Abstract
In mammalian and yeast systems, methyltransferases have been implicated in the regulation of diverse processes, such as protein-protein interactions, protein localization, signal transduction, RNA processing, and transcription. The Cucumber mosaic virus (CMV) 1a protein is essential not only for virus replication but also for movement. Using a yeast two-hybrid system with tobacco plants, we have identified a novel gene encoding a methyltransferase that interacts with the CMV 1a protein and have designated this gene Tcoi1 (tobacco CMV 1a-interacting protein 1). Tcoi1 specifically interacted with the methyltransferase domain of CMV 1a, and the expression of Tcoi1 was increased by CMV inoculation. Biochemical studies revealed that the interaction of Tcoi1 with CMV 1a protein was direct and that Tcoi1 methylated CMV 1a protein both in vitro and in vivo. The CMV 1a binding activity of Tcoi1 is in the C-terminal domain, which shows the methyltransferase activity. The overexpression of Tcoi1 enhanced the CMV infection, while the reduced expression of Tcoi1 decreased virus infectivity. These results suggest that Tcoi1 controls the propagation of CMV through an interaction with the CMV 1a protein.
Protein methylation is a posttranslational modification by which a methyl group from S-adenosylmethionine (AdoMet) is added to a protein. In eukaryotes, proteins can be methylated on the side chain nitrogens of arginine, lysine, and histidine residues or on the carboxyl groups of proteins (1). Whereas N-methylation on nitrogen or oxygen atoms of Lys and Arg side chains, particularly on the same histone tails that are acetylated, is more commonly reported, C-, O-, and S-methylations of protein side chains have also been reported previously (53). The C-methylation of arginine and glutamine side chains has been detected by X-ray analyses of the structures of methyl coenzyme M reductases from methanogenic bacteria (15).
As other posttranslational modifications, protein methylation is involved in regulating protein-protein interactions, resulting in a plethora of effects during key cellular events that include the regulation of transcription (32, 48), stress response, ageing and protein repair (8), nuclear transport (47), and ion channel function (52), as well as cytokine signaling (42).
Furthermore, protein methylation has been shown to be important for virus replication and the infectivity of various viruses. Herpes simplex virus replication is regulated partly by the methylation of the RNA binding domain (BD) in the Herpes simplex virus ICP27 protein (39). In vaccinia virus, the inhibition of AdoMet-dependent protein methylation reactions results in decreased virus replication (4, 28). Arginine methylation of the adenovirus E1B 55-kDa protein-associated protein E1B-AP5 has also been shown previously to be required for efficient adenovirus replication (31). Furthermore, in a small form of hepatitis delta virus delta antigen that can transactivate hepatitis delta virus RNA replication, methylation at arginine residues of the RNA binding motif is required to support RNA replication (34). For human immunodeficiency virus (HIV), various reports implicate the involvement of methylation in virus replication regulation (5, 18, 24, 25, 30, 50, 55). For example, adenosine analogues have been shown previously to have antiviral activity (18), and treatment with adenosine periodate, a general inhibitor of many methyltransferases (MTs), increases virus production but reduces virus infectivity (55).
Cucumber mosaic virus (CMV) is a type member of the genus Cucumovirus in the family Bromoviridae. CMV particles are icosahedral and contain a tripartite, positive-sense RNA genome with components designated RNAs 1 to 3 (44). CMV RNAs 1 and 2 each encode a protein involved in the replication of the viral genome, 1a protein and 2a polymerase protein, respectively (6, 44). RNA 1, encoding sequence motifs conserved in MTs (40, 41, 45) and helicases (Hels) (17, 19, 22), and RNA 2, encoding the polymerase, have been shown to be associated in a membrane-bound RNA-dependent RNA polymerase (RdRp) (21) constituting the CMV replicase. The active CMV replicase consists of both 1a and 2a polymerase proteins, as well as one or more host factors. RNA 2 has an additional open reading frame encoding a protein called 2b (14), which participates in host-specific virus accumulation, the suppression of posttranscriptional gene silencing, and virulence determination (38). RNA 3 encodes the 3a movement protein (MP), which is crucial for the movement of viral RNA from cell to cell (13). The MP also binds to single-stranded nucleic acids in vitro (33, 51). RNA 4 is a subgenomic RNA derived from the 3′ half of RNA 3 and is involved in the synthesis of viral coat protein (CP) (46). The CMV 1a protein has been implicated not only in replication, but also in the regulation of systemic infections (43). Thus, the identification of cellular proteins that are able to interact with and regulate the CMV 1a protein may constitute the first step toward understanding the mechanisms underlying CMV intracellular propagation or other functions of this protein.
To search for cellular components that interact with the CMV 1a protein, yeast two-hybrid screening experiments were conducted by using CMV 1a as bait. A novel protein containing an MT domain was isolated, and its gene was designated Tcoi1 (tobacco CMV 1a-interacting protein 1). This report presents in vitro and in vivo biochemical evidence which shows the interaction of the 1a protein of CMV with Tcoi1. It was also shown that CMV 1a protein was methylated by Tcoi1. In addition, the reduction in the accumulation of CMV RNAs in antisense-Tcoi1 transgenic plants implied that Tcoi1 may affect CMV replication and/or spread.
MATERIALS AND METHODS
Yeast two-hybrid screening.
The MatchMaker II GAL4 two-hybrid system was purchased from Clontech Laboratories (Palo Alto, CA). The pACT2 activation domain (AD) expression vector was used to make a cDNA library with the RNA extracted from CMV-infected tobacco leaves. The library was screened using the pAS2-1::CMV 1a BD fusion vector as bait. Saccharomyces cerevisiae strain Y190 (MATa HIS3 lacZ trp1 leu2 cyhr2) or HF7c (MATa HIS3 lacZ trp1 leu2) was transformed with the two fusion vectors as directed by the manufacturer's instructions. The transformed yeast cells were selected on minimal synthetic dropout medium that lacked Leu, Trp, and His. To eliminate false-positive clones, the synthetic dropout medium plates were supplemented with 50 mM (Y190) or 5 mM (HF7c) 3-amino-1,2,4-triazole. Yeast cells were permitted to grow at 30°C for 8 to 12 days and subjected to the colony-lift filter assay, which monitored β-galactosidase activity using 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) as the substrate. Putative positive clones (Leu+ Trp+ His+) identified by this assay were confirmed to be positive by retransformation. Plasmids were isolated from each Leu+ Trp+ His+ LacZ+ transformant by using a yeast plasmid isolation kit (Clontech). Escherichia coli XL1-Blue cells (Stratagene) were then transformed with the plasmids by electroporation. The bacterially propagated pACT2 fusion plasmid was isolated and analyzed.
Northern blot analysis.
For Northern blot analysis, total RNA was prepared from the tobacco leaves as described by Ausubel et al. (2). Fifteen micrograms of total RNA was electrophoresed in a 1.0% agarose gel containing 6% formaldehyde in MOPS (morpholinepropanesulfonic acid) buffer (pH 7.0) and transferred onto a Nytran Plus membrane. The RNA analysis was repeated three times, and samples from two plants per treatment were collected. Northern blot hybridizations were carried out with 32P-labeled probes. After hybridization, the membranes were washed twice with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at room temperature for 10 min each time, once with 0.1× SSC containing 0.1% sodium dodecyl sulfate (SDS) at room temperature for 10 min, and once with 0.1× SSC containing 0.1% SDS at 65°C for 5 min. The membranes were dried and exposed to X-ray film or directly visualized with the BAS 2500 phosphorimager (using Fuji photo film). Filters were stripped in 0.1× SSC-0.2% SDS at 80°C for 20 min when reprobing was required. For the quantification of the CMV RNA band intensities, the CMV RNA 4 bands on Northern blots were quantified using a phosphorimager and ImageQuant software and the values were normalized by the intensities of the wild-type bands in the corresponding lanes and expressed as log2 ratios.
RT-PCR analysis.
