Abstract
The portal vertex of herpesvirus capsids serves as the conduit through which DNA is inserted during the assembly process. In herpes simplex virus (HSV), the portal is composed of 12 copies of the UL6 gene product, pUL6. Previous results identified a domain in the major capsid scaffold protein, ICP35, required for interaction with pUL6 and its incorporation into capsids formed in vitro (G. P. Singer et al., J. Virol. 74:6838-6848, 2005). In the current studies, pUL6 and scaffold proteins were found to coimmunoprecipitate from lysates of both HSV-infected cells and mammalian cells expressing scaffold proteins and pUL6. The coimmunoprecipitation of pUL6 and scaffold proteins was precluded upon deletion of codons 143 to 151 within UL26.5, encoding ICP35. While wild-type scaffold proteins colocalized with pUL6 when transiently coexpressed as viewed by indirect immunofluorescence, deletion of UL26.5 codons 143 to 151 precluded this colocalization. A recombinant herpes simplex virus, vJB11, was generated that lacked UL26.5 codons 143 to 151. A virus derived from this mutant but bearing a restored UL26.5 was also generated. vJB11 was unable to cleave or package viral DNA, whereas the restored virus packaged DNA normally. vJB11 produced ample numbers of B capsids in infected cells, but these lacked normal levels of pUL6. The deletion in UL26.5 also rendered pUL6 resistant to detergent extraction from vJB11-infected cells. These data indicate that, as was observed in vitro, amino acids 143 to 151 of ICP35 are critical for (i) interaction between scaffold proteins and pUL6 and (ii) incorporation of the HSV portal into capsids.
Herpes simplex virus (HSV) procapsids, like those of all herpesviruses, comprise two interconnected protein shells: an internal hollow sphere composed of more than 2,000 copies of the protein ICP35 or VP22a and an external shell composed primarily of 955 copies of the major capsid protein VP5 (1, 22, 28). Because of its importance in ensuring proper capsid assembly of the outer shell and its eventual loss from DNA-containing capsids, the internal shell has also been referred to as the capsid scaffold.
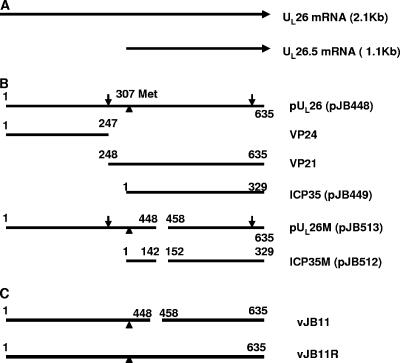
ICP35 is the product of the UL26.5 open reading frame, which is contained entirely within another gene, UL26 (Fig. 1) (15). UL26 encodes a serine protease (termed the maturational protease) that is essential for efficient viral replication (6, 10, 14). Approximately 87 copies of the encoded protein, pUL26, are present within the internal spherical shell of procapsids (20). During procapsid maturation, the UL26-encoded protease cleaves itself between amino acids 247 and 248 to yield two proteins: the N-terminal VP24 containing the protease and VP21, which is largely identical to ICP35 except for an N-terminal extension of 59 amino acids (Fig. 1) (5, 8, 37). In a separate event, the protease also cleaves ICP35 and VP21 to release 25 amino acids from their C termini (8). Because these C termini interact with the outer shell in the procapsid, it has been proposed that the cleavage serves to remove structural constraints within the spherical procapsid that preclude maturation of the outer shell (16, 31, 36). This proteolytic cleavage-triggered event results in a dramatic morphological change in the outer shell, from roughly spherical in the procapsid to the icosahedral shape of mature capsids (11, 34). The maturation of the shell likely coincides with DNA packaging in which concatameric viral DNA is cleaved into genomic lengths and inserted into the capsid.
FIG. 1.
Schematic diagram of wild-type and mutant scaffold-encoding genes and their proteins. A. Diagram of UL26 and UL26.5 RNAs. The direction of transcription is indicated by arrows. B. Schematic diagram of scaffold proteins colinear with the diagram in panel A. The identities of the individual proteins are indicated to the right. Plasmids encoding these are indicated in parentheses. Vertical arrows indicate sites cleaved by the VP24 protease. A vertical triangle indicates the position of the start codon of ICP35 relative to the UL26 open reading frame. A gap in a horizontal line indicates the absence of those sequences in the designated protein. Codon numbers corresponding to N and C termini are indicated above each diagrammed protein. C. Schematic diagram of the UL26 open reading frame of mutant (vJB11) or wild-type (vJB11R) viruses constructed and analyzed in this study.
Remarkably, each functional herpesvirus capsid incorporates a single portal or portal vertex into its outer shell for the purposes of DNA packaging and expulsion. In HSV capsids, the portal consists of a dodecamer of the UL6 gene product (pUL6) (18, 35). How each capsid is prevented from incorporating more than one portal is unknown, but based on studies of bacteriophage portal vertices, it seems likely that the initial portal/shell subassembly acts as a nidus around which the rest of the capsid forms (17). Thus, once the capsid shell continues to expand outward from the original nidus, other portals or their subunits are excluded from incorporation. It has been shown that the portal protein and ICP35 interact in vitro and that amino acids 143 to 151 of ICP35 are critical for incorporation of portal protein into capsids assembled in vitro (19, 27). The current studies were undertaken to investigate the role of this domain in interaction with portal protein in HSV-infected cells and incorporation of the portal into HSV type 1 (HSV-1) capsids produced by such cells.
MATERIALS AND METHODS
Viruses and cells.
Rabbit skin cells and CV1 cells were obtained from the American Type Culture Collection and maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% newborn calf serum, 100 U penicillin per ml, and 100 μg of streptomycin per ml. Flp-In-CV1 cell lines were purchased from Invitrogen and were grown in DMEM supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 100 μg/ml zeocin. CV26.5 and CV26 cell lines described in this paper were cultured in DMEM supplemented with 10% fetal bovine serum, 100 U/ml of penicillin, 100 μg/ml of streptomycin, and 200 μg/ml of hygromycin B. HSV-1 strain F [HSV-1(F)] and UL6 null virus derived from HSV-1 strain 17 were described previously (9, 23).
Plasmids.
PCR amplicons of UL26.5 or UL26 were cloned into the HindIII and EcoRV sites located in the multiple cloning site of expression vector pcDNA3, and the resulting constructs were designated pJB449 and pJB448, respectively. To delete codons 143 to 151 from UL26.5, or codons 449 to 457 from UL26, two-step PCR was performed. The resultant PCR amplicons were cloned into pcDNA5/FRT at the HindIII and EcoRV sites, and the resulting plasmids were designated pJB512 and pJB513, respectively. To construct pJB471 and pJB480, pJB449 and pJB448 were digested with HindIII and EcoRV, and the resulting fragments were subcloned into the HindIII and EcoRV sites of pcDNA5/FRT. All plasmid constructs were confirmed by DNA sequencing (data not shown) and immunoblotting after transient expression in mammalian cells.
Plasmid pCAGGS-nlsCre, expressing Cre recombinase, was a gift from Michael Kotlikoff, Cornell University. Plasmids pBAD-I-SceI, containing the gene encoding the Saccharomyces cerevisiae I-SceI endonuclease, and pEPkan-S, containing aphAI (encoding kanamycin resistance) adjacent to an I-SceI restriction site, were gifts from Nikolaus Osterrieder, Cornell University.
Construction of complementing cell lines.
