Abstract
Koi herpesvirus (KHV) is the causative agent of a lethal disease in koi and common carp. In the present study, we describe the cloning of the KHV genome as a stable and infectious bacterial artificial chromosome (BAC) clone that can be used to produce KHV recombinant strains. This goal was achieved by the insertion of a loxP-flanked BAC cassette into the thymidine kinase (TK) locus. This insertion led to a BAC plasmid that was stably maintained in bacteria and was able to regenerate virions when permissive cells were transfected with the plasmid. Reconstituted virions free of the BAC cassette but carrying a disrupted TK locus (the FL BAC-excised strain) were produced by the transfection of Cre recombinase-expressing cells with the BAC. Similarly, virions with a wild-type revertant TK sequence (the FL BAC revertant strain) were produced by the cotransfection of cells with the BAC and a DNA fragment encoding the wild-type TK sequence. Reconstituted recombinant viruses were compared to the wild-type parental virus in vitro and in vivo. The FL BAC revertant strain and the FL BAC-excised strain replicated comparably to the parental FL strain. The FL BAC revertant strain induced KHV infection in koi carp that was indistinguishable from that induced by the parental strain, while the FL BAC-excised strain exhibited a partially attenuated phenotype. Finally, the usefulness of the KHV BAC for recombination studies was demonstrated by the production of an ORF16-deleted strain by using prokaryotic recombination technology. The availability of the KHV BAC is an important advance that will allow the study of viral genes involved in KHV pathogenesis, as well as the production of attenuated recombinant candidate vaccines.
Common carp (Cyprinus carpio carpio) is the most widely cultivated fish for human consumption mainly in Asia, Europe, and the Middle East (3). In contrast, the koi (Cyprinus carpio koi) subspecies is cultivated as an expensive, beautiful, and colorful pet fish for personal pleasure or competitive showing, especially in Japan but also worldwide (3). Recently, koi herpesvirus (KHV) was identified as the cause of mass mortality among koi and common carp in Israel, the United States, and Germany (7, 21, 22). The intensive culture of common carp, koi shows, and international trading have unfortunately contributed to the rapid global spread of highly contagious and extremely virulent KHV disease (19, 27, 35). Since its emergence, KHV has caused severe financial and economic losses in both koi and common carp culture industries worldwide (20, 34).
The genome of KHV comprises a linear double-stranded DNA sequence of ∼295 kb (2, 24), similar to that of cyprinid herpesvirus 1 (41) but larger than those of other Herpesviridae members, which generally range from 125 to 240 kb. The sequence of the KHV genome revealed a significant number of original DNA sequences with no homology to any other known viral sequences. Moreover, it contains highly divergent DNA sequences encoding polypeptides which resemble those of several other double-stranded DNA viruses, such as other herpesviruses, poxviruses, iridoviruses, and other large DNA viruses (24, 41).
Since the first isolation of KHV, an increasing number of studies have been devoted to the virus. They have reported data related to viral gene content (2, 4, 13, 14, 24-26, 41), pathogenesis (12, 13, 33, 38), epidemiology (24, 32), the diagnosis of KHV infection (1, 4, 14, 15, 17, 18, 37), and control (31, 34). However, no information on the roles of individual KHV genes in the biology of KHV infection or in pathogenesis has been published to date. Two reasons can explain this lacuna. Firstly, the KHV genome sequence has been published only very recently (2). Secondly, prolonged KHV cultivation in vitro leads to the spontaneous attenuation of the virus, making the production of KHV recombinants by classical homologous recombination in eukaryotic cells difficult (34).
Recently, the manipulation of large herpesvirus genomes has been facilitated by the use of bacterial artificial chromosome (BAC) vectors (6, 40). These vectors allow the stable maintenance and efficient mutagenesis of the viral genome in Escherichia coli, followed by the reconstitution of progeny virions by the transfection of permissive eukaryotic cells with the BAC plasmid. Several herpesviruses have been successfully propagated as infectious BAC clones. The 235-kb genome of human cytomegalovirus is to date the largest herpesvirus genome which has been BAC cloned (6).
BAC cloning is an obvious approach to avoid the problems in the production of KHV recombinants described above. However, the large size of the KHV genome and its abundant repetitive sequence content (2) are two intrinsic features of KHV that may render its BAC cloning difficult.
In the present study, we describe for the first time the cloning of the KHV genome as a stable and infectious BAC clone. Several recombinant strains were derived from the BAC clone by using homologous recombination in eukaryotic cells and prokaryotic recombination technology. The comparison of these recombinant strains in vivo revealed that thymidine kinase (TK) gene disruption led to the partial attenuation of KHV and that the deletion of ORF16, encoding a putative G protein-coupled receptor (GPCR), did not affect KHV virulence. The availability of the KHV BAC is an important advance that will allow the study of viral genes involved in KHV pathogenesis, as well as the production of attenuated recombinant candidate vaccines.
MATERIALS AND METHODS
Cells and virus.
Cyprinus carpio brain (CCB) cells (30) were cultured in minimum essential medium (Invitrogen) containing 4.5 g of glucose (d-glucose monohydrate; Merck)/liter and 10% fetal calf serum (FCS). Cells were cultured at 25°C in a humid atmosphere containing 5% CO2. The KHV FL strain was isolated from a kidney of a fish that died from KHV infection (CER, Marloie, Belgium). FL stands for François Lieffrig, who isolated the strain.
BAC cloning of KHV.