Total RNA was isolated from 4-week-old tobacco leaves by the method of Ausubel et al. (2). For the reverse transcription-PCR (RT-PCR) analysis, cDNA was synthesized from DNase I-treated total RNAs by using Moloney murine leukemia virus reverse transcriptase (Promega) and random hexamer primers. PCRs (94°C for 3 min; 25 cycles of 94°C for 1 min, 57°C for 45 s, and 72°C for 2 min; and 72°C for 7 min) were performed for Tcoi1 (primers, 5′-AACACGCAAATGGCTTTCAGTTTCAAG-3′ and 5′-TATGGGGGGAAAATTAGTAAGGAACGGT-3′), PR-1 (primers, 5′-GCGAAAACCTAGCTTGGGGAAG-3′ and 5′-TATATAACGTGAAATGGACGC-3′), the CMV 3′ untranslated region (primers, 5′-TTCTGTGTTTTCCAGAACC-3′ and 5′-GACAGGATCCACGCGTGGTCTCCTTTTGGA-3′), and L25 (primers, 5′-TGCAATGAAGAAGATTGAGGACAACA-3′ and 5′-CCATTCAA GTGTATCTAGTAACTCAAATCCAAG-3′). Unless otherwise specified, all experiments reported in this paper were performed at least three times, with similar results.
In vitro transcription and translation.
The pGADT7 plasmids containing the coding sequences for CMV 1a, the CMV 1a MT domain alone (CMV 1a-MT), or the CMV 1a Hel domain alone (CMV 1a-Hel) under the control of the T7 promoter were used as templates in coupled in vitro transcription-translation reactions with the TNT wheat germ system (Promega). Proteins were synthesized with [35S]Met (Amersham Pharmacia Biotech) for 1 h.
Preparation of recombinant proteins.
All the glutathione S-transferase (GST) fusion proteins were expressed in E. coli and purified by standard procedures. In brief, cells harboring GST or GST fusion expression plasmids were induced with 1 mM isopropyl-1-β-d-thiogalactopyranoside (IPTG) for 2 to 3 h at 30 or 37°C. Cells were washed with phosphate-buffered saline (PBS), resuspended in lysis buffer, and sonicated. Supernatants obtained by centrifugation were loaded onto a glutathione-agarose column (1 by 10 cm). The column was washed with 5 column volumes of lysis buffer. The bead-bound proteins were eluted with lysis buffer containing 10 mM reduced glutathione. Purified proteins were stored at −70°C.
BRET.
Gaussia luciferase (RLuc) and yellow fluorescent protein (YFP) open reading frames were inserted into the SacI site of the modified pCAMBIA2300 vector (CAMBIA) containing the 35S promoter and the Nos terminator. The CMV 1a cDNA was fused upstream of the RLuc cDNA, and cDNA for Tcoi1, the truncation form Tcoi1d3, or the Tcoi1 MT domain alone (Tcoi1-MT) was fused upstream of the YFP gene. RLuc cDNA was obtained from Prolume Ltd. (Pinetop, AZ). For in vivo assays, luminescence activity and the yellow/blue ratio were measured by following previously described procedures (49). Emission was measured using an injector-equipped plate reader spectrofluorometer (Fluostar Optima) at the wavelengths of 475 and 535 nm, corresponding to the maxima of the emission spectra for RLuc and YFP, respectively. Bioluminescence resonance energy transfer (BRET) measurements were taken at 0.4- to 0.5-s intervals.
Immunoprecipitation.
For the Tcoi1-gene fluorescent protein (GFP) gene construct, a fragment was generated by PCR using the primers Tcoi1-5′ (5′-CCGGATCCCATGGCTTTCAGTTTCAAG-3′) and Tcoi1Nonstop (5′-AAGGATCCAGTAAGGAACGGTAGC-3′). The Tcoi1-GFP gene fusion construct was generated by positioning the coding region of Tcoi1 cDNA, without the termination codon, in frame relative to the end of the GFP gene corresponding to the N terminus of soluble modified GFP. The construct comprised the cauliflower mosaic virus 35S-mGFP4-NosT region from pBIN 35S-mGFP4 cloned into the high-copy-number pUC118 plasmid, and site-directed mutagenesis was carried out (10, 11). In the case of the CMV 1a-hemagglutinin (HA) construct, two repeats encoding the HA epitope (YPYDVPDYA) were inserted into the SacI site of the modified pCAMBIA2300 vector (CAMBIA) containing the 35S promoter and the Nos terminator. The CMV 1a cDNA was then fused upstream of the double HA epitope sequence. The fusion constructs were introduced into Arabidopsis thaliana protoplasts (26). Following this introduction, immunoprecipitation experiments were performed as previously described (9). The membrane was then analyzed by a Fujix LAS3000 bioimaging analyzer.
In vitro MT assays.
MT assays were carried out as previously reported (35, 36). Briefly, a soluble cell extract or purified GST-Tcoi1 proteins were allowed to react with methyl acceptors GST-CMV 1a-MT, GST-CMV 1a-Hel, or GST-CMV 2a in the presence of 0.25 μCi of S-adenosyl-l-[methyl-3H]methionine as a methyl donor for 2 h at 30°C. The reaction products were resolved by SDS-polyacrylamide gel electrophoresis (PAGE). The gels were fixed and treated with an amplifier (Amersham-Pharmacia Biotech) for 15 min, dried, and then exposed to X-ray film at −80°C for 5 to 14 days. The gels were then analyzed by using a Fujix BAS 1000 phosphorimager.
In vivo MT assays.
For in vivo MT assays of CMV 1a, the CMV 1a-HA and Tcoi1-GFP or Tcoi1-MT-GFP fusion constructs were introduced into Arabidopsis protoplasts, prepared from whole seedlings by the polyethylene glycol-mediated transformation procedure (26). After 48 h, the incubated cells were harvested, rinsed with PBS, and incubated in 700 μl of lysis buffer (100 mM Tris-HCl [pH 8], 250 mM NaCl, 1% NP-40, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, Roche protease inhibitors) on ice for 30 min. After centrifugation for 15 min in a microcentrifuge, a 20-μl bed volume of protein A-Sepharose and 2 μl of polyclonal anti-HA antibody were added to the supernatant and the mixture was incubated overnight at 4°C with rocking. The Sepharose beads were then washed three times with lysis buffer, and bound proteins were eluted with 40 μl of 2% SDS. The total eluate was loaded onto a 12% polyacrylamide gel and transferred onto a polyvinylidene difluoride membrane. CMV 1a proteins were identified by Western blotting using monoclonal mono-/dimethylarginine antibody (Abcam). The blot was developed with horseradish peroxidase-conjugated secondary antibodies and evaluated using ECL chemiluminescence detection (Amersham Biosciences).
Analysis of transgenic plants with Tcoi1 in the sense or antisense orientation.
Plasmids used in the transformation of tobacco plants were constructed with sense- and antisense-orientated full-length Tcoi1 cDNA that was cloned into a polylinker site of a binary vector, pMBP2 (20). The constructs were introduced into Nicotiana tabacum cv. Samsun NN by using Agrobacterium tumefaciens-mediated transformation as described previously (23).
Detection of the CMV CP by ELISA.
Tobacco leaves inoculated with CMV were harvested at 9 days postinoculation (dpi). The leaves were transferred into a microcentrifuge tube and ground with a disposable grinder in 200 μl of extraction buffer (0.1 M Tris-HCl with 1% sodium sulfite, pH 7.4). The total protein concentrations were determined using Bio-Rad protein assay reagent (Hercules, CA). The enzyme-linked immunosorbent assay (ELISA) plates were activated overnight at 4°C with protein extract (45 μg). The plates were washed with a 0.01 M Tris-HCl solution containing 0.85% NaCl and 0.05% Tween 20 at pH 7.4. The plates were then blocked for 1 h with 1% bovine serum albumin in 0.1 M Tris-HCl with 0.85% NaCl at pH 7.4. After the addition of antigens, the anti-CMV antibody was diluted to 1:500, and 200 μl of the dilution was applied to each plate for 2 h. After the plates were washed, 100-μl aliquots of alkaline phosphatase goat anti-rabbit immunoglobulin G (Sigma) diluted to 1:2,000 were added to the plates for 2 h. The plate contents were developed with 100 μl of a substrate solution containing p-nitrophenyl phosphate (Sigma; 1 mg/ml) in 0.2 M carbonate buffer, pH 9.8, and were measured at A405 in an ELISA reader (Bio-Rad).
Nucleotide sequence accession number.
The nucleotide sequence determined in this study has been deposited in the GenBank database under accession number AY391749.
RESULTS
Identification of Tcoi1, a protein that interacts with CMV 1a protein, by yeast two-hybrid screening.