UL26.5- or UL26-expressing cell lines were constructed by using the Flp-In-CV1 system (Invitrogen) according to the manufacturer's protocol and as described previously (13). Briefly, either pJB471 or pJB480 was cotransfected with plasmid pOG44, containing Flp recombinase under the constitutive cytomegalovirus promoter/enhancer, into an engineered cell line (Flp-In-CV1). Correct insertion of the shuttle vector caused simultaneous loss of zeocin resistance and gain of hygromycin resistance. After recombination, cells resistant to hygromycin were selected by growth in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 200 μg/ml hygromycin B.
Construction of recombinant viruses.
Production and characterization of a bacterial artificial chromosome (BAC) containing the entire HSV-1(F) genome was described previously (29). Recombinant viruses were constructed by En Passant mutagenesis, a two-step Red-mediated recombinant system described by B. K. Tischer et al. (32). The details of the procedure, and their use in construction of recombinant HSV-1 genomes were also described previously (32, 39). The primers for production of a PCR amplicon for eventual deletion of codons 143 to 151 from the UL26.5 gene in the HSV-1(F)-containing BAC were as follows: forward, TCCTACTGCGACCAGGACGAACCGGACGCGGACTACGCGCCGCGCGGGGTCGACTAGGGATAACAGGGTAATCGATTT; reverse, GCGGGCCGCGCGCCGGGAGTCGACCCCGCGCGGCGCGTAGTCCGC GTCCGGTTCGCCAGTGTTACAACCAATTAACC.
The expected mutation in the BAC DNA was confirmed by DNA sequencing, and the resulting recombinant BAC was designated bJB11. Purified bJB11 DNA was cotransfected with a Cre expression plasmid (see above) into CV26.5 cells expressing UL26.5. The presence of viable recombinant virus was indicated by plaque formation, and the resulting virus was subjected to two further rounds of plaque purification. The genotype of the recombinant virus, designated vJB11, was confirmed by PCR and DNA sequencing, whereas the viral phenotype was characterized as described below in Results. To repair the mutated UL26.5 gene, rabbit skin cells were cotransfected with vJB11 viral DNA and the UL26-containing plasmid pJB448. The virus arising from homologous recombination was able to form plaques on rabbit skin cells and was designated vJB11R. The genotype of vJB11R was confirmed by DNA sequencing, immunoblotting, and Southern blot analyses (data not shown).
Immunoprecipitation and immunoblotting.
The immunoprecipitation and immunoblotting procedures were performed essentially as described previously (12). Briefly, CV1 cells were either transfected with expression plasmids containing UL26, UL26.5, or UL6 or were infected with wild-type or recombinant viruses. At 24 h after transfection or 18 h after infection, the cells were washed with cold phosphate-buffered saline (PBS) and lysed in cold radioimmunoprecipitation (RIPA) buffer containing 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, 1 mM EDTA, and 1× protease inhibitor cocktail (Roche). Mouse anti-ICP35 monoclonal antibody (1:200 dilution; MCA406; AbD seroTEC) was used for pUL26 and ICP35 immunoprecipitation, whereas pUL6-specfic rabbit polyclonal serum (1:100 dilution) (30) was used to immunoprecipitate pUL6. Immune complexes, RIPA buffer-solublized clarified lysates, total lysates solublized in 1% sodium dodecyl sulfate (SDS) and β-mercaptoethanol, and in some experiments SDS-denatured purified capsids were separated on SDS-polyacrylamide gels and transferred to nitrocellulose membranes for immunoblotting.
Immunoblots were probed with anti-ICP35 monoclonal antibodies diluted 1:2,000 and/or anti-pUL6 polyclonal antiserum diluted 1:1,000. The bound immunoglobulins were revealed by reaction with horseradish peroxidase-conjugated anti-immunoglobulin antibodies and visualized by enhanced chemiluminescence (Amersham Pharmacia Biotech). Where applicable, the image intensities of specific bands on immunoblots were quantified with an LAS-3000 Mini Fujifilm imaging system (Fuji Photo Film Co. Ltd).
Immunofluorescence microscopy.
Transfection and immunofluorescence microscopy were performed as described previously (39). Briefly, transfected or infected Hep2 cells on glass coverslips were washed with PBS, fixed with 3% (wt/vol) paraformaldehyde in PBS, and permeabilized with 0.2% Triton X-100. Anti-pUL6 rabbit antiserum was preadsorbed against acetone-dehydrated lysates of Hep2 cells infected with a UL6 null virus (23); the preadsorbed antiserum was diluted 1:100 and reacted with the permeabilized cells. The cells were also reacted with mouse antiscaffold monoclonal antibody (MCA406; AbDSeroTEC) diluted 1:250. After extensive washing, the coverslips were reacted with goat anti-rabbit immunoglobulin G (IgG) conjugated to Alexa Fluor 488 (green) and goat anti-mouse IgG conjugated to Alexa Fluor 568 (red). Both conjugates were diluted 1:200. After washing, the cells were visualized using a conventional Nikon Eclipse E600 fluorescence microscope, with a 60× Plan Apo objective. Images were captured with a cooled SensiCam charge-coupled-device camera.
Capsid purification.
CV1 cell monolayers from three 850-cm2 roller bottles were infected with either HSV-1(F) or mutant viruses at a multiplicity of infection (MOI) of 5 PFU/cell. The cells were harvested 18 hours later and washed with cold PBS. Cell pellets were suspended in 25 ml of lysis buffer (1 mM dithiothreitol [DTT], 1 mM EDTA, 20 mM Tris pH 7.6, 500 mM NaCl, 1% Triton X-100, and protease inhibitor), sonicated briefly, and precleared by spinning at 10,000 × g for 15 min. The precleared lysates were pelleted through a 5-ml 35% sucrose cushion in TNE buffer (20 mM Tris-HCl [pH 7.6], 500 mM NaCl, 1 mM EDTA), in an SW28 ultracentrifuge tube at 24,000 rpm for 1 h. The pellets were resuspended in TNE buffer and applied to 20% to 50% sucrose gradients in SW41 ultracentrifuge tubes followed by centrifugation at 24,500 rpm for 1 h. After centrifugation, the light-refracting B capsid band was removed with a Pasteur pipette. Purity of capsid preparations was evaluated by transmission electron microscopy and negative staining (data not shown).
Electron microscopy.
CV1 cells were infected with wild-type or recombinant viruses at 5.0 PFU per cell. Fourteen hours later, the cells were fixed with 2.5% glutaraldehyde (EM Sciences) in 0.1 M sodium cacodylate buffer, pH 7.4, for 90 min. Samples were rinsed three times with cacodylate buffer and then fixed with 2% OsO4 in cacodylate buffer at room temperature for 2 h. Cells were rinsed again three times and then dehydrated through a series of increasing ethanol concentrations followed by acetone. After dehydration, the cells were infiltrated with Epon-Araldite as described previously (2). Thin sections cut with a diamond knife were counterstained with uranyl acetate and Reynold's lead citrate and then examined in a Philips 201 electron microscope as previously described (24). Conventional negatives were digitally scanned and converted to positive images in Adobe Photoshop.
Southern blotting.
Approximately 2 × 106 CV1 cells were mock infected or infected with HSV-1(F), vJB11, or vJB11R. At 18 h postinfection, viral DNA was extracted, digested with BamHI, separated on 0.8% agarose gels, and transferred to nylon membranes as described previously (38). The bound DNA was UV cross-linked to the membrane and hybridized with [32P]dCTP-labeled BamHI P fragment of HSV-1(F) DNA at 42°C for 24 h. The membrane was washed twice with 1× SSC (0.15 M NaCl plus 0.015 M sodium citrate) (25) at 42°C for 15 min each time and once with 0.1× SSC, 0.1% SDS for 1 h at 64°C and then fluorographed by exposure to X-ray film at −80°C in the presence of intensifying screens.