Firstly, a 1,137-bp DNA fragment corresponding to the TK open reading frame (ORF; ORF55) and ORF56 of the KHV genome was amplified by PCR using KHV FL DNA as a template. The following primers were used for the amplification: the forward primer TKfw (5′-ATGGCTATGCTGGAACTGGTG-3′) and the reverse primer TKrev (5′-CTCAACAGGGAAGAGTGGCG-3′), corresponding to nucleotides 1 to 21 of the KHV TK ORF and nucleotides 279 to 297 of ORF56 (GenBank accession no. for the KHV genome, DQ177346), respectively. The amplification product was sequenced and TA cloned into the pGEM-T Easy vector (Promega), resulting in pGEMT-TK (Fig. 1A). A BAC cassette was released by PmeI digestion of the pBeloBACModified-EGFPNeo vector (11) and then ligated into the RsrII site of the pGEMT-TK vector, resulting in the pGEMT-TKBAC vector (Fig. 1A), in which the BAC cassette is flanked by KHV sequences. These KHV homologous sequences were exploited to produce the KHV FL BAC strain by homologous recombination in eukaryotic cells (Fig. 1B). Briefly, freshly seeded CCB cells were infected with KHV at a multiplicity of infection (MOI) of 0.5 PFU/cell. After an incubation period of 2 h, cells were transfected with circular pGEMT-TKBAC by using Lipofectamine Plus (Invitrogen). Four days postinfection (pi), cell supernatant was harvested and inoculated onto confluent CCB cell monolayers (106 cells per 9.5 cm2) in the presence of G418 (final concentration of 500 μg/ml). This step was repeated three times, leading to infected cultures containing predominantly the KHV FL BAC recombinant strain. This viral preparation was inoculated onto freshly seeded CCB cells at a MOI of 1 PFU/cell. The circularized form of the viral BAC recombinant genome was extracted 20 h pi as described previously (29), and 2 μg of DNA was introduced into E. coli DH10B cells (Invitrogen) by electroporation (at 2,250 V, 132 Ω, and 40 μF) as described elsewhere (36). Electroporated cells were plated immediately onto solid-Luria-Bertani medium plates supplemented with chloramphenicol (17 μg/ml). Note that it is crucial at this stage to avoid liquid preculture in order to avoid the preferential growing of bacteria containing incomplete KHV BAC plasmids.
FIG. 1.
Schematic representation of the strategy used to produce the infectious KHV FL BAC plasmid. (A) The genome of the KHV FL strain, flanked by two TRs (the left TR [LTR] and the right TR [RTR]), is shown at the top. A loxP-flanked BAC cassette was inserted into the RsrII sites of the TK ORF of the pGEMT-TK vector, resulting in pGEMT-TKBAC. (B) Flow chart of steps performed to produce the KHV FL BAC plasmid, to control its infectivity, and to demonstrate the possibility of removing the loxP-flanked BAC cassette from the genome of reconstituted virus or to produce a wild-type revertant strain derived from the FL BAC plasmid.
Reconstitution of infectious virus from the KHV FL BAC plasmid.
Permissive CCB cells were transfected with the FL BAC plasmid by using Lipofectamine Plus (Invitrogen) in order to produce the FL BAC recovered strain. To produce a wild-type revertant strain derived from the BAC, CCB cells were cotransfected with the FL BAC plasmid and the pGEMT-TK vector (molar ratio, 1:75). Seven days posttransfection, viral plaques negative for enhanced green fluorescent protein (EGFP) expression were picked and enriched by three successive rounds of plaque purification. Similarly, to reconstitute virions with the BAC cassette excised from the viral genome, CCB cells were cotransfected with the FL BAC plasmid and the pEFIN3-NLS-Cre vector, encoding Cre recombinase fused to a nuclear localization signal (16) (molar ratio, 1:70).
Southern blotting.
Southern blot analysis was performed as described previously (28). Several probes were used. The TK probe was produced by PCR using the TKfw and TKrev primers described above and the KHV FL genome as a template. The terminal repeat (TR) probe corresponded to nucleotides 3817 to 4228 of the left TR and nucleotides 276494 to 276905 of the right TR of the KHV genome. The BAC probe was released from the pBeloBACModified-EGFPNeo vector by PmeI digestion. The ORF16 probe was produced by PCR using ORF16fw and ORF16rev primers corresponding to nucleotides 1 to 50 and 1027 to 1077 of KHV ORF16, respectively.
Indirect immunofluorescence staining.
CCB cells were fixed and permeabilized with acetone-ethanol (50:50, vol/vol) for 10 min at −20°C. Immunofluorescence staining (incubation and washes) was performed in phosphate-buffered saline containing 10% FCS. Samples were incubated at 25°C for 45 min with mouse monoclonal antibody 8G12 raised against an unidentified KHV antigen expressed in the nuclei of infected cells. After three washes, samples were incubated at 25°C for 30 min with Alexa Fluor 568-conjugated goat anti-mouse immunoglobulin G (heavy and light chains [GAM 568; 2 μg/μl; Molecular Probes]) as the secondary conjugate.
Microscopy analysis.
Epifluorescence microscopy analysis was performed with a DMIRBE microscope (Leica) equipped with a DC 300F charge-coupled device camera (Leica) as described previously (39).
Multistep growth curves.
Triplicate cultures of CCB cells were infected at a MOI of 0.5 PFU/cell. After an incubation period of 2 h, cells were washed with phosphate-buffered saline and then overlaid with Dulbecco's modified essential medium (Invitrogen) containing 4.5 g of glucose/liter and 10% FCS. The supernatants of infected cultures were harvested at successive intervals after infection, and the amount of infectious virus was determined by plaque assays with CCB cells as described previously (9).
Production of the KHV FL BAC recombinant plasmid by galK positive selection of bacteria.
A KHV FL BAC recombinant plasmid with the deletion of ORF16 (encoding a putative GPCR) was produced using galK positive selection of bacteria as previously described (42). The recombination fragment consisted of a galactokinase gene (galK) flanked by 50-bp sequences corresponding to the beginning and the end of KHV ORF16. This fragment was produced by PCR using the pgalK vector (42) as a template, the forward primer 16galfw (5′-ATGAAACCTCTGGGTCTTTTTGTTTCTGTGCTCGGGCTGCTTGCCCTGTCCCTGTTGACAATTAATCATCGGCA-3′), and the reverse primer 16galrev (5′-TCATAGGACGCCATCGGTTGAGTTCGCTGCGGCTGCGACTCCCAGTCCTCTCAGCACTGTCCTGCTCCTT-3′). Primer 16galfw consisted of nucleotides 1 to 50 of KHV ORF16 and nucleotides 1 to 24 of the pgalK vector (42). The reverse primer 16galrev consisted of nucleotides 1027 to 1077 of KHV ORF16 and nucleotides 1212 to 1231 of the pgalK vector (42).
Induction of KHV disease in fish.
