antiviral activity was noticed, indicating that TBZE-029 does not hamper early (attachment, entry, or uncoating) or late (assembly or release) events. In parallel, we monitored the single-cycle kinetics of intracellular viral RNA synthesis by means of RT-qPCR (Fig. 3A and B). From panel A it is evident that viral RNA is first detected intracellularly after 3 h p.i., and panel B reveals that a very sudden, steep increase in intracellular viral RNA levels occurs between 4 and 7 h p.i. This increase in viral RNA levels reaches a plateau at 7 to 8 h p.i. The gradual/steep loss of TBZE-029 activity coincides with the time points where a gradual/steep increase in intracellular viral RNA is detected.
FIG. 3.
Time of drug addition. TBZE-029 (80 μM) was added to CVB3-infected cell cultures at 1-hour intervals, starting at 1 h before infection. At 8 h p.i., intra- and extracellular viral RNA was collected and quantified by means of real-time RT-qPCR. (Insets A and B) Kinetics of a single viral replication cycle of CVB3 in the absence of drug (intracellular viral RNA monitored by RT-qPCR).
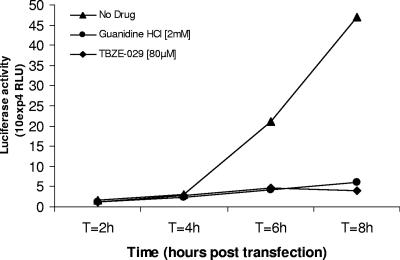
TBZE-029 inhibits accumulation of viral positive-strand RNA.
To study the effect of TBZE-029 on viral positive-strand RNA accumulation, we used a chimeric subgenomic replicon (pCB53/T7-Luc) in which the P1 region of pCB53/T7 is replaced by the firefly luciferase gene (70). It was previously shown that the activity of the reporter protein following transfection correlates with the levels of RNA that accumulate intracellularly (70). The presence of TBZE-029 at a concentration of 80 μM almost completely inhibited accumulation of viral positive-strand RNA, whereas a ∼50-fold increase in luciferase activity was detected in the absence of the drug (Fig. 4). GuaHCl (2 mM) was included as a control and also completely inhibited viral RNA accumulation.
FIG. 4.
Analysis of viral RNA accumulation with subgenomic replicon pCB3/T7-Luc. Accumulation of viral (+)RNA was monitored by transfecting BGM cells grown to confluence (in the presence or absence of TBZE-029) with RNA derived from a chimeric subgenomic replicon (pCB53/T7-Luc). In this replicon, the P1 region of pCB53/T7 is replaced by the firefly luciferase gene. At the indicated times posttransfection, the luciferase activity was quantified and expressed in (relative) light units (RLU).
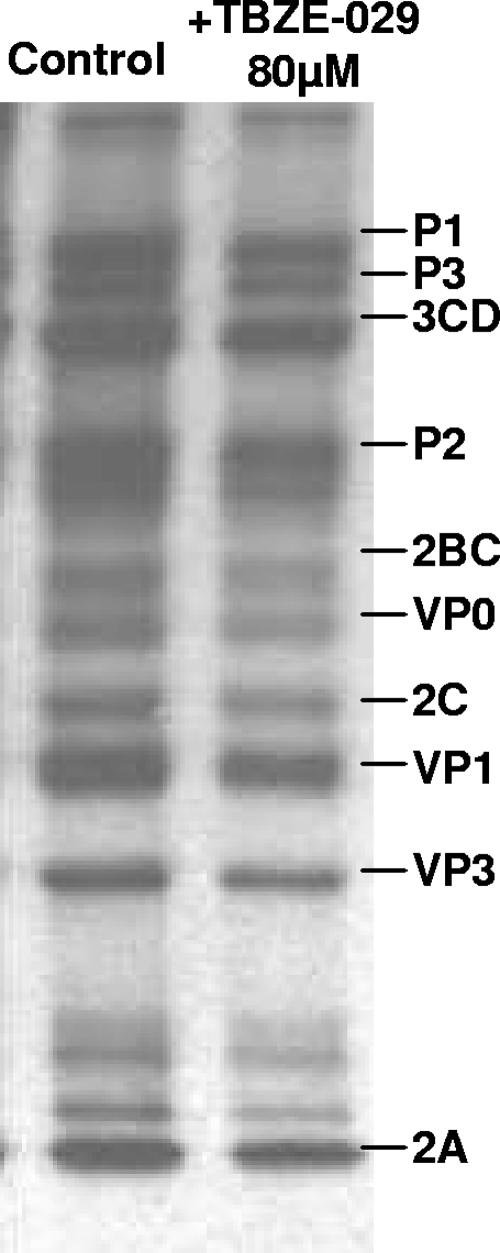
TBZE-029 does not affect polyprotein processing.
As shown using the CVB3-luc replicon, TBZE-029 inhibits accumulation of viral (+)RNA. Theoretically, this could be due to adverse effects of the drug on viral protein synthesis or polyprotein processing. To exclude this possibility, the effect of TBZE-029 on in vivo polyprotein synthesis and/or processing was monitored. At 5 h 45 min p.i., infected-cell cultures were pulse-labeled for 15 min with [35S]Met in the presence (80 μM) or absence of the compound after starvation for methionine for 30 min. From Fig. 5 it is evident that the viral polyprotein processing pattern remains unaffected in the presence of TBZE-029. This excludes the possibility that the deleterious effects of TBZE-029 on viral (+)RNA accumulation are due to an altered rate of protein translation or a general defect in polyprotein processing.
FIG. 5.
