Abstract
Gamma interferon receptor α (IFN-γRα) is stable but posttranslationally modified in herpes simplex virus 1(F) [HSV-1(F)]-infected cells. Studies with antibody directed to the phosphorylation site indicate that IFN-γRα is phosphorylated by the US3 kinase. The modification is abolished in cells infected with ΔUS3, ΔUL13, or Δ(US3/UL13) mutant virus. Transcripts of the IFN-γ-dependent genes do not accumulate in cells transduced with the US3 protein kinase and treated with IFN-γ. In contrast, the accumulation of IFN-γ-dependent gene transcripts is suppressed in cells infected with the wild-type virus, in cells infected with the ΔUS3 mutant virus, and to a lesser extent in the ΔUL41 virus-infected cells. The accumulation of IFN-γ-dependent gene transcripts in ΔUL41-infected cells could be due at least in part to a significant delay and reduction in the accumulation of the US3 protein. The results suggest that the expression of IFN-γ-dependent genes is blocked independently by the degradation of IFN-γ-dependent gene transcripts—a function of the virion host shutoff RNase—and by posttranslational modification of the IFN-γRα protein.
Viruses expend a significant portion of their genome to thwart alpha/beta and gamma interferons (IFN-α/β and -γ) from blocking their replication (22, 34, 54, 64). In the case of herpes simplex viruses (HSVs), at least four viral proteins block IFN and IFN-related pathways. Thus, the RNase expressed by the virion host shutoff protein encoded by the UL41 gene blocks the repopulation of the short-lived components of the IFN signaling pathway, Jak1 and Stat2 (8). ICP0, a product of the α0 gene, blocks interferon regulatory factor 3 (IRF3) and IRF7 and also degrades sumoylated forms of promyelocytic leukemia protein (PML) (4, 23, 24, 36, 37, 39, 40). In cells lacking PML, the signaling pathways of both IFN-α and -γ are blocked (9). Lastly, in infected cells, protein kinase R (PKR) is activated at early times after infection (10). The activated PKR phosphorylates the α subunit of translation initiation factor 2 (eIF-2α) which, in turn, shuts off protein synthesis. The viral protein γ134.5 nullifies the effect of activated PKR by recruiting protein phosphatase 1α to dephosphorylate eIF-2α (25). Lastly, US11, a γ2 protein made very late in infection, also has the capacity to block the activation of PKR (6, 7, 38). In this report, we show that viral protein kinases (PKs), and especially the US3 PK, also play a role in viral defense against IFNs by blocking IFN-γ-dependent gene expression. Relevant to this report are the following characteristics.
(i) The sole member of the type II IFNs, IFN-γ, signals through the IFN-γ receptor complex (IFN-γR) which consists of at least two chains, the ligand-binding IFN-γRα chains and the signal-transducing IFN-γRβ chains (1, 57). It has been suggested that a third (AF-2) and a fourth protein (AF-3) may also be involved in signal transduction of the IFN-γ receptor complex (11, 12, 58).
(ii) IFN-α and IFN-γ signaling activate distinct variants of the Jak-Stat pathway (59), resulting in the activation of two different sets of genes regulated by specific promoter sequences, the IFN-stimulated response elements and the γ-activated sequences, respectively. IFN-γ induces major histocompatibility complex I (MHC I) and MHC II expression; Th1 response; antiviral state; the activation of microbicidal effector functions, such as the NADPH-dependent phagocyte oxidase system; immunomodulation; and leukocyte trafficking (53).
(iii) Among these IFN-γ-responsive genes are monokine induced by IFN-γ (Mig) (16, 17, 19, 65) and complement components C4 (2, 21) and p11 (26). Mig, also called CXCL9, is a member of the platelet factor 4-interleukin-8 cytokine family and has been implicated in the host response to viral infections and tumor immunity. Mig is preferentially activated by IFN-γ, instead of by type I IFNs, possibly through the γ-responsive element in the Mig promoter region (65). Recent reports have suggested that the lipopolysaccharide-induced toll-like receptor 4 ligand NF-κB acts in synergy with IFN-γ-induced STAT to regulate the immune response by influencing specific chemokines, including Mig (31, 47). Another effect of IFN-γ is the upregulation of complement component C4; this was specific for IFN-γ since the effect was abolished by a monoclonal antibody directed against IFN-γ (21). Complement components are mainly synthesized in the liver but are also produced in monocytes, macrophages, fibroblasts, endothelial cells, and epithelial cells (5, 27, 30, 60, 63). Complement proteins function in host defense so that extracellular pathogens undergo receptor-mediated phagocytosis. The p11 protein, also known as S-100A10 or calpactin I light chain, is a member of the S-100 protein family but does not bind calcium (20). p11 is a natural ligand of annexin II. Functional analysis indicated that two γ-activated sequences are important for the induction of p11 promoter by IFN-γ (26).
The US3 PK performs multiple functions in the infected cells. Briefly, US3 blocks apoptosis induced by both viral and cellular gene products, enables the localization of UL34 and UL31 to the nucleus, disrupts the nuclear lamina, and enables the egress of capsids from the nucleus (3, 18, 33, 41, 42, 46, 51, 52, 55). In addition, US3 PK mediates the phosphorylation of histone deacetylases 1 and 2 and acts as a helper in enabling the expression of genes introduced into cells by transduction (45). The US3 PK is autophosphorylated, i.e., phosphorylated both by cellular kinases and by the viral UL13 PK (29, 44, 50). Lastly, the target site of the US3 PK is similar or identical to that of PKA (3). Antibody to the substrate sequence of PKA reacts with the sites phosphorylated by the US3 PK.
The virion host shutoff protein encoded by the UL41 gene is an endoribonuclease with the specificity of RNase A (61, 62). It degrades mRNAs selectively, early in infection (15, 56). In earlier studies it has been shown to play a role in blocking IFNs (8).
In this report, we show that IFN-γRα is posttranslationally modified in HSV-1(F)-infected cells. The modification is abolished in cells infected with ΔUS3 or Δ(US3/UL13) mutant virus and, to a large extent, also in cells infected with the ΔUL13 mutant virus. In cells transduced with the US3 protein kinase, the IFN-γ-dependent genes are not activated in the presence of IFN-γ. In contrast, in infected cells, the accumulation of IFN-γ-dependent gene transcripts is suppressed in the absence of either US3 PK or the ΔUL41-infected cells. The results suggest that the accumulation of IFN-γ-dependent gene transcripts in infected cells is subject to downregulation by a number of factors, including US3 PK and virion host shutoff proteins.
MATERIALS AND METHODS
Cells and viruses.