Specific-pathogen-free koi carp, with an average weight of 7 g, were kept in 60-liter tanks at 24°C. Several groups of fish, each comprising 10 carp (with the exception of mock-infected groups, which consisted of 13 carp), were kept in separate tanks. Koi carp were infected by intraperitoneal (IP) injection with 0.1 ml containing 3 × 102 PFU. The viral inoculums were titrated before inoculation and back titrated after inoculation to ensure that the doses were equivalent among groups. The control group (mock infected) was injected with culture medium under the same conditions. Fishes were examined daily for clinical signs of KHV disease, and dead fishes were removed. The animal study was accredited by the local ethics committee of the University of Liège (Belgium).
Detection of KHV genome by PCR.
DNA was extracted from tissues of fish by using the QIAamp DNA mini kit (Qiagen). PCR amplification was performed using 25 ng of total DNA as a template and the TKfw-TKrev and ORF16fw-ORF16rev primer pairs described above.
RESULTS
Cloning of the KHV genome in E. coli.
The goal of the present study was to clone the genome of KHV as a stable and infectious BAC plasmid. When we started this project, very few sequences from KHV were available. The TK locus was one of the few KHV genes to have been sequenced. This locus was selected for the insertion of the BAC cassette, as TKs encoded by herpesviruses and poxviruses have been shown previously to be dispensable for viral growth in vitro (8a, 30a).
The strategy depicted in Fig. 1 was used for the BAC cloning of KHV. This approach required as a first step the production of a recombinant strain called KHV FL BAC. The molecular structure of this strain was confirmed by a combined SacI restriction endonuclease and Southern blotting approach (Fig. 2). In the parental FL strain, the TK ORF was contained in a DNA fragment of approximately 5.2 kb. In the FL BAC strain, as a consequence of the BAC cassette insertion into the TK locus, the TK sequence was distributed into two fragments of approximately 5.3 and 9.1 kb (Fig. 2). Sequencing of the regions used to target homologous recombination confirmed that the FL BAC strain had the correct molecular structure (data not shown).
FIG. 2.
Structural analysis of the KHV FL BAC plasmid and derived strains. (A) Schematic representation of some of the fragments generated by SacI enzymatic restriction. The genomes of the KHV FL and KHV FL BAC revertant strains are shown at the top. TK, BAC, and TR probes are indicated by bold horizontal lines. Fragment sizes in kilobases are indicated. Note that this cartoon is not drawn to scale. LTR, left TR; RTR, right TR. (B) The KHV FL BAC plasmid and the genomes of the KHV FL, FL BAC, FL BAC recovered, FL BAC-excised, and FL BAC revertant strains were analyzed by SacI restriction (agarose gel, first panel) and further tested by Southern blotting using probes corresponding to the TK ORF (second panel), the BAC cassette (third panel), or the TRs (fourth panel). Black and white arrowheads and open arrowheads indicate restriction fragments containing the TK ORF and the BAC cassette, respectively. Gray arrowheads indicate restriction fragments hybridizing with the TR probe. Marker sizes (MS) are indicated on the left.
Next, we tried to clone circular intermediates of the FL BAC genome into bacteria by classical approaches that we have used successfully in the past for the BAC cloning of other herpesviruses (9, 11, 16). Surprisingly, despite the screening of more than 500 independent clones, we were unable to select a single clone carrying the FL BAC genome. The BAC plasmids obtained generated heterogeneous restriction profiles with only a few bands corresponding to the expected restriction profile (data not shown). Due to the large size of the KHV genome, one may postulate that bacteria carrying incomplete KHV BAC plasmids may have a selective advantage over bacteria carrying a full-length KHV BAC clone and, consequently, that the liquid culture of transformed bacteria (performed before the plating of the bacteria onto solid medium) may lead to the selection of bacteria with BAC plasmids comprising only part of the KHV genome. To test this hypothesis, E. coli cells were plated immediately onto solid Luria-Bertani medium after electroporation. This approach led to BAC plasmids comprising most of the restriction fragments found in the FL BAC strain genome. However, only 1 to 2% of the clones exhibited a restriction profile comparable to that of the FL BAC strain genome. One of these correct clones was characterized by a combined SacI restriction endonuclease and Southern blotting approach (Fig. 2). Due to the circularization of the genome in bacteria, the two bands encompassing the extremities of the left and the right terminal repeats (7.1 and 14.1 kb) present in the FL and FL BAC strains were missing in the FL BAC plasmid (Fig. 2). However, the restriction profile of the FL BAC plasmid did not reveal the expected fused fragment of 21.2 kb (Fig. 2). This observation suggested that the BAC contains a single copy of the terminal repeat. To test this hypothesis, SacI profiles were analyzed by Southern blotting using the TR probe. This analysis revealed the presence of two bands (7.1 and 11.3 kb) in the FL and FL BAC strain profiles and only a single band (11.3 kb) in the FL BAC plasmid profile (Fig. 2). These results demonstrate that the FL BAC plasmid contains only a single copy of the terminal repeat.
Stability of the KHV genome in E. coli.
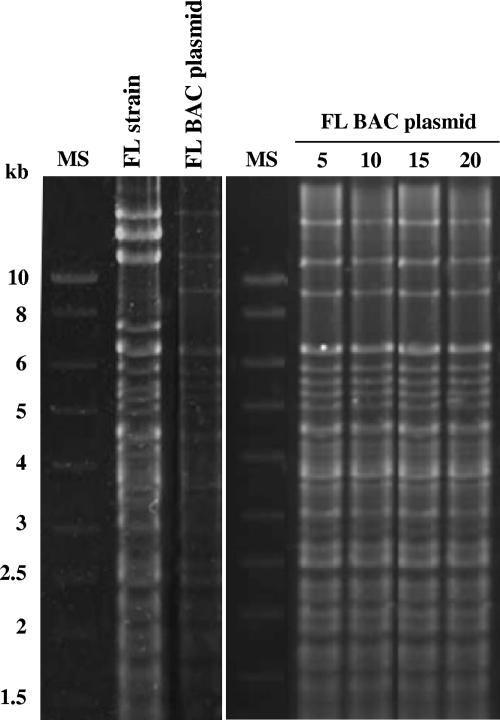
BAC plasmids are usually propagated in bacteria carrying a recA mutation that minimizes recombination. However, the large size and the complex structure of the KHV genome may lead to relative instability of the FL BAC plasmid (25). To assess the stability of the KHV genome as a BAC, bacteria containing the FL BAC plasmid were serially cultured for 20 consecutive days (about 130 generations). After various periods of culture, the BAC plasmids were isolated and characterized by SacI endonuclease digestion (Fig. 3). No difference among plasmids grown for various periods of time was observed, demonstrating a high level of stability of the KHV genome in E. coli.