Effect of TBZE-029 on polyprotein processing. At 5 h 45 min p.i., BGM cells grown to confluence in 24-well plates were pulse-labeled for 15 min with [35S]Met after starvation in methionine-free medium for 30 min, either in the presence (80 μM) or absence of TBZE-029. Cells were lysed, and translation products were analyzed by SDS-polyacrylamide gel electrophoresis.
Resistance to TBZE-029 maps to a short sequence flanking domain C in nonstructural protein 2C.
To identify the precise viral target of TBZE-029, drug-resistant CVB3 was generated. Initially, we tried to recover resistant clones following agar (containing TBZE-029) overlay of infected-cell cultures, but this resulted in complete inhibition of plaque formation and hence, in this particular case, appeared to be not an ideal method to isolate resistant virus. Therefore, we grew virus in liquid medium, in the presence of increasing concentrations of compound, starting at 0.5× EC50. To prevent the generation of resistant clones deriving from one common predecessor, three independent serial passages were carried out. After 15 passages, three highly resistant virus pools that replicated effectively in the presence of 330 μM TBZE-029 were obtained. These virus pools were subjected to two rounds of plaque purification in the presence of TBZE-029, after which six single clones were picked and sequenced. Each of these clones carried three amino acid substitutions (A224V, I227V, and A229V) in protein 2C, immediately downstream of the conserved NTPase domain C, except for clone 5, which carried mutation A224L instead of A224V (Table 1). Clones 2 and 6 carried an additional mutation, K107R in protein 2C and S229T in protein 3D, respectively. Since mutations 2C K107R and 3D S299T each occurred only once in different clones, they were not further considered. To characterize the actual contribution of the A224V, I227V, and A229V mutations to the drug resistance phenotype, seven recombinant viruses were generated, containing either one, two, or three mutations (Table 2). To this end, site-directed mutagenesis was performed with plasmid p53CB3/T7 and RNA transcripts were transfected into Vero cells. Two to 3 days p.i., all cultures exhibited full CPE, except for those infected by clones 3 (A229V) and 5 (A224V and A229V). Since for other 2C inhibitors, drug-dependent phenotypes that could be rescued by addition of GuaHCl have been described previously (32, 66), constructs 3 and 5 were transfected in the presence of 400 μM GuaHCl. Remarkably, 3 to 4 days p.i. full CPE was noted in both cultures. When cultures exhibited extensive CPE, culture flasks were freeze-thawed three times, supernatant was collected, and infectious virus was quantified by titration. The mutant viruses were designated clones 1 to 7 (Table 2), and three of these (clones 4, 6, and 7) were sequenced to verify that no other mutations accumulated after transfection. Subsequently, each clone was evaluated for its susceptibility to TBZE-029. Introduction of the two single mutations at positions 224 and 227 (clones 1 and 2) proved not sufficient to confer the TBZE-029 resistance phenotype. When these mutations were combined, however, a high-TBZE-029-resistance phenotype (clone 4) was generated. Unlike single mutations at positions 224 and 227, a single mutation at position 229 (clone 3) resulted in a transcript that was infectious only in the presence of GuaHCl. This dependence phenotype was also observed when an additional mutation at position 224 was introduced (clone 5). Surprisingly, the dependence phenotype was “rescued” when the 229 mutation was combined with the mutation at position 227, leading to a resistant, but nondependent virus (clone 6). The combination of the three mutations resulted, as expected, in a high-resistance phenotype (clone 7).
TABLE 1.
Amino acid mutations identified in six CVB3 clones following selection for TBZE-029-resistant variants
| CVB3 protein | Amino acid substitution | Nucleotide
|
Clone(s) | |
|---|---|---|---|---|
| Substitution | Position | |||
| 2C | K107R | AAG→AGG | 3620 | 2 |
| 2C | A224V/L | GCA→GTA/TTA | 3971/3970 | 1, 2, 3, 4, 6/5 |
| 2C | I227V | ATT→GTT | 3979 | 1, 2, 3, 4, 5, 6 |
| 2C | A229V | GCT→GTT | 3986 | 1, 2, 3, 4, 5, 6 |
| 3D | S299T | TCA→ACA | 6064 | 6 |
TABLE 2.
Resistance pattern of recombinant CVB3 carrying 2C mutations A224V, I227V, and A229V and cross-resistance pattern with other antienterovirus compoundsa
| Clone | 2C mutationb
|
Mean EC50 (μM) of indicated antiviral molecule ± SDc
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| A224V | I227V | A229V | 2C inhibitors
|
Non-2C inhibitors
|
|||||
| TBZE-029 | HBB | MRL-1237 | Guanidine | Enviroxime | Ruprintrivir | ||||
| WTd | 7.2 ± 3.1 | 55 ± 2 | 15 ± 6 | 272 ± 5 | 0.18 ± 0.03 | 4.7 ± 1.0 | |||
| 1 | X | 9.3 ± 0.9 | 56 ± 4 | 12 ± 2 | 1,681 ± 15 | 0.18 ± 0.03 | 4.4 ± 0.5 | ||
| 2 | X | 10 ± 0 | 59 ± 3 | 199 ± 27 | 286 ± 19 | 0.22 ± 0.02 | 4.3 ± 0.4 | ||
| 3 | X | Dep | Dep | Dep | Dep | ||||
| 4 | X | X | >330 | 283 ± 18 | >314 | 1,687 ± 60 | 0.17 ± 0.05 | 4.4 ± 0.4 | |
| 5 | X | X | Dep | Dep | Dep | Dep | |||
| 6 | X | X | 210 ± 72 | 270 ± 23 | >314 | 1,657 ± 60 | 0.14 ± 0.01 | 4.3 ± 0.6 | |
| 7 | X | X | X | >330 | >446 | >314 | >10,468 | 0.20 ± 0.01 | 2.4 ± 1.5 |
| Res poole | >330 | >446 | 202 ± 12 | >10,468 | 0.47 ± 0.17 | 4.3 ± 0.9 | |||
Mutations at residues 224, 227, and 229 in protein 2C were introduced, either alone or combined, in a full-length cDNA of CVB3. In vitro transcription, followed by transfection, led to seven recombinant viruses that were evaluated for their susceptibility to TBZE-029 and reference compounds.
