Abstract
We have constructed a recombinant herpes simplex virus type 1 (HSV-1) that simultaneously encodes selected structural proteins from all three virion compartments—capsid, tegument, and envelope—fused with autofluorescent proteins. This triple-fluorescent recombinant, rHSV-RYC, was replication competent, albeit with delayed kinetics, incorporated the fusion proteins into all three virion compartments, and was comparable to wild-type HSV-1 at the ultrastructural level. The VP26 capsid fusion protein (monomeric red fluorescent protein [mRFP]-VP26) was first observed throughout the nucleus and later accumulated in viral replication compartments. In the course of infection, mRFP-VP26 formed small foci in the periphery of the replication compartments that expanded and coalesced over time into much larger foci. The envelope glycoprotein H (gH) fusion protein (enhanced yellow fluorescent protein [EYFP]-gH) was first observed accumulating in a vesicular pattern in the cytoplasm and was then incorporated primarily into the nuclear membrane. The VP16 tegument fusion protein (VP16-enhanced cyan fluorescent protein [ECFP]) was first observed in a diffuse nuclear pattern and then accumulated in viral replication compartments. In addition, it also formed small foci in the periphery of the replication compartments which, however, did not colocalize with the small mRFP-VP26 foci. Later, VP16-ECFP was redistributed out of the nucleus into the cytoplasm, where it accumulated in vesicular foci and in perinuclear clusters reminiscent of the Golgi apparatus. Late in infection, mRFP-VP26, EYFP-gH, and VP16-ECFP were found colocalizing in dots at the plasma membrane, possibly representing mature progeny virus. In summary, this study provides new insights into the dynamics of compartmentalization and interaction among capsid, tegument, and envelope proteins. Similar strategies can also be applied to assess other dynamic events in the virus life cycle, such as entry and trafficking.
The herpes simplex virus type 1 (HSV-1) virion consists of three different compartments, capsid, tegument, and envelope. The icosahedral capsid has a diameter of 125 nm and contains the virus genome, a double-stranded DNA of 152 kbp. The structural basis of the capsid are the 162 capsomers, which include 150 hexons and 12 pentons (47). The capsomers are connected in groups of three by a complex formed with two copies of VP23 and one copy of VP19c (47, 54, 68). The hexons are composed of six molecules of the major capsid protein VP5. Eleven of the 12 pentons are composed of five molecules of VP5, while 1 of the 12, the so-called portal, is a cylindrical structure of 12 molecules of UL6 (46). Also involved in capsid assembly, but not physical components of the capsids, are the scaffold polypeptides VP22a, VP21, and the serine protease, VP24, which is required for capsid maturation (9, 26, 38, 51). Six copies of VP26, a 12-kDa polypeptide encoded by the UL35 gene, occupy the tips of each hexon and thus decorate the surface of the capsid (42, 69). Although not essential for virus replication in tissue culture, VP26 was demonstrated to be important for infectious virus production in trigeminal ganglia (12). VP26 is a protein expressed later in the virus replication cycle after the onset of DNA replication and has been demonstrated to have multiple phosphorylated forms (43). VP26 has been shown to be recruited in an ATP-dependent manner after pro-capsid formation (8). As it lacks a nuclear localization signal (NLS), it must form complexes with NLS-containing proteins, such as VP5 and VP22a, in order to specifically accumulate in the nucleus (52, 60).
The virus capsid is surrounded by an amorphous layer, the so-called tegument. The tegument contains at least 15 virus-encoded proteins in various copy numbers which play important structural and functional roles during infection (32). One major structural component of the tegument is VP16, a 54-kDa protein encoded by the UL48 gene (63). Although VP16 is not essential for viral DNA replication, its structural role in the tegument is essential. Recombinants of HSV-1 that lack the UL48 gene show impaired replication, a defect in DNA packaging, and the absence of infectious virus progeny (63). VP16 is responsible for transcriptional regulation of immediate-early (IE) genes and is also involved in the modulation of the activities of early and late virus genes (7, 48, 49). VP16 has been shown to coimmunoprecipitate with virion host shutoff protein (55), to cross-link into complexes with gB, gD, and gH (70), and to copurify with UL47 (67) and with VP22 (16). Due to its involvement in linking capsid and future envelope-associated tegument proteins during virion formation, VP16 is absolutely required for assembly of infectious virus (63) and plays essential roles in viral maturation and egress (24, 44).
The tegument is surrounded by the viral envelope, which is a lipid membrane of cellular origin that contains at least 11 different viral glycoproteins. The glycoproteins are the major antigenic determinants for the host-specific recognition, and they are involved in cell entry, cell-to-cell spread, and immune evasion. Glycoprotein H, the product of the UL22 gene, is a 110-kDa protein which is essential for infectivity (14) and membrane fusion, but not for receptor binding (22, 25). Glycoprotein H must be coexpressed with gL in order for both proteins to be properly processed, folded, and transported to the virion envelope as well as the infected cell surface (33). The gH/gL complex plays essential roles in viral penetration, cell-to-cell spread, and syncytium formation (53). Both glycoproteins are conserved among the herpesviruses, although some differences regarding assembly, structure, and intracellular transport of the heterocomplex exist between individual herpesviruses (27, 33, 34, 50). Only very recently was it shown that the simultaneous deletion of gH and gB results in a severe deficit in nuclear egress leading to the accumulation of virions in the perinuclear space, whereas the deletion of gH or gB alone did not lead to a significant defect (19). One strategy to investigate mechanisms of viral infection, replication, and assembly is the fusion of viral proteins with autofluorescent proteins (reviewed in reference 5). Many of the HSV-1 proteins have been shown to maintain their functional activity when fused with foreign polypeptides. Among these, a green fluorescent protein (GFP)-VP26 fusion was demonstrated to be incorporated into intranuclear capsids and mature virions, where it was capable of interacting with VP5 while retaining its autofluorescence (13). GFP-VP26 retained its biological activity during the replication cycle, as the recombinant virus replicated at a rate comparable to that of wild-type (wt) virus (13). A recombinant HSV-1 that expressed VP16 fused to GFP (VP16-GFP) also showed normal replication kinetics and incorporation of the fusion protein into the virion (35). Similarly, fusion of enhanced yellow fluorescent protein (EYFP) with gH did not markedly alter gH functions, as the recombinant virus was replication competent and showed stable autofluorescence (39). The EYFP-gH fusion protein formed a stable heterocomplex with gL and was incorporated into the virion envelope as well as cellular membranes (39).
In this study, we have constructed a recombinant HSV-1 that simultaneously encodes the VP26 capsid protein fused with monomeric red fluorescent protein (mRFP), the VP16 tegument protein fused with enhanced cyan fluorescent protein (ECFP), and the gH envelope glycoprotein fused with EYFP. This triple-fluorescent recombinant HSV-1, rHSV-RYC, was replication competent and incorporated the autofluorescent fusion proteins into all three virion compartments. Confocal laser scanning microscopy (CLSM) of living infected cells revealed new insights into the organization and dynamics of HSV-1 infection and into the interactions between HSV-1 virion proteins. To our knowledge this is the first report of the construction and live analysis of a recombinant virus encoding three different autofluorescent fusion proteins.
MATERIALS AND METHODS
Cell culture and virus.
BHK, Vero, and Vero 2-2 cells (56) were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Penicillin G at 100 units/ml, 100 μg/ml streptomycin, and 0.25 μg/ml amphotericin B were added to all cell culture media. For culturing Vero 2-2 cells, 500 μg/ml G418 was included in addition.
HSV-1 strain F as well as the recombinant viruses were grown, and titers were determined in Vero cells.
Construction of recombinant viruses.
Recombinant HSV-1 encoding one (rHSV-R or rHSV-48Y), two (rHSV-RYor rHSV-RC), or three (rHSV-RYC or rHSV-RYC/2) different virion proteins fused with autofluorescent proteins were generated by homologous recombination in Escherichia coli SW102 and galK selection/counterselection (62) using a bacterial artificial chromosome (BAC)-cloned HSV-1 strain F genome (pYEbac102; kindly provided by Y. Kawaguchi, Tokyo Medical and Dental University, Japan) (59).
Electroporation and galK positive/negative selection.
To prepare electrocompetent bacteria, 500 μl of an overnight culture of E. coli SW102/YEbac102 was diluted in 25 ml LB medium containing 12.5 μg/ml chloramphenicol in a 50-ml conical flask and grown at 32°C. When the optical density at 600 nm reached 0.6, 10 ml of the culture was transferred to another 50-ml conical flask and incubated at 42°C in a shaking water bath. After exactly 15 min, the culture was briefly cooled on ice, transferred into two 15-ml tubes, and pelleted for 5 min at 5,000 rpm and 0°C. The supernatant was removed, and the pellet was resuspended in 1 ml ice-cold H2O by gently swirling the tube on ice. Then, 9 ml of ice-cold H2O was added, and the cells were pelleted again; this step was repeated once more. After the second washing and centrifugation step, the supernatant was removed, and the pellet (approximately 50 μl) was kept on ice until electroporated with PCR product (targeting cassettes). In a first step (galK selection), a PCR product (see below) that contained the E. coli galK gene flanked by 50 nucleotides of sequence homology to either side of the targeting locus on the virus genome (VP26, gH, and VP16) (Fig. 1) was electroporated into 25 μl of electrocompetent E. coli SW102/YEbac102 cells in a 0.1-cm cuvette (Bio-Rad, Hercules, CA) at 25 μF, 1.75 kV, and 200 Ω. After electroporation, the bacteria were grown in 1 ml LB medium for 1 h at 32°C and then washed twice with 1× M9 salts (37 mM Na2HPO4, 22 mM KH2PO4, 19 mM NaCl) as follows: the culture was pelleted at 13,000 rpm for 15 s, resuspended in 1× M9 salts, and pelleted again. The washing step was repeated once more. After the second wash, the supernatant was removed and the pellet was resuspended in 1× M9 salts before 100-μl aliquots of serial dilutions (1:10, 1:100, and 1:1,000) were plated on galactose minimal medium plates (62) supplemented with 12.5 μg/ml chloramphenicol to select Gal+ recombinant colonies. After 2 to 3 days of incubation at 32°C, colonies were picked and streaked on MacConkey galactose indicator plates to obtain single bright pink/red Gal+ colonies. One or two of these colonies were picked to prepare electrocompetent bacteria for the second recombination step, the in-frame introduction of autofluorescent protein-coding sequences into VP26, gH, or VP16 sequences and galK counterselection. For this, electrocompetent E. coli SW102 cells containing the galK-modified YEbac102 were prepared as described above and electroporated with PCR products (targeting cassettes; see below) containing coding sequences of autofluorescent proteins flanked by the same 50 nucleotides present on the galK PCR product that target the DNA to homologous sequences in the virus genome (VP26, gH, and VP16). After electroporation, the bacteria were recovered in 10 ml LB medium for 4.5 h at 32°C and washed twice with 1× M9 salts, and serial dilutions were plated on minimal medium plates containing glycerol as carbon source, leucine, biotin, and 2-deoxy-galactose (DOG; Acros Organics, Geel, Belgium) for selection against galK (62). After 3 days of incubation at 32°C, colonies were picked, and BAC DNA was isolated and characterized by restriction endonuclease and Southern analysis.
FIG. 1.