FIG. 3.
Stability of the FL BAC plasmid in E. coli. DH10B cells containing the FL BAC plasmid were passaged every day at a ratio of 1:100 (vol/vol) for 20 consecutive days. On the indicated days, BAC DNA from the culture was prepared. Finally, BAC DNA samples collected at various intervals were compared with parental FL strain and FL BAC strain DNA by SacI digestion and agarose gel electrophoresis. Marker sizes (MS) are indicated on the left.
Reconstitution of infectious virus from the FL BAC plasmid.
The usefulness of a herpesvirus BAC clone requires the ability to reconstitute infectious particles from the BAC plasmid. Consequently, we tested whether infectious particles could be produced by the transfection of CCB cells with the FL BAC plasmid (Fig. 1B). Six days posttransfection, viral syncytia expressing EGFP were detected. SacI restriction analysis of the DNA of reconstituted virus (the FL BAC recovered strain) revealed a restriction profile identical to the pattern observed for the FL BAC strain (Fig. 2). These data demonstrate that the BAC is able to regenerate the entire genome and infectious particles even if it includes only a single terminal repeat. To excise the BAC cassette from the genomes of reconstituted virions, CCB cells were cotransfected with the FL BAC plasmid and a Cre recombinase-expressing vector (Fig. 1B). The deletion of the BAC cassette was monitored by the disappearance of EGFP fluorescence and by a combined restriction endonuclease and Southern blotting approach (Fig. 2). The cre-loxP-mediated excision of the BAC cassette left a sequence of 172 bp in the TK ORF, leading to a SacI restriction fragment slightly larger than the corresponding wild-type fragment (Fig. 2). The 172-bp sequence consists of one loxP site (34 bp) and the sequences of the BAC cassette upstream (126 bp) and downstream (12 bp) of the loxP sites. Due to this insertion of a 172-bp foreign sequence into the TK ORF, the FL BAC-excised strain expressed a truncated form of TK corresponding to the first 185 amino acids (aa) of the wild-type protein (217 residues). Finally, to generate a revertant strain, CCB cells were cotransfected with the FL BAC plasmid and the pGEMT-TK vector (Fig. 1B). A revertant recombinant was selected on the basis of the nonexpression of EGFP. Restriction analysis revealed a profile identical to the pattern observed for the parental wild-type FL strain (Fig. 2).
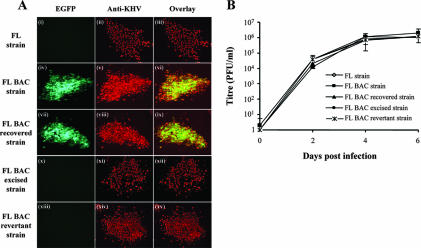
Additional characterization of FL BAC-derived strains in cell cultures was performed. Firstly, microscopic examination of immunostained viral syncytia did not reveal differences among recombinants (Fig. 4A). Secondly, in order to investigate the putative effects of the recombination processes on viral growth in vitro, all recombinant strains were compared using a multistep growth assay (Fig. 4B). All viruses tested exhibited similar growth curves (P ≤ 0.05), leading to the conclusion that TK disruption does not affect KHV replication in vitro and that the KHV genome can support a large insertion (of at least 9.2 kb) despite its large size.
FIG. 4.
Characterization of KHV strains derived from the FL BAC plasmid. (A) Epifluorescence analysis of KHV syncytia. CCB cells were infected (MOI of 0.1 PFU/cell) with FL, FL BAC, FL BAC recovered, FL BAC-excised, and FL BAC revertant strains and were overlaid with Dulbecco's modified essential medium containing 10% FCS and 0.6% (wt/vol) carboxymethyl cellulose (Sigma) to obtain isolated syncytia. Seven days pi, syncytia were revealed by indirect immunofluorescent staining using monoclonal antibody 8G12 and GAM 568 as the primary and secondary antibodies, respectively. The three horizontal panels in each set represent analyses of the same syncytium. Panels i, iv, vii, x, and xiii and panels ii, v, viii, xi, and xiv were analyzed for EGFP and GAM 568 fluorescent emissions, respectively. The merged EGFP and Alexa signals are shown in panels iii, vi, ix, xii, and xv. The side of each panel corresponds to 10 μm of the specimen. (B) Replication kinetics of KHV recombinant strains were compared with those of the parental KHV FL strain as described in Materials and Methods. The data presented are the means ± standard errors of triplicate measurements.
Production of an FL BAC recombinant plasmid by mutagenesis in bacteria with galK positive selection.
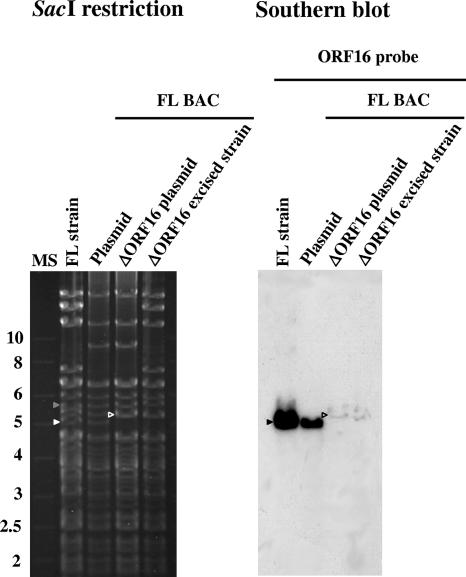
The usefulness of a BAC clone for recombination studies relies on the possibility to use it for the production of recombinants by prokaryotic mutagenesis methods. To test the usefulness of the FL BAC clone, we produced a KHV recombinant strain with the deletion of ORF16 (encoding a putative GPCR) by using galK positive selection of bacteria. The molecular structure of the recombinant plasmid was confirmed by a combined SacI restriction endonuclease and Southern blotting approach (Fig. 5). In the parental FL strain and in the KHV FL BAC plasmid, ORF16 was contained in a DNA fragment of approximately 4.8 kb, whereas in the KHV FL BAC ΔORF16 plasmid, the corresponding fragment had a size of approximately 5 kb due to the deletion of most of ORF16 and the insertion of the galK cassette. This band encompassing the galK cassette is slightly visible on the Southern blot due to the short ORF16 nucleotide sequence left after deletion. Next, to reconstitute virions and to excise the BAC cassette from the genome of the recombinant plasmid, CCB cells were cotransfected with the KHV FL BAC ΔORF16 plasmid and a Cre recombinase-expressing plasmid. The deletion of the BAC cassette was monitored by the disappearance of EGFP fluorescence (data not shown) and by a combined restriction endonuclease and Southern blotting approach (Fig. 5). As described earlier, the cre-loxP-mediated deletion of the BAC cassette leaves a sequence of 172 bp disrupting the TK ORF. Consequently, the FL BAC ΔORF16-excised strain has a disrupted TK locus and a deletion of ORF16.