X indicates the presence of the mutation.
Data were determined by inhibition of CPE formation. Dep, clones that were dependent on GuaHCl in order to generate infectious virus upon transfection. Data are from at least three independent experiments. Boldface indicates resistance to the tested drug.
WT, wild type.
Res pool, resistant pool of CVB3, obtained after serial passaging in the presence of TBZE-029 and prior to plaque purification.
TBZE-029-resistant virus exhibits cross-resistance with other 2C inhibitors.
Next, the susceptibilities of the various mutants to inhibition by other 2C inhibitors (GuaHCl, MRL-1237, and HBB) were studied. Hydantoin was not included, since it lacked antiviral activity against wild-type CVB3 (data not shown). For HBB, a pattern of resistance comparable to that for TBZE-029 was observed. This pattern was almost identical to that for MRL-1237, except in the case of clone 2 (carrying a single mutation at position 227), which was susceptible to inhibition by TBZE-029 and HBB but highly resistant to MRL-1237. Also for GuaHCl, the observed antiviral sensitivity phenotypes were similar to those for TBZE-029, with the exception of that of clone 1, carrying a single amino acid mutation at position 224. This clone was sensitive to inhibition by TBZE-029, MRL-1237, and HBB but proved highly resistant to GuaHCl. Two compounds with a target that is different from 2C were included as well: enviroxime (targeting protein 3A) and the 3C-protease inhibitor rupintrivir. As expected, none of the constructed mutants had a reduced sensitivity to these two reference compounds.
Phenotypic characterization of mutant 6 (I227V, A229V).
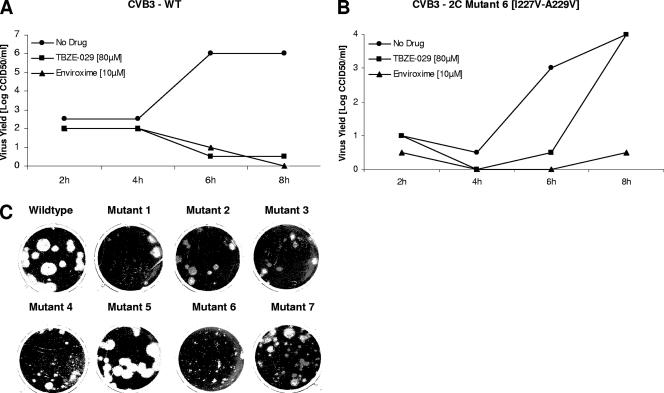
Clone 6, carrying the two most important mutations for resistance (I227V and A229V), was selected for further characterization of a single growth cycle. Both mutant and wild-type viruses were used for infection at an MOI of 1, and growth kinetics were defined by end point titration. At 8 h p.i., recombinant mutant 6 yielded virus titers that were ∼2 logs lower than those of wild-type CVB3, which indicates that the mutations at positions 227 and 229 in protein 2C reduce viral fitness (Fig. 6A and B). As expected, mutant 6 produced markedly smaller plaques than wild-type CVB3 (Fig. 6C), which reflects slower growth kinetics in multiple cycles. As expected, only mutant 6 could replicate to normal titers in the presence of TBZE-029 (80 μM), whereas 10 μM of enviroxime inhibited both mutant and wild-type virus (Fig. 6A and B). Inspection of the plaque phenotypes produced by the other recombinant viruses showed that only mutant 5 was able to produce plaques similar to those produced by wild-type virus, whereas the other viruses generated plaques that had a small or intermediate size compared to those of wild-type virus (Fig. 6C).
FIG. 6.
Phenotypic characterization of wild-type CVB3 and TBZE-029-resistant mutant 6 (I227V, A229V). Confluent Vero monolayers in 24-well plates were infected with wild-type (WT) (A) or mutant (B) virus at an MOI of 1 for 30 min at 37°C. The cells were washed and further cultured in the presence or absence of TBZE-029. At the indicated times postinfection, cells were disrupted by three cycles of freezing and thawing and viral titers were determined by end point titration. (C) Plaque phenotypes of wild-type CVB3 and recombinant clones.
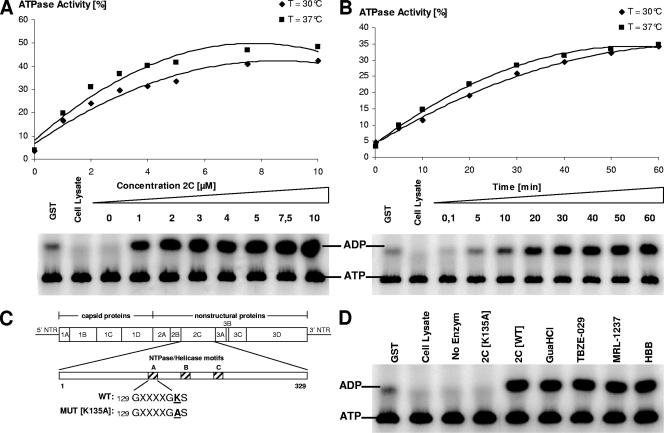
The ATPase activity of 2C is not inhibited by TBZE-029.