CMV 1a fused in frame to the GAL4 DNA BD was used as the bait in a yeast two-hybrid system. A library of N. tabacum cDNA fused to the GAL4 AD sequence and expressed from the pACT2 vector was screened. Of the 1.5 × 106 transformants screened, 368 colonies initially grew on plates devoid of Leu, Trp, and His. When these 368 clones were retransformed and subjected to a colony-lift filter assay, 14 clones were observed to exhibit β-galactosidase activity (data not shown). Three clones were shown to contain the same cDNA insert, varying only in the length upstream of the polyadenylation sequence. This cDNA insert was designated Tcoi1, and the longest form of the insert, ∼900 bp, was further characterized.
The interaction between CMV 1a and Tcoi1 was confirmed using additional plasmid combinations and constructs to test for the autonomous activation of the HIS3 and lacZ reporter genes, for possible direct interactions with the GAL4 BD or AD, and for potential artifacts caused by high-level expression of the encoded GAL4 fusion protein (Table 1). Neither pAS2-1::CMV 1a (bait) nor pACT2::Tcoi1 (prey) alone activated the HIS3 or lacZ reporter gene. Tcoi1 (expressed from pACT2::Tcoi1) did not interact with the GAL4 BD itself, nor did CMV 1a (expressed from pAS2-1::CMV 1a) interact with the GAL4 AD. The CMV 1a protein has two functional domains: a putative MT domain and a putative Hel domain. To test which domain of CMV 1a interacted with Tcoi1, each domain was fused to the GAL4 BD. Tcoi1 was observed to interact with CMV 1a-MT (the region from amino acids 1 to 456 of the CMV 1a protein) but not with CMV 1a-Hel (amino acids 646 to 993). These results were confirmed by quantitative β-galactosidase assays using a very sensitive luminescence-based detection system. The β-galactosidase activity of cotransformants expressing CMV 1a and Tcoi1 was about 44% of that observed in the positive control (carrying pVA3-1 and pTD1-1). Cotransformants expressing CMV 1a-MT and Tcoi1 showed slightly higher activity levels than cotransformants expressing CMV 1a and Tcoi1. Additionally, the interaction of Tcoi1 with other CMV-encoded proteins, which are 2a, MP, and CP, was tested. The Tcoi1 did not interact with any other CMV-encoded proteins (data not shown). Therefore, the Tcoi1 appeared to interact specifically and strongly with CMV 1a through the MT domain of CMV 1a.
TABLE 1.
Interaction of Tcoil and CMV 1a in yeast cells
| GAL4 BD vector | GAL4 AD vector | Result for yeast cells with indicated vector(s) in assay(s) for:
|
|
|---|---|---|---|
| His | β-Galactosidase activityd | ||
| pAS2-1::CMV 1a | Nonea | − | − |
| Nonea | pACT2::Tcoil | − | − |
| pAS2-1::CMV 1a | pACT2 | − | − |
| pAS2-1 | pACT2::Tcoi1 | − | − |
| pAS2-1::CMV 1a | pACT2::Tcoil1 | + | + (33,000 ± 1,000) |
| pAS2-1::CMV 1a-MTb | pACT2::Tcoi1 | + | + (35,000 ± 2,000) |
| pAS2-1::CMV 1a-Helc | pACT2::Tcoil1 | − | − |
No coexpressed vector.
Encodes the MT domain of CMV 1a protein.
Encodes the Hel domain of CMV 1a protein.
The colony-lift filter assay and a liquid culture assay with o-nitrophenyl-β-d-galactopyranoside as the substrate were used to evaluate β-galactosidase activity. +, blue colonies resulting from positive β-galactosidase activity; −, white colonies resulting from negative β-galactosidase activity (same as that in the buffer control). Values are means ± standard deviations and are expressed as Miller units.
Cloning and sequence analysis of Tcoi1 cDNA reveals an MT domain.
Sequence analysis of the 900-bp Tcoi1 cDNA insert identified an open reading frame of 874 nucleotides in frame with the GAL4 AD sequence and showed this cDNA to be an incomplete clone. By using this 900-bp fragment for the screening of the λ-GEM11 N. tabacum cDNA library, we identified two hybridizing plaques, each of which contained a single insert that included a genomic sequence identical to the sequence of the Tcoi1 cDNA clone. A comparison of the Tcoi1 cDNA sequences and the 900-bp fragment showed that the latter appeared to lack the 456 nucleotides of the coding region corresponding to the N terminus of Tcoi1. We then used 5′ and 3′ rapid amplification of cDNA ends to confirm this finding and to clone full-length Tcoi1 cDNA from N. tabacum poly(A)-containing RNA. Clones for a full-length cDNA sequence of 1,355 nucleotides, predicted to encode a protein of 370 amino acids, were obtained. This cDNA contained the 1,100-nucleotide complete Tcoi1 coding sequence, flanked by a 24-nucleotide 5′ untranslated region and a 221-nucleotide 3′ untranslated region (Fig. 1A).
FIG. 1.
Comparison of Tcoi1 nucleotide and protein sequences. (A) Nucleotide sequence of Tcoi1 cDNA and corresponding deduced amino acid sequence. The MT domain is underlined, and motif II and motif III of AdoMet-dependent MTs are marked in italics. (B) Alignment of the predicted Tcoi1 amino acid sequence with those of other UbiE-like C-MT family members. Sequences from A. thaliana (At1g23360), E. coli, and S. cerevisiae are compared. The alignments were generated from DNAStar MegAlign by using the PAM 250 table and the Jotun Hein method. The shaded residues match the consensus within two distance units.
Sequence analysis of Tcoi1 revealed that it was an uncharacterized gene predicted to encode a novel protein. The position-specific iterative Basic Local Alignment Search Tool (PSI-BLAST) predicted residues 188 to 345 of Tcoi1 to contain an MT domain belonging to the UbiE-like C-MT family, the members of which are involved in carbon methylation reactions in the biosynthesis of ubiquinone and menaquinone. As shown in Fig. 1B, the primary sequence of Tcoi1 was only distantly related to those of characterized UbiE MT family members, including the A. thaliana UbiE (33% identity), E. coli UbiE (19.8% identity), and S. cerevisiae UbiE (13.6% identity). Two (motif II and motif III) of the three motifs of sequence similarity present in AdoMet-dependent MTs were present within the predicted Tcoi1 MT domain (27).
Genomic organization and tissue-specific expression of Tcoi1.
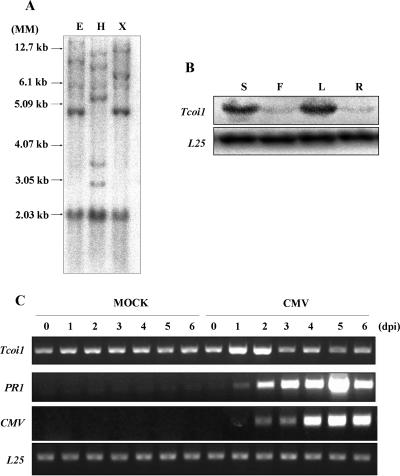
Southern blot analysis carried out with a full-length Tcoi1 probe revealed four or five fragments that hybridized to the probe under medium-stringency conditions (Fig. 2A). This result indicates that the Tcoi1 gene is present as a small gene family in the tobacco genome.
FIG. 2.
Southern blot analysis and comparison of Tcoi1 expression patterns in various organs and in tissues from CMV-Kor- and mock-inoculated plants. (A) Tobacco genomic DNA was digested with EcoRI (E), HindIII (H), or XbaI (X), and digestion products were separated on 0.8% agarose gel. After being transferred onto a Nytran Plus membrane, the blot was hybridized with a 32P-labeled full-length Tcoi1 cDNA probe under conditions of medium stringency. Autoradiograms were visualized with a Fuji BAS 2500 phosphorimager. DNA size standards (MM) are shown at the left. (B) Accumulation of Tcoi1 transcripts in different organs. Lanes: S, stem; F, flower; L, leaf; and R, root. Tcoi1 transcripts were monitored by Northern blot analysis using the Tcoi1-specific 3′ untranslated region as a probe. The transcript level corresponding to ribosomal protein L25 was included as an internal standard for RNA quantity evaluation. (C) RT-PCR analysis of Tcoi1 genes upon CMV-Kor inoculation. Total RNA was extracted from leaf tissues at 0, 1, 2, 3, 4, 5, and 6 dpi. Tissue was isolated from the leaves of either CMV- or mock-inoculated plants. The conserved 3′ ends of CMV RNAs and PR-1 were detected as a positive control for CMV inoculation. As an internal standard for cDNA quantity evaluation, the level of L25 was monitored.