FIG. 5.
Structural analysis of the KHV FL BAC galK recombinant plasmid. The KHV FL BAC plasmid, the derived FL BAC ΔORF16 plasmid, and the genome of the KHV FL BAC ΔORF16-excised strain were analyzed by SacI restriction (agarose gel) and further tested by Southern blotting using a probe corresponding to ORF16. The KHV FL strain was used as a control. White and black arrowheads and open arrowheads indicate restriction fragments containing ORF16 and the galK cassette, respectively. The gray arrowhead indicates a restriction fragment containing the TK ORF. Marker sizes (MS) in kilobases are indicated on the left.
Pathogenicities of FL BAC-derived strains in koi carp.
The strains derived from the FL BAC plasmid described above facilitated the testing of the effect of TK disruption (in FL BAC-excised and FL BAC revertant strains) and the effect of TK disruption and ORF16 deletion (in the FL BAC ΔORF16-excised strain) on the virulence of KHV. To address the effect of TK disruption on KHV virulence, the parental FL, FL BAC excision, and FL BAC revertant strains were compared by IP inoculation of naïve koi carp (Fig. 6A). The parental FL strain induced all the clinical signs associated with KHV disease, including apathy, the folding of the dorsal fin, increased mucus secretions, suffocation, erratic swimming, and the loss of equilibrium. The FL strain induced a mortality rate of 80%. At necropsy, the discoloration of gill filaments, herpetic skin lesions, and necrotic nephritis were observed for most fishes. In comparison to the FL parental strain, the FL BAC-excised strain exhibited a partially attenuated phenotype characterized by the production of similar clinical signs and lesions but with reduced intensities. Consistent with the attenuation observed, the mortality rate of fishes infected with the FL BAC-excised strain was reduced to 40%. Importantly, the virulence of the FL BAC revertant strain was similar to that of the parental FL strain.
FIG. 6.
Cumulative survival rates of carp infected with FL BAC plasmid-derived strains. (A) On day 0, four groups, each consisting of 10 koi carp (with the exception of mock-infected groups, consisting of 13 carp), were inoculated by IP injection with mock-infected culture medium and culture medium containing 3 × 102 PFU of FL, FL BAC-excised, and FL BAC revertant strains. On day 32 pi, surviving fishes were challenged by IP injection with the parental FL strain. (B) On day 0, four groups, each consisting of 10 koi carp (with the exception of mock-infected groups, consisting of 13 carp), were inoculated by IP injection with mock-infected culture medium and culture medium containing 3 × 102 PFU of FL, FL BAC-excised, and FL BAC ΔORF16-excised strains. On day 27 pi, surviving fishes were challenged by IP injection with the parental FL strain. Percentages of surviving carp are expressed according to days pi. The results presented are representative of three independent experiments.
The effect of TK disruption and ORF16 deletion was assessed in the same way, by IP inoculation of naïve koi carp. The parental FL strain and the FL BAC-excised strain were used as controls (Fig. 6B). Fishes infected with the parental FL strain developed KHV disease as described above (80% mortality). In comparison to the parental strain, the FL BAC excision and FL BAC ΔORF16-excised strains exhibited partially attenuated phenotypes, inducing 30 and 40% mortality, respectively. This result suggests that ORF16, encoding a putative GPCR, does not contribute to KHV virulence significantly.
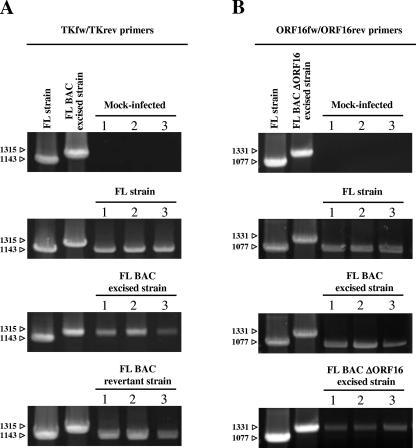
To control the infection of all groups of fish with the correct viral strain and to exclude any possibility of virus spread among tanks, PCR assays were performed on three randomly selected dead fishes from each infected group and three mock-infected fishes randomly selected before the challenge (Fig. 7). PCRs performed with the TKfw/TKrev (Fig. 7A) or ORF16fw/ORF16rev (Fig. 7B) primers confirmed that all samples from infected groups contained the KHV genome, while the sizes of the amplicons excluded the possibility of viral spread among the groups of fishes.
FIG. 7.
PCR detection and characterization of KHV genomes recovered from infected dead carp. (A) DNA was extracted from the intestines of three mock-infected carp (selected randomly before the challenge) and from three dead carp from each of the groups infected with the FL, FL BAC-excised, and FL BAC revertant strains. PCRs were performed with the TKfw/TKrev pair of primers. FL strain DNA and FL BAC-excised strain DNA were used as controls. (B) DNA was extracted from the intestines of mock-infected carp and from dead carp infected with the FL, FL BAC-excised, and FL BAC ΔORF16-excised strains. PCRs were performed with the ORF16fw/ORF16rev pair of primers. FL strain DNA and FL BAC ΔORF16-excised strain DNA were used as controls. The images are photographs of agarose gels. Numbers on the left of each gel are marker sizes.
Finally, on day 32 (Fig. 6A) or day 27 (Fig. 6B) pi, the fishes that survived the primary inoculation were challenged by IP injection with the parental FL strain. Fishes were monitored until day 47. Independently of the strain used for the primary infection, none of the fishes died after the challenge or exhibited clinical signs of disease. In contrast, the challenge of the mock-infected control group led to mortality rates of 60% (Fig. 6A) and 70% (Fig. 6B) by day 15 postchallenge.