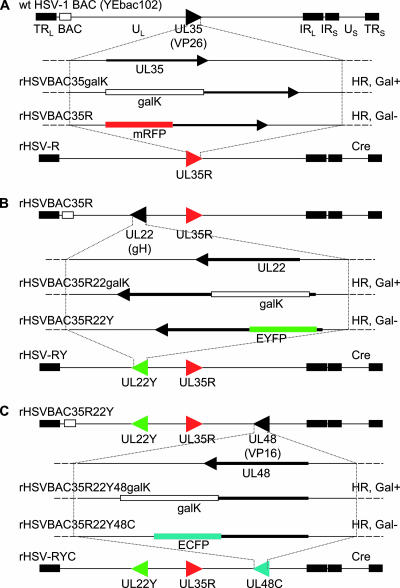
Construction of rHSV-RYC. (A) Representation of the HSV-1 BAC (YEbac102) genome structure, showing the region containing the UL35 gene. The galK expression cassette was inserted into the UL35 gene through homologous recombination (HR) and selection for Gal+ recombinants (rHSVBAC35galK). GalK was then replaced by mRFP coding sequences by HR and counterselected for Gal− recombinants (rHSVBAC35R). The same procedure was used for the fusion of EYFP with the UL22 gene on the rHSVBAC35R genome (B) and for the fusion of ECFP with the UL48 gene on rHSVBAC35R22Y (C). The BAC sequences were removed by the Cre/loxP recombination system (Cre), resulting in the recombinants HSV-1 rHSV-R, rHSV-RY, and rHSV-RYC. TRL, terminal repeat of the long segment; UL, unique long segment; IRL, internal repeat of the long segment; IRS, internal repeat of the short segment; US, unique short segment; TRS, terminal repeat of the short segment.
PCR amplification and purification of targeting cassettes.
Phusion polymerase (Finnzymes, Espoo, Finland) and the following primers were used to amplify the targeting cassettes: (i) mRFP-VP26 fusion, galK selection, ul35-galK-fw, 5′-ACAGCCCTCCCGACCGACACCCCCATATCGCTTCCCGACCTCCGGTCCCG CCTGTTGACAATTAATCATCGGCA-3′, and ul35-galK-rev, 5′-CCAAGCGCCCGGACGCTATCGGTGGTAACGGTGCTGGGGCGGTGAAATTGTCAGCACTGTCCTGCTCCTT-3′; mRFP-VP26 fusion, galK counterselection, ul35-rfp-fw, 5′-ACAGCCCTCCCGACCGACACCCCCATATCGCTTCCCGACCTCCGGTCCCGATGGCCTCCTCCGAGGACGTC-3′, and ul35-rfp-rev, 5′-CCAAGCGCCCGGACGCTATCGGTGGTAACGGTGCTGGGGCGGTGAAATTGGGCGCCGGTGGAGTGGCGGCC-3′; (ii) EYFP-gH fusion, galK selection, UL22-galK-fw, 5′-GTTATTATTTTGGGCGCTGCGTGGGGTCAGGTCCACGACTGGACTGAGCAGCCTGTTGACAATTAATCATCGGCA-3′, and UL22-galK-rev, 5′-CGTGTCGCGCCAGTACATGCGGTCCATGCCCAGGCCATCCAAAAACCATGGTCAGCACTGTCCTGCTCCTT-3′; EYFP-gH fusion, galK counterselection, UL22-EYFP-fw, 5′-GTTATTATTTTGGGCGCTGCGTGGGGTCAGGTCCACGACTGGACTGAGCAGGTGAGCAAGGGCGAGGAGCTGTTC-3′ and UL22-EYFP-rev, 5′-CGTGTCGCGCCAGTACATGCGGTCCATGCCCAGGCCATCCAAAAACCATGGCTTGTACAGCTCGTCCATGCCGAG-3′; (iii) VP16-ECFP fusion, galK selection, UL48-galK-fw, 5′-TTCGAGTTTGAGCAGATGTTTACCGATGCCCTTGGAATTGACGAGTACGGTCCTGTTGACAATTAATCATCGGCA-3′, and UL48-galK-rev, 5′-GGTGACGGGAGGGGAAAACCCAGACGGGGGATGCGGGTCCGGTCGCGCCCCTCAGCACTGTCCTGCTCCTT-3′; VP16-ECFP/EYFP fusion, galK counterselection, UL48-ECFP-fw, 5′-TTCGAGTTTGAGCAGATGTTTACCGATGCCCTTGGAATTGACGAGTACGGTGTGAGCAAGGGCGAGGAGCTGTTC-3′, and UL48-ECFP-rev, 5′-GGTGACGGGAGGGGAAAACCCAGACGGGGGATGCGGGTCCGGTCGCGCCCCTTACTTGTACAGCTCGTCCATGCC-3′. Sequence portions shown in italics are homologous to the galK gene. The following plasmids (10 ng) were used as templates: pgalK for all galK selection/targeting cassettes (obtained from S. Warming, National Cancer Institute, Frederick, MD) (62), pcDNA-mRFP-N1 (obtained from U. F. Greber, University of Zurich, Zurich, Switzerland) for the VP26-mRFP targeting cassette, and pECFP-N1 and pEYFP-N1 for amplification of VP16-ECFP/EYFP and gH-EYFP targeting cassettes, respectively. The PCR conditions were as follows: 94°C for 15 s, 60°C for 30 s, and 72°C for 1 min, for 30 cycles. After completion of the reaction, DpnI (10 U; New England Biolabs, Allschwil, Switzerland) was added for digestion of the template for 2 h. The DpnI-digested reaction mix was run on a 1% agarose gel, and the PCR product was purified using a QIAQuick PCR purification kit (Qiagen, Hombrechtikon, Switzerland) followed by ethanol precipitation. The DNA was resuspended in 40 μl H2O, and an aliquot of 2 to 5 μl (10 to 30 ng) was used for electroporation.
Excision of BAC sequences and isolation of recombinant HSV-1.
To excise the BAC DNA backbone and isolate recombinant viruses, 1.2 × 106 Vero 2-2 cells (56) per 6-cm tissue culture plate were cotransfected with 0.2 μg of plasmid p116, which expresses Cre recombinase with an NLS (kindly provided by K. Tobler, University of Zurich, Zurich, Switzerland), and 2 μg of CsCl gradient-purified recombinant HSV-1 BAC DNA using Lipofectamine (Invitrogen). After 2 to 3 days of incubation at 37°C, the supernatant was harvested and plaque purified twice, and the excision of the BAC sequences was verified by PCR.
Virus replication assays.
For the determination of growth kinetics, Vero cells were inoculated at a multiplicity of infection (MOI) of 0.1 or 5 PFU per cell. After 2 h of incubation at 37°C, 5% CO2, the cultures were washed three times with PBS and then incubated with DMEM containing 2% FBS. Samples (cell culture medium and cells) were removed after 0, 12, 24, 36, and 48 h. The cell culture medium was removed from the cells and serially diluted to determine the titers on Vero cells. The titers of cell-associated virus were determined on Vero cells inoculated with serial dilutions of supernatants of cells prepared by three cycles of freezing and thawing, followed by centrifugation at 1,900 × g.
Determination of particle/PFU ratios.
To determine the particle counts, virus stocks with known titers (PFU/ml) were mixed with 204-nm-diameter latex beads (Agar Scientific, Essex, United Kingdom) of known concentration, adsorbed onto 300-mesh parlodion- and carbon-coated copper electron microscope grids for 5 min at room temperature (RT), briefly washed with H2O, and negatively stained with 2% sodium phosphotungstate, pH 7.4, for 1 min at RT. Samples were examined in a transmission electron microscope (CM12; Philips, Eindhoven, The Netherlands), and the relative numbers of virus particles and latex beads were determined, which allowed us to calculate the absolute numbers of virus particles in the virus stocks.
Purification of virions.
Virions were purified from BHK or Vero cells infected at an MOI of 0.1 PFU with either wt HSV-1, rHSV-RYC, or rHSV-RYC/2. When the cytopathic effect (CPE) was complete, the cultures were frozen and thawed three times, and cell debris was removed by centrifugation for 10 min at 2,600 × g and 4°C. Virions were purified through 60%, 30%, and 10% sucrose (in PBS) gradients in Beckman Ultra-Clear 25- by 89-mm centrifuge tubes, which were centrifuged for 2 h at 28,000 rpm and 4°C using a Beckman SW28 rotor. The interface between the 30% and 60% sucrose layers was collected, diluted in PBS, and ultracentrifuged for 1 h at 25,000 rpm and 4°C. Following resuspension of the pellet in Hanks' buffered saline solution, the virion stocks were frozen in a dry ice-ethanol bath and stored at −80°C.
Immunoprecipitation, SDS-PAGE, Western analysis, and silver staining.
Vero cells (4 × 105 cells per well in a 12-well plate) were mock infected or infected with either wt HSV-1, rHSV-RY, rHSV-RYC, or rHSV-RYC/2 at an MOI of 1 PFU. When CPE was almost complete (between 24 and 48 h postinfection [p.i.]), the cells were washed with cold PBS and prepared for immunoprecipitation or directly lysed with sodium dodecyl sulfate (SDS) loading buffer, boiled for 5 min, and analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting using the antibodies listed below. For immunoprecipitation, the cells were lysed with 200 μl of EBC170 lysis buffer (50 mM Tris pH 8.0, 170 mM NaCl, 0.5% NP-40) supplemented with one tablet of protease inhibitor cocktail (Complete, mini, EDTA-free; Roche Diagnostics, Rotkreuz, Switzerland) per 10 ml. The cell extract was collected and immunoprecipitated with the gH-specific monoclonal antibody (MAb) LP11 (kindly provided by A. Minson and H. Browne, University of Cambridge, United Kingdom) (6) diluted 1:30 in EBC170. After 1 h at 4°C, complexes were allowed to attach to protein A-Sepharose beads. After washing the beads four times with EBC170 buffer, the LP11 MAb complexes were eluted from the beads by adding SDS loading buffer. The samples were boiled for 5 min, and proteins were separated by SDS-PAGE and transferred onto nylon membranes (Protran; Whatman, Bottmingen, Switzerland). Nonspecific reactions were blocked by incubating the membranes for 1 h with PBS containing 5% skimmed milk and 0.3% Tween 20. Membranes were then incubated for 1 h with antibodies against VP26 (rabbit polyclonal antibody [PAb] diluted 1:1,000 in PBS, 0.3% Tween 20 [PBS-T]; kindly provided by A. Helenius, ETH Zurich, Switzerland), VP16 (MAb LP1, diluted 1:500 in PBS-T; kindly provided by A. Minson and H. Browne, University of Cambridge, United Kingdom) (41), GFP (MAb JL-8BD, diluted 1:8,000 in PBS-T; Clontech, Saint-Germain-en-Laye, France), ICP4 (MAb, diluted 1:10,000 in PBS-T; Advanced Biotechnologies, Columbia, MD), ICP8 (MAb clone 10A3, diluted 1:10,000 in PBS-T; Abcam, Cambridge, United Kingdom), and VP22 (rabbit PAb AGV031, diluted 1:10,000 in PBS-T; kindly provided by G. Elliott, Marie Curie Research Institute, Oxted, United Kingdom). After washing, the membranes were incubated with rabbit anti-mouse immunoglobulin G (IgG)-horseradish peroxidase (HRP) (1:10,000 in PBS-T; Sigma-Aldrich, Buchs, Switzerland) or goat anti-rabbit IgG-HRP (1:10,000 in PBS-T; Sigma-Aldrich, Buchs, Switzerland). Target proteins were visualized by enhanced chemiluminescence (ECL Western blotting analysis system; GE Healthcare, Zurich, Switzerland) and autoradiography (Lumi-film chemiluminescent detection film; Roche Diagnostics, Rotkreuz, Switzerland). A molecular weight standard (BenchMark prestained ladder; GIBCO, Invitrogen, Basel, Switzerland) was used to determine the sizes of the protein bands. Silver staining of gels was performed using the Bio-Rad silver stain kit (Bio-Rad, Hercules, CA) according to the manufacturer's manual.
CLSM.