HEL and U2OS cells were obtained from the American Type Culture Collection (Manassas, VA) and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Insect cell line Sf9 (Spodoptera frugiperda) was obtained from Pharmingen and maintained in Grace's medium supplemented with 10% fetal bovine serum. HSV-1(F) is the prototype HSV-1 strain used in this laboratory (14). The mutant viruses lacking UL41 (R2621), US3 (R7041), or UL13 (R7356) or the virus lacking both US3 and UL13 (R7353) was described elsewhere (43, 48, 49). Baculoviruses expressing wild-type US3 and a kinase-inactive mutant have been described elsewhere (45, 46).
Antibodies and reagents.
Rabbit polyclonal antibodies against IFN-γRα and IFN-γRβ were purchased from Santa Cruz Biotechnology (catalog nos. sc-700 and sc-30012) and used at a dilution of 1:1,000. Rabbit polyclonal antibody against PKA-phosphorylated (Ser/Thr) (PKA-P) substrates (catalog no. 9621; Cell Signaling Technology) was used at a dilution of 1:1,000. Mouse monoclonal antibody against actin (A4700; Sigma, St. Louis, MO) was used at a dilution of 1:1,000. The US3 rabbit polyclonal antibody (34) was used at a dilution of 1:1,000. Recombinant human IFN-γ (catalog no. RDI-3002) was purchased from Research Diagnostics, Inc.
Extraction of surface proteins.
Cell monolayer cultures were rinsed twice with ice-cold phosphate-buffered saline (PBS) and reacted with 1 mg/ml sulfosuccinimidyl-6-[biotin-amido]hexanoate (sulfo-NHS-LC-biotin) reagent (Pierce) at 4°C for 30 min. The cells were then rinsed three times with 100 mM glycine in PBS to quench and remove the biotin reagent. The cells were then harvested by scraping and lysed in binding buffer (0.1% sodium dodecyl sulfate [SDS], 1% NP-40, 0.5% sodium deoxycholate, fresh protease inhibitor) at 4°C, followed by brief sonication. Immobilized streptavidin agarose beads (catalog no. 53117; Pierce) were rinsed in binding buffer twice and reacted overnight with the lysate at 4°C. The streptavidin-bound complex was rinsed five times with binding buffer. The sample was then eluted with 8 M guanidine-HCl and subjected to electrophoresis.
Immunoblotting.
Cells were collected and lysed in lysis buffer (100 mM NaCl, 50 mM Tris-HCl, pH 7.5, 0.5% NP-40, 0.5% sodium deoxycholate, and protease inhibitor mixture). The proteins were separated on a denaturing 10% polyacrylamide gel and electrically transferred to a nitrocellulose membrane. The membrane was blocked for 1 h at room temperature with 5% nonfat dry milk in PBS and then reacted overnight at 4°C with the designated primary antibody diluted in PBS containing 1% bovine serum albumin. The membrane was then rinsed three times with 0.2% Tween 20 in PBS (PBS-T) and then exposed to secondary antibody at room temperature for 1.5 h. The secondary antibody was conjugated with alkaline phosphatase (Bio-Rad) or to goat anti-mouse or rabbit (Sigma) conjugated to peroxidase. The antibodies were diluted 1:3,000 in PBS-T containing 5% nonfat dry milk. To develop peroxidase-conjugated secondary antibody, the immunoblot was reacted with enhanced chemiluminescence Western blotting detection reagents, according to the manufacturer's instructions (Amersham Pharmacia). Alternatively, membranes were incubated with alkaline phosphatase-conjugated antibody, followed by development as suggested by the manufacturer's protocol (Amersham Pharmacia).
Immunoprecipitation with IFN-γRα antibody.
HEL cells were mock infected or exposed to 5 PFU of HSV-1(F) or the ΔUS3, ΔUL13, or Δ(US3/UL13) mutant virus per cell for 12 h. The cells were lysed in lysis buffer, and the lysates were incubated with 50 μl of protein A agarose for 3 h. After centrifugation, the supernatant fluid was incubated with 10 μl of rabbit polyclonal anti-IFN-γRα antibody at 4°C overnight. On the next day, the same amount of protein A agarose (50 μl) was added to the lysates and they were rotated at 4°C for 3 h. The immunocomplexes bound to agarose were rinsed five times with lysis buffer, eluted in SDS protein sample buffer, separated by 10% SDS-polyacrylamide gel electrophoresis, and finally immunoblotted with PKA-P substrate antibody as described above.
Treatment with IFN-γ and total RNA extraction.
U2OS cells were transduced with MTS, K220N mutant, or wild-type US3 baculoviruses at 1 PFU/cell for 18 h in 199V medium supplemented with 1% calf serum, followed by induction with 1,000 U/ml human IFN-γ for 3 h. Alternatively, Hel cells or U2OS cells were mock infected or infected with HSV-1(F) or the ΔUS3or ΔUL41 mutant virus at 10 PFU per cell for 12 h before induction with 1,000 U/ml human IFN-γ for 3 h. Total RNA was extracted as previously described (15). Total RNA was extracted using Trizol reagent (Invitrogen), followed by DNase treatment (Ambion), phenol-chloroform extraction (Ambion), and ethanol precipitation to remove possible DNA contamination.
Northern blot analyses.
Total RNA (10 μg) was loaded onto 1% denaturing formaldehyde gel and probed with random hexanucleotide-primed 32P-labeled specific probe after transfer onto a nylon membrane (Ambion). The Mig fragment was amplified from a human brain cDNA library (Invitrogen) by using primers MIG-143F (GCACCAACCAAGGGACTATC) and MIG-500B (TATGCCATCCTCCTTTGGAA). The C4 fragment was amplified using primers 5′ TAAGAGCAGACTCTTGGCCA and 3′ TGAGTGCCATACTCCTGGAG. The p11 fragment was amplified using primers P11-61F (ACCACACCAAAATGCCATCT) and P11-379B (CTGCTCATTTCTGCCTACTT). These fragments were then labeled with [32P]αdCTP by using a Prime-a-Gene labeling system (Promega). The membranes were prehybridized for 2 h at 42°C in ULTRAhyb buffer (Ambion) with 200 μg/ml of denatured salmon sperm DNA (Stratagene) and then hybridized overnight with the 32P-labeled probe. The membranes were then rinsed twice for 5 min at 42°C with a solution containing 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) plus 0.1% SDS and twice at 42°C with a solution containing 0.1× SSC and 0.1% SDS. The membranes were then exposed to phosphoimager screening, followed by scanning for signal detection. The membranes were stripped in boiling 0.5% SDS and reacted with a second probe as described above.
RESULTS
Both total and cell surface IFN-γRαs are posttranslationally modified in cells infected with wild-type HSV-1(F).