DISCUSSION
KHV is the etiological agent of an emerging disease which is highly contagious and extremely virulent and has a high mortality rate (25). Since the discovery of KHV in 1996, an increasing number of studies have been devoted to KHV. However, to date no information on the roles of individual KHV genes in the biology of KHV infection or in pathogenesis has been published. Similarly, there is a lack of safe and efficacious attenuated recombinant vaccines for the control of KHV disease. These lacunas are a consequence of the difficulty of generating KHV recombinant viruses by classical homologous recombination in eukaryotic cells. This problem can be circumvented by the use of BAC cloning technology (6, 40).
In the present study, we describe for the first time the cloning of the KHV genome as a stable and infectious BAC clone. The KHV BAC clone had several interesting features: (i) it was stable when propagated in bacteria, even over long periods of culture corresponding to approximately 130 generations; (ii) it was infectious, as demonstrated by its ability to generate infectious virions after the transfection of permissive cells; (iii) the BAC cassette could be excised from the genome of reconstituted virus; (iv) the insertion of a large DNA sequence into the KHV genome did not affect the ability of KHV to replicate in vitro; (v) importantly, the replication of the FL BAC revertant strain was comparable to that of the FL strain, and the FL BAC revertant strain induced KHV disease in koi carp that was indistinguishable from that induced by the virulent parental strain; and (vi) finally, the usefulness of the KHV BAC clone for recombination studies was demonstrated by the production of an ORF16-deleted strain by prokaryotic recombination technology.
Even if the primary goal of the present study was not to investigate the role of KHV TK in pathogenesis, the recombinants derived from the FL BAC clone allowed us to do so. The FL BAC-excised strain encoding a truncated form of TK exhibited a partially attenuated phenotype in carp (Fig. 6). Four hypotheses may explain the partial attenuation observed. A first hypothesis may be the existence of a KHV enzyme that may partially compensate for TK gene deletion. Viral and cellular TKs have been classified into two types which differ in several respects (5). Type I TKs have higher molecular masses, typically around 40 kDa, and are active as homodimers. This subfamily contains herpesvirus TKs (with the exception of KHV TK) and also human mitochondrial TK. The herpes simplex virus type 1 TK is the viral prototype of this group. It is a multifunctional enzyme that possesses kinase activities normally performed by three separate cellular enzymes. It phosphorylates deoxythymidine and deoxyuridine, as does human TK, and deoxycytidine, as does human deoxycytidine kinase, and acts as a thymidylate kinase, as does human TMP kinase (TMPK) (8). TKs of type II include those from Poxviridae such as vaccinia virus and variola virus, as well as the human cytosolic TK. Type II TKs have smaller polypeptide chains than type I TKs, being ∼25 kDa, but form homotetramers. Moreover, type II TKs have much narrower substrate specificities than type I TKs and phosphorylate only deoxyuridine and/or deoxythymidine. Based on the relatively small size and the nucleotide binding motif of KHV TK, it can be postulated that this TK belongs to type II (10). In poxviruses, the narrower substrate specificities of type II TKs are compensated for by a TMPK gene. Interestingly, the recent sequencing of the KHV genome has revealed the presence of a TMPK ORF (ORF140) (2). It is attractive to speculate that the encoded enzyme may at least partially compensate for the deletion of the KHV TK gene. In support of this hypothesis, it has been shown previously that the replacement of the herpes simplex virus type 1 TK ORF by a human TMPK gene renders the recombinant virus partially competent for replication in mouse sensory ganglia and reactivation from latency upon explant (8). Further studies are required to determine KHV TK and TMPK enzymatic activities and to determine how these enzymes contribute to the pathogenesis in the natural host.
Secondly, the partial attenuation observed with the FL BAC-excised strain may result from residual TK activity expressed by the truncated protein encoded by the FL BAC-excised strain. This hypothesis is very unlikely. Indeed, several studies of herpesviruses and poxviruses have demonstrated previously that the C-terminal region of TK is essential for its activity (23). For example, it has been demonstrated previously that the last 10 residues of the 607-aa-long Epstein-Barr virus TK are essential for its activity (23). In comparison to Epstein-Barr virus TK, KHV TK is rather small, consisting of only 217 aa, among which only the first 185 residues are expressed by the FL BAC-excised strain. A third hypothesis to explain the partial attenuation observed with the FL BAC-excised strain may be that the removal of KHV TK function readily results in a partial-attenuation phenotype in the absence of functional complementation from another virus gene. Finally, a fourth hypothesis may be that host TK may partially replace the eliminated KHV TK.
The usefulness of the KHV BAC clone for recombination studies was demonstrated by the production of an ORF16-deleted strain by using prokaryotic recombination technology. In vivo, the strain induced a mortality rate comparable to that induced by the FL BAC-excised strain, suggesting that ORF16 does not contribute significantly to KHV virulence under the conditions used (Fig. 6B).
In conclusion, this study is the first to report the BAC cloning of a herpesvirus genome as large as that of KHV. The availability of a KHV BAC is an important advance that will allow the study of viral genes involved in KHV pathogenesis, as well as the production of safe and efficacious multiattenuated recombinant candidate vaccines to control KHV infection.
Acknowledgments
The CCB cell line developed by M. Neukirch was obtained through the courtesy of D. Steinhagen. L. Gillet and B. Dewals are postdoctoral researchers of the Fond National de la Recherche Scientifique. V. Stalin Raj is a postdoctoral fellow of the University of Liège.
This work was supported by a grant from the University of Liège (Crédit d'Impulsion).
Footnotes
Published ahead of print on 12 March 2008.