A GST-CVB3 protein 2C fusion was expressed and purified. A concentration- and time-dependent ATPase activity for protein 2C was demonstrated (Fig. 7A and B). Based on these data, protein 2C was used at 3 μM for 30 min at 37°C in subsequent experiments. To verify the specificity of the enzyme activity, an active-site mutant of 2C (K135A) was expressed and purified. It was reported earlier that mutation of the first conserved lysine residue in NTPase motif A (GXXXXGKS) dramatically reduces or abolishes ATPase activity (60, 65), which we could confirm for CVB3 protein 2C (Fig. 7C and D). We next wanted to study whether the antiviral activity of TBZE-029 was due to inhibition of the ATPase activity of 2C. However, even at a concentration as high as 100 μM of TBZE-029, no inhibitory effect on the ATPase activity was noted (Fig. 7C). Likewise, none of the other 2C inhibitors, including GuaHCl (2 mM), were able to inhibit the ATPase activity of 2C.
FIG. 7.
ATPase activity of CVB3 protein 2C and effect of 2C-targeting compounds on enzyme activity. The ATPase activity of CVB3 GST-fused protein 2C was detected using [α-32P]ATP as a substrate. The enzymatic activity was assessed in the presence of different concentrations of protein 2C (A) or as a function of time (B). (C) Generation of an active-site mutant of protein 2C. (D) ATPase activity was assessed in the presence of TBZE-029 and other reported 2C inhibitors at 100 μM (or 2 mM for GuaHCl).
DISCUSSION
Protein 2C is among the most conserved proteins in the picornavirus family. This protein contains three conserved motifs that are characteristic of superfamily III (SFIII) of the NTPase/helicase proteins (30). Motif A (GXXXXGKS) is most likely involved in the binding of the phosphate moieties of the NTP substrate, whereas motif B (two Asp residues preceded by a β-strand) may fix the Mg2+ involved in the cleavage of the NTP γ-phosphate (66). Domain C consists of an Asn preceded by a run of hydrophobic residues. To date, no helicase activity could be demonstrated for picornavirus 2C (57). Helicases are molecular motors that use the energy of ATP hydrolysis to unidirectionally translocate along nucleic acids and separate (unwind) the complementary strands of the nucleic acid duplex (34). In recent years, important progress in determining the structure and function of SFIII helicases has been made (35, 40, 44). In vitro ATPase activity was demonstrated for the purified 2C proteins of poliovirus (45, 57), echovirus (37), human parechovirus (60), and coxsackievirus B3 (this study). Studies have revealed the presence of, apart from 2C's NTPase domain, an N-terminal amphipathic helix (48), a membrane-binding region (22), two RNA-binding domains (58), and a zinc-binding cysteine-rich motif (50). Moreover, 2C was shown to be involved in a multitude of processes required for viral replication, including virus uncoating (41), host cell membrane binding and rearrangement (16, 63, 64), formation of the viral cytoplasmic replication vesicles (1, 16, 22, 62-64), RNA binding and RNA synthesis (4, 5, 7, 71), and possibly encapsidation (67, 68).
Here we describe the antiviral activity of the thiazolobenzimidazole TBZE-029, which we recently reported as an inhibitor of enterovirus replication (19). TBZE-029 inhibited the release of infectious virus progeny in a dose-dependent manner. It blocked viral RNA synthesis without affecting viral polyprotein processing. These findings are corroborated by time of drug addition studies, indicating that TBZE-029 exerts its activity at a stage that coincides with the replication of viral RNA.
To identify the precise target of TBZE-029, drug-resistant CVB3 was generated. Genotyping of this virus led to the identification of three amino acid mutations at positions 224, 227, and 229 in protein 2C that were present in all clones. These residues are located in a region which is just downstream of motif C and have been predicted to form a loop between a beta strand and an alpha helix (64, 66). Reintroduction of these mutations, either alone or combined, into a full-length clone of CVB3 generated seven recombinant clones, which were further characterized. Single amino acid mutations at positions 224 and 227 were not sufficient to confer resistance to TBZE-029, but highly resistant virus was obtained when the amino acid mutation at position 227 was combined with the mutations at either position 224 or 229 or both. Moreover, two transcripts (clones 3 [A229V] and 5 [A224V and A229V]) were dependent on GuaHCl to become infectious upon transfection. Clone 6 (I227V and A229V) showed delayed single-cycle growth kinetics and produced markedly smaller plaques than wild-type virus. In fact, only mutant 5 (exhibiting a GuaHCl dependence phenotype) was able to produce plaques similar to those of wild-type virus.
Interestingly, previous studies with the antiviral compound HBB in echovirus 9 reported that altered sensitivity to this drug mapped, similarly to our findings, to the Ile and Ala residues at positions 227 and 229, respectively (32, 39). In particular, the I227L mutation, either alone or combined with the A229V mutation, conferred resistance to HBB. Akin to the observations in the current study, a single mutation of Ala at position 229, however, led to a noninfectious RNA transcript, which could be rescued by the addition of HBB or GuaHCl (32, 39). Studies of cross-resistance to HBB, using the recombinant TBZE-029-resistant coxsackievirus clones, showed an identical pattern of resistance (Table 2). These observations suggest that TBZE-029 and HBB probably have similar mechanisms of action.