To examine the steady-state transcript levels of the Tcoi1 gene in various plant organs, total RNA was extracted from stems, flowers, leaves, and roots and Northern blot analysis was performed with a Tcoi1-specific probe. Tcoi1 gene transcripts accumulated in the stems and leaves but were not detected in flowers and roots (Fig. 2B).
Expression patterns of Tcoi1 upon CMV inoculation.
The expression pattern of the Tcoi1 gene was monitored using RT-PCR analyses of tobacco plants after CMV inoculation. Total RNA from CMV-inoculated leaves was extracted at designated time points. A mock inoculation treatment was also included as a control to eliminate any possible wounding-induced Tcoi1 gene expression that may arise from rubbing with Carborundum. As shown in Fig. 2C, there were no detectable changes in Tcoi1 transcript accumulation in mock-inoculated leaves. However, in CMV-inoculated leaves, Tcoi1 transcripts were induced at 1 dpi and began to decrease by 3 days after CMV inoculation. The band intensities observed for the CMV-inoculated plants were 2.5 to ∼3 times higher than those observed for the mock-inoculated leaves. The expression of CMV viral genes was investigated by the detection of the conserved 3′ ends of CMV RNAs as a positive control for CMV inoculation. PR-1 gene expression patterns were also monitored in control experiments, as the PR-1 gene has been previously reported to be induced during susceptible responses to CMV (54). As an internal standard for cDNA quantity evaluation, the expression level of ribosomal protein gene L25 was compared to the results obtained. These results indicated that Tcoi1 was induced in response to CMV infection.
Determination of the CMV 1a binding regions of Tcoi1.
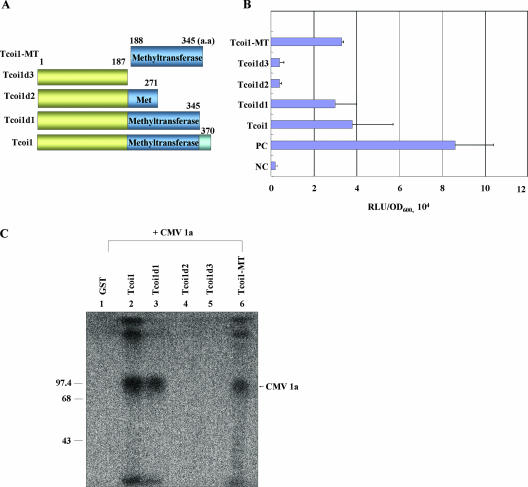
To determine the region in Tcoi1 that serves as a BD for CMV 1a, full-length Tcoi1 and various N- and C-terminal deletion constructs were tested for their interaction with full-length CMV 1a in a yeast two-hybrid system (Fig. 3A and B). Proper expression of the prey proteins was verified by protein gel analysis (data not shown). Some truncation proteins (Tcoi1d2 and Tcoi1d3) showed a complete loss of interaction, while others (Tcoi1d1 and Tcoi1-MT) still interacted with CMV 1a. From these results, it can be concluded that the MT domain of Tcoi1 was sufficient for the interaction with CMV 1a in the yeast two-hybrid system.
FIG. 3.
Identification of the region of Tcoi1 that is necessary for the interaction with CMV 1a protein and in vitro interaction analysis. (A) The names of the truncated constructs and the positions of amino acids (aa) are indicated at the left and above, respectively. (B) Analysis of interaction between different parts of Tcoi1 and CMV 1a protein in the yeast two-hybrid system. The interaction strength was scored by liquid assays using a sensitive luminescence detection system (Clontech). The values are the means ± standard deviations (n = 3) and are expressed as relative light units (RLU) normalized to the cell content (optical density at 600 nm [OD600]). In the evaluation of each interaction, three separate transformed colonies were assayed, and each of the assays was performed in triplicate. PC, positive control; NC, negative control. (C) In vitro GST pulldown assay of the GST-conjugated products of full-length and truncated Tcoi1 clones and in vitro-translated CMV 1a protein. [35S]Met-labeled CMV 1a was incubated with 10 μg of GST (negative control), GST-Tcoi1, GST-Tcoi1d1, GST-Tcoi1d2, GST-Tcoi1d3, or GST-Tcoi1-MT in the presence of glutathione-agarose beads. Precipitates from the binding mixture were subjected to SDS-PAGE, and proteins were visualized by autoradiography. Numbers on the left are molecular size markers.
To confirm the interaction between CMV 1a and the MT domain of Tcoi1, we pursued an in vitro GST pulldown approach. GST fusion proteins with each N- and C-terminal truncation form of Tcoi1 were produced and expressed in E. coli. Intact GST (as a negative control) and GST fusion constructs were readily purified from E. coli (data not shown). For the binding assay, [35S]Met-labeled 1a protein was obtained in a eukaryotic in vitro translation system that synthesized the protein directly from the 1a cDNA clone under the control of the T7 promoter. Each of the purified GST fusion proteins was incubated with the radiolabeled CMV 1a. The mixture was permitted to interact with glutathione-agarose beads at a low temperature, and eluants from the beads were analyzed by SDS-PAGE. The eluted GST-Tcoi1, GST-Tcoi1d1, and GST-Tcoi1-MT fusion proteins trapped by the glutathione-agarose beads appeared to have formed complexes with the CMV 1a polypeptide, as shown by the three respective bands resolved by SDS-PAGE (Fig. 3C, lanes 2, 3, and 6). In contrast, no significant interaction was observed when the CMV 1a polypeptide was incubated with the GST control protein or the GST-Tcoi1d2 or GST-Tcoi1d3 fusion protein (Fig. 3C, lanes 1, 4, and 5). Additionally, Tcoi1-MT was observed to interact with CMV 1a-MT but not with CMV 1a-Hel (data not shown). These results indicate that the intermolecular association between CMV 1a and Tcoi1 or its derivatives that contain the MT domain occurs effectively in vitro.
Direct in vivo interaction of the CMV 1a protein and Tcoi1.
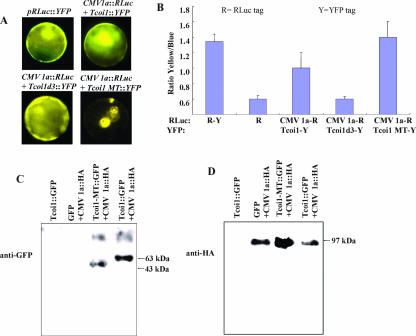
Results obtained from interaction studies within the yeast system and in vitro systems prompted us to investigate whether such interactions occur in planta. The BRET technique offers the advantage of detecting protein-protein interaction in intact living cells. We used this technique to monitor such interactions in real time. Pairs of tagged proteins (i.e., CMV 1a-RLuc with Tcoi1-YFP, Tcoi1d3-YFP, or Tcoi1-MT-YFP) were coexpressed in Arabidopsis protoplasts. At first, the expression of YFP-tagged constructs was observed with a fluorescence microscope (Fig. 4A). The fluorescence of Tcoi1-YFP and Tcoi1d3-YFP fusion proteins accumulated throughout the interiors of the cells, indicating that the proteins were distributed within the cytosol at high concentrations. The Tcoi1-MT-YFP protein also was detected in the cytoplasmic compartment as a ring structure. BRET was measured at 0.4- to 0.5-s intervals. BRET levels for the CMV 1a-RLuc and Tcoi1-YFP protein pair and the CMV 1a-RLuc and Tcoi1-MT-YFP protein pair were highest at 2 min but declined thereafter. On the other hand, BRET was suppressed in presence of the CMV 1a-RLuc and Tcoi1d3-YFP protein pair (as shown in Fig. 4B). pRLuc::YFP and pRLuc were used for the positive control and the negative control, respectively (Fig. 4B, first and second bars).
FIG. 4.
In vivo interaction tests of CMV 1a and Tcoi1, Tcoi1d3, or Tcoi1-MT. (A) Fluorescence micrographs of Arabidopsis protoplasts coexpressing RLuc and YFP fusions. (B) BRET protein interaction analysis of Arabidopsis protoplasts. The yellow/blue ratio in living tissue after the transient coexpression of the indicated fusion proteins was measured. The letters R and Y indicate the positions of the RLuc and YFP tags, respectively. pRLuc::YFP (R-Y) and pRLuc (R) were used as the positive control and the negative control, respectively. The data were averaged from the results for three replicates. Error bars represent the standard deviations. (C and D) Arabidopsis protoplasts were cotransformed with plasmids encoding GFP and CMV 1a-HA, Tcoi1-MT-GFP and CMV 1a-HA, or Tcoi1-GFP and CMV 1a-HA. As a control, protoplasts were transformed with the plasmid encoding Tcoi1-GFP. The immunoprecipitation of the protoplast extracts was performed with anti-HA antiserum. The immunoprecipitated proteins were subjected to Western blot analysis with anti-GFP (C) or anti-HA (D) antibody.