REFERENCES
- 1.Adkison, M. A., O. Gilad, and R. P. Hedrick. 2005. An enzyme linked immunosorbent assay (ELISA) for detection of antibodies to the koi herpesvirus (KHV) in the serum of koi Cyprinus carpio. Fish Pathol. 4053-62. [Google Scholar]
- 2.Aoki, T., I. Hirono, K. Kurokawa, H. Fukuda, R. Nahary, A. Eldar, A. J. Davison, T. B. Waltzek, H. Bercovier, and R. P. Hedrick. 2007. Genome sequences of three koi herpesvirus isolates representing the expanding distribution of an emerging disease threatening koi and common carp worldwide. J. Virol. 815058-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balon, E. K. 1995. Origin and domestication of the wild carp, Cyprinus carpio: from Roman gourmets to the swimming flowers. Aquaculture 1293-48. [Google Scholar]
- 4.Bercovier, H., Y. Fishman, R. Nahary, S. Sinai, A. Zlotkin, M. Eyngor, O. Gilad, A. Eldar, and R. P. Hedrick. 2005. Cloning of the koi herpesvirus (KHV) gene encoding thymidine kinase and its use for a highly sensitive PCR based diagnosis. BMC Microbiol. 513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black, M. E., and D. E. Hruby. 1990. Quaternary structure of vaccinia virus thymidine kinase. Biochem. Biophys. Res. Commun. 1691080-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borst, E. M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 738320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bretzinger, A., T. Fischer-Scherl, R. Oumouma, R. Hoffmann, and U. Truyen. 1999. Mass mortalities in koi, Cyprinus carpio, associated with gill and skin disease. Bull. Eur. Assoc. Fish Pathol. 19182-185. [Google Scholar]
- 8.Chen, S. H., W. J. Cook, K. L. Grove, and D. M. Coen. 1998. Human thymidine kinase can functionally replace herpes simplex virus type 1 thymidine kinase for viral replication in mouse sensory ganglia and reactivation from latency upon explant. J. Virol. 726710-6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a.Coen, D. M., M. Kosz-Vnenchak, J. G. Jacobson, D. A. Leib, C. L. Bogard, P. A. Schaffer, K. L. Tyler, and D. M. Knipe. 1989. Thymidine kinase-negative herpes simplex virus mutants establish latency in mouse trigeminal ganglia but do not reactivate. Proc. Natl. Acad. Sci. USA 864736-4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costes, B., M. Thirion, B. Dewals, J. Mast, M. Ackermann, N. Markine-Goriaynoff, L. Gillet, and A. Vanderplasschen. 2006. Felid herpesvirus 1 glycoprotein G is a structural protein that mediates the binding of chemokines on the viral envelope. Microbes Infect. 82657-2667. [DOI] [PubMed] [Google Scholar]
- 10.Coupar, B. E., S. G. Goldie, A. D. Hyatt, and J. A. Pallister. 2005. Identification of a Bohle iridovirus thymidine kinase gene and demonstration of activity using vaccinia virus. Arch. Virol. 1501797-1812. [DOI] [PubMed] [Google Scholar]
- 11.Dewals, B., C. Boudry, L. Gillet, N. Markine-Goriaynoff, L. de Leval, D. M. Haig, and A. Vanderplasschen. 2006. Cloning of the genome of Alcelaphine herpesvirus 1 as an infectious and pathogenic bacterial artificial chromosome. J. Gen. Virol. 87509-517. [DOI] [PubMed] [Google Scholar]
- 12.Dishon, A., A. Perelberg, J. Bishara-Shieban, M. Ilouze, M. Davidovich, S. Werker, and M. Kotler. 2005. Detection of carp interstitial nephritis and gill necrosis virus in fish droppings. Appl. Environ. Microbiol. 717285-7291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilad, O., S. Yun, M. A. Adkison, K. Way, N. H. Willits, H. Bercovier, and R. P. Hedrick. 2003. Molecular comparison of isolates of an emerging fish pathogen, koi herpesvirus, and the effect of water temperature on mortality of experimentally infected koi. J. Gen. Virol. 842661-2667. [DOI] [PubMed] [Google Scholar]
- 14.Gilad, O., S. Yun, K. B. Andree, M. A. Adkison, A. Zlotkin, H. Bercovier, A. Eldar, and R. P. Hedrick. 2002. Initial characteristics of koi herpesvirus and development of a polymerase chain reaction assay to detect the virus in koi, Cyprinus carpio koi. Dis. Aquat. Org. 48101-108. [DOI] [PubMed] [Google Scholar]
- 15.Gilad, O., S. Yun, F. J. Zagmutt-Vergara, C. M. Leutenegger, H. Bercovier, and R. P. Hedrick. 2004. Concentrations of a koi herpesvirus (KHV) in tissues of experimentally infected Cyprinus carpio koi as assessed by real-time TaqMan PCR. Dis. Aquat. Org. 60179-187. [DOI] [PubMed] [Google Scholar]
- 16.Gillet, L., V. Daix, G. Donofrio, M. Wagner, U. H. Koszinowski, B. China, M. Ackermann, N. Markine-Goriaynoff, and A. Vanderplasschen. 2005. Development of bovine herpesvirus 4 as an expression vector using bacterial artificial chromosome cloning. J. Gen. Virol. 86907-917. [DOI] [PubMed] [Google Scholar]
- 17.Gray, W. L., L. Mullis, S. E. LaPatra, J. M. Groff, and A. Goodwin. 2002. Detection of koi herpesvirus DNA in tissues of infected fish. J. Fish Dis. 25171-178. [Google Scholar]
- 18.Gunimaladevi, I., T. Kono, M. N. Venugopal, and M. Sakai. 2004. Detection of koi herpesvirus in common carp, Cyprinus carpio L., by loop-mediated isothermal amplification. J. Fish Dis. 27583-589. [DOI] [PubMed] [Google Scholar]
- 19.Haenen, O. L. M., K. Way, S. M. Bergmann, and E. Ariel. 2004. The emergence of koi herpesvirus and its significance to European aquaculture. Bull. Eur. Assoc. Fish Pathol. 24293-307. [Google Scholar]
- 20.Hedrick, R. P. 1996. Movement of pathogens with the international trade of live fish: problems and solutions. Rev. Sci. Tech. 15523-531. [DOI] [PubMed] [Google Scholar]
