Abstract
During the assembly of human immunodeficiency virus type 1 (HIV-1) particles, the tetraspanin CD63 can be incorporated into the viral membrane. Indeed, cell surface tetraspanin microdomains that include CD63 have been proposed as sites for virus release. In addition, antibodies against CD63 can inhibit HIV infection of macrophages. In this cell type, HIV assembles into intracellularly sequestered plasma membrane domains that contain several other tetraspanins, including CD81, CD9, and CD53. CD63 is recruited to this domain following HIV infection. Together, these observations suggest that CD63 may have some function in the assembly of infectious virus particles and/or the infectivity of assembled virions. Here we have used RNA interference to knock down CD63 expression in monocyte-derived primary macrophages. We show that in the absence of CD63, HIV assembly is quantitatively comparable to that seen in CD63-expressing macrophages and that virus assembly occurs on compartments positive for CD81, CD9, and CD53. Moreover, the infectivity of macrophage-derived virus is unaffected by the loss of CD63. Together, our results indicate that at least in tissue culture, CD63 expression is not required for either the production or the infectivity of HIV-1.
During the assembly of human immunodeficiency virus type 1 (HIV-1) particles, a number of tetraspanins can be incorporated into the viral membrane (6, 23, 27). In particular, CD63, a tetraspanin found primarily on the internal membranes of late endosomal multivesicular bodies (MVBs), is efficiently incorporated into viral particles (23, 27, 30, 35). Although the cellular function of CD63 remains unclear, a number of studies have suggested that it may have a role in virus release and/or infectivity. Early studies showed that the density of CD63 is increased on HIV-infected H9 and blood-derived mononuclear cells compared to its density on uninfected control cells (20, 21). Subsequently, cell surface tetraspanin microdomains that include CD63 were proposed as sites for virus release (3, 25). In addition, antibodies against CD63, or a recombinant protein containing the large extracellular loop of CD63, can inhibit HIV infection of monocyte-derived macrophages (MDM) (11, 33). A particularly intriguing interaction with CD63 has previously been observed in HIV-infected MDM (8). In this cell type, ultrastructural analyses have shown that HIV assembles into intracellularly sequestered plasma membrane domains that contain several tetraspanins, including CD81, CD9, CD53, and CD63 (8). These domains appear to be associated with a complex array of membranes that, in uninfected cells, contain CD81, CD9, and CD53, but not CD63, i.e., following HIV infection and during the assembly of HIV particles, CD63 becomes detectable in this assembly domain and is incorporated into the membrane of budding viruses (8).
Thus, CD63 is closely associated with HIV assembly, particularly in MDM, where virus infection causes CD63 to accumulate in the assembly compartments and significant amounts of CD63 are incorporated into the viral membrane. This finding suggests that CD63 may have a role in facilitating HIV assembly and/or modulating the infectivity of virus particles. To investigate this suggestion, we have studied MDM in which CD63 expression was reduced by using RNA interference (RNAi). MDM lacking CD63 could be infected by HIV-1, but HIV production and the infectivity of this MDM-derived virus were not affected by the loss of CD63.
MATERIALS AND METHODS
Reagents and antibodies.
Tissue culture medium and supplements were obtained from Invitrogen (Paisley, United Kingdom), and tissue culture plastic was from TPP (Trasadingen, Switzerland). All chemicals were from Sigma-Aldrich (Dorset, United Kingdom), unless stated otherwise. The mouse monoclonal antibody (MAb) against CD63 (1B5, immunoglobulin G2b [IgG2b]) has previously been described (10). Anti-CD81 (M38, IgG1) was provided by F. Berditchevski (University of Birmingham, United Kingdom), and anti-CD53 (MEM-53, IgG1) and anti-CD9 (MCA469GA, IgG2b) were from Abcam Ltd. (Cambridge, United Kingdom) and Serotec (Oxford, United Kingdom), respectively. Anti-clathrin heavy chain (clone 23, IgG1), anti-LAMP-1 (H4A3, IgG1), and anti-LAMP-2 (H4B4, IgG1) antibodies were from BD Biosciences Europe (Erembodegem, Belgium), and anti-vesicular stomatitis virus G (VSV-G) (P5D4, IgG1) was from T. Kreis, University of Geneva, Geneva (Switzerland). MAbs to p24/p55 (MAbs 38:96K and EF7, both mouse IgG1, ARP365 and 366, respectively) and Env (MAbs b12, EVA3065 and 37.1.1, ARP372) were obtained through the NIBSC Centralised Facility for AIDS Reagents supported by the EU Programme EVA and the UK Medical Research Council. The rabbit antiserum UP595 against HIV p17 was provided by M. Malim (Guy's, King's, and St. Thomas' School of Medicine, London, United Kingdom). Alexa Fluor-labeled fluorescent secondary antibodies were from Invitrogen. Protein A-gold (PAG) reagents were from the EM Lab, Utrecht University, The Netherlands.
Cells and infections.
Peripheral blood mononuclear cells (PBMCs) were prepared from buffy coats from healthy donors (National Blood Service, Essex, United Kingdom), and adherent monocytes were isolated as described previously (27). Monocytes were differentiated to MDM in complete medium (RPMI 1640, 100 U/ml penicillin, 0.1 mg/ml streptomycin, and 10% human AB serum; PAA Laboratories GmbH, Pasching, Austria) by culturing with 10 ng/ml of macrophage colony-stimulating factor (R&D Systems, Oxon, United Kingdom) for 2 days after plating and then without macrophage colony-stimulating factor until required.
HIV-1BaL was originally obtained from R. Shattock (St. George's Hospital, London, United Kingdom). A first virus stock was prepared by the infection of PBMCs (grown in RPMI 1640 with penicillin, streptomycin, 10% fetal calf serum and 20 U/ml interleukin-2 [R&D Systems] and after treatment with 0.5 mg/ml Phaseolus vulgaris phytohemagglutinin for 3 days). PBMC-grown HIV-1BaL was passaged once through MDM to make an MDM-derived HIV-1 stock. Titers for virus stocks were determined on NP2-CD4-CCR5 cells as described previously (27). For experiments, MDM were infected with 3 focus-forming units (FFU)/cell of HIV-1BaL in a volume of 0.6 to 1 ml. During the next day, the medium was topped up to 3 ml and then changed 8 h later. Alternatively, at 7 days after isolation, MDM were infected with 3 FFU/cell HIV-1BaL by spinoculation at 2.5 krpm for 2 h at room temperature (RT) and cultured for various times as indicated. TAK-779 (50 μM; NIH Aids Research & Reference Reagents Program) was added to MDM medium when required to prevent subsequent reinfection.
Infectivity assay.
Single-cycle infectivities were determined by challenging TZM-β-galactosidase (β-gal) indicator cells (obtained from J. Martin-Serrano, Guy's, King's and St. Thomas' School of Medicine, London, United Kingdom) (34) with cell-free virus-containing supernatants, as described previously (2). Briefly, 2 × 104 cells/well in 24-well plates were infected with virus preparations containing 3 to 5 ng of p24 by spinoculation, as described above, and the induced expression of β-gal activity in cell lysates was measured 24 h later by using the Galacto-Star system from Applied Biosystems (Foster City, CA). β-gal activity was expressed as FFU per milliliter calibrated against known infectivity values from an infectivity assay on NP2-CD4-CCR5, as described previously (27). The number of infectious units per particle was calculated by assuming there are 5,000 Gag molecules per virion (4).
Knockdown of CD63 by siRNA.