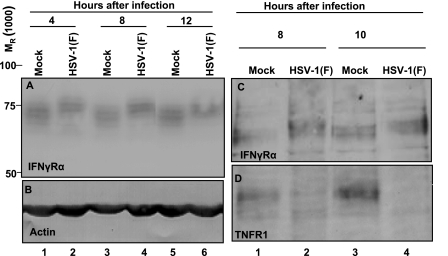
We report two series of experiments. In the first, HEL cells were mock infected or exposed to 5 PFU of wild-type virus per cell. The cells were harvested at 4, 8, or 12 h after infection, solubilized, subjected to electrophoresis in denaturing gels, transferred to a nitrocellulose membrane, and reacted with either anti-IFN-γRα (Fig. 1A) or anti-actin antibody (Fig. 1B). The results seen in Fig. 1A show that in cells infected with the wild-type virus, the total IFN-γRα protein formed prominent bands that migrated more slowly than the bands in mock-infected cells. The change in the electrophoretic mobility occurred as early as 4 h after infection.
FIG. 1.
Modification of total and cell surface IFN-γRα protein by HSV-1(F) infection. (A and B) HEL cells were mock infected or infected with 5 PFU of HSV-1(F) per cell for 4, 8, or 12 h. Total cell lysates were separated on 10% denaturing polyacrylamide gels and immunoblotted with anti-IFN-γRα antibody (A) or anti-actin antibody (B). Relative molecular weights are shown on the left. (C and D) HEL cells were mock infected or exposed to 5 PFU of HSV-1(F) per cell. The cells were harvested after 8 or 10 h of infection. Surface proteins were extracted as described in Materials and Methods, separated on a 10% denaturing polyacrylamide gel, and reacted with anti-IFN-γRα antibody (C) or anti-TNF-R1 antibody (D).
In the second series of experiments, the cells were either mock infected or exposed to virus as described above. They were harvested at 8 or 10 h after infection, and the surface proteins were extracted as described in Materials and Methods, subjected to electrophoresis in denaturing gel, transferred to a nitrocellulose membrane, and reacted with antibody to either IFN-γRα or tumor necrosis factor receptor 1 (TNF-R1). The results shown in Fig. 1C indicate that the cell surface IFN-γRα protein was also subject to posttranslational modification and migrated more slowly than the protein present on the surface of mock-infected cells. The experiment shown in Fig. 1D served as a positive control; elsewhere we reported that TNF-αR1 has a short half-life and that in infected cells, the receptor turns over and is not repopulated as a consequence of the action of the RNase encoded by the UL41 gene (35). As illustrated in this figure, the amounts of IFN-γRα protein remained quite stable throughout the test interval (12 h), in contrast to the TNF-R1.
The posttranscriptional modification of IFN-γRα protein is mediated by the viral PKs.
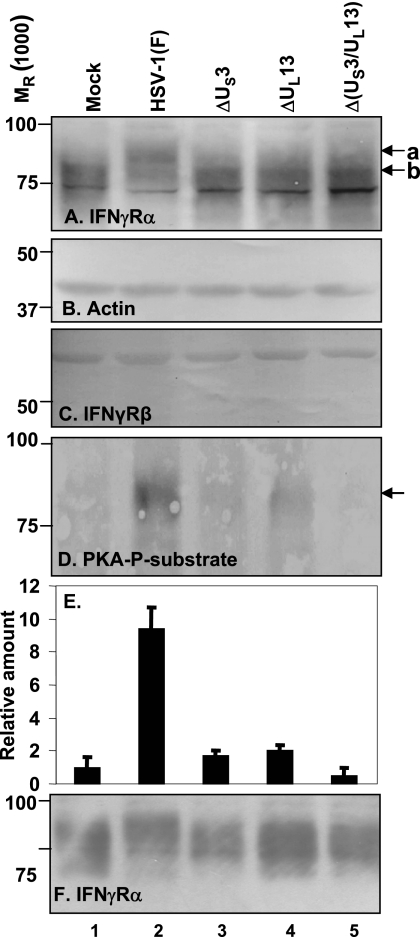
To test whether the viral PKs encoded by US3 and UL13 mediated the change in the electrophoretic mobility of the IFN-γRα protein and whether the IFN-γRβ protein was similarly modified, HEL cells were mock infected or exposed to 5 PFU of HSV-1(F) or the ΔUS3, ΔUL13, or Δ(US3/UL13) mutant virus per cell. The cells were harvested at 8 h after infection, and lysates prepared as described in Materials and Methods were subjected to electrophoresis in denaturing gels and probed with antibodies against IFN-γRα (Fig. 2A), IFN-γRβ (Fig. 2C), or actin (Fig. 2B). In this series of experiments, as expected on the basis of the results presented in Fig. 1, the IFN-γRα contained in HSV-1(F)-infected cells exhibited a slower electrophoretic mobility than that of mock-infected cells or cells infected with the mutant viruses (Fig. 2A). The electrophoretic mobility of IFN-γRα protein in the lysates of cells infected with the ΔUS3 or Δ(US3/UL13) mutant virus was largely similar to that of mock-infected cells. We noted a small amount of slowly migrating protein in the lysates of ΔUL13 or Δ(US3/UL13), but this material was also present in lysates of mock-infected cells. In contrast, there was no change in the electrophoretic mobility or amount of the IFN-γRβ protein (Fig. 2C). These experiments suggest that in infected cells, the posttranslational modification of the IFN-γRα protein requires the presence of both the US3 and UL13 PK.
FIG. 2.
Modification of IFN-γRα protein in HEL cells infected with HSV-1 mutant viruses. HEL cells were mock infected or infected with 5 PFU of HSV-1(F), ΔUS3, ΔUL13, or Δ (US3/UL13) double-deletion virus per cell for 8 h. Total cell lysates were separated on 10% denaturing polyacrylamide gel and immunoblotted with anti-IFN-γRα antibody (A), anti-actin antibody (B), or anti-IFN-γRβ antibody (C). In the experiment whose results are shown in panels D and E, cells were infected with designated viruses for 12 h. The cells were lysed and immunoprecipitated with anti-IFN-γRα antibody as described in Materials and Methods. Protein samples were run on a 10% SDS-polyacrylamide gel and reacted with antibody to PKA-P substrate (D) or IFN-γRα antibody (F). Relative amount of PKA-P substrate normalized against IFN-γRα (F) is shown in panel E. Error bars show standard deviations. a and b indicate posttranslationally modified and unmodified forms of IFN-γRα protein, respectively. Relative molecular weights are shown on the left.
The objective of the second experiment illustrated in Fig. 2 was to determine whether the IFN-γRα protein is phosphorylated by the US3 kinase. Earlier studies (3) have shown that the US3 kinase phosphorylates sites identical to those of PKA and that antibody to those sites reacts with the sites phosphorylated by the US3 PK. However, in infected cells, the phosphorylation of PKA-P substrates is due largely, if not exclusively, to the US3 PK (46). The design of this experiment is similar to that described above for Fig. 2A. Total cell lysates were immunoprecipitated with anti-IFN-γRα antibody. One set of electrophoretically separated proteins was reacted with antibody to the PKA-P substrate (Fig. 2D). The other set was reacted with antibody to the IFN-γRα protein. The amounts of protein reacting with antibodies were quantified with the aid of a General Dynamics Storm phosphorimager. The amount of protein reacting with the antibody to the PKA-P substrate (Fig. 2D) was normalized with respect to the total protein, as shown in Fig. 2E, and with respect to the amount of antibody reacting with PKA-P substrate in the lysates of mock-infected cells. The results shown in Fig. 2F indicate that the IFN-γRα protein was extensively phosphorylated by the US3 PK in cells infected with the wild-type virus, but not in cells infected with mutant viruses.