- 21.Hedrick, R. P., O. Gilad, S. Yun, J. Spangenberg, R. Marty, M. Nordhausen, M. Kebus, H. Bercovier, and A. Eldar. 2000. A herpesvirus associated with mass mortality of juvenile and adult koi, a strain of common carp. J. Aquat. Anim. Health. 1244-55. [DOI] [PubMed] [Google Scholar]
- 22.Hedrick, R. P., R. Marty, M. Nordhausen, M. Kebus, H. Bercovier, and A. Eldar. 1999. An herpesvirus associated with mass mortality of juvenile and adult koi Cyprinus carpio. Fish Health Newsl. 27(no. 3)7. [DOI] [PubMed] [Google Scholar]
- 23.Hsu, T. Y., M. W. Liu, Y. R. Chang, C. Y. Pai, M. Y. Liu, C. S. Yang, and J. Y. Chen. 1996. Functional analysis of C-terminal deletion mutants of Epstein-Barr virus thymidine kinase. J. Gen. Virol. 771893-1899. [DOI] [PubMed] [Google Scholar]
- 24.Hutoran, M., A. Ronen, A. Perelberg, M. Ilouze, A. Dishon, I. Bejerano, N. Chen, and M. Kotler. 2005. Description of an as yet unclassified DNA virus from diseased Cyprinus carpio species. J. Virol. 791983-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ilouze, M., A. Dishon, and M. Kotler. 2006. Characterization of a novel virus causing a lethal disease in carp and koi. Microbiol. Mol. Biol. Rev. 70147-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishioka, T., M. Yoshizumi, S. Izumi, K. Suzuki, H. Suzuki, K. Kozawa, M. Arai, K. Nobusawa, Y. Morita, M. Kato, T. Hoshino, T. Iida, K. Kosuge, and H. Kimura. 2005. Detection and sequence analysis of DNA polymerase and major envelope protein genes in koi herpesviruses derived from Cyprinus carpio in Gunma prefecture, Japan. Vet. Microbiol. 11027-33. [DOI] [PubMed] [Google Scholar]
- 27.Liu, H., X. Shi, L. Gao, and Y. Jiang. 2002. Study on the aetiology of koi epizootic disease using the method of nested-polymerase chain reaction assay (nested-PCR). J. Huazhong Agric. Univ. 21414-418. [Google Scholar]
- 28.Markine-Goriaynoff, N., L. Gillet, O. A. Karlsen, L. Haarr, F. Minner, P. P. Pastoret, M. Fukuda, and A. Vanderplasschen. 2004. The core 2 beta-1,6-N-acetylglucosaminyltransferase-M encoded by bovine herpesvirus 4 is not essential for virus replication despite contributing to post-translational modifications of structural proteins. J. Gen. Virol. 85355-367. [DOI] [PubMed] [Google Scholar]
- 29.Morgan, R. W., J. L. Cantello, and C. H. McDermott. 1990. Transfection of chicken embryo fibroblasts with Marek's disease virus DNA. Avian Dis. 34345-351. [PubMed] [Google Scholar]
- 30.Neukirch, M., K. Böttcher, and S. Bunnajrakul. 1999. Isolation of a virus from koi with altered gills. Bull. Eur. Assoc. Fish Pathol. 19221-224. [Google Scholar]
- 30a.Panicali, D., and E. Paoletti. 1982. Construction of poxviruses as cloning vectors: insertion of the thymidine kinase gene from herpes simplex virus into the DNA of infectious vaccinia virus. Proc. Natl. Acad. Sci. USA. 794927-4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perelberg, A., A. Ronen, M. Hutoran, Y. Smith, and M. Kotler. 2005. Protection of cultured Cyprinus carpio against a lethal viral disease by an attenuated virus vaccine. Vaccine 233396-3403. [DOI] [PubMed] [Google Scholar]
- 32.Perelberg, A., M. Smirnov, M. Hutoran, A. Diamant, Y. Bejerano, and M. Kotler. 2003. Epidemiological description of a new viral disease afflicting cultured Cyprinus carpio in Israel. Isr. J. Aquaculture 555-12. [Google Scholar]
- 33.Pikarsky, E., A. Ronen, J. Abramowitz, B. Levavi-Sivan, M. Hutoran, Y. Shapira, M. Steinitz, A. Perelberg, D. Soffer, and M. Kotler. 2004. Pathogenesis of acute viral disease induced in fish by carp interstitial nephritis and gill necrosis virus. J. Virol. 789544-9551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ronen, A., A. Perelberg, J. Abramowitz, M. Hutoran, S. Tinman, I. Bejerano, M. Steinitz, and M. Kotler. 2003. Efficient vaccine against the virus causing a lethal disease in cultured Cyprinus carpio. Vaccine 214677-4684. [DOI] [PubMed] [Google Scholar]
- 35.Sano, M., T. Ito, J. Kurita, T. Yanai, N. Watanabe, M. Satoshi, and T. Iida. 2004. First detection of koi herpesvirus in cultured common carp Cyprinus carpio in Japan. Fish Pathol. 39165-168. [Google Scholar]
- 36.Smith, G. A., and L. W. Enquist. 2000. A self-recombining bacterial artificial chromosome and its application for analysis of herpesvirus pathogenesis. Proc. Natl. Acad. Sci. USA 974873-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soliman, H., and M. El-Matbouli. 2005. An inexpensive and rapid diagnostic method of Koi Herpesvirus (KHV) infection by loop-mediated isothermal amplification. Virol. J. 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.St.-Hilaire, S., N. Beevers, K. Way, R. M. Le Deuff, P. Martin, and C. Joiner. 2005. Reactivation of koi herpesvirus infections in common carp Cyprinus carpio. Dis. Aquat. Org. 6715-23. [DOI] [PubMed] [Google Scholar]
- 39.Vanderplasschen, A., and G. L. Smith. 1997. A novel virus binding assay using confocal microscopy: demonstration that the intracellular and extracellular vaccinia virions bind to different cellular receptors. J. Virol. 714032-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wagner, M., Z. Ruzsics, and U. H. Koszinowski. 2002. Herpesvirus genetics has come of age. Trends Microbiol. 10318-324. [DOI] [PubMed] [Google Scholar]
- 41.Waltzek, T. B., G. O. Kelley, D. M. Stone, K. Way, L. Hanson, H. Fukuda, I. Hirono, T. Aoki, A. J. Davison, and R. P. Hedrick. 2005. Koi herpesvirus represents a third cyprinid herpesvirus (CyHV-3) in the family Herpesviridae. J. Gen. Virol. 861659-1667. [DOI] [PubMed] [Google Scholar]
- 42.Warming, S., N. Costantino, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33e36. [DOI] [PMC free article] [PubMed] [Google Scholar]