Another benzimidazole derivative, MRL-1237, was also recently reported to select for mutations in poliovirus 2C (61). Of 15 mutants, 14 were shown to carry a single amino acid mutation at position 164, conferring high resistance to both MRL-1237 and guanidine. The other mutant carried a mutation at position 120 but exhibited only weak resistance to MRL-1237 and was not cross-resistant to guanidine. None of the mutations described for MRL-1237 were observed in TBZE-029-resistant coxsackievirus. However, when the constructed coxsackievirus mutants in this study were evaluated for cross-resistance to MRL-1237, a substantial overlap in the resistance profile was noticed. In fact, all the constructed mutants, except for the one carrying a single amino acid mutation at position 224, showed cross-resistance to MRL-1237 (Table 2). This suggests that the antiviral activity of MRL-1237 is also dependent on interactions within this short region in 2C (224-227-229). However, the precise interactions between protein 2C and MRL-1237 must differ from those between 2C and TBZE-029 or HBB, since amino acid 227 has a greater impact on the resistance profile of MRL-1237 than on those of TBZE-029 and HBB.
The most extensively studied inhibitor of 2C is probably GuaHCl (14, 17, 42, 56). Several studies with guanidine-resistant poliovirus (52-54, 66) led to the conclusion that guanidine resistance can be attributed mainly to two mutations in a loop adjacent to motif B, designated “class N” or “class M” mutations. The first class involves a mutated Asn at position 179 and the second class a mutated Met at residue 187. Besides these “main” mutations, guanidine-resistant poliovirus was shown to carry some additional mutations that were located at residue 143, 227, or 233. It should be emphasized that residue 227 is also involved in the resistance of echovirus to HBB (39) and, as reported here, of coxsackievirus to TBZE-029. Shimizu et al. (61) demonstrated that poliovirus carrying either of the guanidine “main” mutations was not only resistant to guanidine but also cross-resistant to MRL-1237.
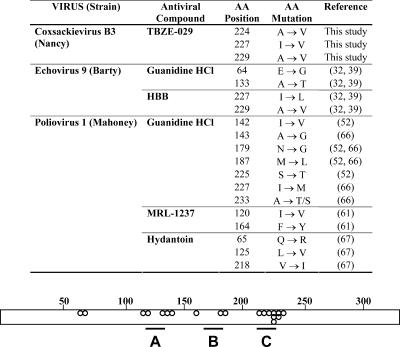
A general important observation is that almost every mutation conferring resistance to any 2C inhibitor reported to date implies almost by definition cross-resistance to GuaHCl, which is not necessarily true the other way around. Similarly, the drug-dependent RNA transcripts, observed when echo- and coxsackievirus 2C proteins were mutated at position 229, became infectious upon transfection only in the presence of GuaHCl. An overview of all reported amino acid mutations involved in resistance to 2C inhibitors reported in this study and the literature to date is presented in Fig. 8. Even though several compounds select for different amino acid mutations at different sites, it is evident that most of the mutations are clustered within each of the three NTPase/helicase domains.
FIG. 8.
Overview of mutations identified in several enteroviruses showing resistance to several 2C inhibitors. The bar represents the 329-amino-acid protein 2C. Dots represent reported amino acid mutations present in virus resistant to 2C inhibitors. A, B, and C represent the conserved NTPase/helicase domains.
In the present study, no inhibition of the in vitro ATPase activity of purified 2C by TBZE-029 was observed. In fact, studies with purified 2C in an in vitro system may not be a valuable model for testing the inhibitory action of drugs, given the complex interactions with membranes and other viral and cellular proteins that have been described for 2C in infected cells (64).
Alternatively, another 2C function may be inhibited. One might speculate that, as the region just outside domain C is a “hot spot” for mutations and as this domain is conserved among the helicase SFIII, the putative helicase function is the target of TBZE-029. Although this is a possible explanation, it should be emphasized again that so far all attempts to demonstrate that purified 2C exerts in vitro helicase activity have failed (57). The CVB3 precursor protein 2BC has previously been shown to homomultimerize (18). Importantly, TBZE-029 did not interfere with 2BC homomultimerization (our unpublished data).
Several questions remain to be addressed. Is TBZE-029 an inhibitor of the enterovirus helicase? What is the molecular interaction between the identified amino acids flanking domain C and TBZE-029? What is the importance of residue 229 since mutation of this residue generates noninfectious RNA transcripts? Unraveling the precise mechanism of action by which TBZE-029 inhibits picornavirus replication may (i) provide insights into the biology of picornavirus replication and (ii) allow the rational design of more-potent analogues as novel inhibitors.
Acknowledgments
This work was supported by the VIZIER integrated project (LSHG-CT-2004-511960) from the European Union 6th PCRDT.
We thank Miette Stuyck, Karen Dalle, Katrin Jäger, and Violaine Lantez for excellent technical assistance. We also thank Arie Gerlof, who kindly provided the Gateway plasmid for His-6-GST fusion.
Footnotes
Published ahead of print on 12 March 2008.