Virus replication assay.
Approximately 2 × 106 cells in 25-cm2 flasks were infected with the viruses indicated in Table 1, below, at a multiplicity of infection of 0.1 PFU/cell. After adsorption for 2 hours at 37°C with shaking, the inocula were removed, and the cells were washed with CBS buffer (40 mM citric acid, 10 mM KCl, 135 mM NaCl, pH 3.0) to remove residual infectivity. The cells were then washed with PBS once and overlaid with 5 ml of DMEM supplemented with 5% newborn calf serum. Twenty-four hours after infection, virus was harvested by three cycles of freezing and thawing, and the infectious yields were determined by plaque assay on the cell monolayers indicated in Table 1.
TABLE 1.
Virus replication assaya
| Virus | Genotype | Cell type infected/cell type used for plaque assay | Titer (PFU/ml) |
|---|---|---|---|
| vJB11 | Deletion of codons 143 to 151 of UL26.5 | Vero/CV26.5b | No viral plaques detected |
| vJB11 | Deletion of codons 143 to 151 of UL26.5 | CV26.5/CV26.5 | 8 × 105 |
| vJB11 | Deletion of codons 143 to 151 of UL26.5 | CV26c/CV26.5 | 6 × 105 |
| vJB11 | Deletion of codons 143 to 151 of UL26.5 | CV26Md/CV26.5 | No plaques detected |
| vJB11 | Deletion of codons 143 to 151 of UL26.5 | CV26.5Me/CV26.5 | No plaques detected |
| vJB11R | Deletion repaired | Vero/Vero | 2.5 × 107 |
| vJB11R | Deletion repaired | CV26.5/Vero | 3 × 107 |
| HSV-1(F) | Wild type | CV26.5/Vero | 2 × 107 |
| HSV-1(F) | Wild type | CV26.5M/Vero | 2.4 × 107 |
| HSV-1(F) | Wild type | CV26M/Vero | 2 × 107 |
Cells were infected with the indicated viruses at 0.1 PFU/cell. Twenty-four hours after infection, virus was harvested by three cycles of freezing and thawing, and infectious yields were determined by plaque assay on the indicated cell monolayers.
CV26.5 cells constitutively express UL26.5.
CV26 cells constitutively express UL26.
CV26M cells constitutively express UL26 lacking codons 449 to 457.
CV26.5M cells constitutively express UL26.5 lacking codons 143 to 151.
RESULTS
Scaffold proteins and portal protein coimmunoprecipitate from infected cells.
Previous results showed that amino acids 143 to 151 of ICP35 were necessary for in vitro interaction with the portal protein pUL6 and incorporation of pUL6 into HSV capsids reconstituted in vitro (27). To determine if these observations reflected an interaction between pUL6 and ICP35 in HSV-infected cells, CV1 cells were mock infected or were infected with wild-type HSV-1(F) or a deletion mutant lacking the UL6 open reading frame (23). The cells were harvested at 18 h postinfection, and clarified lysates were subjected to immunoprecipitation with mouse IgG or antibodies directed against ICP35 and other scaffold proteins. After purification and electrophoretic separation of immune complexes, the presence of pUL6 and ICP35 was determined by immunoblotting with pUL6-specfiic or ICP35-specific antibodies. As a control, the relative amounts of the respective proteins in the lysate were also determined by immunoblotting.
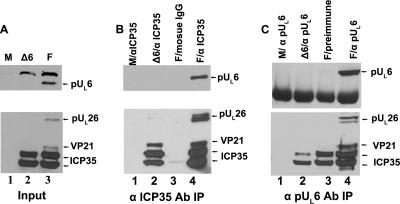
As shown in Fig. 2, the ICP35-specific antibody readily immunoprecipitated ICP35 from lysates of cells infected with the UL6 deletion mutant and ICP35, pUL26, and VP21 from cells infected with HSV-1(F). In the control reactions, ICP35 was not expressed in mock-infected cells, and mouse IgG immunoprecipitated only barely detectable amounts of ICP35. Importantly, the scaffold-specific antibody coimmunoprecipitated pUL6 from lysates of cells infected with HSV-1(F), indicating that these proteins interacted in the infected cell lysate.
FIG. 2.
Interaction between pUL6 and ICP35 in virus-infected cells detected by coimmunoprecipitation. CV1 cells were mock infected (M) or infected with 5 PFU per cell of HSV-1(F) (lanes labeled F) or a UL6 null mutant (lanes labeled Δ6). At 18 h postinfection, proteins were extracted with RIPA buffer, and the soluble fraction was subjected to immunoprecipitation with scaffold-specific monoclonal antibody MCA406 (designated αICP35) or pUL6-specific antibody (designated αpUL6). (A) Sample of the lysate of mock-infected or infected cells. (B) Approximately 40-fold more lysate as in panel A was subjected to immunoprecipitation with scaffold-specific antibody. Immunoprecipitated proteins were separated on an SDS-polyacrylamide gel, transferred to a nitrocellulose membrane, and detected by immunoblotting with anti-pUL6 (upper panel) or antiscaffold (lower panel) antibodies. (C) Approximately 40-fold more lysate as for panel A was reacted with pUL6-specific antibody, and an immunoblot of immunoprecipitated material was probed with antibodies directed against pUL6 (upper panel) or scaffold proteins (lower panel). The positions of the indicated proteins are indicated by arrows to the right of each panel. Ab, antibody; IP, immunoprecipitation.
To confirm the interaction between pUL6 and scaffold proteins, a reciprocal immunoprecipitation was performed by reacting the cell lysates with pUL6-specfic antibody followed by immunoblotting the electrophoretically separated purified immune complexes with scaffold-specific antibody. As shown in Fig. 2C, pUL6 was readily immunoprecipitated from lysates infected with HSV-1(F) but not from lysates of mock-infected cells, cells infected with the UL6 null virus, or HSV-1(F)-infected cells reacted with preimmune antibody.
Reaction with pUL6-specific antibody also caused coimmunoprecipitation of pUL26, ICP35, and VP21. The latter migrates slower than ICP35 due to the presence of 59 additional amino acids (Fig. 1B). ICP35 was brought down nonspecifically inasmuch as ICP35 was present in immunoprecipitation mixtures containing (i) pUL6-specific antibody reacted with lysates of cells infected with the UL6 null virus and (ii) preimmune antibody reacted with lysates of wild-type virus-infected cells. Thus, densitometric analysis indicated that the amounts of ICP35 coimmunoprecipitated with pUL6 antibody from lysates of cells infected with HSV-1(F) were increased 1.5-fold over that obtained upon reaction with preimmune antibody and 5-fold more than was immunoprecipitated from lysates of cells infected with the UL6 null virus. Perhaps more convincing for the specificity of the interaction were the observations that (i) coimmunoprecipitation of detectable levels of VP21 by pUL6-specific antibody required pUL6 (i.e., it was not observed in lysates of cells infected with the UL6 null mutant reacted with the pUL6-specific antibody) and did not occur upon reaction of lysates of cells infected with HSV-1(F) with preimmune antibody, and (ii) pUL26 was specifically coimmunoprecipitated with pUL6-specific antibody but not preimmune antibody. We could not assess the specificity of the interaction with pUL26, inasmuch as little was detected in the lysates of cells infected with the UL6 null mutant. Nevertheless, taken with the data shown in Fig. 1B, these data indicate that pUL6 can interact directly or indirectly with ICP35, pUL26, and VP21 in infected cells. The coimmunoprecipitation of these scaffold proteins by pUL6 antibody might be expected inasmuch as ICP35 and VP21 share considerable sequence identity (Fig. 1) and interact with one another (26).