Furthermore, the in vivo interaction between CMV 1a and Tcoi1 was confirmed via coimmunoprecipitation experiments using cotransformed Arabidopsis protoplasts in which Tcoi1-MT or Tcoi1 was fused with GFP and CMV 1a was tagged with HA epitopes (Fig. 4C). After the homogenization of the cotransformed Arabidopsis protoplasts, the extract was pulled down using polyclonal anti-HA antibody. The pellet was washed with PBS and then analyzed using a Western blot technique, and the blot was developed with polyclonal GFP antibody. As shown in Fig. 4C, the molecular mass bands were detected at approximately 43.0 and 63.0 kDa, respectively, the estimated sizes of the recombinant Tcoi1-MT-GFP and Tcoi1-GFP proteins. No significant interactions in the extracts from Arabidopsis protoplasts transformed with a plasmid expressing Tcoi1-GFP only or GFP and CMV 1a-HA were detected (Fig. 4C). In the control experiment, the correct synthesis of the HA-tagged CMV 1a was verified via Western blotting, in which development was achieved with polyclonal HA antibody (Fig. 4D). Additionally, we verified that anti-HA serum did not precipitate Tcoi1-GFP alone (Fig. 4D). This result indicated that Tcoi1 and CMV 1a formed a protein complex in plant cells through the MT domain of Tcoi1.
In vitro and in vivo MT activity of Tcoi1.
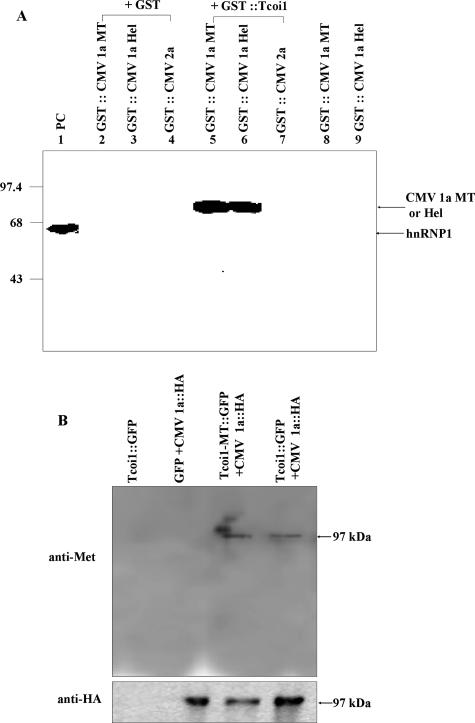
We next wanted to demonstrate the MT activity of Tcoi1 and determine whether CMV 1a protein could serve as a substrate for Tcoi1 in vitro by incubating either recombinant GST-CMV 1a-MT or GST-CMV 1a-Hel with purified GST-Tcoi1 and [3H]AdoMet in PBS (pH 7.4) for up to 24 h at 25°C. In addition, we wanted to assess the substrate specificity of Tcoi1 by using GST-CMV 2a as a negative control for the Tcoi1 substrate. Surprisingly, both CMV 1a-MT and CMV 1a-Hel were methylated under these conditions (Fig. 5A, lanes 5 and 6). The CMV 2a protein was not methylated (Fig. 5A, lane 7), as was predicted since CMV 2a did not interact with Tcoi1 in the yeast two-hybrid assay. When we tested whether GST protein itself had MT activity with GST-CMV 1a-MT, GST-CMV 1a-Hel, or GST-CMV 2a, no signals were detected (Fig. 5A, lanes 2, 3, and 4). Additionally, both GST-CMV 1a-MT and GST-CMV 1a-Hel did not have auto-MT activity (Fig. 5A, lanes 8 and 9). The arginine MT (PRMT) using heterogeneous nuclear RNA protein 1 (hnRNP1) as a substrate was employed as a positive control for the MT reaction (Fig. 5A, lane 1).
FIG. 5.
In vitro and in vivo methylation of CMV 1a by Tcoi1. (A) Methylation of the CMV 1a MT domain and the CMV 1a Hel domain by purified Tcoi1. The protein MT assay was carried out with equal amounts (10 μg) of the indicated purified proteins in the presence of 0.25 μCi of S-adenosyl-l-[methyl-3H]methionine. Lane 1, PRMT1 and hnRNP1 (as a positive control [PC]). The reaction products were separated by SDS-10% PAGE and visualized by autoradiography. The hnRNP1 (59-kDa), GST-CMV 1a-MT (77-kDa), and GST-CMV 1a-Hel (75-kDa) bands are indicated by arrows. Molecular size markers (in kilodaltons) are indicated on the left. (B) Arabidopsis protoplasts were cotransformed with plasmids encoding the indicated proteins. As a control, protoplasts were transformed with the plasmid encoding Tcoi1-GFP. The immunoprecipitation of the protoplast extracts was performed with anti-HA antiserum. The immunoprecipitated proteins were subjected to Western blot analysis with anti-mono-/dimethylarginine (anti-Met) (upper panel) or anti-HA (lower panel) antibody.
Next, we also tested the Tcoi1 MT activity in vivo using cotransformed Arabidopsis protoplasts in which the Tcoi1-MT-GFP or Tcoi1-GFP and CMV 1a-HA constructs were used (Fig. 5B). After the homogenization of the cotransformed Arabidopsis protoplasts, the extract was pulled down using a polyclonal anti-HA antibody. The pellet was washed with PBS and then analyzed by using a Western blot that was developed with monoclonal mono-/dimethylarginine antibody. As shown in the upper panel in Fig. 5B, the molecular mass band was detected at approximately 97 kDa, the estimated size of the recombinant CMV 1a-HA, in the extract from Arabidopsis protoplasts cotransformed with plasmids expressing CMV 1a-HA and Tcoi1-MT-GFP or Tcoi1-GFP. No significant bands for the extracts from Arabidopsis protoplasts transformed with a plasmid expressing Tcoi1-GFP only or GFP and CMV 1a-HA were detected (Fig. 5B). In the control experiment, the correct synthesis of the HA-tagged CMV 1a was verified via Western blotting, in which development was achieved with polyclonal HA antibody (Fig. 5B). Additionally, we verified that anti-HA serum did not precipitate Tcoi1-GFP alone (Fig. 5B). These results indicate that Tcoi1 has MT activity to methylate CMV 1a protein not only in vitro but also in vivo.
Reduced susceptibility to CMV infection in antisense-oriented-Tcoi1 transgenic tobacco plants.
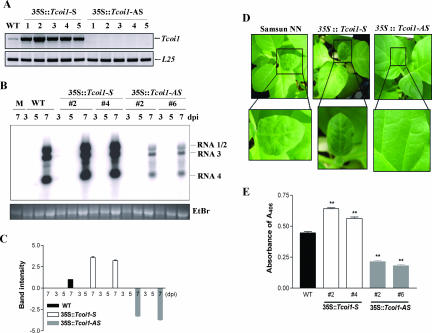
To further understand the biological function of Tcoi1 in vivo, we generated both sense- and antisense-oriented-Tcoi1 transgenic tobacco plants. All experiments using transgenic plants were performed in triplicate using the second-generation (T2) descendants of independent T0 lines that had a single copy of Tcoi1. At first, we compared the levels of RNA expression in transgenic plants and control plants by using RT-PCR. A low level of endogenous Tcoi1 gene expression in untransformed plants was apparent, and the expression level in transgenic plants harboring the sense Tcoi1 gene (sense transgenic plants) was higher than that in the control plants. The expression level in control plants, in turn, was higher than that in transgenic plants harboring the antisense Tcoi1 gene (antisense transgenic plants) (Fig. 6A). We examined whether Tcoi1 transgenic tobacco plants exhibited altered responses against CMV infection compared to those of wild-type plants. Four leaves from each of 10 6-leaf-stage transgenic T2 plants per independent line and 10 untransformed control plants of cultivar Samsun NN were inoculated with the CMV Korean strain (CMV-Kor) for the assessment of virus infectivity.
FIG. 6.