REFERENCES
- 1.Aldabe, R., and L. Carrasco. 1995. Induction of membrane proliferation by poliovirus proteins 2C and 2BC. Biochem. Biophys. Res. Commun. 20664-76. [DOI] [PubMed] [Google Scholar]
- 2.Baltera, R. F., Jr., and D. R. Tershak. 1989. Guanidine-resistant mutants of poliovirus have distinct mutations in peptide 2C. J. Virol. 634441-4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee, R., and A. Dasgupta. 2001. Interaction of picornavirus 2C polypeptide with the viral negative-strand RNA. J. Gen. Virol. 822621-2627. [DOI] [PubMed] [Google Scholar]
- 4.Banerjee, R., A. Echeverri, and A. Dasgupta. 1997. Poliovirus-encoded 2C polypeptide specifically binds to the 3′-terminal sequences of viral negative-strand RNA. J. Virol. 719570-9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banerjee, R., W. Tsai, W. Kim, and A. Dasgupta. 2001. Interaction of poliovirus-encoded 2C/2BC polypeptides with the 3′ terminus negative-strand cloverleaf requires an intact stem-loop b. Virology 28041-51. [DOI] [PubMed] [Google Scholar]
- 6.Banerjee, R., M. K. Weidman, A. Echeverri, P. Kundu, and A. Dasgupta. 2004. Regulation of poliovirus 3C protease by the 2C polypeptide. J. Virol. 789243-9256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barton, D. J., and J. B. Flanegan. 1997. Synchronous replication of poliovirus RNA: initiation of negative-strand RNA synthesis requires the guanidine-inhibited activity of protein 2C. J. Virol. 718482-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bedard, K. M., and B. L. Semler. 2004. Regulation of picornavirus gene expression. Microbes Infect. 6702-713. [DOI] [PubMed] [Google Scholar]
- 9.Bergamini, G., T. Preiss, and M. W. Hentze. 2000. Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA 61781-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berrow, N. S., K. Bussow, B. Coutard, J. Diprose, M. Ekberg, G. E. Folkers, N. Levy, V. Lieu, R. J. Owens, Y. Peleg, C. Pinaglia, S. Quevillon-Cheruel, L. Salim, C. Scheich, R. Vincentelli, and D. Busso. 2006. Recombinant protein expression and solubility screening in Escherichia coli: a comparative study. Acta Crystallogr. Sect. D Biol. Crystallogr. 621218-1226. [DOI] [PubMed] [Google Scholar]
- 11.Bienz, K., D. Egger, and L. Pasamontes. 1987. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology 160220-226. [DOI] [PubMed] [Google Scholar]
- 12.Bienz, K., D. Egger, T. Pfister, and M. Troxler. 1992. Structural and functional characterization of the poliovirus replication complex. J. Virol. 662740-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bienz, K., D. Egger, M. Troxler, and L. Pasamontes. 1990. Structural organization of poliovirus RNA replication is mediated by viral proteins of the P2 genomic region. J. Virol. 641156-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caliguiri, L. A., and I. Tamm. 1968. Action of guanidine on the replication of poliovirus RNA. Virology 35408-417. [DOI] [PubMed] [Google Scholar]
- 15.Chimirri, A., S. Grasso, P. Monforte, A. Rao, M. Zappala, A. M. Monforte, C. Pannecouque, M. Witvrouw, J. Balzarini, and E. De Clercq. 1999. Synthesis and biological activity of novel 1H,3H-thiazolo[3,4-a]benzimidazoles: non-nucleoside human immunodeficiency virus type 1 reverse transcriptase inhibitors. Antivir. Chem. Chemother. 10211-217. [DOI] [PubMed] [Google Scholar]
- 16.Cho, M. W., N. Teterina, D. Egger, K. Bienz, and E. Ehrenfeld. 1994. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology 202129-145. [DOI] [PubMed] [Google Scholar]
- 17.Crowther, D., and J. L. Melnick. 1961. Studies of the inhibitory action of guanidine on poliovirus multiplication in cell cultures. Virology 1565-74. [DOI] [PubMed] [Google Scholar]
- 18.de Jong, A. S., I. W. Schrama, P. H. Willems, J. M. Galama, W. J. Melchers, and F. J. van Kuppeveld. 2002. Multimerization reactions of coxsackievirus proteins 2B, 2C and 2BC: a mammalian two-hybrid analysis. J. Gen. Virol. 83783-793. [DOI] [PubMed] [Google Scholar]
- 19.De Palma, A. M., W. Heggermont, P. Leyssen, G. Purstinger, E. Wimmer, E. De Clercq, A. Rao, A. M. Monforte, A. Chimirri, and J. Neyts. 2007. Anti-enterovirus activity and structure-activity relationship of a series of 2,6-dihalophenyl-substituted 1H,3H-thiazolo[3,4-a]benzimidazoles. Biochem. Biophys. Res. Commun. 353628-632. [DOI] [PubMed] [Google Scholar]
- 20.De Palma, A. M., I. Vliegen, E. De Clercq, and J. Neyts. Selective inhibitors of picornavirus replication. Med. Res. Rev., in press. [DOI] [PubMed]
- 21.Echeverri, A., R. Banerjee, and A. Dasgupta. 1998. Amino-terminal region of poliovirus 2C protein is sufficient for membrane binding. Virus Res. 54217-223. [DOI] [PubMed] [Google Scholar]
- 22.Echeverri, A. C., and A. Dasgupta. 1995. Amino terminal regions of poliovirus 2C protein mediate membrane binding. Virology 208540-553. [DOI] [PubMed] [Google Scholar]