Scaffold proteins and portal protein interact in the absence of other HSV proteins.
To test whether ICP35 and pUL26 were sufficient to interact with pUL6 in the absence of other viral proteins in mammalian cells, ICP35 and pUL26 were transiently coexpressed with pUL6, and lysates of the cells were subjected to immunoprecipitation with pUL6 and scaffold-specific antibodies. The presence of pUL6, pUL26, VP21, and ICP35 in immunoprecipitated material was then determined by immunoblotting.
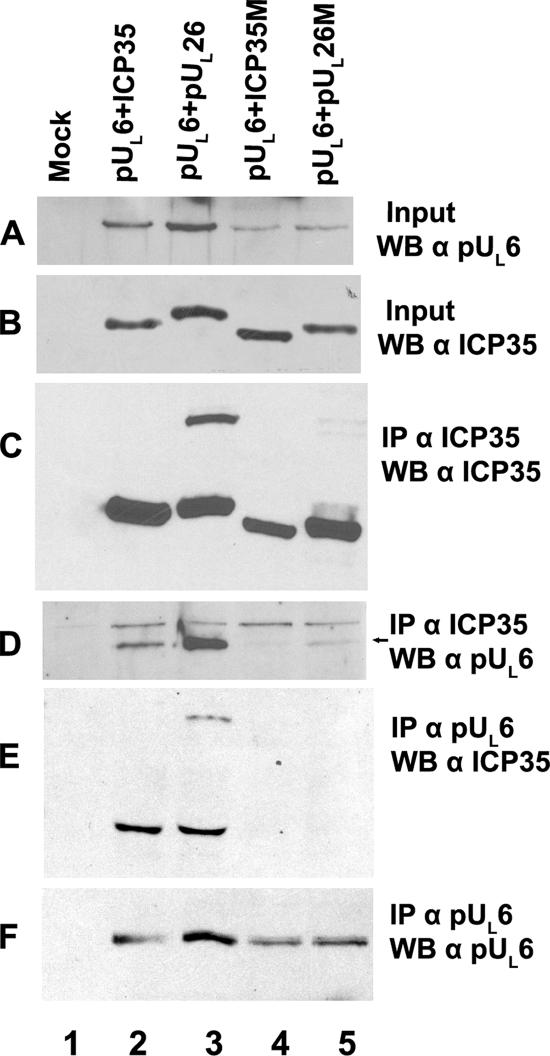
As shown in Fig. 3, expression of UL26 caused production of VP21, which migrated slower than ICP35 (Fig. 3B, lane 3). Similar to the results in infected cells (Fig. 2), scaffold-specific antibody immunoprecipitated transiently expressed ICP35, VP21, and pUL26 (Fig. 3C, lanes 2 and 3). More importantly, transiently coexpressed pUL6, VP21, pUL26, and ICP35 were readily coimmunoprecipitated by both the antiscaffold and anti-pUL6 antibodies (Fig. 3D and E, lanes 2 and 3).
FIG. 3.
Interaction between portal and scaffold proteins in the absence of other viral proteins. CV1 cells were mock transfected (Mock) or were cotransfected with plasmids expressing pUL6 and wild-type ICP35, wild-type pUL26, mutant ICP35 (ICP35M), or mutant pUL26 (pUL26M). The mutant plasmids are diagrammed in Fig. 1. Cell lysates (A and B) or proteins from 40-fold more lysate immunoprecipitated with antiscaffold antibody (designated αICP35) (C and D) or anti-pUL6 antibody (αpUL6) (E and F) were transferred to nitrocellulose. The transferred proteins were probed with scaffold-specific (B, C, and E) or pUL6-specific (A, D, and F) antibodies. Bound immunoglobulins were revealed by enhanced chemiluminescence. An arrow to the right of panel D indicates the position of the pUL6-specific band, which is located below a nonspecific band in that panel. IP, immunoprecipitation with the indicated antibody; WB, Western blotting with the indicated antibody.
Domain in scaffold proteins critical for interaction with portal protein.
Given that a robust immunoprecipitation assay was now available, we next wanted to determine whether a previously identified pUL6 interaction domain within scaffold proteins was necessary for coimmunoprecipitation of scaffold and portal. To test this possibility, plasmids (i) encoding mutant ICP35 lacking amino acids 143 to 151 or (ii) lacking the corresponding region of otherwise-full-length pUL26 were coexpressed with pUL6 and analyzed in the coimmunoprecipitation assay described above. As shown in Fig. 3, lanes 4 and 5, removal of codons 143 to 151 precluded detectable coimmunoprecipitation of ICP35, VP21, and pUL26 by pUL6-specific antibody (Fig. 3E). Using cross-reacting bands (Fig. 3D) and transiently expressed ICP35 as loading controls, it was noted by densitometric scan that around twofold less pUL6 was present in the clarified lysates of cells coexpressing mutant ICP35 and pUL26, as opposed to lysates of cells expressing wild-type versions of these proteins (Fig. 3A, compare lanes 2 to 5). Further analyses indicated that this observation reflected less pUL6 in the soluble fraction, as opposed to a decrease in overall expression of pUL6 (data not shown). Taking into account the reduced amounts of soluble pUL6, mutant ICP35 and pUL26 still coimmunoprecipitated fivefold less efficiently with pUL6-specific antibody compared to the amounts of coimmunoprecipitated wild-type proteins (Fig. 3D, lane 2 versus lane 4 and lane 3 versus lane 5). Thus, the reduced coimmunoprecipitation of pUL6 with mutant scaffold proteins is not solely a consequence of lowered solubility of pUL6. We therefore conclude that the region in scaffold proteins corresponding to amino acids 143 to 151 in ICP35 is critical for the interaction with the portal protein in mammalian cells.
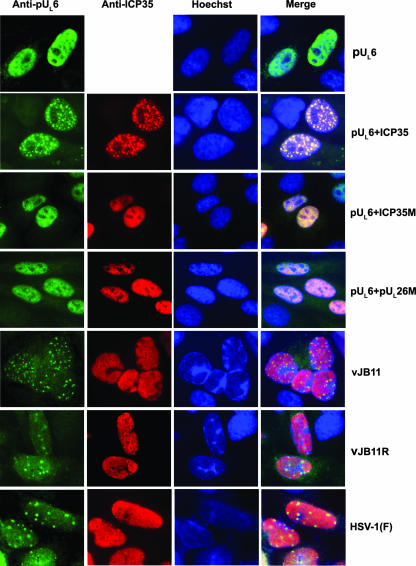
It was also of interest to determine whether wild-type and mutant scaffold proteins affected distribution of pUL6 within cells. Cells were therefore transfected with equal amounts of plasmids encoding pUL6 alone or together with plasmids encoding wild-type ICP35 or mutant ICP35 or pUL26. As shown in Fig. 4, pUL6 localized in a diffuse pattern throughout the nucleus when expressed alone. In contrast to this observation, some pUL6 localized in a punctate pattern when coexpressed with either wild-type ICP35 (Fig. 4) or pUL26 (data not shown). Moreover, these intranuclear puncta contained ICP35/UL26 immunoreactivity, as viewed by the yellow color on merging the respective images of scaffold or portal immunostaining. In contrast, coexpression of pUL26/VP21 (not shown) or ICP35 lacking the putative pUL6 interaction domain (designated pUL26 M and ICP35M in Fig. 4) caused pUL6 to localize in a diffuse intranuclear pattern. Thus, codons 143 to 151 of ICP35 and 449 to 457 of UL26 were necessary for the ICP35- or pUL26-mediated alteration of the localization of pUL6. This is consistent with the conclusion that these domains of ICP35 and pUL26/VP21 are critical for interaction with pUL6.