Expression of Tcoi1 gene in transgenic tobacco plants and comparison of CMV RNA levels and patterns of CMV CP accumulation and symptom development in wild-type and transgenic plants. (A) RT-PCR comparison of the transgenic T2 tobacco plants and the untransformed control plants. As an internal standard for cDNA quantification, the level of L25 transcripts was also monitored. WT, wild type; 35S::Tcoi1-S, sense-Tcoi1 transgenic plants; 35S::Tcoi1-AS, antisense-Tcoi1 transgenic plants. (B) The levels of CMV RNA accumulation in the transgenic T2 plants, along with those in the control plants, were monitored by Northern blotting using the CMV RNA 3′ untranslated region as a probe on the indicated days after inoculation. The uninoculated upper leaves from sense-Tcoi1 and antisense-Tcoi1 plants and wild-type plants, as indicated, were monitored at 3, 5, and 7 dpi and then harvested for RNA analysis. The positions of the bands representing CMV RNAs 1, 2, 3, and 4 are shown to the right of the panel. The RNA loading control was stained with ethidium bromide (EtBr). M, molecular size marker. (C) The intensities of the bands for CMV RNA 4 on Northern blots were quantified. Boxes and error bars represent the means and standard deviations of results from three independent experiments examining relative levels of viral RNA accumulation, normalized by the intensities of the wild-type bands in the corresponding lanes. Values are expressed as log2 ratios. (D) Comparison of the CMV-Kor infection symptoms in uninoculated upper leaves of transgenic and untransformed plants at 9 dpi. The lower panels show higher magnifications of the boxed regions. (E) Comparison of CMV-Kor CP accumulation patterns in uninoculated upper leaves at 9 dpi. The accumulation of CMV CP was measured via ELISA, and the results were expressed as the absorbance at 405 nm (A405) per 45 μg of total protein (n = 30 plants). The data were analyzed by analysis of variance. Error bars indicate the standard deviations, and asterisks indicate significant differences compared with the wild-type control (P < 0.001).
At various time points after inoculation, the levels of accumulation of CMV viral RNAs were determined by RNA hybridization with the end of RNA 3 corresponding to the C terminus of the product as a probe. The CMV RNA accumulation levels in inoculated leaves of untransformed plants and transgenic plants were similar (data not shown). Next, the accumulation pattern of virus-related RNA in uninoculated upper leaves was investigated. The accumulation of CMV RNAs in untransformed plants and sense transgenic plants was detectable at 7 dpi. The levels of CMV RNAs in sense transgenic plants were higher than those in untransformed plants. In contrast, CMV RNA levels were reduced in antisense transgenic plants compared to the levels in untransformed plants (Fig. 6B). The intensities of the bands for CMV RNA 4 on Northern blots were quantified using a phosphorimager and ImageQuant software and were normalized by the intensities of the wild-type bands in the corresponding lanes (Fig. 6C). The band intensities for sense transgenic plants were 2.5 to ∼3 times higher than those observed for the untransformed plants, and the band intensities for antisense transgenic plants were 5.5 to ∼6 times lower than those observed for the untransformed plants. We also observed the patterns of symptom development in uninoculated upper leaves. As shown in Fig. 6D, typical symptoms of CMV infection were evident in untransformed plants within 9 dpi and were slightly more severe but did not occur earlier in sense transgenic plants, whereas under the same conditions, the antisense transgenic plants exhibited no visible signs of virus infection. The accumulation of CMV CP in the uninoculated upper leaves at 9 dpi was assessed by ELISA. The degree of CMV CP accumulation in sense transgenic plants was 27 to ∼45% higher than that in untransformed plants, and that in antisense transgenic plants was 52 to 59% lower than that in untransformed plants (Fig. 6E). However, 15 days after virus infection, the symptoms were also detected in antisense transgenic plants. These results suggested that the replication and/or spread of CMV was compromised in antisense-Tcoi1 transgenic plants.
DISCUSSION
We identified a novel protein MT, Tcoi1, based on its specific interaction with CMV 1a protein. This interaction was shown to cause the methylation of CMV 1a protein both in vitro and in vivo. The finding that CMV infectivity was enhanced, although only in uninoculated upper leaves, in transgenic tobacco plants that overexpressed Tcoi1 and reduced in antisense-Tcoi1 transgenic plants suggested that protein methylation was crucial in regulating the function of CMV 1a protein, thereby facilitating virus replication and movement.
Tcoi1 and CMV 1a were shown to interact both in vitro, in the absence of other proteins, and in vivo by using four independent assays: the classic yeast two-hybrid system, a GST pulldown assay, BRET analysis, and coimmunoprecipitation from Arabidopsis protoplasts (Table 1; Fig. 3 and 4). In addition, the expression of Tcoi1 was increased upon CMV infection (Fig. 2C), and when Tcoi1 was expressed from a YFP vector, it accumulated in the cytosolic compartments of the Arabidopsis protoplasts (Fig. 4A). Thus, Tcoi1 is a cytosolic protein in plant cells, and this finding supports its role in protein methylation modification.
It has been reported previously that PRMTs, including CARM1 (PRMT4), share a highly conserved domain encompassing the MT activity (7). In addition to its MT activity, this homology domain is also responsible for the formation of homodimers or large homo-oligomers (58). We also found that the MT domain of Tcoi1 formed a homodimer in yeast cells (data not shown). Additionally, the MT domain of Tcoi1 was important for interaction with CMV 1a protein (Fig. 3), and this domain showed MT activity against CMV 1a protein (Fig. 5). Although Tcoi1 interacted with CMV 1a-MT, but not with CMV 1a-Hel, Tcoi1 methylated both the MT and Hel domains in vitro (Fig. 5A). When we predicted the site of protein methylation modification in CMV 1a by using a Web tool, MeMo (http://www.bioinfo.Tsinghua.edu.cn/∼tigerchen/memo.html), there were three possible sites (amino acids 32, 111, and 182) of arginine methylation in the MT domain of CMV 1a and one site (amino acid 659) in the Hel domain of CMV 1a. Thus, it is likely that Tcoi1 interacted with CMV 1a through the MT domain of CMV 1a and then methylated both the MT and Hel domains.
Posttranslational modifications of proteins expand the structural and functional diversity of the proteome (53). But how the posttranslational modifications of CMV 1a are involved in the life cycle of CMV in plants is not currently understood. A postulated mechanism includes the regulation of CMV 1a MT or Hel domain activity by a posttranslational modification such as CMV 1a protein methylation through interaction with Tcoi1. It was previously reported that the yeast Coq5 C-MT regulates the stability of other polypeptides involved in coenzyme Q synthesis (3). Additionally, Invernizzi et al. described the novel methylation in HIV type 1 (HIV-1) Rev's arginine-rich motif by PRMT6 (24). PRMT6 binds HIV-1 Rev and decreases the stability of Rev in cells. Furthermore, the methylated Rev was shown previously to bind the Rev-responsive element in HIV-1 RNAs (12) poorly and is associated with attenuated Rev-Rev-responsive element-dependent export of viral transcripts from the nucleus to the cytoplasm. Similar to Coq5 and PRMT6, Tcoi1 may participate in regulating CMV 1a stability. Moreover, methylation may regulate the processing and/or assembly of the CMV 1a protein, as has been suggested previously for the regulation of the HIV-1 Gag protein processing (55). In that report, the authors mentioned differences in the compositions and sizes of virion proteins in the presence and absence of adenosine periodate, a finding consistent with perturbed virion assembly when methylation events were inhibited, leading ultimately to the significant decrease in HIV-1 infectivity.
The interaction of CMV 1a and 2a was important for the formation of the replication complex, and the phosphorylation of the CMV replicase, protein 2a, inhibited the 1a-2a protein interaction (29). We found that, relative to those of wild-type plants, CMV RNA levels in systemically infected leaves of sense-Tcoi1 transgenic plants were increased and those of antisense-Tcoi1 transgenic plants were decreased (Fig. 6B). However, the CMV RNA levels in inoculated leaves were unchanged between wild-type plants and transgenic plants expressing sense and antisense full-length Tcoi1 cDNA. Despite the role of CMV 1a in RNA replication, Tcoi1 does not seem to have a significant effect on CMV RNA replication in single cells or on the initial cell-to-cell spread of CMV infection in inoculated leaves. Thus, CMV 1a methylation is unlikely to regulate the 1a-2a interaction. Furthermore, Tcoi1 appears to affect long-range systemic spread (Fig. 6B, D, and E), which is often controlled by induced host defenses (16). Additionally, the inhibition of systemic spread may represent a threshold effect, changing the outcome of the race between virus spread and host defense by slightly accelerating host defense induction or slightly slowing early CMV spread out of inoculated leaves. While the results are consistent with Tcoi1's affecting CMV infection by methylating 1a, we cannot exclude the possibility that Tcoi1 action on another viral or cellular protein(s) causes or contributes to the effects on systemic infection.