- 23.Egger, D., N. Teterina, E. Ehrenfeld, and K. Bienz. 2000. Formation of the poliovirus replication complex requires coupled viral translation, vesicle production, and viral RNA synthesis. J. Virol. 746570-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eggers, H. J. 1976. Successful treatment of enterovirus-infected mice by 2-(alpha-hydroxybenzyl)-benzimidazole and guanidine. J. Exp. Med. 1431367-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eggers, H. J., and I. Tamm. 1961. Spectrum and characteristics of the virus inhibitory action of 2-(alpha-hydroxybenzyl)-benzimidazole. J. Exp. Med. 113657-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eggers, H. J., and I. Tamm. 1963. Inhibition of enterovirus ribonucleic acid synthesis by 2-(alpha-hydroxy-benzyl)-benzimidazole. Nature 1971327-1328. [Google Scholar]
- 27.Eggers, H. J., and I. Tamm. 1963. Synergistic effect of 2-(alpha-hydroxybenzyl)-benzimidazole and guanidine on picornavirus reproduction. Nature 199513-514. [DOI] [PubMed] [Google Scholar]
- 28.Gorbalenya, A. E., and E. V. Koonin. 1989. Viral proteins containing the purine NTP-binding sequence pattern. Nucleic Acids Res. 178413-8440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorbalenya, A. E., E. V. Koonin, A. P. Donchenko, and V. M. Blinov. 1988. A conserved NTP-motif in putative helicases. Nature 33322. [DOI] [PubMed] [Google Scholar]
- 30.Gorbalenya, A. E., E. V. Koonin, and Y. I. Wolf. 1990. A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett. 262145-148. [DOI] [PubMed] [Google Scholar]
- 31.Guenther, B., R. Onrust, A. Sali, M. O'Donnell, and J. Kuriyan. 1997. Crystal structure of the delta′ subunit of the clamp-loader complex of E. coli DNA polymerase III. Cell 91335-345. [DOI] [PubMed] [Google Scholar]
- 32.Hadaschik, D., M. Klein, H. Zimmermann, H. J. Eggers, and B. Nelsen-Salz. 1999. Dependence of echovirus 9 on the enterovirus RNA replication inhibitor 2-(alpha-hydroxybenzyl)-benzimidazole maps to nonstructural protein 2C. J. Virol. 7310536-10539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwai, M., H. Yoshida, K. Matsuura, T. Fujimoto, H. Shimizu, T. Takizawa, and Y. Nagai. 2006. Molecular epidemiology of echoviruses 11 and 13, based on an environmental surveillance conducted in Toyama Prefecture, 2002-2003. Appl. Environ. Microbiol. 726381-6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iyer, L. M., D. D. Leipe, E. V. Koonin, and L. Aravind. 2004. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 14611-31. [DOI] [PubMed] [Google Scholar]
- 35.James, J. A., A. K. Aggarwal, R. M. Linden, and C. R. Escalante. 2004. Structure of adeno-associated virus type 2 Rep40-ADP complex: insight into nucleotide recognition and catalysis by superfamily 3 helicases. Proc. Natl. Acad. Sci. USA 10112455-12460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirschke, D. L., T. F. Jones, S. C. Buckingham, A. S. Craig, and W. Schaffner. 2002. Outbreak of aseptic meningitis associated with echovirus 13. Pediatr. Infect. Dis. J. 211034-1038. [DOI] [PubMed] [Google Scholar]
- 37.Klein, M., H. J. Eggers, and B. Nelsen-Salz. 1999. Echovirus 9 strain barty non-structural protein 2C has NTPase activity. Virus Res. 65155-160. [DOI] [PubMed] [Google Scholar]
- 38.Klein, M., H. J. Eggers, and B. Nelsen-Salz. 2000. Echovirus-9 protein 2C binds single-stranded RNA unspecifically. J. Gen. Virol. 812481-2484. [DOI] [PubMed] [Google Scholar]
- 39.Klein, M., D. Hadaschik, H. Zimmermann, H. J. Eggers, and B. Nelsen-Salz. 2000. The picornavirus replication inhibitors HBB and guanidine in the echovirus-9 system: the significance of viral protein 2C. J. Gen. Virol. 81895-901. [DOI] [PubMed] [Google Scholar]
- 40.Li, D., R. Zhao, W. Lilyestrom, D. Gai, R. Zhang, J. A. DeCaprio, E. Fanning, A. Jochimiak, G. Szakonyi, and X. S. Chen. 2003. Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen. Nature 423512-518. [DOI] [PubMed] [Google Scholar]
- 41.Li, J. P., and D. Baltimore. 1990. An intragenic revertant of a poliovirus 2C mutant has an uncoating defect. J. Virol. 641102-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loddo, B., W. Ferrari, G. Brotzu, and A. Spanedda. 1962. In vitro inhibition of infectivity of polio viruses by guanidine. Nature 19397-98. [DOI] [PubMed] [Google Scholar]
- 43.Lum, L. C., K. B. Chua, P. C. McMinn, A. Y. Goh, R. Muridan, S. A. Sarji, P. S. Hooi, B. H. Chua, and S. K. Lam. 2002. Echovirus 7 associated encephalomyelitis. J. Clin. Virol. 23153-160. [DOI] [PubMed] [Google Scholar]
- 44.Mancini, E. J., D. E. Kainov, J. M. Grimes, R. Tuma, D. H. Bamford, and D. I. Stuart. 2004. Atomic snapshots of an RNA packaging motor reveal conformational changes linking ATP hydrolysis to RNA translocation. Cell 118743-755. [DOI] [PubMed] [Google Scholar]
- 45.Mirzayan, C., and E. Wimmer. 1994. Biochemical studies on poliovirus polypeptide 2C: evidence for ATPase activity. Virology 199176-187. [DOI] [PubMed] [Google Scholar]
- 46.Padalko, E., D. Nuyens, A. De Palma, E. Verbeken, J. L. Aerts, E. De Clercq, P. Carmeliet, and J. Neyts. 2004. The interferon inducer Ampligen [poly(I)-poly(C12U)] markedly protects mice against coxsackie B3 virus-induced myocarditis. Antimicrob. Agents Chemother. 48267-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patick, A. K. 2006. Rhinovirus chemotherapy. Antivir. Res. 71391-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paul, A. V., A. Molla, and E. Wimmer. 1994. Studies of a putative amphipathic helix in the N-terminus of poliovirus protein 2C. Virology 199188-199. [DOI] [PubMed] [Google Scholar]