FIG. 4.
Conventional indirect immunofluorescence of pUL6 and ICP35 in transfected and infected Hep2 cells. Cells were transfected with pUL6 alone or together with plasmids expressing wild-type ICP35, mutant ICP35 (ICP35M), or mutant pUL26 (pUL26M). Alternatively, cells were infected with the indicated viruses (bottom three rows). Cells were fixed and permeabilized at 24 h posttransfection or 18 h postinfection, and localization of pUL6 (left column) and scaffold proteins (middle left column) was determined by indirect immunofluorescence microscopy. Primary antibodies were mouse monoclonal antiscaffold antibody (anti-ICP35) or rabbit antiserum directed against pUL6 (anti-pUL6). Bound antibodies were revealed by reaction with Alexa Fluor 488-labeled goat anti-rabbit IgG (green) and Alexa Fluor 568-labeled goat anti-mouse IgG (red). Nuclei were identified by Hoechst staining (middle right column). The images of the same panel are merged in the rightmost column. All images were recorded with a charge-coupled-device camera.
The portal protein interaction domain of scaffold proteins is essential for viral replication, DNA cleavage/packaging, and incorporation of the portal into capsids.
Because some results in transient assays can be misleading, we constructed a recombinant virus, lacking codons 143 to 151 of UL26.5, through the genetic manipulation of an HSV-1(F) genome maintained in a recombinant BAC. Moreover, an additional recombinant virus was constructed in which this lesion was restored to wild-type sequences. The genotypes of the deletion and restored viruses, designated vJB11 and vJB11R, respectively, were confirmed by DNA sequencing, restriction length polymorphism, and Southern blot analyses (data not shown).
To characterize the effect(s) of codons 143 to 151 of UL26.5 or the equivalent region of UL26 on HSV-1 replication, normal Vero cells or CV1 cells (derived from Vero cells) engineered to express wild-type or mutant (lacking codons 143 to 151 of UL26.5 or its equivalent in UL26) UL26 or UL26.5 were infected with HSV-1(F), vJB11, or vJB11R, and yields of infectious virus at 24 h postinfection were determined on permissive cell lines. The results, shown in Table 1, were as follows:
(i) vJB11 was unable to produce infectious virus or appreciable viral plaques in noncomplementing Vero cells, whereas vJB11R produced infectious virus to levels comparable to those of wild-type HSV-1(F) virus. In the cell lines expressing either wild-type UL26.5 or UL26, the virus was able to form plaques and replicate, but viral yields were reduced approximately 30- to 50-fold relative to production of wild-type or restored virus in these cells.
(ii) vJB11 was unable to replicate or form appreciable plaques on cell lines expressing mutant UL26 or UL26.5.
(iii) Both the wild-type HSV-1(F) virus and the genetically restored virus vJB11R were able to replicate to wild-type levels on all cell lines tested, including cells expressing mutant UL26 and mutant UL26.5. These data indicate that the mutation did not confer an obvious dominant negative effect that would have inhibited wild-type virus replication.
Together, these data indicate that codons 143 to 151 of ICP35, and their equivalent in UL26, are essential for viral replication. The data also indicate that the UL26 mutation in vJB11 was solely responsible for the inability to replicate on normal cells.
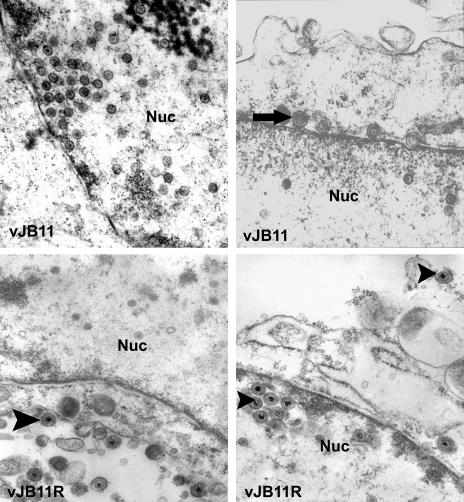
To more precisely determine the stage of virus replication precluded by the mutation in vJB11, cells were infected with this virus or vJB11R, 14 hours later they were fixed and embedded, and thin sections were examined by electron microscopy. As shown in Fig. 5, the nuclei of cells infected with vJB11 contained numerous viral capsids, but these capsids lacked electron-dense cores indicative of the presence of packaged viral DNA. Some enveloped capsids accumulated in the perinuclear space between the inner and outer nuclear membranes (upper right panel), but such capsids were not observed in the extracellular space, nor in the cytoplasm free of the nuclear membrane. In contrast, cells infected with the restored virus contained ample enveloped capsids within virions in the cytoplasm and extracellular space (Fig. 5), and most of these contained electron-dense cores, suggesting that they contained viral DNA. These data suggest that the lesion in UL26.5 of vJB11 precludes viral DNA packaging and capsid egress from the nucleus but does not preclude assembly of B-like capsids.
FIG. 5.
Ultrastructure of Vero cells infected with mutant and restored viruses. Vero cells were infected with viruses vJB11 (top panels) or vJB11R (bottom panels) and were fixed and permeabilized at 14 h after infection. The cells were subsequently embedded in Epon and stained with uranyl acetate and OsO4. Thin sections were viewed in a Phillips electron microscope. Images from conventional negatives were digitally scanned and rendered positive with Adobe Photoshop. Enveloped B capsids are indicated with an arrow. Arrowheads indicate virions containing viral DNA. Nuc, nucleus. As a size standard, HSV capsids are 125 nm in diameter.
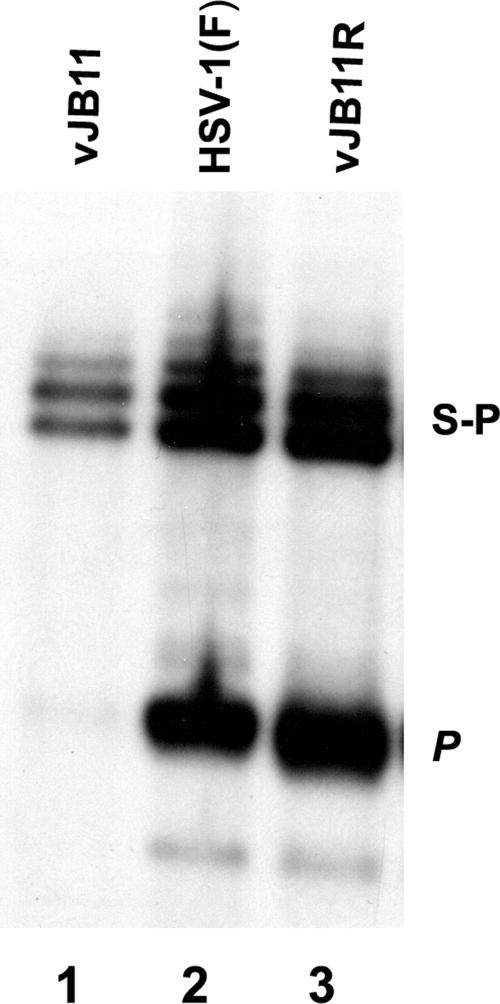
To determine whether the deletion of codons 143 to 151 of UL26.5 affected viral DNA cleavage, viral DNA was purified from cells infected with vJB11, vJB11R, or HSV-1(F) and digested with BamHI. The DNA fragments were then separated by electrophoresis, transferred to a nylon membrane, and probed with radiolabeled BamHI restriction fragment P, which contains sequences located at genomic ends (33). As shown in Fig. 6, viral DNA from cells infected with vJB11 contained the S-P fragment, which is present in both linear and concatameric viral DNA, but lacked the BamHI P fragment indicative of genomic ends. In contrast, the BamHI P fragment was readily detectable in viral DNA purified from cells infected with the restored virus vJB11R and wild type HSV-1(F). Together, these data indicate that codons 143 to 151 of UL26.5 are essential for cleavage and packaging of viral DNA.