Small interfering RNA (siRNA) was used to knock down the expression of CD63 in MDM. Two oligonucleotides targeting the CD63-specific sequences, 5′-GTTCTTGCTCTACGTCCTC-3′ (7) and 5′-GGAGAACTATTGTCTTATG-3′ (16), were purchased from Ambion (Austin, TX). A scrambled oligonucleotide with the sequence 5′-AATTCTCCGAACGTGTCACGT-3′ was purchased from Qiagen (Crawley, United Kingdom) and was used as a negative control. MDM, differentiated for 7 or 8 days, were nucleofected with 4 μg of scrambled or CD63-specific siRNAs by using a macrophage nucleofection kit (Amaxa Biosystems, Köln, Germany) according to the manufacturer's instructions. The levels of CD63 expression were subsequently analyzed by immunofluorescence and Western blotting.
Immunofluorescence.
For immunofluorescence staining of whole cells, MDM were fixed by adding an equal volume of double-strength fixative, 6% paraformaldehyde (TAAB, Aldermaston, United Kingdom), to the culture medium, for 30 min at RT. After being washed, free aldehyde groups were quenched with 50 mM NH4Cl and the cells were permeabilized for 1 h in blocking buffer (0.1% saponin, 0.5% bovine serum albumin [BSA] in phosphate-buffered saline [PBS] containing 10 μg/ml purified human IgG). Cells were stained for 1 h with primary antibodies diluted in blocking buffer, washed, and incubated for 1 h with appropriate combinations of fluorescent secondary antibodies. Appropriate controls to exclude cross-reactions were included. The cell nuclei were stained with Hoechst 33258, and the coverslips were mounted in Mowiol (Merck Biosciences Ltd., Beeston, Nottingham, United Kingdom) and examined directly or stored at −20°C. Staining was analyzed with a Nikon Optiphot-2 microscope equipped with an MRC Bio-Rad 1024 confocal laser scanner and a planapochromat 60× objective lens. Images were processed using Adobe Photoshop 8 or ImageJ.
For immunofluorescence staining of cryosections, 0.5-μm sections from cell samples prepared for cryosection electron microscopy (EM) were placed on glass slides, quenched in 50 mM glycine-50 mM NH4Cl, stained with antibodies diluted in PBS with 1% BSA, and mounted in Mowiol. Sections were examined with an Axioskop microscope (Carl Zeiss Ltd., Welwyn Garden City, Hertfordshire, United Kingdom) fitted with Plan-Neofluar oil immersion objectives. Images were recorded with a Hamamatsu Orca C4742-95 charge-coupled device camera controlled by Openlab 3.1.7 software (Improvision Ltd., Coventry, United Kingdom), converted to TIFF files, adjusted for brightness and contrast, and assembled into montages by using Adobe Photoshop 8.
EM.
Human MDM differentiated for 7 days were nucleofected with siRNA against CD63 or scrambled oligonucleotides. Two days later, half of the cells were infected with HIV-1BaL and the other half were left as uninfected controls. At 10 days postinfection (dpi), cells were fixed in 4% paraformaldehyde in 0.1 M sodium phosphate buffer, pH 7.4, embedded in gelatin, and frozen for cryosectioning as described previously (8, 29). Ultrathin (50-nm) cryosections were stained with mouse IgG1 MAb against LAMP-1 (H4A3) or HIV p24/p55 (38:96K and EF7), a rabbit anti-mouse bridging antibody (DakoCytomation, Ely, United Kingdom), and 15 nm PAG. Sections were fixed in 1% glutaraldehyde for 10 min, quenched in 50 mM glycine-50 mM NH4Cl in PBS and incubated with the IgG1 MAb P5D4 against the VSV-G protein to block any IgG1-reactive sites on the bridging antibody. After a further round of fixation in 1% glutaraldehyde (which destroys protein A binding sites) and requenching as above, sections were stained with the anti-CD63 antibody 1B5 (IgG2b) and 5 nm PAG. Sections were embedded in uranyl acetate in methylcellulose as described previously (29) and examined with a CM10 transmission electron microscope (FEI Company UK Ltd., Cambridge, United Kingdom), and images were recorded onto Kodak SO-163 electron image film. Negatives were digitized with a FlexTight Precision II rotating drum charge-coupled device scanner (Imacon), and TIFF images were acquired with the ColorFlex 1.9.5 FlexTight Interface software. Images were adjusted for brightness and contrast, and figures were assembled with Adobe Photoshop 8.
p24 assay.
Supernatants from HIV-1BaL-infected cells were centrifuged for 8 min at 1,200 rpm to remove contaminating cells, and p24 levels in the supernatants were then assayed by enzyme-linked immunosorbent assay (AIDS Vaccine Program, NCI-Frederick Cancer Research and Development Center, Frederick, MD) by following the manufacturer's instructions.
Western blotting.
For Western blot analysis, 2 × 106 MDM were lysed in cell lysis buffer (150 mM NaCl, 1.0% NP-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 50 mM Tris-Cl) with protease inhibitors (Roche Diagnostic, Basel, Switzerland) at 4°C. The lysates were centrifuged at 8,000 rpm for 5 min, separated on 10% SDS-polyacrylamide gel electrophoresis gels under nonreducing conditions, transferred to nitrocellulose membranes (Protran, SLS, Nottingham, United Kingdom) at 10 V for 30 min, quenched for 20 min at RT with blocking buffer (PBS-0.1% Tween-milk 5%), and then immunoblotted for CD63, Gag, Env (MAb 37.1.1), or clathrin for 1 h at RT. Subsequently, the blots were washed three times for 5 min with PBS-0.1% Tween and the membranes were then incubated with a goat anti-mouse horseradish peroxidase conjugate purchased from Perbio (Tattenhall, Cheshire, United Kingdom) for 1 h at RT. After three washes of 5 min with PBS-0.1% Tween, membranes were incubated for 5 min in Supersignal West Femto luminol (Perbio). Signals were detected with Hyperfilm ECL from Amersham Biosciences (Little Chalfont, United Kingdom), and films were scanned and quantified using Quantity One software from Bio-Rad (Hemel Hempstead, Herts, United Kingdom).
Proviral load analysis.
An Alu-long terminal repeat (LTR)-based real-time nested-PCR assay was used to quantify integrated HIV-1BaL in control and CD63 knockdown MDM as described previously (5). Genomic DNA concentrations were normalized with real-time PCR quantification of the β-globin gene, adapted from a previous study (1). Levels of HIV-1 DNA and the β-globin gene were determined by reference to standard curves generated by serial dilutions of purified PCR amplification products and are expressed in arbitrary units.
Virus immunoprecipitation.
For immunoprecipitation, we followed a procedure described previously (27). Briefly, HIV-1BaL cell-free supernatants were diluted to 1 × 106 FFU/ml in PBS containing 3% (wt/vol) BSA and incubated at 37°C for 1 h with antibodies against CD63, VSV-G, or Env (MAb b12; all MAbs were at 10 μg/ml in a final volume of 100 μl). Pansorbin cells (formalin-fixed Staphylococcus aureus; Calbiochem) were washed and blocked for 1 h at RT in PBS containing 3% (wt/vol) BSA and then used to capture the virus antibody mixture for 1 h at RT. Pansorbin cell-virus complexes were sedimented by centrifugation at 4,000 rpm for 30 min at 4°C. The supernatants, containing the unprecipitated viruses, were used to infect TZM-β-gal indicator cells as described above. Infection levels were assessed by measuring β-gal activity in the TZM-β-gal cell lysates after 24 h.
RESULTS
CD63 knockdown in MDM.