Interestingly, CARM1 has been shown previously to repress CREB-dependent cellular gene expression (57). The methylation of the CBP/p300 KIX domain by CARM1 blocks CREB activation by disabling the interaction between KIX and the KID domain of CREB. Furthermore, arginine methylation of HIV-1 Tat protein was shown previously to negatively affect Tat-Tat transactivation region-cyclin T1 ternary complex formation and diminish cyclin T1-dependent Tat transcriptional activation (56). Thus, the methylation of CMV 1a protein may alter the interactions of CMV 1a proteins with other host proteins, leading to either the increase or the decrease of the level of CMV infection.
Arabidopsis plants with a disruption of the menG gene encoding 2-phytyl-1,4-naphthoquinone MT show significantly reduced growth and chlorophyll contents that are caused by changes in photosystem I stability or turnover, and the limitation in functional photosystem I complexes results in the overreduction of photosystem II under high light levels (37). Antisense-Tcoi1 transgenic plants showed slightly reduced leaf size compared to the leaf size of the untransformed plants (data not shown). Therefore, Tcoi1 may be one of the genes that regulate photosynthesis.
Although the exact significance of the Tcoi1-CMV 1a interaction and the methylation of CMV 1a is unclear at this stage and the site of the CMV 1a protein methylation is yet to be identified, the idea that protein methylation plays a role in virus multiplication is attractive. Therefore, the identification of the methylated residues of the CMV 1a will improve our understanding of such novel modes of CMV viral protein regulation and will further illuminate the subtle complexities of protein structure and function.
In summary, we have discovered the function of a previously uncharacterized novel MT, Tcoi1, and our data support the conclusions that Tcoi1 and CMV 1a interact directly and that this interaction is specific, physiologically relevant, and important for CMV systemic movement and CMV infection symptom development.
Acknowledgments
This work was supported by the Science Research Center-Engineering Research Center program (Plant Signaling Network Research Center) of the Ministry of Science and Technology-Korea Science and Engineering Foundation (grant no. R11-2003-008-02001-0). B.-K. Ham was supported by a postdoctoral fellowship from the Korea Science and Engineering Foundation, and the scholarships of M. J. Kim and S. U. Huh were provided by the BK21 program of Korea.
Footnotes
Published ahead of print on 5 March 2008.
REFERENCES
- 1.Aletta, J. M., T. R. Cimato, and M. J. Ettinger. 1998. Protein methylation: a signal event in post-translational modification. Trends Biochem. Sci. 2389-91. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1995. Short protocols in molecular biology. Wiley, New York, NY.
- 3.Baba, S. W., G. I. Belogrudov, J. C. Lee, P. T. Lee, J. Strahan, J. N. Shepherd, and C. F. Clarke. 2004. Yeast Coq5 C-methyltransferase is required for stability of other polypeptides involved in coenzyme Q biosynthesis. J. Biol. Chem. 27910052-10059. [DOI] [PubMed] [Google Scholar]
- 4.Borchardt, R. T., B. T. Keller, and U. Patel-Thombre. 1984. Neplanocin A. A potent inhibitor of S-adenosylhomocysteine hydrolase and of vaccinia virus multiplication in mouse L929 cells. J. Biol. Chem. 2594353-4358. [PubMed] [Google Scholar]
- 5.Boulanger, M. C., C. Liang, R. S. Russell, R. Lin, M. T. Bedford, M. A. Wainberg, and S. Richard. 2005. Methylation of Tat by PRMT6 regulates human immunodeficiency virus type 1 gene expression. J. Virol. 79124-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buck, K. W. 1996. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 47159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, D., N. Yadav, R. W. King, M. S. Swanson, E. J. Weinstein, and M. T. Bedford. 2004. Small molecule regulators of protein arginine methyltransferases. J. Biol. Chem. 27923892-23899. [DOI] [PubMed] [Google Scholar]
- 8.Clarke, S. 2003. Aging as war between chemical and biochemical processes: protein methylation and the recognition of age-damaged proteins for repair. Ageing Res. Rev. 2263-285. [DOI] [PubMed] [Google Scholar]
- 9.Crofts, A. J., N. Leborgne-Castel, M. Pesca, A. Vitale, and J. Denecke. 1998. BiP and calreticulin form an abundant complex that is independent of endoplasmic reticulum stress. Plant Cell 10813-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis, S. J., and R. D. Viestra. 1996. Soluble derivatives of green fluorescent protein (GFP) for use in Arabidopsis thaliana. Weeds World 343-48. [Google Scholar]
- 11.Davis, S. J., and R. D. Viestra. 1998. Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol. Biol. 36521-528. [DOI] [PubMed] [Google Scholar]
- 12.Dayton, A. I. 2004. Within you, without you: HIV-1 Rev and RNA export. Retrovirology 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding, B., Q. Li, L. Nguyen, P. Palukaitis, and W. J. Lucas. 1995. Cucumber mosaic virus 3a protein potentiates cell-to-cell trafficking of CMV RNA in tobacco plants. Virology 207345-353. [DOI] [PubMed] [Google Scholar]
- 14.Ding, S.-W., B. J. Anderson, H. R. Haase, and R. H. Symons. 1994. New overlapping gene encoded by the cucumber mosaic virus genome. Virology 198593-601. [DOI] [PubMed] [Google Scholar]
- 15.Ermler, U., W. Grabarse, S. Shima, M. Goubeaud, and R. K. Thauer. 1997. Crystal structure of methyl-coenzyme M reductase: the key enzyme of biological methane formation. Science 2781457-1462. [DOI] [PubMed] [Google Scholar]
- 16.Gale, M., Jr., and E. M. Foy. 2005. Evasion of intracellular host defence by hepatitis C virus. Nature 436939-945. [DOI] [PubMed] [Google Scholar]
- 17.Gorbalenya, A. E., E. V. Koonin, A. P. Donchenko, and V. M. Blinov. 1989. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 174713-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gordon, R. K., K. Ginalski, W. R. Rudnicki, L. Rychlewski, M. C. Pankaskie, J. M. Bujnicki, and P. K. Chiang. 2003. Anti-HIV-1 activity of 3-deaza-adenosine analogs. Inhibition of S-adenosylhomocysteine hydrolase and nucleotide congeners. Eur. J. Biochem. 2703507-3517. [DOI] [PubMed] [Google Scholar]
- 19.Habili, N., and R. H. Symons. 1989. Evolutionary relationship between luteovirus and other RNA viruses based on sequence motifs in their putative RNA polymerase and nucleic acid helicases. Nucleic Acids Res. 179543-9555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han, S.-J., H. Y. Cho, J.-S. You, Y.-W. Nam, E. K. Park, J.-S. Shin, Y. I. Park, W. M. Park, and K.-H. Paek. 1999. Gene silencing-mediated resistance in transgenic tobacco plants carrying potato virus Y coat protein gene. Mol. Cells 9376-383. [PubMed] [Google Scholar]
- 21.Hayes, R. J., and K. W. Buck. 1990. Complete replication of a eukaryotic virus RNA in vitro by a purified RNA-dependent RNA polymerase. Cell 63363-368. [DOI] [PubMed] [Google Scholar]
- 22.Hodgman, T. C. 1988. A new superfamily of replicative proteins. Nature 33322-23. [DOI] [PubMed] [Google Scholar]
- 23.Hoekema, A., P. Hirsch, P. J. J. Hooykaas, and R. Schilperoort. 1983. A binary plant vector strategy based on separation of vir- and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature 303179-180. [Google Scholar]