- 49.Paul, A. V., J. Peters, J. Mugavero, J. Yin, J. H. van Boom, and E. Wimmer. 2003. Biochemical and genetic studies of the VPg uridylylation reaction catalyzed by the RNA polymerase of poliovirus. J. Virol. 77891-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pfister, T., K. W. Jones, and E. Wimmer. 2000. A cysteine-rich motif in poliovirus protein 2C (ATPase) is involved in RNA replication and binds zinc in vitro. J. Virol. 74334-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pfister, T., and E. Wimmer. 1999. Characterization of the nucleoside triphosphatase activity of poliovirus protein 2C reveals a mechanism by which guanidine inhibits poliovirus replication. J. Biol. Chem. 2746992-7001. [DOI] [PubMed] [Google Scholar]
- 52.Pincus, S. E., D. C. Diamond, E. A. Emini, and E. Wimmer. 1986. Guanidine-selected mutants of poliovirus: mapping of point mutations to polypeptide 2C. J. Virol. 57638-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pincus, S. E., H. Rohl, and E. Wimmer. 1987. Guanidine-dependent mutants of poliovirus: identification of three classes with different growth requirements. Virology 15783-88. [DOI] [PubMed] [Google Scholar]
- 54.Pincus, S. E., and E. Wimmer. 1986. Production of guanidine-resistant and -dependent poliovirus mutants from cloned cDNA: mutations in polypeptide 2C are directly responsible for altered guanidine sensitivity. J. Virol. 60793-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reed, S. E., and H. Muench. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 27493-497. [Google Scholar]
- 56.Rightsel, W. A., J. R. Dice, R. J. McAlpine, E. A. Timm, I. W. McLean, Jr., G. J. Dixon, and F. M. Schabel, Jr. 1961. Antiviral effect of guanidine. Science 134558-559. [DOI] [PubMed] [Google Scholar]
- 57.Rodriguez, P. L., and L. Carrasco. 1993. Poliovirus protein 2C has ATPase and GTPase activities. J. Biol. Chem. 2688105-8110. [PubMed] [Google Scholar]
- 58.Rodriguez, P. L., and L. Carrasco. 1995. Poliovirus protein 2C contains two regions involved in RNA binding activity. J. Biol. Chem. 27010105-10112. [DOI] [PubMed] [Google Scholar]
- 59.Rotbart, H. A. 2002. Treatment of picornavirus infections. Antivir. Res. 5383-98. [DOI] [PubMed] [Google Scholar]
- 60.Samuilova, O., C. Krogerus, I. Fabrichniy, and T. Hyypia. 2006. ATP hydrolysis and AMP kinase activities of nonstructural protein 2C of human parechovirus 1. J. Virol. 801053-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shimizu, H., M. Agoh, Y. Agoh, H. Yoshida, K. Yoshii, T. Yoneyama, A. Hagiwara, and T. Miyamura. 2000. Mutations in the 2C region of poliovirus responsible for altered sensitivity to benzimidazole derivatives. J. Virol. 744146-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suhy, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 748953-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Teterina, N. L., K. Bienz, D. Egger, A. E. Gorbalenya, and E. Ehrenfeld. 1997. Induction of intracellular membrane rearrangements by HAV proteins 2C and 2BC. Virology 23766-77. [DOI] [PubMed] [Google Scholar]
- 64.Teterina, N. L., A. E. Gorbalenya, D. Egger, K. Bienz, and E. Ehrenfeld. 1997. Poliovirus 2C protein determinants of membrane binding and rearrangements in mammalian cells. J. Virol. 718962-8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Teterina, N. L., K. M. Kean, A. E. Gorbalenya, V. I. Agol, and M. Girard. 1992. Analysis of the functional significance of amino acid residues in the putative NTP-binding pattern of the poliovirus 2C protein. J. Gen. Virol. 731977-1986. [DOI] [PubMed] [Google Scholar]
- 66.Tolskaya, E. A., L. I. Romanova, M. S. Kolesnikova, A. P. Gmyl, A. E. Gorbalenya, and V. I. Agol. 1994. Genetic studies on the poliovirus 2C protein, an NTPase. A plausible mechanism of guanidine effect on the 2C function and evidence for the importance of 2C oligomerization. J. Mol. Biol. 2361310-1323. [DOI] [PubMed] [Google Scholar]
- 67.Vance, L. M., N. Moscufo, M. Chow, and B. A. Heinz. 1997. Poliovirus 2C region functions during encapsidation of viral RNA. J. Virol. 718759-8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Verlinden, Y., A. Cuconati, E. Wimmer, and B. Rombaut. 2000. The antiviral compound 5-(3,4-dichlorophenyl) methylhydantoin inhibits the post-synthetic cleavages and the assembly of poliovirus in a cell-free system. Antivir. Res. 4861-69. [DOI] [PubMed] [Google Scholar]
- 69.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wessels, E., D. Duijsings, R. A. Notebaart, W. J. Melchers, and F. J. van Kuppeveld. 2005. A proline-rich region in the coxsackievirus 3A protein is required for the protein to inhibit endoplasmic reticulum-to-Golgi transport. J. Virol. 795163-5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wimmer, E., C. U. Hellen, and X. Cao. 1993. Genetics of poliovirus. Annu. Rev. Genet. 27353-436. [DOI] [PubMed] [Google Scholar]