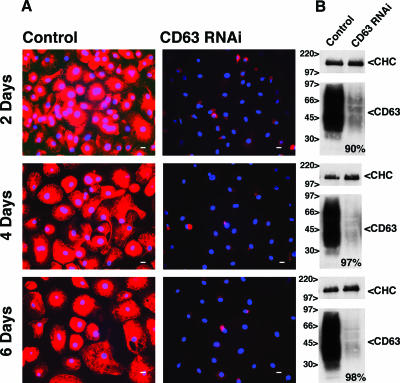
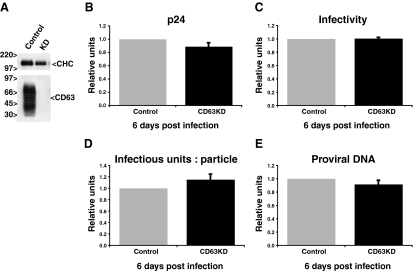
RNAi-mediated reduction of CD63 expression has previously been shown to modulate the cell surface expression of H+K+-ATPase (7). We therefore used the same target sequence to reduce CD63 expression in MDM. Seven days after monocyte isolation, siRNA was introduced into cultured MDM by nucleofection. Immunofluorescence staining demonstrated that within 2 days of treatment with CD63 siRNA, CD63 had disappeared from most cells in the culture (Fig. 1A, top panel). This result was confirmed by Western blotting of cell lysates, which revealed a reduction in the cellular CD63 levels of about 90% (Fig. 1B, top panel). A further reduction of CD63 expression was seen at 4 and 6 days postnucleofection (97 and 98% of control cells, respectively) (Fig. 1A and B, middle and bottom panels), and low CD63 expression levels were maintained for up to 2 weeks (see below). Similar efficiencies of CD63 knockdown were seen consistently with MDM obtained from different donors.
FIG. 1.
CD63 knockdown in MDM. Seven days after monocyte isolation (day 0), MDM were nucleofected with scrambled or CD63-specific siRNAs. CD63 knockdown was assessed by immunofluorescence and by Western blot analysis 2, 4, and 6 days after nucleofection. (A) Permeabilized cells were stained with anti-CD63, followed by Alexa Fluor 594 anti-mouse and Hoechst dye for immunofluorescence analysis. Scale bars = 10 μm. (B) For Western blot analysis, 2 × 106 MDM were solubilized in lysis buffer. Cell proteins were separated in nonreducing buffer on 10% SDS-polyacrylamide gel electrophoresis gels and immunoblotted for CD63 and clathrin heavy chain (CHC). CD63 runs as a broad smear due to glycosylation (18). The blots were quantified using Bio-Rad Quantity One software, and band intensities were normalized against clathrin. The loss of CD63 is expressed as a percentage of the CD63 levels in control cells (bottom right in the blots).
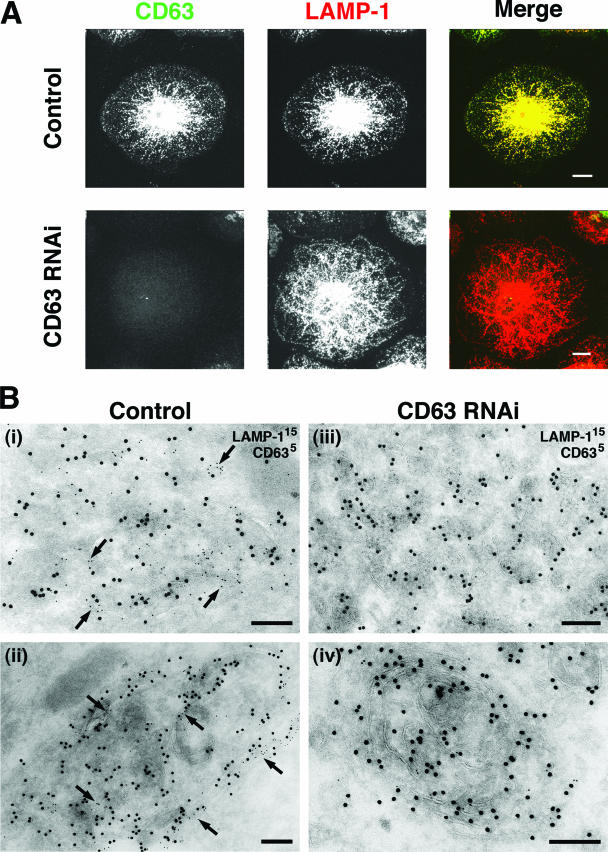
Since CD63 is a late endosome/lysosome marker that colocalizes with the lysosomal membrane proteins LAMP-1 and LAMP-2 in MDM (8), we investigated whether CD63 knockdown would affect the distribution and/or morphology of lysosomes in these cells. As observed previously, CD63 is primarily located in an extensive network of tubulovesicular organelles in MDM that costained almost completely with LAMP-1 (Fig. 2A, top panels) (8). In cells lacking CD63, the overall LAMP-1 distribution was indistinguishable from that seen in the controls (Fig. 2A, lower panels), indicating that CD63 is not required for the maintenance of the late endosomal/lysosomal morphology. Similarly, CD63 knockdown did not affect the distribution of LAMP-2 (data not shown). The lack of an effect of CD63 depletion on lysosome morphology was also confirmed by EM immunolabeling. Ultrathin cryosections of MDM with or without CD63 knockdown were colabeled for LAMP-1 and CD63. Control cells showed the tubulovesicular late endosome/lysosome structures that have previously been described for these cells (8) as well as larger, more electron-dense organelles with lamellar membranes that were strongly labeled for both LAMP-1 and CD63 (Fig. 2B, panels i and ii). Morphologically similar structures were observed in the CD63 knockdown MDM, but these structures were labeled only for LAMP-1 (Fig. 2B, panels iii and iv). This analysis supports the view that CD63 is not required to maintain late endosomal/lysosomal morphology in MDM.
FIG. 2.
Distribution of lysosomal markers in CD63 knockdown MDM. MDM were nucleofected with scrambled or CD63-specific siRNAs and returned to culture for 2 to 12 days. (A) Cells were fixed 6 days after nucleofection, permeabilized, and stained with MAbs against CD63, followed by anti-mouse IgG2b Alexa Fluor 488, and against LAMP-1, followed by anti-mouse IgG1 Alexa Fluor 594. Images show projections of 22 and 23 confocal sections (0.5 μm thick) for the control and CD63 RNAi samples, respectively. Scale bars = 10 μm. Observations similar to those above were made 2 and 4 days after nucleofection. (B) EM immunolabeling of control (panels i and ii) or CD63 knockdown MDM (panels iii and iv) 12 days after nucleofection. Ultrathin cryosections were double labeled with MAbs against LAMP-1/PAG (15 nm) and CD63/PAG (5 nm). Arrows indicate CD63 staining. Scale bars = 200 nm.
HIV localizes to the tetraspanin-enriched compartment in CD63 knockdown MDM.