FIG. 6.
Southern blot of viral DNA digested with BamHI. Approximately 2 × 106 CV1 cells were infected with the indicated viruses, and viral DNA was extracted, digested with BamHI, transferred to a nylon membrane (0.45 μm), and hybridized with radiolabeled BamHI P fragment of HSV-1(F) DNA. Bound DNA was visualized by autoradiography. The positions of the S-P fragment corresponding to junctions of the S and L component in concatameric DNA and the P fragment indicative of genomic end fragments are indicated.
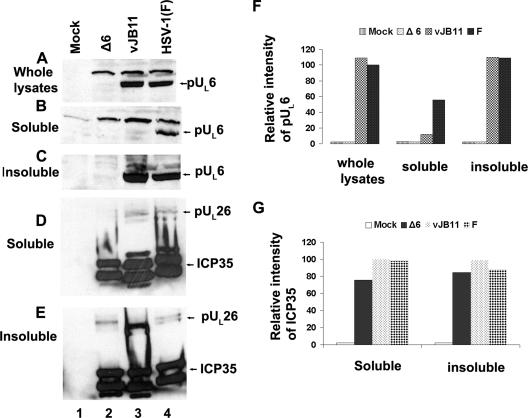
The next set of experiments was designed to determine the effects of the deletion of codons 143 to 151 of UL26.5 on interactions with the UL6-encoded portal protein in infected cells and to determine if this mutation precluded portal incorporation into capsids. As a first step, the solubility of pUL6 and ICP35 in cells infected with vJB11 was compared with the solubility in cells infected with HSV-1(F). As shown in Fig. 7, whereas ICP35 was solubilized in RIPA buffer from cells infected with wild-type HSV-1(F) or vJB11, pUL6 was virtually undetected in the soluble fractions of cells infected with vJB11. This was the case despite the observations that pUL6 was (i) readily detected in whole lysates and insoluble fractions of cells infected with vJB11 and (ii) readily solubilized from cells infected with HSV-1(F) by extraction in RIPA buffer. Although the insolubility of pUL6 precluded meaningful coimmunoprecipitation experiments, it argued that the region corresponding to amino acids 143 to 151 of ICP35 was necessary for the normal biochemical conformation and/or behavior of pUL6 in infected cells. This interpretation is consistent with the possibility that the scaffold and portal proteins directly or indirectly interact with one another in these cells.
FIG. 7.
Immunoblots of soluble or insoluble cell lysates from vJB11-infected cells. Approximately 2 × 106 CV1 cells were mock infected or were infected with pUL6 null virus, vJB11, or HSV-1(F). At 18 hours postinfection, cells were washed with cold PBS and solubilized in either (i) 800 μl of SDS sample buffer (whole-cell lysates) (A) or in 400 μl of RIPA buffer (B to E). After extensive clarification of the RIPA-fractionated material, proteins in the supernatant (soluble) (B and D) or pellet (insoluble) (C and E) were denatured in SDS, separated on an SDS-polyacrylamide gel, transferred to a nitrocellulose membrane, and probed with pUL6 (A, B, and C) or scaffold-specific antibodies (D and E). Bound immunoglobulins were revealed by reaction with appropriate secondary antibodies and enhanced chemiluminescence. (F) The chemiluminescent intensities corresponding to the position of the pUL6-containing band in each lane of panels A, B, and C were determined using an LAS-3000 Mini Fujifilm imaging system and are reported as a percentage of that obtained in panel A, lane 4. (G) Similar to panel F, but intensities of ICP35 signals were normalized to that in panel D, lane 4.
Because localization of pUL6 was altered upon transient coexpression with ICP35 (Fig. 4), experiments were performed to determine whether the mutation in ICP35 altered pUL6 localization in infected cells. Cells were infected with vJB11, vJB11R, or HSV-1(F) and were fixed, permeabilized, and reacted with antibodies directed against pUL6 or ICP35. As shown in Fig. 4, pUL6 appeared in a punctate pattern in the nuclei of cells infected with all three viruses (Fig. 4, bottom three rows), whereas scaffold proteins localized in a smooth pattern throughout the nucleus except in areas of dense chromatin revealed by staining with Hoechst dye. Some pUL6-containing foci lacked obvious scaffold protein-specific staining at their centers, whereas scaffold and pUL6 colocalized at the edges of these “holes” in the otherwise-smooth scaffold-specific staining pattern. This feature was not specific to any particular virus used in the experiment. Thus, although pUL6 solubility was greatly affected by the deletion of codons in UL26 or ICP35, the distribution of pUL6 in infected cells was not noticeably affected by this mutation. Given the difference in appearance of pUL6 in transient expression versus infection, we conclude that whereas ICP35 is necessary and sufficient to localize pUL6 into punctate intranuclear foci in uninfected cells, viral functions other than those encoded by UL26.5 or UL26 dramatically affect the distribution of pUL6 in infected cells.
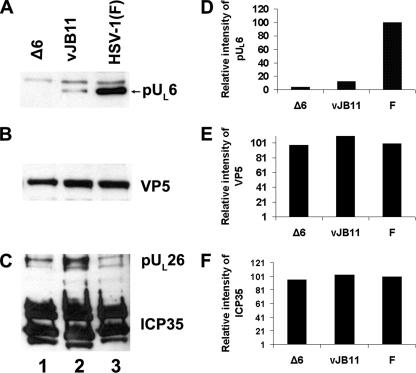
To more directly assess the effects of the mutation in UL26/UL26.5 on portal protein incorporation into capsids, B capsids were purified from cells infected with vJB11, the UL6 deletion virus, or HSV-1(F). The capsid-associated proteins were then denatured in SDS, electrophoretically separated, and probed with antibodies directed against scaffold proteins and pUL6. As loading controls, the immunoblots were also probed with antibodies directed against scaffold proteins and the major external capsid protein VP5. As shown in Fig. 8, for a given amount of VP5, vJB11 incorporated approximately 20-fold less pUL6 per capsid on average than did wild-type HSV-1(F) virus. These data indicate that codons 143 to 151 of UL26.5 are critical for incorporation of pUL6 into HSV capsids.
FIG. 8.