- 24.Invernizzi, C. F., B. Xie, S. Richard, and M. A. Wainberg. 2006. PRMT6 diminishes HIV-1 Rev binding to and export of viral RNA. Retrovirology 393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishida, T., A. Hamano, T. Koiwa, and T. Watanabe. 2006. 5′ long terminal repeat (LTR)-selective methylation of latently infected HIV-1 provirus that is demethylated by reactivation signals. Retrovirology 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin, J. B., Y. A. Kim, S. J. Kim, S. H. Lee, D. H. Kim, G. W. Cheong, and I. Hwang. 2001. A new dynamin-like protein, ADL6, is involved from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 131511-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kagan, R. M., and S. Clarke. 1994. Widespread occurrence of three sequence motifs in diverse S-adenosylmethionine-dependent methyltransferases suggests a common structure for these enzymes. Arch. Biochem. Biophys. 310417-427. [DOI] [PubMed] [Google Scholar]
- 28.Keller, B. T., and R. T. Borchardt. 1987. Adenosine dialdehyde: a potent inhibitor of vaccinia virus multiplication in mouse L929 cells. Mol. Pharmacol. 31485-492. [PubMed] [Google Scholar]
- 29.Kim, S. H., P. Palukaitis, and Y. I. Park. 2002. Phosphorylation of cucumber mosaic virus RNA polymerase 2a protein inhibits formation of replicase complex. EMBO J. 212292-2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwak, Y. T., J. Guo, J., S. Prajapati, K. J. Park, R. M. Surabhi, B. Miller, P. Gehrig, and R. B. Gaynor. 2003. Methylation of SPT5 regulates its interaction with RNA polymerase II and transcriptional elongation properties. Mol. Cell 111055-1066. [DOI] [PubMed] [Google Scholar]
- 31.Kzhyshkowska, J., E. Kremmer, M. Hofmann, H. Wolf, and T. Dobner. 2004. Protein arginine methylation during lytic adenovirus infection. Biochem. J. 383259-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, D. Y., C. Teyssier, B. D. Strahl, and M. R. Stallcup. 2005. Role of protein methylation in regulation of transcription. Endocr. Rev. 26147-170. [DOI] [PubMed] [Google Scholar]
- 33.Li, Q., and P. Palukaitis. 1996. Comparison of the nucleic acid- and NTP-binding properties of the movement protein of cucumber mosaic cucumovirus and tobacco mosaic tobamovirus. Virology 21671-79. [DOI] [PubMed] [Google Scholar]
- 34.Li, Y. J., M. R. Stallcup, and M. M. Lai. 2004. Hepatitis delta virus antigen is methylated at arginine residues, and methylation regulates subcellular localization and RNA replication. J. Virol. 7813325-13334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin, W. J., J. D. Gary, M. C. Yang, S. Clarke, and H. R. Herschman. 1996. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J. Biol. Chem. 27115034-15044. [DOI] [PubMed] [Google Scholar]
- 36.Linder, P., P. F. Lasko, M. Ashburner, P. Leroy, P. J. Nielsen, K. Nishi, J. Schnier, and P. P. Slonimski. 1989. Birth of the D-E-A-D box. Nature 337121-122. [DOI] [PubMed] [Google Scholar]
- 37.Lohmann, A., M. A. Schöttler, C. Bréhélin, F. Kessler, R. Bock, E. B. Cahoon, and P. Dörmann. 2006. Deficiency in phylloquinone (vitamin K1) methylation affects prenyl quinone distribution, photosystem I abundance, and anthocyanin accumulation in the Arabidopsis AtmenG mutant. J. Biol. Chem. 28140461-40472. [DOI] [PubMed] [Google Scholar]
- 38.Lucy, A. P., H. S. Guo, W. Li, and S. W. Ding. 2000. Suppression of post-transcriptional gene silencing by a plant viral protein localized in the nucleus. EMBO J. 191672-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mears, W. E., and S. A. Rice. 1996. The RGG box motif of the herpes simplex virus ICP27 protein mediates an RNA-binding activity and determines in vivo methylation. J. Virol. 707445-7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mi, S., and V. Stollar. 1991. Expression of Sindbis virus nsP1 and methyltransferase activity in Escherichia coli. Virology 184423-427. [DOI] [PubMed] [Google Scholar]
- 41.Mi, S., R. Durbin, H. V. Huang, C. M. Rice, and V. Stollar. 1989. Association of Sindbis virus RNA methyltransferase activity with the nonstructural protein nsP1. Virology 170385-391. [DOI] [PubMed] [Google Scholar]
- 42.Mowen, K. A., B. T. Schurter, J. W. Fathman, M. David, and L. H. Glimcher. 2004. Arginine methylation of NIP45 modulates cytokine gene expression in effector T lymphocytes. Mol. Cell 15559-571. [DOI] [PubMed] [Google Scholar]
- 43.Palukaitis, P., and F. Garcia-Arenal. 2003. Cucumoviruses. Adv. Virus Res. 62241-323. [DOI] [PubMed] [Google Scholar]
- 44.Palukaitis, P., M. J. Roossinck, R. G. Dietzgen, and R. I. B. Francki. 1992. Cucumber mosaic virus. Adv. Virus Res. 41281-348. [DOI] [PubMed] [Google Scholar]
- 45.Rozanov, M. N., E. V. Koonin, and A. E. Gorbalenya. 1992. Conservation of the putative methyltransferase domain: a hallmark of the ‘Sindbis-like’ supergroup of positive-strand RNA viruses. J. Gen. Virol. 732129-2134. [DOI] [PubMed] [Google Scholar]
- 46.Schwinghamer, M. W., and R. H. Symons. 1975. Fractionation of cucumber mosaic virus RNA and its translation in a wheat embryo cell-free system. Virology 63252-262. [DOI] [PubMed] [Google Scholar]
- 47.Smith, W. A., B. T. Schurter, F. Wong-Staal, and M. David. 2004. Arginine methylation of RNA helicase determines its subcellular localization. J. Biol. Chem. 27922795-22798. [DOI] [PubMed] [Google Scholar]
- 48.Stallcup, M. R., J. H. Kim, C. Teyssier, Y. H. Lee, H. Ma, and D. Chen. 2003. The roles of protein-protein interactions and protein methylation in transcriptional activation by nuclear receptors and their coactivators. J. Steroid Biochem. Mol. Biol. 85139-145. [DOI] [PubMed] [Google Scholar]
- 49.Subramanian, C., J. Woo, X. Cai, X. Xu, S. Servick, C. H. Johnson, A. Nebenfuhr, and A. G. von Arnim. 2006. A suite of tools and application notes for in vivo protein interaction assays using bioluminescence resonance energy transfer (BRET). Plant J. 48138-152. [DOI] [PubMed] [Google Scholar]
- 50.Tanaka, J., T. Ishida, B. I. Choi, J. Yasuda, T. Watanabe, and Y. Iwakura. 2003. Latent HIV-1 reactivation in transgenic mice requires cell cycle-dependent demethylation of CREB/ATF sites in the LTR. AIDS 17167-175. [DOI] [PubMed] [Google Scholar]
- 51.Vaquero, C., Y.-C. Liao, J. Nahring, and R. Fischer. 1997. Mapping of the RNA-binding domain of the cucumber mosaic virus movement protein. J. Gen. Virol. 782095-2099. [DOI] [PubMed] [Google Scholar]
- 52.Vemuri, R., and K. D. Philipson. 1988. Protein methylation inhibits Na+-Ca2+ exchange activity in cardiac sarcolemmal vesicles. Biochim. Biophys. Acta 939503-508. [DOI] [PubMed] [Google Scholar]
- 53.Walsh, C. T., S. Garneau-Tsodikova, and G. J. Gatto, Jr. 2005. Protein posttranslational modifications: the chemistry of proteome diversifications. Angew. Chem. Int. Ed. Engl. 447342-7372. [DOI] [PubMed] [Google Scholar]
- 54.Whitham, S. A., S. Quan, H. S. Chang, B. Cooper, B. Estes, T. Zhu, X. Wang, and Y. M. Hou. 2003. Diverse RNA viruses elicit the expression of common sets of genes in susceptible Arabidopsis thaliana plants. Plant J. 33271-283. [DOI] [PubMed] [Google Scholar]
- 55.Willemsen, N. M., E. M. Hitchen, T. J. Bodetti, A. Apolloni, D. Warrilow, S. C. Piller, and D. Harrich. 2006. Protein methylation is required to maintain optimal HIV-1 infectivity. Retrovirology 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xie, B., C. F. Invernizzi, S. Richard, and M. A. Wainberg. 2007. Arginine methylation of the human immunodeficiency virus type 1 Tat protein by PRMT6 negatively affects Tat interactions with both cyclin T1 and the Tat transactivation region. J. Virol. 814226-4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu, W., H. Chen, K. Du, H. Asahara, M. Tini, B. M. Emerson, M. Montminy, and R. M. Evans. 2001. A transcriptional switch mediated by cofactor methylation. Science 2942507-2511. [DOI] [PubMed] [Google Scholar]
- 58.Zhang, X., L. Zhou, and X. Cheng. 2000. Crystal structure of the conserved core of protein arginine methyltransferase PRMT3. EMBO J. 193509-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]