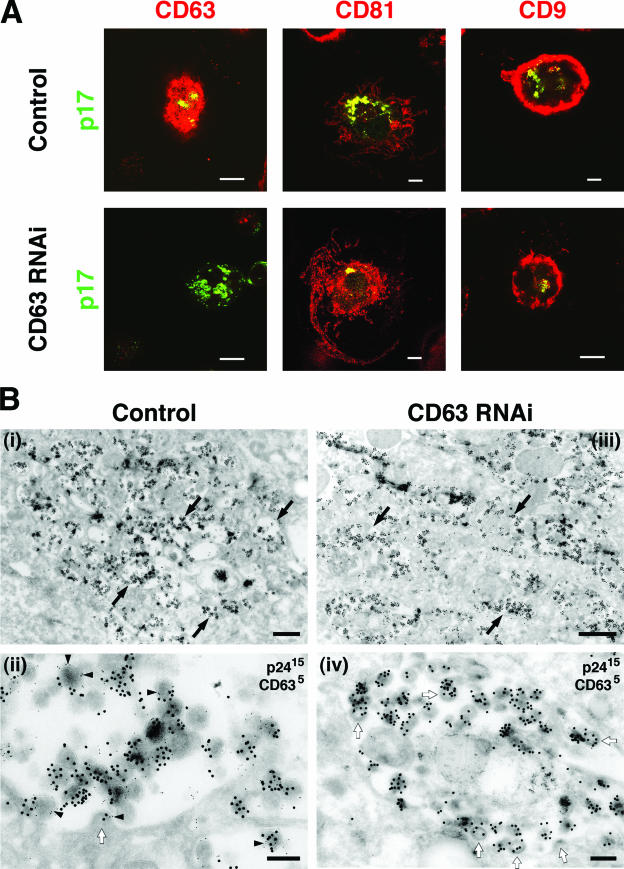
To determine whether MDM lacking CD63 can be infected with HIV, MDM were nucleofected with siRNA against CD63 and 2 days later were exposed to HIV-1BaL. After 10 days, the cells were fixed and the presence and distribution of HIV was assessed by immunofluorescence with anti-p17 (MA) antibodies. HIV staining was readily detected in these cells, showing that MDM lacking CD63 can be infected by HIV-1 (see below). Previous studies have indicated that HIV assembly in MDM occurs in a complex, intracellularly located, plasma membrane-linked compartment containing the tetraspanins CD81, CD9, and CD53 (8). Accordingly, double staining showed that labeling for HIV p17 (MA) was associated with intracellular CD81- and CD9-containing structures (Fig. 3A, top row). In addition, the virus-containing compartment also labeled for CD63 (Fig. 3A, top row, left panel). In the CD63 knockdown MDM, p17 labeling also colocalized with CD81, CD9 (Fig. 3A, bottom row), or CD53 (data now shown), even though CD63 was almost completely absent from these cells (Fig. 3A, bottom row, left panel).
FIG. 3.
CD63 is not required for HIV-1 targeting to the assembly compartment. MDM were nucleofected with scrambled or CD63-specific siRNA as described in the text. Two days later, the cells were infected with HIV-1BaL. At 10 dpi the cells were analyzed by immunofluorescence or by EM. (A) For immunofluorescence analysis, cells were fixed, permeabilized, and stained with mouse MAbs against the tetraspanin CD63, CD81, or CD9, followed by Alexa Fluor 594 anti-mouse (red), and costained with rabbit anti-HIV-1 p17 (MA) followed by Alexa Fluor 488 anti-rabbit (green). Single confocal sections are shown. Areas where the markers colocalize appear yellow. Scale bars = 10 μm. (B) For EM analysis, ultrathin cryosections of control (panels i and ii) and CD63 knockdown MDM (panels iii and iv) were double labeled with MAbs against the HIV capsid protein p24 and PAG (15 nm) and with CD63/PAG (5 nm). Black arrows in panels i and iii indicate a virus-filled vacuole. The higher-magnification views in panels ii and iv show mature virus particles as well as immature virions or buds (white arrows). CD63 is incorporated into the HIV particles (arrowheads in panel ii). Scale bars, 1 μm (panels i and iii) and 200 nm (panels ii and iv).
When HIV-infected MDM were examined by immunolabeling EM, HIV particles were observed to accumulate in a complex array of apparently intracellular structures (Fig. 3B, panel i), which have previously been described as connected to the cell surface (8). The assembling virus particles were strongly labeled with antibodies to the HIV capsid protein p24. In addition, CD63 staining was associated with the virus assembly compartment and CD63 could be detected in the viral membranes and being incorporated into budding virions (Fig. 3B, panel ii) (27). In CD63 siRNA-treated cells, virus particles accumulated in similar intracellular compartments and both immature and mature virus particles of sizes and morphologies similar to those seen in control cells were labeled with anti-p24 antibodies. However, these virus-containing compartments and viruses did not label for CD63 (Fig. 3B, panel iv).
Together, these data indicate that CD63 knockdown MDM can be infected with HIV and can assemble mature virus particles in compartments that are morphologically indistinguishable from the virus assembly compartments seen in untreated cells. The compartments contain CD81, CD9, and CD53. Thus, CD63 does not appear to be required for the assembly of mature virus particles or the organization of the assembly compartments in MDM.
CD63 is not required for the production of infectious HIV-1 from MDM.
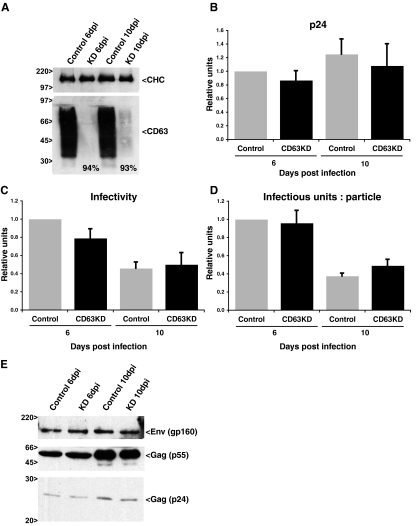
Although the morphology of uninfected and HIV-infected MDM was not affected following CD63 knockdown, and the cells appeared healthy and viable for periods in excess of 12 days after CD63 knockdown, it is possible that the amount of virus production and the quality of the virus particles was affected by the loss of CD63. To investigate this possibility, MDM were infected with HIV-1BaL 2 days after nucleofection with scrambled oligonucleotides or siRNA against CD63. At this time point, a nearly complete reduction in the level of CD63 expression was obtained (Fig. 1, top panels). The infected MDM were cultured for several days to allow virus to accumulate. At 5 and 9 dpi, the medium was changed. Twenty-four hours later, supernatants were collected, centrifuged to remove cells and debris, and analyzed for their p24 content and infectious virus levels. In parallel, cell lysates were analyzed by Western blotting with antibodies against CD63, Env, and Gag (p24 and p55).
At both 6 and 10 dpi, CD63 expression in the siRNA-treated cells was almost undetectable by Western blotting (Fig. 4A), indicating that the siRNA-mediated knockdown of CD63 was stable for long periods. Assays of p24 levels in the 6- and 10-dpi supernatants showed that MDM treated with CD63 siRNA produced similar amounts of virus to control cells, with virus release at 10 dpi slightly higher than that at 6 dpi. (Fig. 4B and Table 1). Levels of infectious viruses in the supernatants were determined in single-cycle infectivity assays on TZM-β-gal indicator cells. The infectious virus levels present in supernatants from control or CD63 knockdown MDM were very similar, although infectivity was reduced by about 50% at 10 dpi compared to the level for the 6-dpi samples (Fig. 4C and Table 1). Likewise, the ratios for infectious units per particle (the number of infectious FFUs divided by the number of virus particles) showed little difference between control and CD63 knockdown MDM-derived samples (Fig. 4D and Table 1).
FIG. 4.
Production of infectious HIV-1 is not affected by CD63 knockdown. MDM were nucleofected with scrambled or CD63-specific siRNA and infected with HIV-1BaL as described in the text. (A) CD63 expression levels were assessed by Western blot analysis at 6 and 10 dpi. The clathrin heavy chain (CHC) was analyzed as a loading control. The percentage of knockdown is shown in the relevant lanes. (B to D) Cell-free supernatants from control and CD63 knockdown (KD) HIV-1BaL-infected MDM were assayed for p24 levels (B) or infectivity (C), and the number of infectious units per particle were calculated (D). Data from experiments with five different donors were averaged and are represented as relative units after normalizing with respect to control values of p24, infectivity, and infectious units per particle at 6 dpi. For panels B through D, bars represent the average relative units, and error bars represent standard errors of the means. (E) Env and Gag (p24 and pr55) expression levels in control and CD63 knockdown HIV-infected MDM at 6 and 10 dpi were assessed by Western blotting.
TABLE 1.