CLSM was performed on a Leica TCS SP2 AOBS confocal laser scanning microscope (Leica Microsystems, Wetzlar, Germany) equipped with an incubation chamber (THE BOX; Live Imaging Services, Reinach, Switzerland), a temperature control device (THE CUBE; Live Imaging Services, Reinach, Switzerland), and a gas mixer (THE BRICK; Live Imaging Services, Reinach, Switzerland). The settings for the individual fluorophores were as follows: 4′,6-diamidine-2′-phenylindole (DAPI), excitation at 405 nm and recording at 415 to 480 nm; ECFP, excitation at 458 nm and recording at 468 to 510 nm; fluorescein isothiocyanate (FITC), excitation at 488 nm and recording at 498 to 570 nm; EYFP, excitation at 514 nm and recording at 520 to 550 nm; mRFP/Atto590/Alexa Fluor 594 (AF594), excitation at 594 nm and recording at 604 to 700 nm. In order to avoid channel overlap, blue and red channels were recorded simultaneously while the yellow/green channels were recorded separately. The images shown in Fig. 7 and 8, below, were deconvolved with a blind deconvolution algorithm using the Huygens Essential 2.6.0p1 software (Scientific Volume Imaging, Hilversum, The Netherlands). Image processing was done with Imaris 5.0.1 (Bitplane AG, Zurich, Switzerland) and Adobe Photoshop CS 8.0.1 software. The detailed procedures were as follows.
FIG. 7.
High-resolution CLSM of large VP26 foci. (A) Vero cells were infected with rHSV-RY at an MOI of 0.3 PFU, fixed at the indicated times postinfection, stained with DAPI, and observed by CLSM with settings specific for DAPI, EYFP (EYFP-gH fusion protein), and mRFP (mRFP-VP26 fusion protein). Images represent single z stacks of the cells. (B) Panels a and c show magnifications of the mRFP-VP26 foci marked with the numbered arrows shown in panel A, while panels b and d show surpass views of three-dimensional reconstructions of the same foci. (C) Vero cells were infected with wt HSV-1 at an MOI of 10 PFU, fixed at 12 h p.i., and stained with the rabbit anti-VP26 PAb VP26/C and a FITC-conjugated secondary antibody. HSV-1 DNA was detected by FISH using an Atto590-labeled probe. Cells were observed by CLSM with settings specific for DAPI, FITC (VP26), and Atto590 (HSV-1 DNA). Images represent single z stacks of the cells.
FIG. 8.
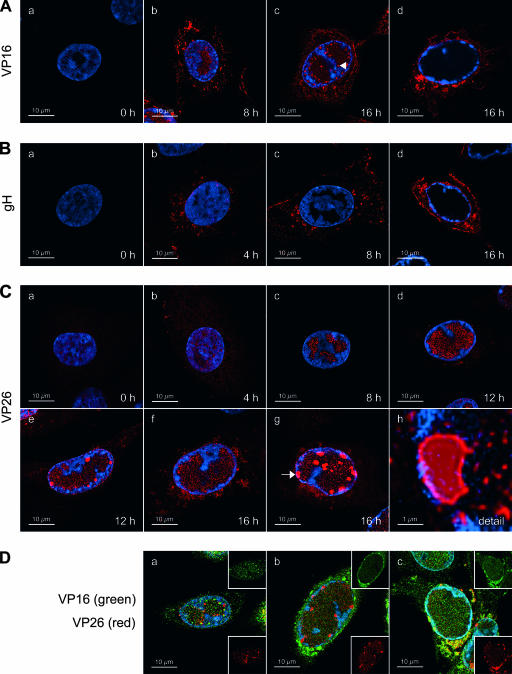
Immunofluorescence staining for VP16, gH, and VP26 in wt HSV-1-infected cells. (A) Immunofluorescence staining for VP16. Vero cells were infected with wt HSV-1 at an MOI of 10 PFU. At the indicated times postinfection, the cells were fixed and stained with the anti-VP16 MAb LP1 and an AF594-conjugated secondary antibody, as well as DAPI. The cells were then observed by CLSM with settings specific for DAPI and AF594. Filled arrowhead, VP16 foci in the periphery of RCs. (B) Immunofluorescence staining for gH. Vero cells were infected and stained as described for panel A, except that the anti-gH MAb LP11 was used. (C) Immunofluorescence staining for VP26. Vero cells were infected and stained as described for panel A, except that the rabbit anti-VP26 PAb VP26/C was used. Panel h shows a magnification of the VP26 focus marked with an arrow. (D) Vero cells were infected as described for panel A, fixed at 16 h p.i., and stained with DAPI, the anti-VP16 MAb LP1 (detected with an AF594-conjugated secondary antibody), and the rabbit anti-VP26 PAb VP26/C (detected with a FITC-conjugated secondary antibody). The cells were then observed by CLSM with settings specific for DAPI, AF594 (VP16, shown in green), and FITC (VP26, shown in red). Images in panels A to D represent single z stacks of the cells.
(i) High-resolution CLSM of live or fixed, infected cells.
Vero cells were seeded on Lab-Tek four-well chambered coverglasses (Nalge Nunc International, Rochester, NY) at 105 cells/well. On the following day, the cells were washed once with cold (4°C) DMEM and incubated for 15 min at 4°C, and the DMEM was replaced with cold (4°C) virus inoculum at the MOIs described in the figure legends. The viruses were then allowed to adsorb to the cells for 1 h at 4°C with gentle shaking. Subsequently, the virus inoculum was replaced with warm (37°C) Iscove's modified Dulbecco's medium (GIBCO, Invitrogen, Basel, Switzerland) supplemented with 25 mM HEPES and 2% FBS, and the cells were incubated at 37°C, 5% CO2. At the indicated times after infection (temperature shift), live cells were observed by CLSM in a humidified atmosphere at 37°C, 5%CO2. Where mentioned in the text below, the cells were fixed with 3.7% formaldehyde in PBS for 15 min at RT prior to microscopy.
(ii) Time-lapse CLSM of live, infected cells.
Vero cells were seeded into 35-mm glass-bottom dishes (MatTek, Ashland, MA) at 5 × 105 cells/dish. On the following day, the cells were infected with rHSV-RY or rHSV-RYC diluted in DMEM at the MOIs described in the text below. The viruses were allowed to adsorb for 2 h at 37°C, 5% CO2. The cells were then washed with PBS, overlaid with Iscove's modified Dulbecco's medium supplemented with 25 mM HEPES and 2% FBS, and incubated at 37°C, 5% CO2. At the indicated time, live cells were observed by CLSM in a humid atmosphere at 37°C, 5% CO2. Images were recorded at intervals of 15 to 25 min.
Immunofluorescence.
Vero cells were seeded on round 12-mm coverglasses in 24-well plates at 105 cells/well. On the following day, the cells were washed with PBS and infected with wt HSV-1 or rHSV-RY diluted in DMEM at the MOIs described in the figure legends. The viruses were allowed to adsorb for 1 to 2 h at 37°C, 5% CO2. The cells were then washed with PBS, overlaid with DMEM supplemented with 2% FBS, and incubated at 37°C, 5% CO2. At the indicated time points, the cells were washed once with cold PBS and fixed with 3.7% formaldehyde in PBS for 15 min at RT, and the fixation was stopped with 0.1 M glycine in PBS for 5 min at RT. Immunofluorescence staining, DAPI staining, and embedding of cells were performed as described previously (29) except that the cells were permeabilized with 0.2% Triton X-100 in PBS for 15 min at RT and that 0.2 mg/ml human IgG (Sigma-Aldrich, Buchs, Switzerland) was included in the blocking solution when cells were stained with antibodies of rabbit origin. Primary antibodies were used at the following dilutions: anti-HSV-1 ICP8 MAb 7381 (kindly provided by R. D. Everett, MRC Virology Unit, Glasgow, United Kingdom), 1:500 (see Fig. 6C, below) or 1:1,000 (see Fig. 6D, below); anti-HSV-1 gH MAb LP11, 1:3; anti-HSV-1 VP16 MAb LP1, 1:3 (see Fig. 8A, below) or 1:200 (see Fig. 8D, below); rabbit anti-HSV-1 VP26 PAb VP26/C (kindly provided by P. Desai, Johns Hopkins University, Baltimore, MD) (12), 1:400. Secondary antibodies were used as follows: F(ab′)2 fragment of goat anti-rabbit IgG(H+L)-AF594 (Molecular Probes, Invitrogen, Basel, Switzerland), 1:200; goat anti-rabbit IgG(H+L)-FITC (Southern Biotechnology, Birmingham, AL), 1:100; goat anti-mouse IgG(H+L)-AF594 (Molecular Probes, Invitrogen, Basel, Switzerland), 1:200; Fab fragment of goat anti-mouse IgG(H+L)-FITC (Jackson ImmunoResearch, West Grove, PA), 1:100.
FIG. 6.
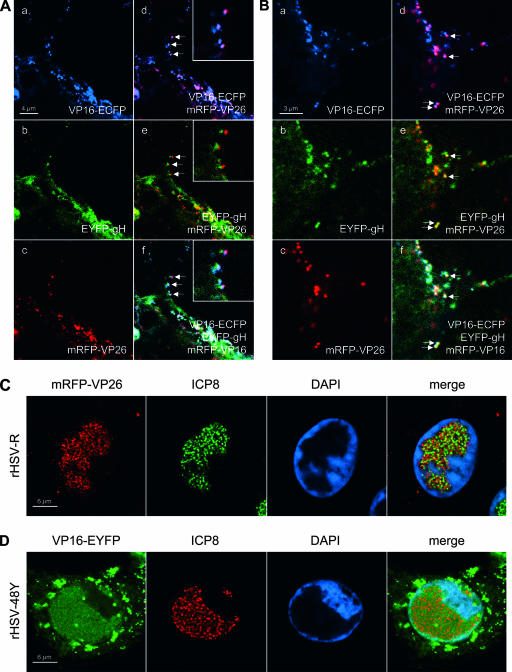
High-resolution CLSM of rHSV-RYC-infected cells and rHSV-R- and rHSV-48Y-infected cells stained for ICP8. (A) Vero cells were infected with rHSV-RYC, and living cells were observed by CLSM as described for Fig. 5. The images show a high magnification of a protrusion of the plasma membrane (top of picture). The arrows point to foci in which all three fusion proteins colocalize. The insets show magnifications of these foci. (B) Vero cells were infected with rHSV-RYC, and fixed cells were observed by CLSM as described for panel A. (C) Vero cells were infected with rHSV-R at an MOI of 10 PFU, fixed at 12 h p.i., and stained with the anti-ICP8 MAb 7381 and a FITC-conjugated secondary antibody, as well as DAPI. The cells were observed by CLSM with settings specific for DAPI, FITC (ICP8), and mRFP (mRFP-VP26 fusion protein). (D) Vero cells were infected with rHSV-48Y at an MOI of 10 PFU and stained as described for panel C, except that an AF594-conjugated secondary antibody was used. The cells were observed by CLSM with settings specific for DAPI, EYFP (VP16-EYFP fusion protein), and AF594 (ICP8). Images in panels A to D represent single z stacks of the cells.
FISH.
Infection with wt HSV-1 and immunofluorescence staining for VP26 were performed as described above. Fluorescent in situ hybridization (FISH) was performed as described previously (18) with the following modifications: (i) cells were fixed with precooled methanol for 10 min at −20°C; (ii) immunofluorescence staining was performed before the hybridization step; (iii) fluorescent labeling of BAC-cloned HSV-1 DNA (YEbac102) was performed with an Atto590 nick translation labeling kit according to the manufacturer's manual (Jena Bioscience, Jena, Germany).
Electron microscopy.