We conclude that both viral kinases are required for the extensive posttranslational modifications of the IFN-γRα protein in infected cells. The results also indicate that the IFN-γRα protein from cells infected with the wild-type virus is phosphorylated by the US3 PK and raise the possibility that extensive phosphorylation of IFN-γRα by US3 requires the presence of the UL13 PK.
In transduced cells, the US3 PK blocks the activation of the IFN-γ pathway.
The experiments described above suggested that the US3 PK mediates a posttranslational modification of the IFN-γRα protein but that this requires the presence of the UL13 PK. This is not unexpected since the UL13 PK has been shown to phosphorylate the US3 PK (29, 44). The US3 PK performs multiple functions, and it is conceivable that the execution of some of the functions is regulated by the UL13 PK. To determine whether these modifications alter the function of the receptor and block the activation of IFN-γ response genes, we used U2OS cells transduced with recombinant baculoviruses. The rationale for this experiment is as follows. As reported elsewhere, the advantages of using baculoviruses for the transduction of mammalian cells stems from the observation that baculoviruses uniformly transduce cells and the expression of the transgene is baculovirus dose dependent. In most cell lines, the expression of the transgene requires inhibitors of histone deacetylases (e.g., sodium butyrate). This is not the case for U2OS cells (45).
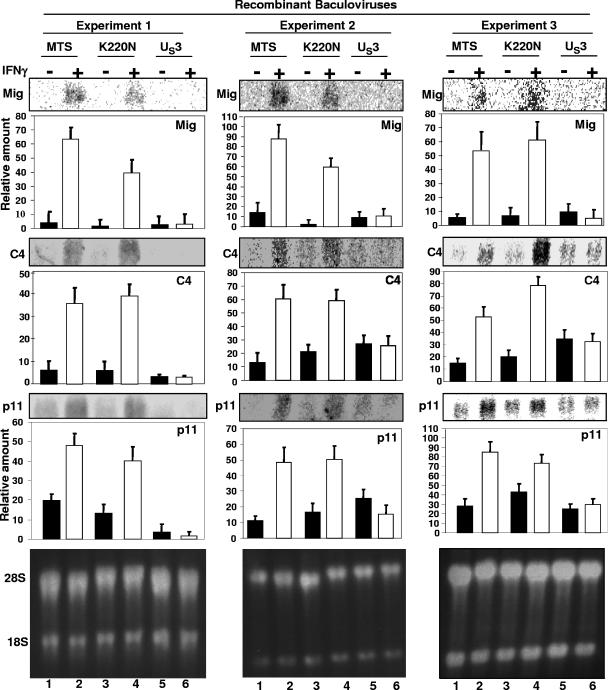
In the experiments reported in this section, U2OS cells were transduced with baculoviruses encoding the US3 PK, an inactive US3 protein carrying the K220N substitution, or an empty vector. After 18 h of incubation, a replicate set of the cell cultures was exposed to 1,000 U of IFN-γ per ml of medium for 3 h. The cells were then harvested, and total mRNAs extracted. Equivalent amounts of RNA were separated on a 1% denaturing formaldehyde gel. The electrophoretically separated mRNAs were assayed by Northern blot analysis with 32P-labeled Mig, C4, or p11 probes and quantified as described in Materials and Methods. The results of three independently performed experiments shown in Fig. 3 indicate that each of the three IFN-γ-dependent genes was activated in cells transduced with the empty vector or K220 mutant gene, but not in cells transduced with the wild-type US3 gene. We conclude from the results of this experiment that in transduced cells, US3 PK blocked the activation of IFN-γ-dependent genes in the absence of other viral proteins.
FIG. 3.
US3 expression by baculovirus blocked the expression of the activation of IFN-γ-dependent genes. U2OS cells were transduced with baculoviruses expressing empty vector MTS, wild-type US3 PK, or the inactive kinase carrying the substitution K220N for 18 h, followed by exposure to human IFN-γ (1,000 U/ml) for 3 h. Total RNA was extracted, and an equivalent amount of total RNA was separated on a 1% denaturing formaldehyde gel and hybridized with 32P-labeled Mig, C4, or p11 probes as described in Materials and Methods. The amounts of mRNA detected by the hybridization with labeled probes were quantified relative to the amounts of ribosomal mRNA shown in the bottom panels. The figure shows the results of three experiments. Error bars show standard deviations. +, present; −, absent.
In infected cells, the inhibition of the activation of IFN-γ-dependent genes depends on the virion host shutoff product of the UL41 gene.
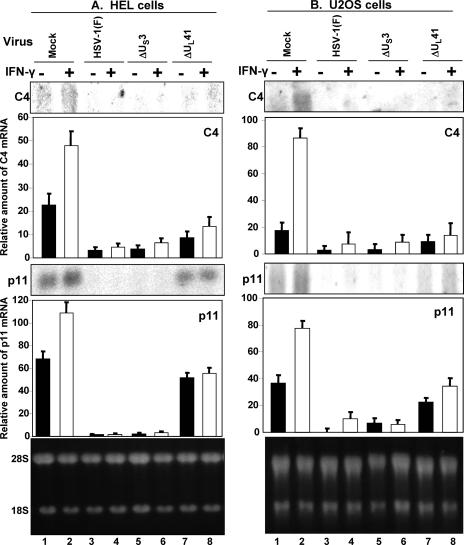
The objectives of this experiment were to determine whether the US3 PK blocked the activation of IFN-γ-dependent genes in infected cells. In this series of experiments, replicate cultures of HEL or U2OS cells were mock infected or exposed to 10 PFU of HSV-1(F), ΔUS3, or ΔUL41 virus per cell. After 12 h of incubation at 37°C, the cultures were replenished with medium containing IFN-γ (1,000 U/ml), and incubation was continued for three more hours. Total RNA was then extracted and separated on a 1% denaturing formaldehyde gel, followed by Northern blot analyses with 32P-labeled C4 or p11 probes. The amounts of RNA were quantified with respect to the amounts of 28S rRNA. The results (Fig. 4) were as follows.
FIG. 4.
Inhibition of IFN-γ pathway in HSV-1-infected cells. HEL or U2OS cells were mock infected or exposed to 10 PFU of HSV-1(F), ΔUS3, or ΔUL41 virus per cell for 12 h, followed by treatment with human IFN-γ (1,000 U/ml) for 3 h. Total RNA was extracted, separated on a 1% denaturing formaldehyde gel, and hybridized with labeled C4 or p11 probes as described in Materials and Methods. The C4 and p11 RNAs accumulating in the mock infected and infected cells were normalized with respect to the ribosomal RNAs shown in the bottom panels. Error bars show standard deviations. +, present; −, absent.