Analysis of infectious virus production from control and CD63 knockdown MDM at 6 and 10 dpi
| dpi | Experiment no. | p24 level (ng/ml) for:
|
Infectivity level (105 FFU/ml) for:
|
Infectious unit per particleb for:
|
|||
|---|---|---|---|---|---|---|---|
| Control | KDa | Control | KD | Control | KD | ||
| 6 | 1 | 6.21 | 7.31 | 5.19 | 5.54 | 0.0167 | 0.0152 |
| 2 | 3.06 | 3.67 | 3.32 | 3.03 | 0.0217 | 0.0165 | |
| 3 | 4.94 | 2.73 | 4.66 | 3.89 | 0.0189 | 0.0284 | |
| 4 | 13.17 | 7.62 | 17.40 | 8.35 | 0.0264 | 0.0219 | |
| 5 | 11.59 | 9.60 | 7.62 | 4.96 | 0.0132 | 0.0103 | |
| 10 | 1 | 10.20 | 13.40 | 3.17 | 4.85 | 0.0062 | 0.0072 |
| 2 | 5.77 | 4.50 | 1.86 | 1.88 | 0.0064 | 0.0083 | |
| 3 | 3.91 | 2.81 | 1.56 | 1.16 | 0.0080 | 0.0083 | |
| 4 | 10.50 | 6.78 | 4.46 | 3.88 | 0.0085 | 0.0114 | |
| 5 | 13.09 | 8.03 | 4.02 | 4.03 | 0.0061 | 0.0100 | |
KD, CD63 knockdown MDM.
The number of infectious units per particle was calculated from the infectivity in FFU/ml and the p24 levels, assuming 5,000 Gag molecules per virion (4).
In order to correlate the levels of virus production in control and CD63-depleted cells with the levels of virus protein in the MDM, cell lysates underwent Western blotting with antibodies against the viral Gag proteins (p55 and p24) or the viral envelope protein (Env). Again, similar levels of Env and Gag proteins were found in lysates from the CD63 knockdown cells compared to the levels in cells nucleofected with scrambled siRNAs (Fig. 4E).
Previous studies have indicated that antibodies to CD63 can inhibit HIV-1 infection at an early step of the virus life cycle, prior to reverse transcription (33). Accordingly, it is possible that the reduced levels of CD63 following siRNA treatment might also influence virus entry. In order to exclude any effects of CD63 knockdown on virus entry, MDM were infected with HIV-1BaL on day 7 of differentiation. Then, 1 day later, the infected MDM were nucleofected with scrambled oligonucleotides or with siRNA against CD63. After nucleofection, the cells were recultured in the presence of 50 μM TAK779, a CCR5 antagonist and HIV entry inhibitor used to block multiple rounds of infection. Four days later (i.e., 5 dpi) the medium was replaced, and 24 h later (at 6 dpi) cell-free supernatants from control and CD63-depleted MDM were harvested and assayed for p24 and infectivity, as described above. Western blot analysis of lysates from the cells confirmed a successful CD63 knockdown (Fig. 5A). The levels of p24 released into the supernatant were similar in control and CD63 siRNA-treated MDM (Fig. 5B and Table 2). Likewise, infectivity assays on the TZM-β-gal indicator cells showed no significant differences in the number of infectious viruses released (Fig. 5C and Table 2). The number of infectious units per particle was also similar in control and CD63 knockdown cells (Fig. 5D and Table 2). In order to confirm similar levels of virus entry in the MDM prior to CD63 knockdown, we measured the proviral DNA levels in CD63-depleted and control MDM. DNA from control and CD63 knockdown cells was extracted at 6 dpi and assayed in an Alu-LTR-based, real-time, quantitative, nested PCR; the proviral levels for each sample were normalized to the genomic DNA content using real-time PCR quantification of the β-globin gene. We confirmed similar proviral loads in the control and CD63-depleted MDM with this experimental protocol (Fig. 5E and Table 2).
FIG. 5.
Production of infectious HIV-1 is not affected in CD63 knockdown MDM with the same levels of infection. MDM were infected with HIV-1BaL 7 days after monocyte isolation (day 0) and nucleofected with scrambled or CD63-specific siRNAs 1 day after infection. Cells were returned to culture in medium containing the entry inhibitor TAK779 to avoid multiple rounds of infection. After 5 days, the medium was changed and virus-containing supernatants were collected 24 h later (i.e., at 6 dpi). (A) CD63 expression levels were assessed by Western blot analysis at 6 dpi. CHC was analyzed as a loading control. KD, CD63 knockdown MDM. (B to D) Cell-free supernatants from CD63 knockdown and control MDM were assayed for p24 levels (B) and infectivity (C), and the number of infectious units per particle were calculated (D). (E) In addition, an Alu-LTR-based real-time nested-PCR assay was used to quantify the integrated HIV DNA levels in control and CD63 knockdown MDM. Data corresponding to four experiments with three different donors were averaged and are represented as relative units after normalization with respect to control values at 6 dpi of p24, infectivity, infectious units per particle, or proviral DNA. For panels B through E, bars represent the average relative units, and error bars represent standard errors of the means.
TABLE 2.
Analysis of infectious virus production from control and CD63 knockdown MDM at 6 dpi
| Experiment no. | p24 level (ng/ml) for:
|
Infectivity level (105 FFU/ml) for:
|
Infectious unit(s) per particleb (10−3)
|
Proviral DNAc (au) for:
|
||||
|---|---|---|---|---|---|---|---|---|
| Control | KDa | Control | KD | Control | KD | Control | KD | |
| 6 | 8.74 | 8.13 | 0.07 | 0.07 | 0.15 | 0.17 | 50.3 | 41.0 |
| 7 | 7.58 | 7.65 | 0.22 | 0.22 | 0.58 | 0.56 | 55.9 | 45.9 |
| 8 | 10.55 | 9.24 | 1.97 | 1.92 | 3.74 | 4.15 | 84.0 | 83.1 |
| 9 | 8.08 | 5.97 | 1.95 | 2.04 | 4.84 | 6.82 | 66.5 | 69.3 |
KD, CD63 knockdown MDM.
The number of infectious units per particle was calculated from the infectivity in FFU/ml and the p24 levels, assuming 5,000 Gag molecules per virion (4).
Proviral DNA is expressed as arbitrary units (au) based on the ratio of the integrated HIV-1 DNA level to the β-globin gene level.
An alternative target sequence for CD63 knockdown does not affect the production of infectious HIV-1.
To ascertain that these results were independent of the target sequence for CD63 knockdown, an alternative oligonucleotide was used to downregulate CD63 levels (16). The reduction of CD63 expression with this siRNA was similar to that obtained with the other target sequence. Again, analyses of the production of infectious HIV-1 showed no differences between control and CD63 knockdown cells. Quantitative PCR also revealed no change in the proviral load in control and CD63-depleted MDM (data not shown). Thus, two different target sequences were both effective in reducing CD63 expression and both had little, if any, effect on the production of infectious HIV-1 from MDM.
Virions from knockdown MDM do not incorporate CD63.