Vero cells were grown on sapphire disks for 2 days prior to infection with rHSV-RYC. After 24 h, cells were fixed with 0.25% glutaraldehyde for 30 min and then frozen in a high-pressure freezer (HPM010; BAL-TEC Inc., Balzers, Liechtenstein) as previously described (37). The sapphire disks carrying the frozen cells were transferred into a freeze-substitution unit (FS 7500; Boeckeler Instruments, Tucson, AZ) precooled to −88°C for substitution with acetone and subsequent fixation with 0.25% glutaraldehyde and 0.5% osmium tetroxide at temperatures between −30°C and +2°C as previously described in detail (64) and embedded in Epon at 4°C. Sections of 50 to 60 nm were analyzed in a transmission electron microscope (CM12; Philips, Eindhoven, The Netherlands) equipped with a slow-scan charge-coupled-device camera (Gatan, Pleasanton, CA) at an acceleration voltage of 100 kV.
RESULTS
Construction of a recombinant HSV-1 encoding mRFP-VP26, VP16-ECFP, and EYFP-gH fusion proteins.
The goal of this study was to construct and characterize a recombinant HSV-1 that simultaneously encodes selected structural proteins from all three virion compartments fused with autofluorescent proteins, in particular, capsid protein VP26 fused with mRFP, tegument protein VP16 fused with ECFP, and envelope glycoprotein H fused with EYFP. This triple-fluorescent recombinant HSV-1 was constructed via homologous recombination in E. coli using a BAC-cloned HSV-1 strain F genome (pYEbac102) (59) and the galK positive/negative selection method (62) as described in Materials and Methods. Briefly, in a first step, a DNA fragment containing the galK expression cassette flanked by homology arms that target the cassette between codons 1 and 5 of the VP26 coding sequences was electroporated into E. coli SW102 cells that contained the pYEbac102 HSV-1 BAC DNA. Gal-positive recombinant bacteria were selected on galactose minimal medium plates. BAC DNA prepared from these clones was characterized by restriction endonuclease analysis (not shown), and one clone (rHSVBAC35galK) was selected for the second recombination step. For the second step, the galK expression cassette was replaced by electroporating a DNA fragment containing the mRFP expression cassette, again flanked by the homology arms that facilitate the in-frame insertion of mRFP coding sequences between codons 1 and 5 of the VP26 coding sequences, into E. coli SW102/rHSVBAC35galK. After galK counterselection with DOG, Gal-negative colonies were picked, BAC DNA was prepared and characterized by restriction endonuclease analysis (not shown), and one clone (rHSVBAC35R; Fig. 1A) was selected for further manipulation. The subsequent fusion of gH with EYFP (rHSVBAC35R22Y; Fig. 1B) and VP16 with ECFP (rHSVBAC35R22Y48C; Fig. 1C) was performed using the same galK positive/negative selection procedure described above and in Materials and Methods. The final recombinant, rHSVBAC35R22Y48C, contained codons 1 to 225 of mRFP fused to codons 5 to 108 of VP26, codons 2 to 239 of EYFP inserted between codons 6 and 9 of gH, and codons 1 to 489 of VP16 fused to codons 2 to 239 of ECFP. Two additional recombinant HSV-1 BACs were constructed that contained either codons 1 to 489 of VP16 fused to codons 2 to 239 of EYFP (rHSVBAC48Y) or codons 1 to 225 of mRFP fused to codons 5 to 108 of VP26 and codons 1 to 489 of VP16 fused to codons 2 to 239 of ECFP (rHSVBAC35R48C). A second triple-fluorescent recombinant HSV BAC, rHSVBAC35R22Y48C/2, was created by introducing codons 2 to 239 of EYFP between codons 6 and 9 of gH in rHSVBAC35R48C. The two triple-fluorescent HSV BACs, rHSVBAC35R22Y48C and rHSVBAC35R22Y48C/2, encode the same fusion proteins, but these were inserted in different orders. To confirm that the recombinations had occurred as expected, BAC DNA was analyzed by restriction endonuclease digestion and Southern blot analysis (not shown). Finally, to rescue recombinant viruses and release the virus genome from the BAC backbone, recombinant HSV-1 BAC DNA was cotransfected with a Cre recombinase-expressing plasmid, p116, into mammalian cells (Fig. 1). Progeny virus was harvested after 2 to 3 days and plaque purified twice, and the excision of the BAC sequences was confirmed by PCR analysis (not shown). The recombinant viruses constructed and used in this study were designated as follows: rHSV-R (mRFP-VP26), rHSV-48Y (VP16-EYFP), rHSV-RY (mRFP-VP26 and EYFP-gH), rHSV-RC (mRFP-VP26 and VP16-ECFP), rHSV-RYC (mRFP-VP26, EYFP-gH, and VP16-ECFP), and rHSV-RYC/2 (mRFP-VP26, EYFP-gH, and VP16-ECFP). The two triple-fluorescent recombinants, rHSV-RYC and rHSV-RYC/2, produced comparable titers and showed identical patterns of fluorescence in infected cells (not shown).
Synthesis of autofluorescent fusion proteins in infected cells.
To verify that the recombinant viruses expressed the fluorescent fusion proteins, Vero cells were mock infected or infected with either wt HSV-1, rHSV-RY, or rHSV-RYC at an MOI of 1 PFU per cell. When the CPE was approximately 90%, cells were harvested and analyzed. The 90% CPE was reached approximately 24 h p.i. for the wt virus and 48 h p.i. for the recombinant virus, consistent with delayed replication kinetics of the recombinant viruses (see Fig. 3, below). Western blotting with a rabbit anti-VP26 PAb (kindly provided by A. Helenius, ETH, Zurich, Switzerland) detected the 12-kDa VP26 protein from wt HSV-1-infected cell lysates. In the lysates of rHSV-RY- or rHSV-RYC-infected cells, the 12-kDa VP26 was not detected but was replaced with a band of approximately 50 kDa, which corresponds to the size expected for the mRFP-VP26 fusion protein (Fig. 2A). The band intensity of the wt 12-kDa VP26 protein was consistently weaker than that of the 50-kDa mRFP-VP26 fusion protein. The reason for this might be that the relatively small VP26 protein adsorbed less well to the nitrocellulose membrane than the larger fusion protein.
FIG. 3.
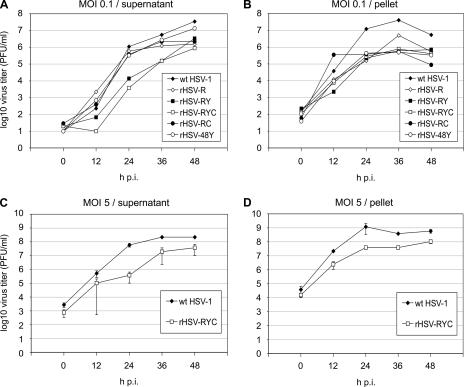
Growth kinetics of recombinant HSV-1 and wt HSV-1. Vero cells were infected with MOIs of 0.1 (A and B) or 5 (C and D) PFU of either a recombinant HSV-1 (rHSV-R, rHSV-RY rHSV-RYC, rHSV-RC, or rHSV-48Y) or wt HSV-1, and progeny virus was harvested from the cell culture medium (A and C) or from the cells (B and D) at 0, 12, 24, 36, and 48 h p.i. Titers are expressed as PFU per ml. The titers represent means from three experiments. Error bars represent standard deviations.
FIG. 2.
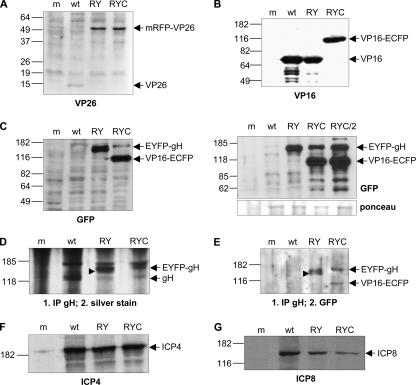
Expression of fluorescent fusion proteins in infected cells. Vero cells were mock infected (m) or infected with either wt HSV-1 (wt), rHSV-RY (RY), rHSV-RYC (RYC), or rHSV-RYC/2 (RYC/2) at an MOI of 1 PFU and harvested when the CPE was approximately 90%. Cell lysates were analyzed by SDS-PAGE followed by Western blotting with antibodies against VP26 (A), VP16 (B), GFP (C), ICP4 (F), and ICP8 (G). For detection of gH and EYFP-gH, cell lysates were immunoprecipitated with a gH-specific antibody followed by SDS-PAGE and silver staining (D) or Western blotting with a GFP-specific antibody (E). Arrows indicate the mRFP-VP26, VP16-ECFP, and EYFP-gH fusion proteins, as well as the wt VP26, VP16, gH, ICP4, and ICP8 proteins. The arrowheads in panels D and E point to a band that likely represents the EYFP-gH precursor. Sizes of molecular weight standards are indicated.
Western blotting with the anti-VP16 MAb LP1 (41) detected the VP16 protein in lysates of cells infected with either wt HSV-1 or rHSV-RY. In rHSV-RYC-infected cell lysates, however, the native VP16 was replaced by a band of approximately 100 kDa, which was also detected by the GFP-specific MAb JL-8BD (Fig. 2B and C). As the GFP-specific antibody reacts not only with GFP and ECFP but also with EYFP, a band of approximately 140 kDa was visible in both rHSV-RY- and rHSV-RYC-infected cell lysates which represented the EYFP-gH fusion protein (Fig. 2C, left panel). Of note is that the levels of the EYFP-gH fusion protein appeared to be higher in rHSV-RY- than in rHSV-RYC-infected cells. We therefore compared the EYFP-gH expression level of rHSV-RYC to that of the independently constructed triple-labeled virus, rHSV-RYC/2, as well as to rHSV-RY. As shown in Fig. 2C (right panel), the ratios between EYFP-gH and VP16-ECFP were comparable for both triple-labeled viruses. In addition, in this experiment, the total amounts of EYFP-gH appeared similar for the double- and triple-labeled viruses. The reason for the fairly low levels of EYFP-gH detected in some experiments therefore appears to be due to variations in the efficiency of infection between different experiments, rather than due to an inherent inability of the triple-colored viruses to express normal amounts of EYFP-gH.
To compare the expression levels of gH from wt HSV-1 and those of EYFP-gH from the recombinant viruses, cell lysates were immunoprecipitated with the gH-specific MAb LP11 (6), which binds to a conformational epitope of gH, followed by SDS-PAGE and silver staining of the gel (Fig. 2D). A band corresponding to the molecular mass of gH (110 kDa) was visible for wt HSV-1 but was replaced with bands corresponding to the molecular mass of EYFP-gH (140 kDa) in the samples of the recombinant viruses. The levels of gH appeared to be higher for wt HSV-1 and for rHSV-RY than for rHSV-RYC. To confirm expression of the EYFP-gH fusion protein, lysates were first immunoprecipitated with the gH-specific MAb LP11 and then analyzed by SDS-PAGE and Western blotting. The GFP-specific antibody detected the approximately 140-kDa EYFP-gH fusion protein in both rHSV-RY- and rHSV-RYC-infected cell lysates (Fig. 2E). Of note is that the levels of EYFP-gH were similar in this setting. Interestingly, in rHSV-RY-infected cell lysates but not in rHSV-RYC-infected cell lysates, a slightly smaller band was also visible, which likely represents the EYFP-gH precursor (30) (Fig. 2E). The accumulation of the EYFP-gH precursor might be a consequence of the higher total levels of EYFP-gH of rHSV-RY resulting in incomplete processing of the precursor, while the lower EYFP-gH levels expressed by rHSV-RYC may be completely processed into the mature form. The GFP-specific antibody also detected the VP16-ECFP fusion protein in rHSV-RYC-infected and LP11-immunoprecipitated cell lysates (Fig. 2E), which is consistent with an earlier finding that gH can coimmunoprecipitate with VP16 (31). An alternative explanation, however, could be that VP16-ECFP and EYFP-gH coimmunoprecipitate because GFP-derived proteins can undergo weak dimerization (57, 65, 66).