(i) Both C4 and p11 mRNAs were present in lysates of mock-infected cells, but the level of mRNA was higher in IFN-γ-treated than in untreated cells.
(ii) The amounts of C4 or p11 mRNA detected in HSV-1(F) or ΔUS3 mutant virus-infected cells were close to the background levels, indicating that in cells infected with the wild-type or ΔUS3 mutant virus, the accumulation of the mRNAs was suppressed.
(ii) Both cell lines infected with the ΔUL41 mutant virus accumulated amounts of p11 mRNA that were lower than those in mock-infected cells but significantly higher than those in cells infected with ΔUS3 mutant virus. A similar, albeit lower increase was observed for C4 mRNA in HEL cells infected with the ΔUL41.
The experiments illustrated in Fig. 4 were reproducible. The failure to accumulate C4 or p11 mRNA in cells infected with the ΔUS3 mutant virus suggested that in infected cells, another gene product may also play a role in blocking the accumulation of IFN-γ-dependent gene response. An obvious candidate is the virion host shutoff RNase encoded by the UL41 gene. We expected, however, that the US3 PK made in UL41 mutant virus-infected cells would block the accumulation of IFN-γ-dependent genes, but the results indicate that the accumulation of these transcripts was reduced but not blocked completely. The central question, then, was whether the US3 PK accumulated in ΔUL41-infected cells.
In ΔUL41-infected cells, the expression of US3 PK is reduced and IFN-γRα is not posttranslationally modified.
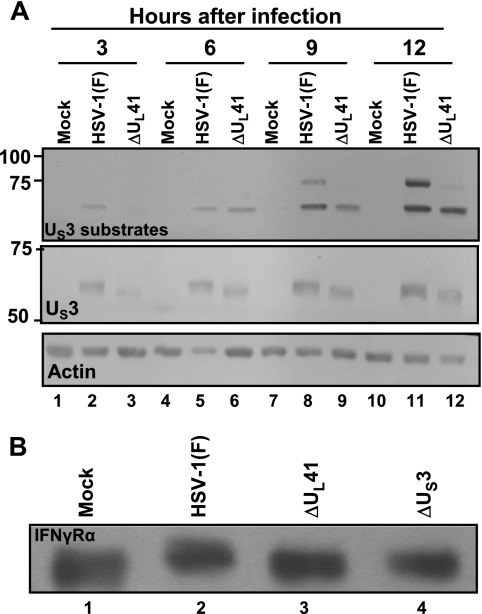
The objective of this series of experiments was to examine the status of the US3 PK in ΔUL41 mutant virus-infected cells. The objective of the first series of experiments was to obtain a rough profile of the activity of the US3 kinase in cells infected with wild-type or ΔUL41 mutant virus. As noted earlier in the text, the substrate specificity of the US3 PK is similar to that of the PKA, and the antibody to the PKA-P substrates reacts with numerous proteins in cells infected with wild-type virus, but not in cells infected with the mutants lacking the gene encoding the US3 PK. In this series of experiments, replicate U2OS cell cultures were mock infected or exposed to 10 PFU of HSV-1(F) or ΔUL41 virus per cell. The cells were harvested at 3, 6, 9, or 12 h after infection, lysed, subjected to electrophoresis in denaturing gels, and reacted with anti-PKA-P substrate antibody, anti-US3 antibody, or anti-actin antibodies. As shown in Fig. 5A, there was a delay in the appearance of phosphorylated proteins reactive with the anti-PKA-P substrate antibody. At the same time there was a delay in the accumulation of US3 protein. Particularly noteworthy is that the electrophoretic mobility of the US3 protein as late as 12 h after infection with ΔUL41 mutant virus was faster than that of the US3 protein accumulating in cells infected with the wild-type virus. The results indicate that the US3 protein accumulating in ΔUL41-infected cells was not posttranslationally processed.
FIG. 5.
Substrate phosphorylation by US3 in HSV-1(F)-infected cells. (A) U2OS cells were mock infected or exposed to 10 PFU of HSV-1(F) or ΔUL41 virus per cell. The cells were harvested at 3, 6, 9, and 12 h after infection. Total cell lysates were electrophoretically separated on a denaturing gel and reacted with antibody to PKA-P substrate or to US3 PK. Molecular size markers in thousands are shown on the left. (B) U2OS cells were mock infected or exposed to 10 PFU of HSV-1(F), ΔUL41, or ΔUS3 virus per cell. The cells were harvested at 12 h after infection. Surface proteins were extracted and immunoblotted with anti-IFN-γRα antibody.
In the second series of experiments, we examined the status of the IFN-γRα in U2OS cells mock infected or exposed to 10 PFU of HSV-1(F) or the ΔUL41 or ΔUS3 mutant virus per cell. After 12 h of incubation, the cell surface proteins were extracted, processed as described in Materials and Methods, and then reacted with the anti-IFN-γRα antibody. The results seen in Fig. 5B show that IFN-γRα was posttranslationally modified in cells infected with the wild-type virus, but not in cells infected with either the ΔUS3 or ΔUL41 mutant virus.
DISCUSSION
The salient features of the results presented in this report center on three observations.
First, unlike TNF-αR1 that has a short half-life and is not repopulated, in cells infected with the wild-type virus, the IFN-γRα protein is stable and could be readily detected on the cell surface as late as 15 h after infection.
Second, IFN-γRα protein is extensively posttranslationally modified in a manner that is dependent on both US3 and UL13 PKs. In the experiment whose results are illustrated in Fig. 2, we have shown that the IFN-γRα protein present in lysates of cells infected with the wild-type virus reacted with the antibody specific for substrates phosphorylated by PKA. Elsewhere we have shown that the substrate specificity of US3 PK is similar if not identical to that of the PKA and that antibody to the phosphorylated US3 substrate reacts with the anti-PKA-P substrate (3). The results therefore strongly suggest that the posttranslational modification of the IFN-γRα protein involves phosphorylation by the US3 PK, but only in the presence of the UL13 PK. The simplest explanation is that the extensive phosphorylation of the IFN-γRα protein is mediated by US3 posttranslationally modified by the UL13 PK. Indeed, there is ample evidence that the US3 PK is phosphorylated by the UL13 PK (29, 44). At this time, we cannot exclude the possibility that the posttranslational modification of the IFN-γRα protein requires prior modification by a process mediated by the UL13 PK.
Lastly, we showed that in transduced cells, US3 PK blocked the accumulation of transcripts of IFN-γ-dependent genes following exposure to IFN-γ. This process did not require the involvement of any other viral protein. The results suggest that US3 modified either the IFN-γRα protein or a component of the downstream signaling pathway to block either the activation of the IFN-γ-dependent genes or the accumulation of the transcripts of these genes.