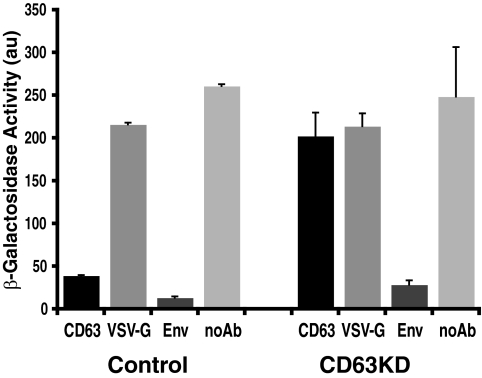
During HIV particle assembly, CD63 is incorporated into the viral envelope (23, 27). EM analysis demonstrated CD63 staining in virus particles in control cells, but CD63 could not be detected in virions in CD63-depleted MDM (Fig. 3B). To confirm the absence of CD63 in viruses derived from cells treated with CD63 siRNA, virus-containing cell-free supernatants were isolated from control and CD63 knockdown MDM and analyzed using a previously characterized immunoprecipitation/immunodepletion assay (27) that detects proteins incorporated into infectious virus particles. The virus-containing supernatants were incubated with antibodies against CD63, or with antibodies against VSV-G (as a negative control) or the viral Env glycoprotein (positive control), and the virus-antibody mixtures were then treated with Pansorbin cells to immunoprecipitate viruses. The infectivity remaining in the supernatant was assessed in TZM-β-gal indicator cells and expressed as β-gal activity (Fig. 6). Almost no infectivity was found in the virus supernatants treated with anti-Env antibody. Virus samples from control cells showed an 82% reduction of infectivity when treated with anti-CD63 antibody compared to samples treated with the anti-VSV-G antibody. High infectivity levels were found in the untreated sample. However, infectivity levels were similar in the supernatants from CD63 knockdown cells treated with anti-CD63 or anti-VSV-G antibodies (Fig. 6). These data confirm that viruses derived from CD63-depleted MDM do not incorporate CD63 into their envelopes.
FIG. 6.
Immunoprecipitation of virus particles from control and CD63 knockdown MDM. Cell-free supernatants from control and CD63 knockdown MDM-derived viruses were incubated at 37°C for 1 h with antibodies against CD63, VSV-G (negative control), and Env (positive control). The virus-antibody mixture was incubated with Pansorbin cells, and viruses were precipitated by centrifugation. The supernatants, containing the unprecipitated viruses, were used to infect TZM-β-gal indicator cells. β-Gal activity was measured 24 h after infection and was expressed as arbitrary units (au). Bars represent averages of values from six different dilutions per sample from a representative experiment. Error bars represent the standard errors of the means. noAb, untreated sample.
Failure to recruit CD63 by an HIV-1BaL stock passaged through MDM.
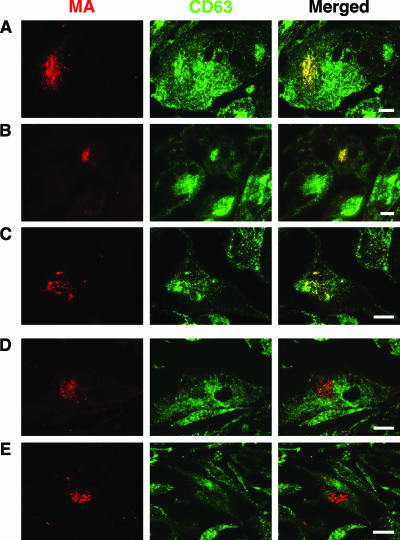
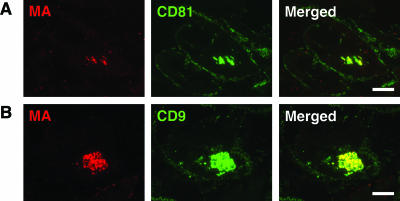
We and others have previously shown that in MDM, HIV-1 accumulates in intracellular structures that label strongly for CD63 (8, 27, 30) and that the virus particles incorporate CD63 into their membranes (8, 27). Interestingly, we observed one stock of HIV-1BaL that lost the ability to recruit CD63 to the virus assembly compartment after it was passaged repeatedly through MDM. This was particularly apparent when HIV-infected MDM preparations from a number of different donors were compared by immunofluorescence staining of semithin cryosections with a rabbit antiserum to HIV-1 MA and a mouse monoclonal antibody against CD63. Sections prepared from various frozen specimens of MDM infected with HIV-1BaL for various times (7 to 20 days) all showed a strong association of CD63 with the virus-containing intracellular compartments (Fig. 7A to C); indeed, the virus-containing areas could be identified in the CD63-stained images as strongly stained structures of irregular shape, often larger than the fine tubulovesicular late endosome/lysosome structures (Fig. 7, middle column). By contrast, even though MDM samples prepared from the serially passaged HIV-1BaL stock showed high levels of infection, there was no accumulation of CD63 in the virus-containing compartments (Fig. 7D and E). Single-color images of the CD63 staining tended to show a void in the area where the virus-containing compartment was located. Nonetheless, the passaged HIV-1BaL still assembled in the tetraspanin-containing, surface-connected intracellular compartment, where it colocalized with CD81 or CD9 (Fig. 8) (8). Despite the lack of recruitment of CD63, viruses from these cells could infect fresh MDM cultures. These results again demonstrate that CD63 is not required for the production of infectious HIV-1 in MDM.
FIG. 7.
Colocalization of the HIV matrix protein with CD63 on HIV-infected MDM. Semithin cryosections from human MDM samples infected with HIVBaL for 7 days (A), 14 days (B), or 20 days (C) were stained with a rabbit antiserum against HIV p17 and a mouse MAb to CD63, followed by anti-rabbit Alexa Fluor 594 and anti-mouse Alexa Fluor 488. Sections were examined with an Axioskop fluorescence microscope. The HIV-containing structures (red, left column) costained strongly with CD63 (green, center column), giving a yellow color in the merged images (right column). This distribution was seen for MDM cultures infected for various lengths of time, in single infected cells or syncytia, and for virus-containing compartments of various sizes and morphologies. By contrast, MDM samples infected with a passaged stock of HIV-1BaL for 8 days (D) or 13 days (E) did not accumulate CD63 in the virus-containing compartment, which appears red even in the merged images (right column). Scale bars = 10 μm.
FIG. 8.
Serially passaged HIV-1BaL assembles in a CD81- and CD9-containing compartment. Semithin cryosections from MDM infected with the passaged stock of HIV-1BaL for 13 days were stained with a rabbit antiserum against HIV p17 and mouse MAbs to CD81 (A) or CD9 (B), followed by anti-rabbit Alexa Fluor 594 and anti-mouse Alexa Fluor 488. The HIV-containing structures (red, left column) costained strongly with CD81 and CD9 (green, center column), giving a yellow color in the merged images (right column). Scale bars = 10 μm.
DISCUSSION
The tetraspanin CD63 (LAMP-3) has previously been described as a late endosome or lysosome marker that colocalizes with the lysosomal glycoproteins LAMP-1 and -2 (22). CD63 is expressed at high levels in late endosomes with the appearance of MVBs, where it is enriched in the intralumenal vesicles, and has previously been found in exosomes, small (50 to 100 nm diameter) vesicles secreted from cells as a result of the fusion of MVBs with the plasma membrane (9). Low levels of CD63 are also found at the plasma membrane. As a tetraspanin, CD63 contains four transmembrane domains and two extracellular loops, of which the second larger loop contains three putative N-glycosylation sites. Short N- and C-terminal domains are located in the cytoplasm, and the C-terminal domain contains sorting information, in particular a GYEVM motif that controls CD63 sorting to late endosomes and lysosomes through interaction with AP-2 and AP-3 adaptor complexes (12, 13, 31). Other adaptors, such as syntenin-1, have also been implicated in CD63 trafficking (17). The functions of CD63 are poorly understood. CD63 has previously been proposed to be a component of tetraspanin-enriched microdomains (TEMs). Together with other tetraspanins (e.g., CD9, CD81, and CD82), CD63 has previously been shown to modulate the migration of dendritic cells, where it may participate actively in the processing of complex antigens through its association with major histocompatibility complex class II molecules (19). There have also been suggestions that CD63 may be involved in T-cell activation concomitant with T-cell receptor triggering (28).