We also compared the accumulation of the HSV-1 IE protein ICP4 and the early protein ICP8 among wt HSV-1 and the recombinants. While ICP4 (Fig. 2F) accumulated to comparable levels in cells infected with either wt HSV-1, rHSV-RY, or rHSV-RYC, the levels of ICP8 (Fig. 2G) were reduced in cells infected with the recombinant viruses. In conclusion, these results demonstrated that the fusion proteins mRFP-VP26, VP16-ECFP, and EYFP-gH were expressed in infected cells with ongoing compromised levels of gene expression.
Growth kinetics of autofluorescent recombinant HSV-1.
We assessed the growth properties of recombinants that expressed the mRFP-VP26 fusion alone or coexpressed two or three of the fusion proteins. In the first set of experiments, Vero cells were infected at a low MOI (0.1 PFU) with either wt HSV-1, rHSV-R, rHSV-48Y, rHSV-RY, rHSV-RC, or rHSV-RYC, and cultures were harvested at 12, 24, 36, and 48 h p.i. as described in Materials and Methods. Virus yields in both cell culture supernatant (Fig. 3A) and cells (Fig. 3B) were titrated separately (PFU/ml). The growth properties of the recombinant viruses compared to wt HSV-1 can be summarized as follows: (i) the kinetics of virus release was delayed for recombinants rHSV-RY and rHSV-RYC, while all the other recombinants showed kinetics similar to that of wt HSV-1 (Fig. 3A); (ii) in the supernatant, the final titers of the recombinant viruses were reduced 2.6-fold (rHSV48Y) to 40-fold (rHSV-RYC) at 48 h p.i; (iii) in the cell pellet, the final titers of the recombinant viruses were reduced 7.7-fold (rHSV-RY) to 62-fold (rHSV-RC) at 48 h p.i. While it appeared that the fusion of EYFP to gH contributed to the delayed kinetics of virion release of rHSV-RY and rHSV-RYC (Fig. 3A), the contribution of the individual fusions to the reduced final titers was less obvious; specifically, the fusion of EYFP to VP16 alone (rHSV-48Y) had the weakest effect on the final titers, and all other recombinants, whether single, double, or triple labeled, showed similar final titers in the supernatants (Fig. 3A).
In a second set of experiments, we compared the growth properties of rHSV-RYC and wt HSV-1 at a high MOI (5 PFU) (Fig. 3C and D). Under these conditions, the growth deficit of the triple-labeled recombinant was less pronounced. Although the delayed kinetics of virus production was still observed, the final titers at 48 h p.i. differed only by a factor of 5.8 in the supernatant and a factor of 5.6 in the pellet. These findings suggest that the growth deficit of rHSV-RYC can be partially overcome by infection at high MOI, which obviates the need for efficient cell-to-cell spread.
To further characterize the growth properties of the recombinant viruses, we determined the particle/PFU ratios of two recombinants, rHSV-RY and rHSV-RYC, as well as wt HSV-1. We observed particle/PFU ratios of 68 ± 49 (mean ± standard deviation) for rHSV-RY, 26 ± 19 for rHSV-RYC, and 1.3 ± 0.9 for wt HSV-1, indicating that the recombinant viruses were on average 20- to 50-fold less infectious than the wt virus, a finding which at least partially explains the reduced titers in the recombinant virus stocks.
In summary, the recombinant viruses showed reduced titers both in the supernatant as well as in the pellet, with more pronounced reductions at low than at high MOI. While the fusion of EYFP to gH seemed to contribute to a delay in the kinetics of virion release, there was no clear correlation between the number of tagged virion proteins and the reduction in titer. Finally, the recombinant viruses did not seem to have a specific deficit at the level of virion egress, since the differences between the wt virus and the recombinants were comparable in the supernatant and the pellet. The data rather suggest that the kinetics of recombinant virus replication was delayed.
Incorporation of autofluorescent fusion proteins into the virion.
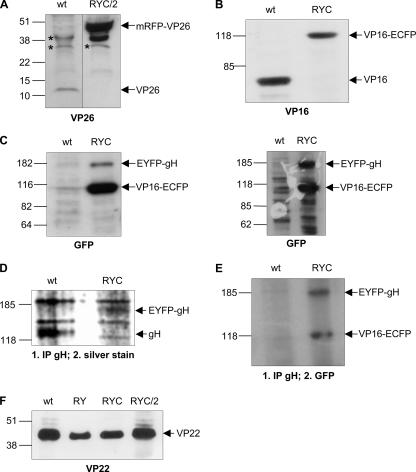
To confirm that all three fusion proteins were incorporated into the virion, wt HSV-1 virions and rHSV-RYC virions were prepared by sucrose gradient centrifugation and analyzed for the presence of the recombinant proteins. Western blotting with the VP26-specific PAb revealed the 12-kDa VP26 in wt HSV-1 virions and the 50-kDa mRFP-VP26 fusion protein in rHSV-RYC virons (Fig. 4A). Similar to the observations made in Fig. 2A, the band intensity of the wt VP26 protein was weaker than that of the mRFP-VP26 fusion protein. As previously mentioned, this difference might be due to different efficiencies of adsorption to the nitrocellulose membrane, since it is unlikely that the fusion protein is incorporated more efficiently into the virions than the wt protein. Similar observations were also made in a previous report (45). In order to obtain a visible band for wt VP26, a doubled amount of wt virions was loaded for the blot shown in Fig. 4A. Western blotting with the VP16-specific MAb revealed the presence of VP16 in the wt virions and VP16-ECFP in the rHSV-RYC virions, the amounts of which appeared comparable (Fig. 4B). The VP16-ECFP fusion protein, but not wt VP16, could also be detected with the GFP-specific MAb (Fig. 4C). As expected, the anti-GFP MAb also detected the EYFP-gH fusion protein (Fig. 4C). As noted in Fig. 2, the ratio of EYFP-gH to VP16-ECFP was quite variable between individual experiments (Fig. 2C, compare left and right panels). To detect gH from wt HSV-1 and EYFP-gH from rHSV-RYC, virion proteins were immunoprecipitated with the gH-specific MAb LP11 followed by SDS-PAGE and silver staining. The results shown in Fig. 4D demonstrate the presence of gH in wt virions and EYFP-gH in rHSV-RYC virions. Again, it appeared that the level of gH was higher in the wt virus than in with the recombinant. As expected, immunoprecipitation of virion lysates with LP11 followed by Western blotting with the anti-GFP MAb detected the EYFP-gH fusion protein but not the wt gH (Fig. 4E). As previously noted (Fig. 2E), VP16-ECFP coimmunoprecipitated with EYFP-gH (Fig. 4E). Staining for the VP22 tegument protein confirmed that comparable amounts of virions were loaded on the gels (Fig. 4F). Taken together, these results demonstrate that all three fusion proteins, mRFP-VP26, VP16-ECFP, and EYFP-gH, are indeed incorporated into the virus particle, although the amounts of incorporated EYFP-gH were lower than those of wt gH.
FIG. 4.
Incorporation of fluorescent proteins into virions. Purified wt HSV-1 (wt), rHSV-RY (RY), rHSV-RYC (RYC), and rHSV-RYC/2 (RYC/2) were analyzed by SDS-PAGE followed by Western blot analysis with antibodies specific for VP26 (A), VP16 (B), GFP (C), and VP22 (F). For detection of gH and EYFP-gH, virion proteins were immunoprecipitated with a gH-specific antibody and analyzed by SDS-PAGE followed by silver staining (D) or Western blotting with a GFP-specific antibody (E). The corresponding fusion proteins mRFP-VP26, VP16-ECFP, and EYFP-gH, as well as the wt VP26, VP16, gH, and VP22 proteins are indicated. Unspecific bands are marked by asterisks. Sizes of molecular weight markers are shown.
Dynamics of compartmentalization of mRFP-VP26, VP16-ECFP, and EYFP-gH in infected cells.
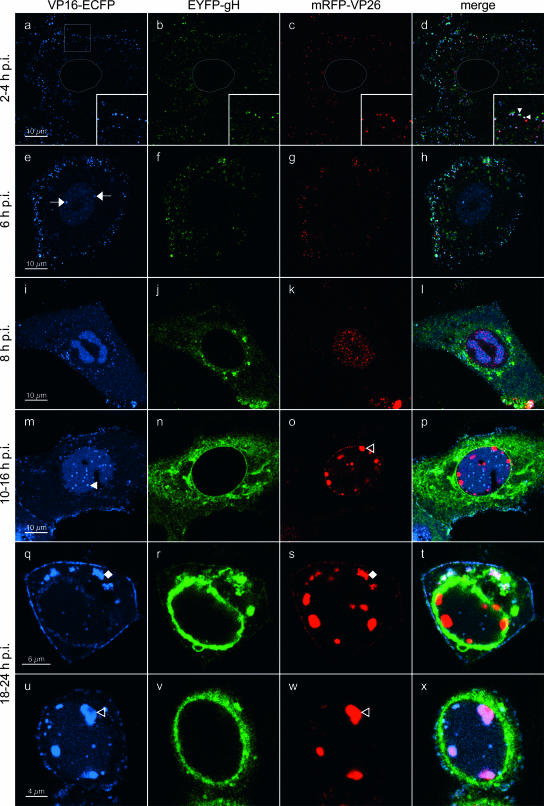
Next, we monitored the localization of mRFP-VP26, VP16-ECFP, and EYFP-gH in infected cells. For this, Vero cells were plated onto chambered coverglasses and infected with rHSV-RYC, and live cells were examined by CLSM at various times after infection. In the first hours postinfection, small fluorescent foci were detected on the surface of the cells and within the cytoplasm (Fig. 5a to d). Some foci show colocalizations of ECFP, EYFP, and mRFP signals, suggesting that they represent intact virions from the virus inoculum (Fig. 5a to d, insets). De novo protein synthesis was first observed at 6 h p.i. for VP16-ECFP, which was found diffusely in the nucleus with accumulation in very early viral replication compartments (RCs) (Fig. 5e and h). At 8 h p.i., VP16-ECFP was present in mature nuclear RCs as well as in the cytoplasm, where it was found in a diffuse pattern with accumulation in some small perinuclear foci (Fig. 5i and l). At this time, expression of EYFP-gH and mRFP-VP26 also became apparent. In particular, EYFP-gH was found diffusely in the cytoplasm with some accumulation in a vesicular pattern around the nucleus (Fig. 5j and l), while mRFP-VP26 was found in small microfoci which were distributed throughout the nucleus with accumulation in viral RCs (Fig. 5k and l). These small microfoci displayed a high mobility within the nucleus (not shown), reflecting active movement of nuclear capsids (21). The localization of the VP26 and VP16 fusion proteins to RCs was confirmed by staining rHSV-R- and rHSV-48Y-infected cells for ICP8, a marker for RCs (Fig. 6C and D). Between 10 and 16 h p.i., VP16-ECFP was predominantly found in the viral RCs as well as in small foci in their periphery. In addition, the cytoplasmic VP16-ECFP accumulation became more intense, with a pronounced recruitment of VP16-ECFP to the plasma membrane, where it was found in a dot-like pattern (Fig. 5m and p). At this stage, EYFP-gH was still prominent in the cytoplasm but now strongly accumulated in the nuclear membrane (Fig. 5n and p). Microfoci of mRFP-VP26 were still observed in the nucleus, but in addition, mRFP-VP26 was also observed in much larger foci which preferentially formed in the periphery of the RCs, but it did not colocalize with the small VP16-ECFP foci (Fig. 5o and p). These large foci have previously been suggested to correspond to sites of capsid assembly, so-called assemblons (13). Between 18 and 24 h p.i., two additional patterns became apparent. First, the majority of the VP16-ECFP was consistently recruited out of the nucleus into the cytoplasm. Specifically, it was found accumulating in an asymmetric, perinuclear pattern reminiscent of the Golgi complex, where it often colocalized with mRFP-VP26 and associated with EYFP-gH (Fig. 5q to t). Second, in a subset of cells, VP16-ECFP was recruited into the large mRFP-VP26 foci in the periphery of the nucleus, while EYFP-gH remained in a pronounced perinuclear pattern (Fig. 5u to x). Finally, at these late time points, the dot-like accumulations of VP16-ECFP at the plasma membrane consistently colocalized with EYFP-gH and mRFP-VP26 (Fig. 5q to t), suggesting that they reflect the accumulation of progeny virus at the plasma membrane. This possibility was further investigated by high-magnification CLSM of the plasma membrane, which, as previously described, often formed pronounced plasma membrane protrusions (35) (Fig. 6A and B). On such plasma membrane protrusions, we observed a very strong association of VP16-ECFP and mRFP-VP26 (Fig. 6A, panel d) as well as a somewhat looser association of EYFP-gH with mRFP-VP26 and VP16-ECFP (Fig. 6A, panels e and f). The looser association of EYFP-gH with mRFP-VP26 and VP16-ECFP is probably due to the fact that we observed living cells and that while the ECFP and the mRFP channels were recorded simultaneously, the EYFP channel was recorded separately, thus explaining the observed differences in the degree of colocalization. Indeed, the association between the three signals was more pronounced in fixed cells (Fig. 6B). These observations, together with the fact that the number of dots increased in the course of infection, support the hypothesis that the dot-like accumulation of virion proteins of three different compartments at the plasma membrane might correspond to accumulating virion progeny. Nevertheless, it cannot be excluded that at least a subset of these foci also represent virions from the inoculum which did not successfully enter the cell and thus remained bound to the plasma membrane.