Analyses of the events occurring in infected cells indicated that US3 may not be the only viral protein whose function it is to block IFN-γ-mediated gene expression. Specifically, we expected that in the absence of the US3 PK, infected cells would respond to IFN-γ by activating IFN-γ-dependent genes. Preliminary experiments indicated that this was not the case and suggested that other viral gene products interfere with the activation of IFN-γ-dependent genes. The most common blocker of the accumulation of cellular transcripts is the RNase encoded by the UL41 gene. In the experiment whose results are shown in this report and in the results of other studies, transcripts encoded by IFN-γ-dependent genes accumulated in IFN-γ-treated cells infected with ΔUL41 mutant virus, but not to the same level as in IFN-γ-treated mock-infected cells. This observation in itself was puzzling, inasmuch as it would have been expected that US3 protein made in ΔUL41-infected cells would block IFN-γ-dependent gene expression. Since this was partly the case, we examined the status of the US3 PK accumulation and function in cells infected with the ΔUL41 mutant. The results showed that in ΔUL41 mutant virus-infected cells, the accumulation of US3 PK was delayed, the US3 substrates were not fully phosphorylated, and the IFN-γRα protein was not posttranslationally processed within the time frame of the studies. In essence, the accumulation of the US3 PK is retarded in the ΔUL41 mutant virus-infected cells.
Our results, then, indicate that HSV-1 blocks the activation of IFN-γ-dependent genes as a result of the functions expressed by at least two proteins, US3 PK and the virion host shutoff RNase. The evidence favoring the host shutoff RNase is based on the results of the studies on infected cells illustrated in Fig. 4. The evidence supporting a role for the US3 protein in blocking IFN-γ is based on the results of studies on cells transduced with the US3 PK and the observation that it mediates a posttranslational modification of the IFN-γRα protein. The role of the UL13 PK is less clear. As indicated above, UL13 PK enhances the posttranslational modification of the IFN-γRα protein by the phosphorylation of US3 PK, by direct phosphorylation of the IFN-γRα protein, or by both (29, 44, 50).
It is noteworthy that Eisemann et al. (13) measured the cell surface expression of IFN-γRα protein by flow cytometry. The authors reported a 50% cell surface loss at 16 h after infection for cells infected with wild-type or ΔUL41 mutant virus. At 24 h after infection, the number of cells exhibiting IFN-γRα on the surface of cells infected with the ΔUL41 mutant virus decreased significantly below that of cells infected with the wild-type virus. In the studies reported here, the modification of IFN-γRα was observed as early as 4 h after infection.
It is tempting to speculate that the RNases encoded by the UL41 gene and the US3 PK act independently at different times after infection to block the expression of IFN-γ-dependent genes. Thus, the UL41 RNase degrades the expression of IFN-γ-dependent genes early in infection. At late times, when the RNase activity of the UL41 protein ceases, the newly synthesized US3 protein inactivates IFN-γRα.
Two other observations presented in this report are worthy of note. First, the results of the experiments shown in Fig. 5 indicate that the activation of IFN-γ-dependent genes in cells infected with the ΔUL41 mutant was lower than that observed in mock-infected cells. One hypothesis that could explain these results is that the US3 PK packaged in virions precluded in part the activation of IFN-γ-dependent genes or that the accumulation of the mRNAs of IFN-γ-dependent genes was in part blocked by ICP27. Second, it has been known for many years that in ΔUL41 mutant virus-infected cells, the expression of late genes is retarded (32). The results of recent studies also indicate that in the absence of the UL41 gene, the localization of ICP4 and ICP0 is altered in a manner similar to that occurring in cells lacking the genes encoding glycoprotein E or I (28). On the basis of the known function of glycoproteins E and I, the possibility exists that in ΔUL41 mutant virus-infected cells, the localization or, more appropriately, the transport of several proteins may be perturbed. A change in the distribution of the US3 PK in ΔUL41 mutant virus-infected cells, coupled with a delay in its synthesis, may explain the failure of surface IFN-γRα protein to be posttranslationally modified.
The studies cited in the introduction have shown that ICP0, virion host shutoff RNase, g34.5, and US11 mediate resistance to IFNs. In this report, we have extended the range of viral functions that specifically target IFN-γ. The major conclusion of this study is that HSV-1 encodes at least two additional functions capable of inhibiting the activation of IFN-γ-dependent genes.
Acknowledgments
These studies were aided by a grant (CA88860) from the National Cancer Institute.
Footnotes
Published ahead of print on 5 March 2008.
REFERENCES
- 1.Aguet, M., Z. Dembic, and G. Merlin. 1988. Molecular cloning and expression of the human gamma interferon receptor. Cell 55273-280. [DOI] [PubMed] [Google Scholar]
- 2.Belt, K. T., M. C. Carroll, and R. R. Porter. 1984. The structural basis of the multiple forms of human complement component C4. Cell 36907-914. [DOI] [PubMed] [Google Scholar]
- 3.Benetti, L., and B. Roizman. 2004. Herpes simplex virus protein kinase US3 activates and functionally overlaps protein kinase A to block apoptosis. Proc. Natl. Acad. Sci. USA 1019411-9416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boutell, C., S. Sadis, and R. D. Everett. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and its isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brooimans, R. A., P. S. Hiemstra, A. A. J. van der Ark, R. B. Sim, L. A. van Es, and M. R. Daha. 1989. Biosynthesis of complement factor H by human umbilical vein endothelial cells: regulation by T cell growth factor and IFN-γ. J. Immunol. 1422024-2030. [PubMed] [Google Scholar]
- 6.Cassady, K. A., and M. Gross. 2002. The herpes simplex virus type 1 US11 protein interacts with protein kinase R in infected cells and requires a 30-amino-acid sequence adjacent to a kinase substrate domain. J. Virol. 762029-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cassady, K. A., M. Gross, and B. Roizman. 1998. The herpes simplex virus US11 protein effectively compensates for the γ134.5 gene if present before activation of protein kinase R by precluding its phosphorylation and that of the α subunit of eukaryotic translation initiation factor 2. J. Virol. 728620-8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chee, A. V., and B. Roizman. 2004. Herpes simplex virus 1 gene products occlude the interferon signaling pathway at multiple sites. J. Virol. 784185-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chee, A. V., P. Lopez, P. P. Pandolfi, and B. Roizman. 2003. Promyelocytic leukemia protein mediates interferon-based anti-herpes simplex virus 1 effects. J. Virol. 777101-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chou, J., J. Chen, M. Gross, and B. Roizman. 1995. Association of a Mr 90,000 phosphoprotein with protein kinase PKR in cells exhibiting enhanced phosphorylation of translation initiation factor eIF-2α and premature shutoff of protein synthesis after infection with γ134.5− mutants of herpes simplex virus 1. Proc. Natl. Acad. Sci. USA 9210516-10520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cook, J. R., S. L. Emanuel, R. J. Donnely, J. Soh, T. M. Mariano, B. Schwartz, S. Rhee, and S. Pestka. 1994. Sublocalization of the human gamma interferon receptor accessory factor gene and characterization of accessory factor activity by yeast artificial chromosomal fragmentation. J. Biol. Chem. 2697013-7018. [PubMed] [Google Scholar]