CD63 has also been implicated in HIV infection. CD63 is incorporated into virus particles (6, 23, 27) and, in model tissue culture cells or in T-cell lines, is present at sites where particles assemble (3, 23, 24, 27). Indeed, it has previously been suggested that HIV buds from TEMs at the surface of these cells (3, 25). In MDM, HIV particles have been shown to accumulate at intracellular sites. Recent studies have shown that nonetheless, these structures are connected to the cell surface (8, 15, 35) and thus represent unusual plasma membrane domains. This intracellular plasma membrane compartment is enriched in the tetraspanins CD81, CD9, and CD53 (8), but CD63 also accumulates in the compartment after HIV infection (8). Furthermore, antibodies against CD63 have been shown to inhibit the infection of MDM by R5 or dual-tropic HIV-1 strains (33) and the recombinant large extracellular domain 2 of CD63 has previously been reported to inhibit HIV infection of MDM (11). Together, these observations suggested that CD63 may have important functions in HIV infection.
To investigate the role of CD63 in HIV assembly and infectivity directly, we set up conditions to efficiently knock down CD63 expression in primary human MDM. CD63 levels were dramatically reduced within 2 days of nucleofection with CD63-specific siRNAs, and the reduced expression was maintained over more than 12 days. Despite this long-term suppression of CD63 expression, the MDM looked normal and there was no obvious effect on cell viability. Immunolabeling studies showed that neither the expression nor the distribution of the lysosomal markers LAMP-1 and -2 was affected by the CD63 knockdown. EM immunolabeling showed that LAMP-1-containing lysosomes, including tubulovesicular structures and larger, more electron-dense multilaminar lysosomes, appeared similar to those seen in CD63-expressing control cells. Thus, CD63 is not required for the maintenance of lysosome structure. Similarly, the distribution of other tetraspanins, including CD81, CD9, and CD53, was not altered in the absence of CD63.
Despite the profound depletion of CD63, we found that HIV could infect MDM lacking CD63 and, moreover, that virus was also produced from these cells. Immunofluorescence and EM studies showed that HIV still assembled in the CD81- and CD9-containing compartment as in wild-type MDM, and the ultrastructure of the compartment and of the budding virions appeared unchanged, although they lacked CD63. Therefore, the CD81/CD9/CD53-containing compartment appears to be stable in the absence of CD63 and HIV assembles normally in cells lacking CD63. Furthermore, quantitative analysis indicated that the depletion of CD63 had no effect on virus production or infectivity. This was surprising, given that anti-CD63 antibodies or soluble CD63 extracellular domain 2 can block HIV infection (11, 33). These reagents mainly affect virus entry, while our study has focused on the role of CD63 in virus assembly. To eliminate possible effects of CD63 knockdown on virus entry, we also infected MDM with HIV-1BaL before treatment of the cells with CD63 siRNA and blocked multiple rounds of infection by including the entry inhibitor TAK779. With this protocol, we also saw no differences in the production of infectious viruses between CD63 knockdown MDM and controls. The same result was obtained when a second siRNA targeting a different region of CD63 was used. Thus, CD63 does not appear to be required for HIV infection of MDM or for the production of infectious HIV-1 in these model tissue culture scenarios.
Further evidence for the dispensability of CD63 for the production of infectious HIV-1 in MDM came from the observation that an HIV-1BaL stock that had been passaged repeatedly through MDM failed to recruit CD63 to the virus assembly compartment. Although these viruses still assembled in the CD81- and CD9-containing tetraspanin compartment, we did not see any colocalization with CD63, regardless of the time in culture or the level of infection. Again, this result suggests that CD63 is not essential for the production of infectious HIV-1. Rather the selection of this virus suggests that in the culture system, there may be a competitive advantage for virions that do not recruit CD63. How CD63 is targeted to the virus assembly compartment and incorporated into budding HIV particles is still unclear, as no specific interaction between CD63 and viral components has been demonstrated to date.
Although CD63-containing, tetraspanin-enriched domains appear to be essential sites for HIV assembly, both at the cell surface in T cells and tissue culture cell lines (25) and in the intracellular plasma membrane-connected domains in MDM (8), CD63 appears not to be essential for HIV infection, for the structure of the HIV assembly platforms or the targeting of viral components to these sites, or for the production of infectious viruses. Hence, other tetraspanins and/or other molecules present in the TEMs, such as the lipid phosphatidylinositol 4,5-bis-phosphate (26, 32), may play a more important role in HIV assembly in MDM. Since there is significant functional overlap and redundancy among the various tetraspanins, further studies will be needed to assess the importance of CD81 and other tetraspanins or their role in HIV infection of other cell types (such as T cells, dendritic cells, or monocytes) and in HIV infection in vivo. Preliminary experiments suggest that MDM treated with siRNA for CD81, which is associated more closely with the HIV assembly compartment (8), also has no significant effect on the assembly of infectious HIV particles (E. Ruiz-Mateos, A. Pelchen-Matthews, and M. Marsh, unpublished data). Better knowledge of how tetraspanins are involved in cell-cell interactions and virus transmission, e.g., via the formation of virological synapses (14), should improve our understanding of HIV transmission and pathogenesis and may shed light on the physiological functions of CD63 as well as other tetraspanins.
Acknowledgments
We thank Aine McKnight and Robin Weiss for access to the containment facilities of the Wohl Virion Centre. We are grateful to the National Institute for Biological Standards and Control Centralised Facility for AIDS Reagents and to F. Beditchevski, T. Kreis, M. Malim, R. Shattock, and J. Martin-Serrano for providing reagents. We also thank Emma Tippett and Kristina Theusner for critical reading of the manuscript.
E. Ruiz-Mateos was supported by Fondo de Investigacion Sanitaria grant CD05/00174 and Redes Telemáticas de Investigación Cooperativa en Salud (RETICS; 2006, Red de SIDA RD06/0006/0021, 2007-2010). A. Pelchen-Matthews, M. Deneka, and M. Marsh were supported by UK Medical Research Council funding to the Cell Biology Unit.
Footnotes
Published ahead of print on 5 March 2008.