FIG. 5.
High-resolution CLSM of living rHSV-RYC-infected cells. Vero cells were infected with an MOI of 18 PFU, and live cells were observed by CLSM with settings specific for ECFP (VP16-ECFP fusion protein), EYFP (EYFP-gH fusion protein), and mRFP (mRFP-VP26 fusion protein). The thin gray lines in panels a to d mark the contours of the nucleus. The insets in panels a to d show a magnification of the sector denoted by the white square. The filled arrowheads within the insets point to colocalizations of ECFP, EYFP, and mRFP signals. Arrows, early RCs; filled triangle, VP16-ECFP foci in periphery of RCs; open triangles, large mRFP-VP26 and VP16-ECFP foci in periphery of nuclei; filled diamonds, asymmetric, perinuclear accumulation of VP16-ECFP and mRFP-VP26. Images represent single z stacks of the cells.
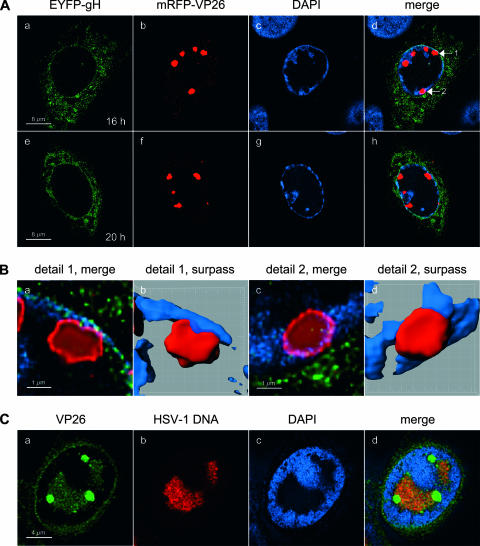
In order to obtain a more detailed picture of the subnuclear distribution of the large mRFP-VP26 foci, we infected cells with rHSV-RY, fixed them at 16 or 20 h p.i., and stained the nuclei with DAPI. CLSM analysis revealed that, at such late time points, the large mRFP-VP26 foci were often located at the very periphery of the nuclei (Fig. 7A) and that the density of mRFP-VP26 within these large foci was not homogenous (Fig. 7B). Rather, mRFP-VP26 accumulated strongly at the rim and displayed only a weak accumulation in the interior, suggesting a hollow structure of these foci (Fig. 7B). In addition, we observed that while some of the VP26 structures were still separated from the nuclear membrane by a thin layer of condensed chromatin (Fig. 7B, panels a and b), others displaced the chromatin to such an extent that they virtually gained access to the nuclear membrane, which was visible by virtue of EYFP-gH fluorescence (Fig. 7B, panels c and d). In order to test if these large VP26 foci corresponded to an accumulation of DNA-filled capsids, we codetected VP26 and HSV-1 DNA in wt HSV-1-infected cells (Fig. 7C). While VP26 and HSV-1 DNA colocalized within the RCs, the large VP26 foci did not contain HSV-1 DNA. Assuming that DNA within virions is accessible for FISH, these data suggest that the large VP26 foci do not correspond to accumulations of DNA-filled capsids.
To ascertain that the distribution of the fluorescent virion proteins corresponded to that of the respective wt proteins, we performed infections with wt HSV-1 and detected VP16, gH, and VP26 by immunofluorescence. CLSM analysis of stained cells revealed that the patterns for all three wt virion proteins were very similar to those observed with the recombinant viruses (Fig. 8A to C). However, it has to be noted that the accumulation of VP16 in large foci at the nuclear periphery and their colocalization with VP26 was not observed in wt HSV-1-infected cells stained with VP26- and VP16-specific antibodies (Fig. 8D). This analysis also demonstrated that the structure of the large VP26 foci was identical to that observed with the mRFP-VP26 fusion protein (Fig. 8C, panel h). Interestingly, the proportion of cells displaying such large VP26 foci appeared to be somewhat smaller in wt HSV-1-infected cells than in cells infected with the recombinant viruses.
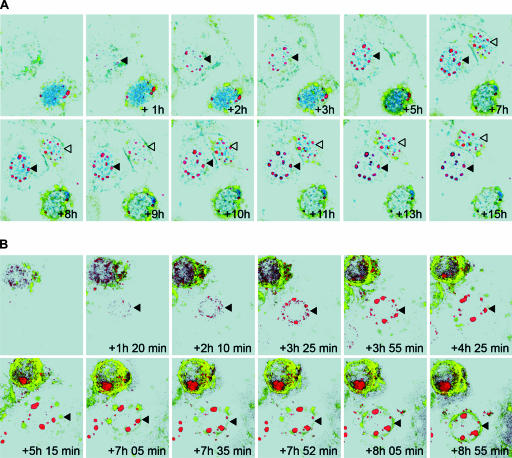
Time-lapse CLSM of cells infected with either rHSV-RYC or rHSV-RY allowed us to obtain a dynamic view of the interaction between VP16, gH, and VP26 in the infected cells (Fig. 9; see also movies S1 to S3 in the supplemental material). Specifically, the image series demonstrated a very dynamic compartmentalization of VP16-ECFP in the course of infection. While VP16-ECFP initially steadily accumulated in the nuclear RCs and later in the small foci in the periphery of the RCs, there was a very rapid and pronounced recruitment of VP16-ECFP out of the nucleus into the cytoplasm and, in some cells, into the large mRFP-VP26 foci at the periphery of the nucleus in a late stage of infection. This redistribution was fairly rapid and generally occurred within 1 to 2 h (Fig. 9A). In addition, the time-lapse series revealed the progressive accumulation of EYFP-gH at the nuclear membrane and in the perinuclear cytoplasmic pattern (Fig. 9A and B). Finally, the analysis demonstrated that the initially small foci of mRFP-VP26 expanded and coalesced to form much larger foci, which were pushed toward the periphery of the nucleus as infection progressed (Fig. 9A and B).
FIG. 9.
Time-lapse CLSM of rHSV-RYC- and rHSV-RY-infected cells. Vero cells were infected with rHSV-RYC (A) or rHSV-RY (B) at MOIs of 2 and 0.3, respectively. Cells that just started to accumulate fluorescent proteins were monitored by CLSM with settings specific for ECFP (VP16-ECFP), EYFP (EYFP-gH), and mRFP (mRFP-VP26). Selected frames at the indicated intervals are shown. Images were processed with Imaris software in the surpass view mode. Arrowheads denote the cells described in the text.
Ultrastructural analysis of rHSV-RYC-infected cells.
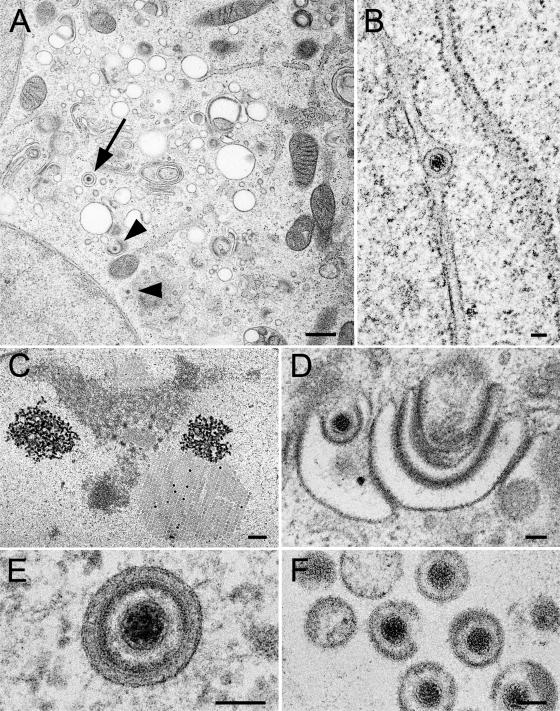
Electron microscopy revealed clear indications for normal maturation of the triple-fluorescent recombinant HSV-1, rHSV-RYC, with identical phenotypes to wt HSV-1 (Fig. 10). These phenotypes include formation of capsids with occasional crystalline-like accumulation within the nucleus (Fig. 10C), budding of capsids at the inner nuclear membrane (Fig. 10B), budding of capsids at Golgi membranes (Fig. 10D), virions within vacuoles (Fig. 10E), and virions in the extracellular space (Fig. 10F).
FIG. 10.
Electron micrographs of Vero cells infected with rHSV-RYC after prefixation at 24 h p.i. followed by freezing and freeze-substitution. (A) Low-power micrograph showing Golgi membranes, virions within a vacuole (arrow), and capsids (arrowheads) within the cytoplasm and budding at Golgi membranes, respectively. (B) Virion within the perinuclear space of the nuclear envelope. (C) Accumulation of capsids in a crystalline manner within the nucleus. (D) Budding capsid at Golgi membranes. (E) Virion within a concentric vacuole. (F) Virions in the extracellular space. Bars, 500 nm (A and C) and 100 nm (B, D, and E).
DISCUSSION
Autofluorescent proteins have been used extensively to study many different biological processes, including the replication cycle of viruses (reviewed in reference 5). Recombinant herpesviruses encoding individual virus proteins fused with autofluorescent proteins have previously been constructed to study several different aspects of the herpesvirus life cycle, including trafficking, assembly, and maturation (11, 13, 15, 17, 20, 35, 39). Here, we report for the first time the construction and time-lapse analysis of a recombinant HSV-1 that simultaneously encodes virus proteins from three different virion compartments, capsid (VP26), tegument (VP16), and envelope (gH), fused with different autofluorescent proteins (mRFP, ECFP, and EYFP, respectively).