- 12.Cook, J. R., V. Jung, B. Schwartz, P. Wang, and S. Pestka. 1992. Structural analysis of the human gamma interferon receptor: specific requirements of a small segment of the intracellular domain for class I MHC antigen induction and antiviral activity. Proc. Natl. Acad. Sci. USA 8911317-11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eisemann, J., P. Mühl-Zürbes, A. Steinkasserer, and M. Kummer. 2008. Infection of mature dendritic cells with herpes simplex virus type 1 interferes with the interferon signaling pathway. Immunobiology 212877-886. [DOI] [PubMed] [Google Scholar]
- 14.Ejercito, P. M., E. D. Kieff, and B. Roizman. 1968. Characterization of herpes simplex virus strains differing in their effects on social behavior of infected cells. J. Gen. Virol. 2357-364. [DOI] [PubMed] [Google Scholar]
- 15.Esclatine, A., B. Taddeo, and B. Roizman. 2004. The UL41 protein of herpes simplex virus mediates selective stabilization or degradation of cellular mRNAs. Proc. Natl. Acad. Sci. USA 10118165-18170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farber, J. M. 1990. A macrophage mRNA selectively induced by gamma interferon encodes a member of the platelet factor 4 family of cytokines. Proc. Natl. Acad. Sci. USA 875238-5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farber, J. M. 1993. HuMIG: a new member of the chemokine family of cytokines. Biochem. Biophys. Res. Commun. 192223-230. [DOI] [PubMed] [Google Scholar]
- 18.Galvan, V., and B. Roizman. 1998. Herpes simplex virus 1 induces and blocks apoptosis at multiple steps during infection and protects cells from exogenous inducers in a cell-type-dependent manner. Proc. Natl. Acad. Sci. USA 953931-3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gasperini, S., M. Marchi, F. Calzetti, C. Laudanna, L. Vicentini, H. Olsen, M. Murphy, F. Liao, J. Farber, and M. A. Cassatella. 1999. Gene expression and production of the monokine induced by IFN-gamma (MIG), IFN-inducible T cell alpha chemoattractant (I-TAC), and IFN-gamma-inducible protein-10 (IP-10) chemokines by human neutrophils. J. Immunol. 1624928-4937. [PubMed] [Google Scholar]
- 20.Gerke, V., and K. Weber. 1985. The regulatory chain in the p36-kd substrate complex of viral tyrosine-specific protein kinases is related in sequence to the S-100 protein of glial cells. EMBO J. 42917-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerritsma, J. S. J., A. F. Gerritsen, M. De Ley, L. A. van Es, and M. R. Daha. 1997. Interferon-γ induces biosynthesis of complement components C2, C4 and factor H by human proximal tubular epithelial cells. Cytokine 9276-283. [DOI] [PubMed] [Google Scholar]
- 22.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 812341-2364. [DOI] [PubMed] [Google Scholar]
- 23.Gu, H., and B. Rroizman. 2003. The degradation of promyelocytic leukemia and Sp100 proteins by herpes simplex virus 1 is mediated by the ubiquitin-conjugating enzyme UbcH5a. Proc. Natl. Acad. Sci. USA 1008963-8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hagglund, R., C. Van Sant, P. Lopez, and B. Roizman. 2002. Herpes simplex virus 1-infected cell protein 0 contains two E3 ubiquitin ligase sites specific for different E2 ubiquitin-conjugating enzymes. Proc. Natl. Acad. Sci. USA 99631-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He, B., M. Gross, and B. Roizman. 1997. The γ134.5 protein of herpes simplex virus 1 complexes with protein phosphoatase 1α to dephosphorylate the α subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. USA 94843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang, X., R. Pawliczak, X. Yao, M. J. Cowan, M. T. Gladwin, M. J. Walter, M. J. Holtzman, P. Madara, C. Logun, and J. H. Shelhamer. 2003. Interferon-γ induces p11 gene and protein expression in human epithelial cells through interferon-γ-activated sequences in the p11 promoter. J. Biol. Chem. 2789298-9308. [DOI] [PubMed] [Google Scholar]
- 27.Johnson, E., and G. Hetland. 1988. Mononuclear phagocytes have the potential to synthesize the complete functional complement systems. Scand. J. Immunol. 27489-493. [DOI] [PubMed] [Google Scholar]
- 28.Kalamvoki, M., J. Qu, and B. Roizman. 2008. Translocation and colocalization of ICP4 and ICP0 in cells infected with herpes simplex virus 1 mutants lacking glycoprotein E, glycoprotein I, or the virion host shutoff product of the UL41 gene. J. Virol. 821701-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato, A., M. Yamamoto, T. Ohno, M. Tanaka, T. Sata, Y. Nishyama, and Y. Kawaguchi. 2006. Herpes simplex virus 1-encoded protein kinase UL13 phosphorylates viral US3 protein kinase and regulates nuclear localization of viral envelopment factors UL34 and UL31. J. Virol. 801476-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katz, Y., and R. C. Strunk. 1988. Synovial fibroblast-like cells synthesize seven proteins of the complement system. Arthritis Rheum. 311365-1370. [DOI] [PubMed] [Google Scholar]
- 31.Kwak, H. B., S. W. Lee, H. M. Jin, H. Ha, S. H. Lee, S. Takeshita, S. Tanaka, H.-M. Kim, H.-H. Kim, and Z. H. Lee. 2005. Monokine induced by interferon-γ is induced by receptor activator of nuclear factor κB ligand and is involved in osteoclast adhesion and migration. Blood 1052963-2969. [DOI] [PubMed] [Google Scholar]
- 32.Laurent, A. M., J. J. Madjar, and A. Greco. 1998. Translational control of viral and host protein synthesis during the course of herpes simplex virus type 1 infection: evidence that initiation of translation is the limiting step. J. Gen. Virol. 792765-2775. [DOI] [PubMed] [Google Scholar]
- 33.Leopardi, R., C. Van Sant, and B. Roizman. 1997. The herpes simplex virus 1 protein kinase US3 is required for protection from apoptosis induced by the virus. Proc. Natl. Acad. Sci. USA 947891-7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levy, D. E., and A. Garcia-Sastre. 2001. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 12143-156. [DOI] [PubMed] [Google Scholar]