REFERENCES
- 1.Bauer, H. M., Y. Ting, C. E. Greer, J. C. Chambers, C. J. Tashiro, J. Chimera, A. Reingold, and M. M. Manos. 1991. Genital human papillomavirus infection in female university students as determined by a PCR-based method. JAMA 265472-477. [PubMed] [Google Scholar]
- 2.Bishop, K. N., R. K. Holmes, and M. H. Malim. 2006. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 808450-8458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Booth, A. M., Y. Fang, J. K. Fallon, J. M. Yang, J. E. Hildreth, and S. J. Gould. 2006. Exosomes and HIV Gag bud from endosome-like domains of the T cell plasma membrane. J. Cell Biol. 172923-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Briggs, J. A., M. N. Simon, I. Gross, H. G. Krausslich, S. D. Fuller, V. M. Vogt, and M. C. Johnson. 2004. The stoichiometry of Gag protein in HIV-1. Nat. Struct. Mol. Biol. 11672-675. [DOI] [PubMed] [Google Scholar]
- 5.Brussel, A., and P. Sonigo. 2003. Analysis of early human immunodeficiency virus type 1 DNA synthesis by use of a new sensitive assay for quantifying integrated provirus. J. Virol. 7710119-10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chertova, E., O. Chertov, L. V. Coren, J. D. Roser, C. M. Trubey, J. W. Bess, Jr., R. C. Sowder II, E. Barsov, B. L. Hood, R. J. Fisher, K. Nagashima, T. P. Conrads, T. D. Veenstra, J. D. Lifson, and D. E. Ott. 2006. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J. Virol. 809039-9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Codina, J., J. Li, and T. D. Dubose, Jr. 2005. CD63 interacts with the carboxy terminus of the colonic H+-K+-ATPase to increase plasma membrane localization and 86Rb+ uptake. Am. J. Physiol. Cell Physiol. 288C1279-C1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deneka, M., A. Pelchen-Matthews, R. Byland, E. Ruiz-Mateos, and M. Marsh. 2007. In macrophages, HIV-1 assembles into an intracellular plasma membrane domain containing the tetraspanins CD81, CD9, and CD53. J. Cell Biol. 177329-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Escola, J. M., M. J. Kleijmeer, W. Stoorvogel, J. M. Griffith, O. Yoshie, and H. J. Geuze. 1998. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem. 27320121-20127. [DOI] [PubMed] [Google Scholar]
- 10.Fraile-Ramos, A., T. N. Kledal, A. Pelchen-Matthews, K. Bowers, T. W. Schwartz, and M. Marsh. 2001. The human cytomegalovirus US28 protein is located in endocytic vesicles and undergoes constitutive endocytosis and recycling. Mol. Biol. Cell 121737-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho, S. H., F. Martin, A. Higginbottom, L. J. Partridge, V. Parthasarathy, G. W. Moseley, P. Lopez, C. Cheng-Mayer, and P. N. Monk. 2006. Recombinant extracellular domains of tetraspanin proteins are potent inhibitors of the infection of macrophages by human immunodeficiency virus type 1. J. Virol. 806487-6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ihrke, G., A. Kyttala, M. R. Russell, B. A. Rous, and J. P. Luzio. 2004. Differential use of two AP-3-mediated pathways by lysosomal membrane proteins. Traffic 5946-962. [DOI] [PubMed] [Google Scholar]
- 13.Janvier, K., and J. S. Bonifacino. 2005. Role of the endocytic machinery in the sorting of lysosome-associated membrane proteins. Mol. Biol. Cell 164231-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jolly, C., and Q. J. Sattentau. 2007. Human immunodeficiency virus type 1 assembly, budding and cell-cell spread in T cells takes place in tetraspanin-enriched plasma membrane domains. J. Virol. 817873-7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jouvenet, N., S. J. Neil, C. Bess, M. C. Johnson, C. A. Virgen, S. M. Simon, and P. D. Bieniasz. 2006. Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol. 4e435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jung, K. K., X. W. Liu, R. Chirco, R. Fridman, and H. R. Kim. 2006. Identification of CD63 as a tissue inhibitor of metalloproteinase-1 interacting cell surface protein. EMBO J. 253934-3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Latysheva, N., G. Muratov, S. Rajesh, M. Padgett, N. A. Hotchin, M. Overduin, and F. Berditchevski. 2006. Syntenin-1 is a new component of tetraspanin-enriched microdomains: mechanisms and consequences of the interaction of syntenin-1 with CD63. Mol. Cell. Biol. 267707-7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis, V., S. A. Green, M. Marsh, P. Vihko, A. Helenius, and I. Mellman. 1985. Glycoproteins of the lysosomal membrane. J. Cell Biol. 1001839-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mantegazza, A. R., M. M. Barrio, S. Moutel, L. Bover, M. Weck, P. Brossart, J. L. Teillaud, and J. Mordoh. 2004. CD63 tetraspanin slows down cell migration and translocates to the endosomal-lysosomal-MIICs route after extracellular stimuli in human immature dendritic cells. Blood 1041183-1190. [DOI] [PubMed] [Google Scholar]
- 20.Meerloo, T., H. K. Parmentier, A. D. Osterhaus, J. Goudsmit, and H. J. Schuurman. 1992. Modulation of cell surface molecules during HIV-1 infection of H9 cells. An immunoelectron microscopic study. AIDS 61105-1116. [DOI] [PubMed] [Google Scholar]
- 21.Meerloo, T., M. A. Sheikh, A. C. Bloem, A. de Ronde, M. Schutten, C. A. van Els, P. J. Roholl, P. Joling, J. Goudsmit, and H. J. Schuurman. 1993. Host cell membrane proteins on human immunodeficiency virus type 1 after in vitro infection of H9 cells and blood mononuclear cells. An immuno-electron microscopic study. J. Gen. Virol. 74129-135. [DOI] [PubMed] [Google Scholar]
- 22.Metzelaar, M. J., P. L. Wijngaard, P. J. Peters, J. J. Sixma, H. K. Nieuwenhuis, and H. C. Clevers. 1991. CD63 antigen. A novel lysosomal membrane glycoprotein, cloned by a screening procedure for intracellular antigens in eukaryotic cells. J. Biol. Chem. 2663239-3245. [PubMed] [Google Scholar]
- 23.Nguyen, D. G., A. Booth, S. J. Gould, and J. E. Hildreth. 2003. Evidence that HIV budding in primary macrophages occurs through the exosome release pathway. J. Biol. Chem. 27852347-52354. [DOI] [PubMed] [Google Scholar]
- 24.Nydegger, S., M. Foti, A. Derdowski, P. Spearman, and M. Thali. 2003. HIV-1 egress is gated through late endosomal membranes. Traffic 4902-910. [DOI] [PubMed] [Google Scholar]
- 25.Nydegger, S., S. Khurana, D. N. Krementsov, M. Foti, and M. Thali. 2006. Mapping of tetraspanin-enriched microdomains that can function as gateways for HIV-1. J. Cell Biol. 173795-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ono, A., and E. O. Freed. 2004. Cell-type-dependent targeting of human immunodeficiency virus type 1 assembly to the plasma membrane and the multivesicular body. J. Virol. 781552-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pelchen-Matthews, A., B. Kramer, and M. Marsh. 2003. Infectious HIV-1 assembles in late endosomes in primary macrophages. J. Cell Biol. 162443-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfistershammer, K., O. Majdic, J. Stockl, G. Zlabinger, S. Kirchberger, P. Steinberger, and W. Knapp. 2004. CD63 as an activation-linked T cell costimulatory element. J. Immunol. 1736000-6008. [DOI] [PubMed] [Google Scholar]
- 29.Raposo, G., M. J. Kleijmeer, G. Posthuma, J. W. Slot, and H. J. Geuze. 1997. Immunogold labeling of ultrathin cryosections: application in immunology, p. 1-11. In L. A. Herzenberg, D. M. Weir, L. A. Herzenberg, and C. Blackwell (ed.), Handbook of experimental immunology. Blackwell Science, Inc., Oxford, United Kingdom.
- 30.Raposo, G., M. Moore, D. Innes, R. Leijendekker, A. Leigh-Brown, P. Benaroch, and H. Geuze. 2002. Human macrophages accumulate HIV-1 particles in MHC II compartments. Traffic 3718-729. [DOI] [PubMed] [Google Scholar]
- 31.Rous, B. A., B. J. Reaves, G. Ihrke, J. A. Briggs, S. R. Gray, D. J. Stephens, G. Banting, and J. P. Luzio. 2002. Role of adaptor complex AP-3 in targeting wild-type and mutated CD63 to lysosomes. Mol. Biol. Cell 131071-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saad, J. S., J. Miller, J. Tai, A. Kim, R. H. Ghanam, and M. F. Summers. 2006. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc. Natl. Acad. Sci. USA 10311364-11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.von Lindern, J. J., D. Rojo, K. Grovit-Ferbas, C. Yeramian, C. Deng, G. Herbein, M. R. Ferguson, T. C. Pappas, J. M. Decker, A. Singh, R. G. Collman, and W. A. O'Brien. 2003. Potential role for CD63 in CCR5-mediated human immunodeficiency virus type 1 infection of macrophages. J. Virol. 773624-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 461896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Welsch, S., O. T. Keppler, A. Habermann, I. Allespach, J. Krijnse-Locker, and H. G. Krausslich. 2007. HIV-1 buds predominantly at the plasma membrane of primary human macrophages. PLoS Pathog. 3e36. [DOI] [PMC free article] [PubMed] [Google Scholar]