Crucial to the construction of the triple-fluorescent recombinant HSV-1 was the availability of a BAC-cloned HSV-1 genome (59), which facilitated the easy manipulation of the virus genome in bacteria. The combination of the HSV-1 BAC with the possibility for both positive and negative selection in bacteria provided by the galK system (62) allowed the serial introduction of the three fusion genes into the virus genome. The reconstitution of recombinant virus progeny and the elimination of the BAC backbone were accomplished following cotransfection of the recombinant HSV-1 BAC DNA with a Cre recombinase-expressing plasmid in mammalian cells (Fig. 1).
Western analysis of infected cells demonstrated that all three fusion proteins were synthesized (Fig. 2). Interestingly, the triple-fluorescent recombinant HSV-1, rHSV-RYC, was replication competent, although with delayed kinetics, and incorporated all three fusion proteins into the different virion compartments. It appeared that the fusion of EYFP to gH was responsible for a delay in the kinetics of virus release (Fig. 3). This is consistent with a previous report describing the fusion of EYFP to the N terminus of gH (39). Although the authors of that study contended that the recombinant virus replicated to titers comparable to those of wt HSV-1, it has to be noted that the wt virus used in their study (strain 17 syn+) reached only a titer of 5.25 × 106 PFU/ml, which is rather low for a wt virus. The fusion of autofluorescent proteins to VP16 and VP26 also seemed to affect virus titers, although there were no indications that fusion to the essential VP16 would have a greater impact on viral replication fitness than fusion to the nonessential VP26. In addition, the analysis did not support the notion that the effects of the individual fusions were additive. Rather it appeared that the addition of at least one autofluorescent fusion protein to a virus protein resulted in a somewhat delayed kinetics of virus production (gH) or reduced final titers (VP16 or VP26). Electron microscopy analysis revealed that the triple-fluorescent recombinant HSV-1 was comparable to wt HSV-1 also at the ultrastructural level (Fig. 10). Moreover, CLSM of infected cells demonstrated that the distribution and compartmentalization of the three autofluorescent fusion proteins encoded by rHSV-RYC were comparable to those observed with either wt HSV-1 or recombinants of HSV-1 that encode autofluorescent proteins fused with VP26, gH, or VP16 individually (Fig. 5 to 8) (13, 35, 39). Finally, the strong dot-like association of autofluorescent fusion proteins of three different virion compartments, VP16-ECFP, mRFP-VP26, and EYFP-gH, at plasma membrane protrusions (Fig. 6A and B) supports the hypothesis that these dots indeed are mature virions and that progeny virions incorporate sufficient amounts of the fluorescent fusion proteins to allow their visualization, although this has to be confirmed on the ultrastructural level. Nevertheless, it would be less convincing to draw such a conclusion with a recombinant HSV-1 that encodes only one or two autofluorescent virion proteins.
The major advantage of our approach is the possibility for simultaneously visualizing the distributions and interactions between different virus proteins and virion compartments on the single cell level. Moreover, our strategy allows study of the dynamics of these events in live cells. For example, mRFP-VP26 and VP16-ECFP colocalized within RCs in the nucleus but, in addition, they both also formed foci in the periphery of the RCs that did not colocalize (Fig. 5). In a previous report it was hypothesized that the VP16 foci in the periphery of the RCs may correspond to sites where capsids acquire VP16 (35). Our experiments, however, suggest that this hypothesis is unlikely. In the course of the infection, the mRFP-VP26 foci expanded and coalesced to form larger foci that relocated to the periphery of the nucleus. In the majority of the cells, VP16-ECFP was redistributed from the nucleus to the cytoplasm late in infection, in a process that occurred very rapidly (generally within 1 to 2 h). Of note is that in a subset of cells, VP16-ECFP was, in addition, recruited into the large mRFP-VP26 foci at the periphery of the nucleus (Fig. 5 and 9; see also movies S1 to S3 in the supplemental material). This phenomenon was, however, not observed in wt HSV-1-infected cells stained with VP16- and VP26-specific antibodies, in which VP26, but not VP16, was consistently observed in the large foci at the nuclear periphery (Fig. 8). In contrast, the colocalization of VP26 and VP16 in the cytoplasm late in infection was readily observed in both rHSV-RYC- and wt HSV-1-infected cells (Fig. 5 and 8). There are two possible explanations for this discrepancy: first, the fusion of ECFP to VP16 may have altered some of its biological properties, leading to its recruitment into the large mRFP-VP26 foci at the nuclear periphery. Second, it is conceivable that VP16 and VP26 similarly colocalized in wt HSV-1-infected cells but that the VP16-specific antibody employed did not detect VP16 when present in the large VP26 foci at the periphery of the nucleus, possibly because of an altered conformation of VP16 in these structures. However, the very late appearance of this colocalization (18 to 24 h p.i.) suggests that it likely does not represent a crucial step in the assembly and egress of progeny virus, since infectious progeny are produced already at earlier time points (i.e., 12 h p.i.) (Fig. 3). The accumulation of VP26 in large nuclear foci was previously documented in cells infected with wt HSV-1 and stained with VP26-specific antibodies, as well as by expression of VP26 fused to GFP. The formation of nuclear VP26 foci was demonstrated to depend on the presence of VP5 and VP22a and to occur in an ATP-dependent manner (8, 11, 13, 52). These VP26 foci were hypothesized to correspond to the sites of capsid assembly, similar to the assemblons described by Ward and coworkers, which contained the capsid proteins ICP35, VP5, and VP19c and formed very late in infection at the periphery of the nuclear RCs (13, 61). Interestingly, Ward et al. (61) detected a partial colocalization between assemblons and VP16 by staining with VP19c- and VP16-specific antibodies. However, this partial colocalization clearly differs from the almost perfect colocalization of VP16-ECFP and mRFP-VP26 in large foci at the nuclear periphery observed in our study (Fig. 5). The ultrastructural correlate of this colocalization remains to be determined. It is possible that the VP26 foci at the nuclear periphery may represent the accumulation of dead-end products, as previously suggested for assemblons (10, 36), to which VP16 is recruited late in infection. This hypothesis is further supported by the finding that these structures do not contain HSV-1 DNA, indicating that they do not correspond to accumulations of DNA-filled capsids (Fig. 7).
In summary, this study demonstrates the feasibility of the construction of a recombinant HSV-1 simultaneously expressing autofluorescent proteins fused to VP16, VP26, and gH. This study sheds light on the spatial and temporal organization of HSV-1 infection at the single-cell level. Specifically, the simultaneous fluorescence labeling of several virion components allowed assessment of the interactions of the different viral proteins in the course of the infection. This approach may be used to address several open questions in HSV-1 biology, for example, the spatial organization of capsid assembly and maturation by fluorescent labeling of several components of the pro-capsid and those of mature capsids. In addition, fluorescently labeled virus proteins may be combined with systems for the live visualization of viral DNA, as previously described for several viruses, such as HSV-1, Epstein-Barr virus, and adeno-associated virus (1, 23, 28, 29, 58), to specifically assess the dynamics of the association of viral proteins with viral DNA. Finally, the simultaneous fluorescent labeling of capsid, tegument, and envelope components may prove useful for the study of virus trafficking, for example, to assess and compare the composition of virions transported in an anterograde versus retrograde direction within axons (2-4, 20, 40).
Supplementary Material
Acknowledgments
We kindly thank U. F. Greber, H. Browne, A. Minson, A. Helenius, P. Desai, Y. Kawaguchi, S. Warming, R. D. Everett, G. Elliott, and K. Tobler for providing reagents and I. Heid for technical assistance.
This work was supported by the Swiss National Science Foundation (grants 3100A0-100195 and 3100AO-112462 to C.F.), the Novartis Foundation for Biomedical Research (grant 06C77 to C.F.), and the National Institutes of Health (grant CA69246 to X.O.B. and C.F.).
Footnotes
Published ahead of print on 12 March 2008.
REFERENCES
- 1.Amon, W., R. E. White, and P. J. Farrell. 2006. Epstein-Barr virus origin of lytic replication mediates association of replicating episomes with promyelocytic leukaemia protein nuclear bodies and replication compartments. J. Gen. Virol. 871133-1137. [DOI] [PubMed] [Google Scholar]
- 2.Antinone, S. E., G. T. Shubeita, K. E. Coller, J. I. Lee, S. Haverlock-Moyns, S. P. Gross, and G. A. Smith. 2006. The herpesvirus capsid surface protein, VP26, and the majority of the tegument proteins are dispensable for capsid transport toward the nucleus. J. Virol. 805494-5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antinone, S. E., and G. A. Smith. 2006. Two modes of herpesvirus trafficking in neurons: membrane acquisition directs motion. J. Virol. 8011235-11240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bearer, E. L., X. O. Breakefield, D. Schuback, T. S. Reese, and J. H. LaVail. 2000. Retrograde axonal transport of herpes simplex virus: evidence for a single mechanism and a role for tegument. Proc. Natl. Acad. Sci. USA 978146-8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandenburg, B., and X. Zhuang. 2007. Virus trafficking—learning from single-virus tracking. Nat. Rev. Microbiol. 5197-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buckmaster, E. A., U. Gompels, and A. Minson. 1984. Characterisation and physical mapping of an HSV-1 glycoprotein of approximately 115 × 103 molecular weight. Virology 139408-413. [DOI] [PubMed] [Google Scholar]
- 7.Campbell, M. E., J. W. Palfreyman, and C. M. Preston. 1984. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J. Mol. Biol. 1801-19. [DOI] [PubMed] [Google Scholar]
- 8.Chi, J. H., and D. W. Wilson. 2000. ATP-dependent localization of the herpes simplex virus capsid protein VP26 to sites of procapsid maturation. J. Virol. 741468-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davison, M. D., F. J. Rixon, and A. J. Davison. 1992. Identification of genes encoding two capsid proteins (VP24 and VP26) of herpes simplex virus type 1. J. Gen. Virol. 732709-2713. [DOI] [PubMed] [Google Scholar]
- 10.de Bruyn Kops, A., S. L. Uprichard, M. Chen, and D. M. Knipe. 1998. Comparison of the intranuclear distributions of herpes simplex virus proteins involved in various viral functions. Virology 252162-178. [DOI] [PubMed] [Google Scholar]
- 11.Desai, P., J. C. Akpa, and S. Person. 2003. Residues of VP26 of herpes simplex virus type 1 that are required for its interaction with capsids. J. Virol. 77391-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desai, P., N. A. DeLuca, and S. Person. 1998. Herpes simplex virus type 1 VP26 is not essential for replication in cell culture but influences production of infectious virus in the nervous system of infected mice. Virology 247115-124. [DOI] [PubMed] [Google Scholar]
- 13.Desai, P., and S. Person. 1998. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J. Virol. 727563-7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desai, P. J., P. A. Schaffer, and A. C. Minson. 1988. Excretion of non-infectious virus particles lacking glycoprotein H by a temperature-sensitive mutant of herpes simplex virus type 1: evidence that gH is essential for virion infectivity. J. Gen. Virol. 691147-1156. [DOI] [PubMed] [Google Scholar]