- 35.Liang, L., and B. Roizman. 2006. Herpes simplex virus 1 precludes replenishment of the short-lived receptor of tumor necrosis factor alpha by virion host shutoff-dependent degradation of its mRNA. J. Virol. 807756-7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin, R., R. S. Noyce, S. E. Collins, R. D. Everett, and K. L. Mossman. 2004. The herpes simplex virus ICP0 RING finger domain inhibits IRF3- and IRF7-mediated activation of interferon-stimulated genes. J. Virol. 781675-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melroe, G. T., L. Silva, P. A. Schaffer, and D. M. Knipe. 2007. Recruitment of activated IRF-3 and CBP/p300 to herpes simplex virus ICP0 nuclear foci: potential role in blocking IFN-β induction. Virology 360305-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mohr, I., and Y. Gluzman. 1996. A herpesvirus genetic element which affects translation in the absence of the viral GADD34 function. EMBO J. 154759-4766. [PMC free article] [PubMed] [Google Scholar]
- 39.Mossman, K. L., P. F. Macgregor, J. J. Rozmus, A. B. Goryachev, A. M. Edwards, and J. R. Smiley. 2001. Herpes simplex virus triggers and then disarms a host antiviral response. J. Virol. 75750-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Müller, S., and A. Dejean. 1999. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J. Virol. 735137-5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Munger, J., A. V. Chee, and B. Roizman. 2001. The US3 protein kinase blocks apoptosis induced by the d120 mutant of herpes simplex virus 1 at a premitochondrial stage. J. Virol. 755491-5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ogg, P. D., P. J. McDonell, B. J. Ryckman, C. M. Knudson, and R. J. Roller. 2004. The HSV-1 US3 protein kinase is sufficient to block apoptosis induced by overexpression of a variety of Bcl-2 family members. Virology 319212-224. [DOI] [PubMed] [Google Scholar]
- 43.Poon, A. P., and B. Roizman. 1997. Differentiation of the shutoff of protein synthesis by virion host shutoff and mutant γ134.5 genes of herpes simplex virus 1. Virology 22998-105. [DOI] [PubMed] [Google Scholar]
- 44.Poon, A. P., and B. Roizman. 2005. The herpes simplex virus 1 ICP22 regulates the accumulation of a shorter mRNA and of a truncated US3 protein kinase that exhibits altered functions. J. Virol. 798470-8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poon, A. P., H. Gu, and B. Roizman. 2006. ICP0 and the US3 protein kinase of herpes simplex virus 1 independently block histone deacetylation to enable gene expression. Proc. Natl. Acad. Sci. USA 1039993-9998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poon, A. P., L. Benetti, and B. Roizman. 2006. US3 and US3.5 protein kinases of herpes simplex virus 1 differ with respect to their functions in blocking apoptosis and in virion maturation and egress. J. Virol. 803752-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Powell, J. D., S. Boodoo, and M. R. Horton. 2004. Identification of the molecular mechanism by which TLR ligation and IFN-γ synergize to induce Mig. Clin. Dev. Immunol. 1177-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Purves, F. C., and B. Roizman. 1992. The UL13 gene of herpes simplex virus 1 encodes the functions for posttranslational processing associated with phosphorylation of the regulatory protein alpha 22. Proc. Natl. Acad. Sci. USA 897310-7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Purves, F. C., R. M. Longnecker, D. P. Leader, and B. Roizman. 1987. Herpes simplex virus 1 protein kinase is encoded by open reading frame US3 which is not essential for virus growth in cell culture. J. Virol. 612896-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Purves, F. C., W. O. Ogle, and B. Roizman. 1993. Processing of the herpes simplex virus regulatory protein alpha 22 mediated by the UL13 protein kinase determines the accumulation of a subset of alpha and gamma mRNAs and proteins in infected cells. Proc. Natl. Acad. Sci. USA 906701-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reynolds, A. E., E. G. Wills, R. J. Roller, B. J. Ryckman, and J. D. Baines. 2002. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 768939-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reynolds, A. E., L. Liang, and J. D. Baines. 2004. Conformational changes in the nuclear lamina induced by herpes simplex virus type 1 require genes UL31 and UL34. J. Virol. 785564-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schroder, K., P. J. Hertzog, T. Ravasi, and D. A. Hume. 2004. Interferon-γ: an overview of signals, mechanisms and functions. J. Leukoc. Biol. 75163-189. [DOI] [PubMed] [Google Scholar]
- 54.Sen, G. C. 2001. Viruses and interferons. Annu. Rev. Microbiol. 55255-281. [DOI] [PubMed] [Google Scholar]
- 55.Simpson-Holley, M., J. D. Baines, R. Roller, and D. M. Knipe. 2004. Herpes simplex virus 1 UL31 and UL34 gene products promote the late maturation of viral replication compartments to the nuclear periphery. J. Virol. 785591-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smiley, J. R. 2004. Herpes simplex virus virion host shutoff protein: immune evasion mediated by a viral RNase? J. Virol. 781063-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soh, J., R. J. Donnelly, S. Kotenko, T. H. Mariano, J. R. Cook, N. Wang, S. Emanuel, B. Schwartz, T. Miki, and S. Pestka. 1994. Identification and sequence of an accessory factor required for activation of the human interferon-γ receptor. Cell 76793-802. [DOI] [PubMed] [Google Scholar]
- 58.Soh, J., R. J. Donnelly, T. M. Mariano, J. R. Cook, B. Schwartz, and S. Pestka. 1993. Identification of a yeast artificial chromosome clone encoding an accessory factor for the human interferon γ receptor: evidence for multiple accessory factors. Proc. Natl. Acad. Sci. USA 908737-8741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67227-264. [DOI] [PubMed] [Google Scholar]
- 60.Strunk, R. C., D. M. Eidlen, and R. J. Mason. 1988. Pulmonary alveolar type II epithelial cells synthesize and secrete proteins of the classical and alternative complement pathways. J. Clin. Investig. 811419-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taddeo, B., and B. Roizman. 2006. The virion host shutoff protein (UL41) of herpes simplex virus 1 is an endoribonuclease with a substrate specificity similar to that of RNase A. J. Virol. 809341-9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taddeo, B., W. Zhang, and B. Roizman. 2006. The U(L)41 protein of herpes simplex virus 1 degrades RNA by endonucleolytic cleavage in absence of other cellular or viral proteins. Proc. Natl. Acad. Sci. USA 1032827-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Warren, H. B., P. Pantazis, and P. F. Davies. 1987. The third component of complement is transcribed and secreted by cultured human endothelial cells. Am. J. Pathol. 1299-13. [PMC free article] [PubMed] [Google Scholar]
- 64.Weber, F., G. Kochs, and O. Haller. 2004. Inverse interference: how viruses fight the interferon system. Viral Immunol. 17498-515. [DOI] [PubMed] [Google Scholar]
- 65.Wong, P., C. W. Severns, N. B. Guyer, and T. M. Wright. 1994. A unique palindromic element mediates gamma interferon induction of mig gene expression. Mol. Cell. Biol. 14914-922. [DOI] [PMC free article] [PubMed] [Google Scholar]