Immunoblot of B capsids probed with anti-pUL6, ICP35, or VP5 antibodies. Capsids were purified from CV1 cells infected with UL6 null virus, vJB11, or HSV-1(F) as detailed in Materials and Methods. B capsids denatured in SDS were either diluted 10-fold (B and C) or were loaded undiluted (A) onto an SDS-polyacrylamide gel through which they were separated by electrophoresis and transferred to a nitrocellulose sheet. The nitrocellulose sheet was probed with pUL6 (A), VP5 (B), or ICP35 (C) specific antibodies, and bound immunoglobulin was revealed by reaction with appropriately conjugated secondary antibodies followed by enhanced chemiluminescence. (D) The relative amounts of pUL6 in lanes 1 to 3 were determined using an LAS-3000 Mini Fujifilm imaging system. The chemiluminescent intensity of each pUL6-containing band in panel A is reported as a percentage of the signal obtained in panel A, lane 3. (E) Similar to panel D except that values reflect chemiluminescent intensities of VP5 in lanes of panel B normalized to that in panel B, lane 3. (F) Similar to panel D, but values reflect the ICP35 specific signals in panel C normalized to panel C, lane 3.
DISCUSSION
The data presented herein using mammalian cells and mutant HSV are largely consistent with conclusions from previous studies using capsids reconstituted in an in vitro capsid assembly system (19, 27). Specifically, the data suggest that the portal protein encoded by pUL6 interacts with internal capsid shell proteins and requires the region corresponding to amino acids 143 to 151 of ICP35 for the interaction. The interaction between portal and scaffold was supported by coimmunoprecipitation of pUL6 with scaffold components (Fig. 3) and alteration of the localization of transiently expressed pUL6 when coexpressed with ICP35 (Fig. 4). In further indirect support of the interaction, the solubility of pUL6 was dependent on the scaffold protein domain, shown to be essential for the interaction as assessed by immunoprecipitation and for incorporation of the portal vertex into capsids (Fig. 8). These observations suggest that early in infection pUL6 requires ICP35, VP21, and/or pUL26 for its normal biochemical behavior, which culminates in its incorporation into the portal vertex. This is consistent with the observation that the portal may act very early in formation of capsids in vitro (21).
Because the inner shell of procapsids contains a mixture of ICP35 and pUL26, and both were coimmunoprecipitated with pUL6-specific antibody, we cannot distinguish whether one or both proteins interact directly with the portal. Although it also coimmunoprecipitated efficiently with pUL6 from lysates of infected cells, the absence of VP21 from procapsids would argue against this protein as the main partner responsible for portal incorporation into capsids.
The data presented herein also indicate that the scaffold domain studied here was essential for viral replication and DNA cleavage and packaging. We assume that the DNA packaging defect is a consequence of aberrant portal incorporation into capsids; thus, there is no conduit through which DNA can be inserted into the preformed capsid. That viral DNA was not cleaved in cells infected with the mutant virus is consistent with previous studies showing that DNA cleavage is dependent on the presence of the portal and intact capsids (7, 23). Thus, DNA cleavage only occurs when enzymes required for DNA cleavage (the viral terminase) and capsids are expressed together, presumably to adopt the proper conformation.
Expression of the mutant scaffold proteins in cell lines did not interfere with replication of wild-type viruses, i.e., it did not confer an obvious dominant negative effect. One possibility to explain this observation is that the mutant proteins were entirely incapable of interaction with the portal protein, arguing that the scaffold domain studied is solely responsible for the interaction with portal protein. We cannot exclude the possibility, however, that the levels of mutant scaffold expressed in the engineered cell lines were too low to efficiently compete with wild-type scaffold proteins. The latter would be expected to be abundantly expressed by the infecting virus. The low level of complementation of vJB11 replication conferred by cell lines expressing the wild-type proteins (Table 1) might also reflect the low levels of wild-type scaffold proteins encoded by the cell lines as opposed to the abundance of mutant proteins encoded by the virus.
We have noted that pUL6 distributes in a diffuse pattern when expressed alone but partly in punctate foci when coexpressed with wild-type scaffold proteins. These distribution patterns differed from that of pUL6 in infected cells, in which pUL6 localized almost exclusively within discrete foci. Thus, proteins other than pUL26, VP21, or ICP35 contribute to the punctate distribution of pUL6 in infected cells. Likely candidates to confer this function include (i) heat shock proteins induced by HSV infection that have been shown to colocalize with pUL6 or (ii) other components of the capsid shell that must eventually directly or indirectly interact with the portal (3, 4). In addition, many regions with intense pUL6 staining did not contain scaffold-specific staining, unlike the case in cells only expressing ICP35 and pUL6. Possibilities to explain this observation are that (i) the pUL6-containing foci contain scaffold proteins but associated epitopes are masked from the reaction with the antiscaffold antibody or (ii) pUL6 and scaffold proteins do not associate in these foci. The latter would argue that these foci do not represent sites in which portals are inserted into capsids. Alternatively, we cannot exclude the possibility that the different levels induced during transient expression versus viral infection could contribute to the different staining patterns.
Acknowledgments
We thank Elizabeth G. Wills for electron microscopy, Gary Whittaker for use of the Nikon florescence microscope, and Arvind Patel for the UL6 deletion virus.
These studies were supported by grant R01 GM 50740 from the National Institutes of Health.
Footnotes
Published ahead of print on 12 March 2008.
REFERENCES
- 1.Baines, J. D., and C. Duffy. 2006. Nucleocapsid assembly and envelopment of herpes simplex virus, p. 175-204. In R. M. Sandri-Goldin (ed.), Alphaherpesviruses: pathogenesis, molecular biology and infection control. Caister Scientific Press, Norfolk, United Kingdom.