- 15.Dohner, K., K. Radtke, S. Schmidt, and B. Sodeik. 2006. Eclipse phase of herpes simplex virus type 1 infection: efficient dynein-mediated capsid transport without the small capsid protein VP26. J. Virol. 808211-8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elliott, G., G. Mouzakitis, and P. O'Hare. 1995. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J. Virol. 697932-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elliott, G., and P. O'Hare. 1999. Live-cell analysis of a green fluorescent protein-tagged herpes simplex virus infection. J. Virol. 734110-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Everett, R. D., and J. Murray. 2005. ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J. Virol. 795078-5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farnsworth, A., T. W. Wisner, M. Webb, R. Roller, G. Cohen, R. Eisenberg, and D. C. Johnson. 2007. Herpes simplex virus glycoproteins gB and gH function in fusion between the virion envelope and the outer nuclear membrane. Proc. Natl. Acad. Sci. USA 10410187-10192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feierbach, B., M. Bisher, J. Goodhouse, and L. W. Enquist. 2007. In vitro analysis of transneuronal spread of an alphaherpesvirus infection in peripheral nervous system neurons. J. Virol. 816846-6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forest, T., S. Barnard, and J. D. Baines. 2005. Active intranuclear movement of herpesvirus capsids. Nat. Cell Biol. 7429-431. [DOI] [PubMed] [Google Scholar]
- 22.Forrester, A., H. Farrell, G. Wilkinson, J. Kaye, N. Davis-Poynter, and T. Minson. 1992. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J. Virol. 66341-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fraefel, C., A. G. Bittermann, H. Bueler, I. Heid, T. Bachi, and M. Ackermann. 2004. Spatial and temporal organization of adeno-associated virus DNA replication in live cells. J. Virol. 78389-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuchs, W., H. Granzow, B. G. Klupp, M. Kopp, and T. C. Mettenleiter. 2002. The UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions. J. Virol. 766729-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fuller, A. O., R. E. Santos, and P. G. Spear. 1989. Neutralizing antibodies specific for glycoprotein H of herpes simplex virus permit viral attachment to cells but prevent penetration. J. Virol. 633435-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibson, W., A. I. Marcy, J. C. Comolli, and J. Lee. 1990. Identification of precursor to cytomegalovirus capsid assembly protein and evidence that processing results in loss of its carboxy-terminal end. J. Virol. 641241-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gillet, L., J. S. May, S. Colaco, and P. G. Stevenson. 2007. Glycoprotein L disruption reveals two functional forms of the murine gammaherpesvirus 68 glycoprotein H. J. Virol. 81280-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glauser, D. L., O. Saydam, N. A. Balsiger, I. Heid, R. M. Linden, M. Ackermann, and C. Fraefel. 2005. Four-dimensional visualization of the simultaneous activity of alternative adeno-associated virus replication origins. J. Virol. 7912218-12230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glauser, D. L., R. Strasser, A. S. Laimbacher, O. Saydam, N. Clement, R. M. Linden, M. Ackermann, and C. Fraefel. 2007. Live covisualization of competing adeno-associated virus and herpes simplex virus type 1 DNA replication: molecular mechanisms of interaction. J. Virol. 814732-4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gompels, U. A., and A. C. Minson. 1989. Antigenic properties and cellular localization of herpes simplex virus glycoprotein H synthesized in a mammalian cell expression system. J. Virol. 634744-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gross, S. T., C. A. Harley, and D. W. Wilson. 2003. The cytoplasmic tail of herpes simplex virus glycoprotein H binds to the tegument protein VP16 in vitro and in vivo. Virology 3171-12. [DOI] [PubMed] [Google Scholar]
- 32.Haarr, L., and S. Skulstad. 1994. The herpes simplex virus type 1 particle: structure and molecular functions. APMIS 102321-346. [DOI] [PubMed] [Google Scholar]
- 33.Hutchinson, L., H. Browne, V. Wargent, N. Davis-Poynter, S. Primorac, K. Goldsmith, A. C. Minson, and D. C. Johnson. 1992. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J. Virol. 662240-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kinzler, E. R., and T. Compton. 2005. Characterization of human cytomegalovirus glycoprotein-induced cell-cell fusion. J. Virol. 797827-7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.La Boissiere, S., A. Izeta, S. Malcomber, and P. O'Hare. 2004. Compartmentalization of VP16 in cells infected with recombinant herpes simplex virus expressing VP16-green fluorescent protein fusion proteins. J. Virol. 788002-8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lamberti, C., and S. K. Weller. 1998. The herpes simplex virus type 1 cleavage/packaging protein, UL32, is involved in efficient localization of capsids to replication compartments. J. Virol. 722463-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leuzinger, H., U. Ziegler, E. M. Schraner, C. Fraefel, D. L. Glauser, I. Heid, M. Ackermann, M. Mueller, and P. Wild. 2005. Herpes simplex virus 1 envelopment follows two diverse pathways. J. Virol. 7913047-13059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu, F. Y., and B. Roizman. 1991. The herpes simplex virus 1 gene encoding a protease also contains within its coding domain the gene encoding the more abundant substrate. J. Virol. 655149-5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lorentzen, E. U., B. R. Eing, W. Hafezi, R. Manservigi, and J. E. Kuhn. 2001. Replication-competent herpes simplex virus type 1 mutant expressing an autofluorescent glycoprotein H fusion protein. Intervirology 44232-242. [DOI] [PubMed] [Google Scholar]
- 40.Luxton, G. W., S. Haverlock, K. E. Coller, S. E. Antinone, A. Pincetic, and G. A. Smith. 2005. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc. Natl. Acad. Sci. USA 1025832-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McLean, C., A. Buckmaster, D. Hancock, A. Buchan, A. Fuller, and A. Minson. 1982. Monoclonal antibodies to three non-glycosylated antigens of herpes simplex virus type 2. J. Gen. Virol. 63297-305. [DOI] [PubMed] [Google Scholar]
- 42.McNabb, D. S., and R. J. Courtney. 1992. Identification and characterization of the herpes simplex virus type 1 virion protein encoded by the UL35 open reading frame. J. Virol. 662653-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McNabb, D. S., and R. J. Courtney. 1992. Posttranslational modification and subcellular localization of the p12 capsid protein of herpes simplex virus type 1. J. Virol. 664839-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mossman, K. L., R. Sherburne, C. Lavery, J. Duncan, and J. R. Smiley. 2000. Evidence that herpes simplex virus VP16 is required for viral egress downstream of the initial envelopment event. J. Virol. 746287-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagel, C. H., K. Dohner, M. Fathollahy, T. Strive, E. M. Borst, M. Messerle, and B. Sodeik. 2008. Nuclear egress and envelopment of herpes simplex virus capsids analyzed with dual-color fluorescence HSV1(17+). J. Virol. 823109-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newcomb, W. W., R. M. Juhas, D. R. Thomsen, F. L. Homa, A. D. Burch, S. K. Weller, and J. C. Brown. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 7510923-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Newcomb, W. W., B. L. Trus, F. P. Booy, A. C. Steven, J. S. Wall, and J. C. Brown. 1993. Structure of the herpes simplex virus capsid. Molecular composition of the pentons and the triplexes. J. Mol. Biol. 232499-511. [DOI] [PubMed] [Google Scholar]
- 48.O'Hare, P., and C. R. Goding. 1988. Herpes simplex virus regulatory elements and the immunoglobulin octamer domain bind a common factor and are both targets for virion transactivation. Cell 52435-445. [DOI] [PubMed] [Google Scholar]
- 49.O'Hare, P., C. R. Goding, and A. Haigh. 1988. Direct combinatorial interaction between a herpes simplex virus regulatory protein and a cellular octamer-binding factor mediates specific induction of virus immediate-early gene expression. EMBO J. 74231-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Omerovic, J., L. Lev, and R. Longnecker. 2005. The amino terminus of Epstein-Barr virus glycoprotein gH is important for fusion with epithelial and B cells. J. Virol. 7912408-12415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Person, S., S. Laquerre, P. Desai, and J. Hempel. 1993. Herpes simplex virus type 1 capsid protein, VP21, originates within the UL26 open reading frame. J. Gen. Virol. 742269-2273. [DOI] [PubMed] [Google Scholar]
- 52.Rixon, F. J., C. Addison, A. McGregor, S. J. Macnab, P. Nicholson, V. G. Preston, and J. D. Tatman. 1996. Multiple interactions control the intracellular localization of the herpes simplex virus type 1 capsid proteins. J. Gen. Virol. 772251-2260. [DOI] [PubMed] [Google Scholar]
- 53.Roop, C., L. Hutchinson, and D. C. Johnson. 1993. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells, and its particles lack glycoprotein H. J. Virol. 672285-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saad, A., Z. H. Zhou, J. Jakana, W. Chiu, and F. J. Rixon. 1999. Roles of triplex and scaffolding proteins in herpes simplex virus type 1 capsid formation suggested by structures of recombinant particles. J. Virol. 736821-6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smibert, C. A., B. Popova, P. Xiao, J. P. Capone, and J. R. Smiley. 1994. Herpes simplex virus VP16 forms a complex with the virion host shutoff protein vhs. J. Virol. 682339-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith, I. L., M. A. Hardwicke, and R. M. Sandri-Goldin. 1992. Evidence that the herpes simplex virus immediate early protein ICP27 acts post-transcriptionally during infection to regulate gene expression. Virology 18674-86. [DOI] [PubMed] [Google Scholar]
- 57.Snapp, E. L., R. S. Hegde, M. Francolini, F. Lombardo, S. Colombo, E. Pedrazzini, N. Borgese, and J. Lippincott-Schwartz. 2003. Formation of stacked ER cisternae by low affinity protein interactions. J. Cell Biol. 163257-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sourvinos, G., and R. D. Everett. 2002. Visualization of parental HSV-1 genomes and replication compartments in association with ND10 in live infected cells. EMBO J. 214989-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tanaka, M., H. Kagawa, Y. Yamanashi, T. Sata, and Y. Kawaguchi. 2003. Construction of an excisable bacterial artificial chromosome containing a full-length infectious clone of herpes simplex virus type 1: viruses reconstituted from the clone exhibit wild-type properties in vitro and in vivo. J. Virol. 771382-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taylor, T. J., M. A. Brockman, E. E. McNamee, and D. M. Knipe. 2002. Herpes simplex virus. Front. Biosci. 7d752-d764. [DOI] [PubMed] [Google Scholar]
- 61.Ward, P. L., W. O. Ogle, and B. Roizman. 1996. Assemblons: nuclear structures defined by aggregation of immature capsids and some tegument proteins of herpes simplex virus 1. J. Virol. 704623-4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Warming, S., N. Costantino, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weinheimer, S. P., B. A. Boyd, S. K. Durham, J. L. Resnick, and D. R. O'Boyle II. 1992. Deletion of the VP16 open reading frame of herpes simplex virus type 1. J. Virol. 66258-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wild, P., E. M. Schraner, H. Adler, and B. M. Humbel. 2001. Enhanced resolution of membranes in cultured cells by cryoimmobilization and freeze-substitution. Microsc. Res. Tech. 53313-321. [DOI] [PubMed] [Google Scholar]
- 65.Yang, F., L. G. Moss, and G. N. Phillips, Jr. 1996. The molecular structure of green fluorescent protein. Nat. Biotechnol. 141246-1251. [DOI] [PubMed] [Google Scholar]
- 66.Zacharias, D. A., J. D. Violin, A. C. Newton, and R. Y. Tsien. 2002. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296913-916. [DOI] [PubMed] [Google Scholar]
- 67.Zhang, Y., D. A. Sirko, and J. L. McKnight. 1991. Role of herpes simplex virus type 1 UL46 and UL47 in alpha TIF-mediated transcriptional induction: characterization of three viral deletion mutants. J. Virol. 65829-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou, Z. H., M. Dougherty, J. Jakana, J. He, F. J. Rixon, and W. Chiu. 2000. Seeing the herpesvirus capsid at 8.5 Å. Science 288877-880. [DOI] [PubMed] [Google Scholar]
- 69.Zhou, Z. H., J. He, J. Jakana, J. D. Tatman, F. J. Rixon, and W. Chiu. 1995. Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nat. Struct. Biol. 21026-1030. [DOI] [PubMed] [Google Scholar]
- 70.Zhu, Q., and R. J. Courtney. 1994. Chemical cross-linking of virion envelope and tegument proteins of herpes simplex virus type 1. Virology 204590-599. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.