- 2.Baines, J. D., A. P. W. Poon, J. Rovnak, and B. Roizman. 1994. The UL15 gene of herpes simplex virus encodes two proteins and is required for cleavage of viral DNA. J. Virol. 688118-8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burch, A. D., and S. K. Weller. 2004. Nuclear sequestration of cellular chaperone and proteasomal machinery during herpes simplex virus type 1 infection. J. Virol. 787175-7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cardone, G., D. C. Winkler, B. L. Trus, N. Cheng, J. E. Heuser, W. W. Newcomb, J. C. Brown, and A. C. Steven. 2007. Visualization of the herpes simplex virus portal in situ by cryo-electron tomography. Virology 361426-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davison, M. D., F. J. Rixon, and A. J. Davison. 1992. Identification of genes encoding two capsid proteins (VP24 and VP26) of herpes simplex type 1. J. Gen. Virol. 732709-2713. [DOI] [PubMed] [Google Scholar]
- 6.Deckman, I. C., M. Hagen, and P. J. McCann III. 1992. Herpes simplex virus type 1 protease expressed in Escherichia coli exhibits autoprocessing and specific cleavage of the ICP35 assembly protein. J. Virol. 667362-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desai, P., N. A. DeLuca, J. C. Glorioso, and S. Person. 1993. Mutations in herpes simplex virus type 1 genes encoding VP5 and VP23 abrogate capsid formation and cleavage of replicated DNA. J. Virol. 671357-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DiIanni, C. L., D. A. Drier, I. C. Deckman, P. J. McCann, F. Liu, B. Roizman, R. J. Colonno, and M. G. Cordingley. 1993. Identification of the herpes simplex virus-1 protease cleavage sites by direct sequence analysis of autoproteolytic cleavage products. J. Biol. Chem. 2682048-2051. [PubMed] [Google Scholar]
- 9.Ejercito, P. M., E. D. Kieff, and B. Roizman. 1968. Characterization of herpes simplex virus strains differing in their effects on social behavior of infected cells. J. Gen. Virol. 2357-364. [DOI] [PubMed] [Google Scholar]
- 10.Gao, M., L. Matusick-Kumar, W. Hurlburt, S. F. DiTusa, W. W. Newcomb, J. C. Brown, P. J. McCann, I. Deckman, and R. J. Colonno. 1994. The protease of herpes simplex virus type 1 is essential for functional capsid formation and viral growth. J. Virol. 683702-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heymann, J. B., N. Cheng, W. W. Newcomb, B. L. Trus, J. C. Brown, and A. C. Steven. 2003. Dynamics of herpes simplex virus capsid maturation visualized by time-lapse cryo-electron microscopy. Nat. Struct. Biol. 10334-341. [DOI] [PubMed] [Google Scholar]
- 12.Jacobson, J. G., K. Yang, J. D. Baines, and F. L. Homa. 2006. Linker insertion mutations in the herpes simplex virus type 1 UL28 gene: effects on UL28 interaction with UL15 and UL33 and identification of a second-site mutation in the UL15 gene that suppresses a lethal UL28 mutation. J. Virol. 8012312-12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang, L., M. Tanaka, Y. Kawaguchi, and J. D. Baines. 2004. Cell lines that support replication of a novel herpes simplex 1 UL31 deletion mutant can properly target UL34 protein to the nuclear rim in the absence of UL31. Virology 32968-76. [DOI] [PubMed] [Google Scholar]
- 14.Liu, F., and B. Roizman. 1991. The herpes simplex virus 1 gene encoding a protease also contains within its coding domain the gene encoding the more abundant substrate. J. Virol. 655149-5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu, F., and B. Roizman. 1991. The promoter, transcriptional unit, and coding sequences of herpes simplex virus 1 family 35 proteins are contained within and in frame with the UL26 open reading frame. J. Virol. 65206-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matusick-Kumar, L., W. W. Newcomb, J. C. Brown, P. J. McCann, W. Hurlburt, S. P. Weinheimer, and M. Gao. 1995. The C-terminal 25 amino acids of the protease and its substrate ICP35 of herpes simplex virus type 1 are involved in the formation of sealed capsids. J. Virol. 694347-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murialdo, H., and A. Becker. 1978. Head morphogenesis of complex double-stranded deoxyribonucleic acid bacteriophages. Microbiol. Rev. 42529-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newcomb, W. W., R. M. Juhas, D. R. Thomsen, F. L. Homa, A. D. Burch, S. K. Weller, and J. C. Brown. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 7510923-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newcomb, W. W., D. R. Thomsen, F. L. Homa, and J. C. Brown. 2003. Assembly of the herpes simplex virus capsid: identification of soluble scaffold-portal complexes and their role in formation of portal-containing capsids. J. Virol. 779862-9871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newcomb, W. W., B. L. Trus, F. P. Booy, A. C. Steven, J. S. Wall, and S. C. Brown. 1993. Structure of the herpes simplex virus capsid molecular composition of the pentons and the triplexes. J. Mol. Biol. 232499-511. [DOI] [PubMed] [Google Scholar]
- 21.Newcomb, W. W., F. L. Homa, and J. C. Brown. 2005. Involvement of the portal at an early step in herpes simplex virus capsid assembly. J. Virol. 7910540-10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newcomb, W. W., B. L. Trus, N. Cheng, A. C. Steven, A. K. Sheaffer, D. J. Tenney, S. K. Weller, and J. C. Brown. 2000. Isolation of herpes simplex virus procapsids from cells infected with a protease-deficient mutant virus. J. Virol. 741663-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel, A. H., F. J. Rixon, C. Cunningham, and A. J. Davison. 1996. Isolation and characterization of herpes simplex virus type 1 mutants defective in the UL6 gene. Virology 217111-123. [DOI] [PubMed] [Google Scholar]
- 24.Salmon, B., C. Cunningham, A. J. Davison, W. J. Harris, and J. D. Baines. 1998. The herpes simplex virus 1 UL17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J. Virol. 723779-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York, NY.
- 26.Sheaffer, A. K., W. W. Newcomb, J. C. Brown, M. Gao, S. K. Weller, and D. J. Tenney. 2000. Evidence for controlled incorporation of herpes simplex virus type 1 UL26 protease into capsids. J. Virol. 746838-6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singer, G. P., W. W. Newcomb, D. R. Thomsen, F. L. Homa, and J. C. Brown. 2005. Identification of a region in the herpes simplex virus scaffolding protein required for interaction with the portal. J. Virol. 79132-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steven, A. C., and P. G. Spear. 1997. Herpesvirus capsid assembly and envelopment, p. 312-351. In W. Chiu, R. M. Burnett, and R. L. Garcea (ed.), Structural biology of viruses. Oxford University Press, New York, NY.
- 29.Tanaka, M., H. Kagawa, Y. Yamanashi, T. Sata, and Y. Kawaguchi. 2003. Construction of an excisable bacterial artificial chromosome containing a full-length infectious clone of herpes simplex virus type 1: viruses reconstituted from the clone exhibit wild-type properties in vitro and in vivo. J. Virol. 771382-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taus, N. S., B. Salmon, and J. D. Baines. 1998. The herpes simplex virus 1 UL17 gene is required for localization of capsids and major and minor capsid proteins to intranuclear sites where viral DNA is cleaved and packaged. Virology 252115-125. [DOI] [PubMed] [Google Scholar]
- 31.Thomsen, D. R., W. W. Newcomb, J. C. Brown, and F. L. Homa. 1995. Assembly of the herpes simplex virus capsid: requirement for the carboxy-terminal twenty-five amino acids of the proteins encoded by the UL26 and UL26.5 genes. J. Virol. 693690-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tischer, B. K., J. von Einem, B. Kaufer, and N. Osterrieder. 2006. Two-step RED-mediated recombination for versatile high-efficiency markerless DNA manipulation in Eschericia coli. BioTechniques 40191-197. [DOI] [PubMed] [Google Scholar]
- 33.Tognon, M., E. Cassai, A. Rotola, and B. Roizman. 1983. The heterogenous regions in herpes simplex virus 1 DNA. Microbiologica 6191-198. [PubMed] [Google Scholar]
- 34.Trus, B. L., F. P. Booy, W. W. Newcomb, J. C. Brown, F. L. Homa, D. R. Thomsen, and A. C. Steven. 1996. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19C and VP23 in assembly. J. Mol. Biol. 263447-462. [DOI] [PubMed] [Google Scholar]
- 35.Trus, B. L., N. Cheng, W. W. Newcomb, F. L. Homa, J. C. Brown, and A. C. Steven. 2004. Structure and polymorphism of the UL6 portal protein of herpes simplex virus type 1. J. Virol. 7812668-12671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Warner, S. C., G. Chytrova, P. Desai, and S. Person. 2001. Mutations in the N-terminus of VP5 alter its interaction with the scaffold proteins of herpes simplex virus type 1. Virology 284308-316. [DOI] [PubMed] [Google Scholar]
- 37.Weinheimer, S. P., P. J. McCann, D. R. O'Boyle, J. T. Stevens, B. A. Boyd, D. A. Drier, G. A. Yamanaka, C. L. DiIanni, I. C. Deckman, and M. G. Cordingley. 1993. Autoproteolysis of herpes simplex virus type 1 protease releases an active catalytic domain found in intermediate capsid particles. J. Virol. 675813-5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang, K., and J. D. Baines. 2006. The putative terminase subunit of herpes simplex virus 1 encoded by UL28 is necessary and sufficient to mediate interaction between pUL15 and pUL33. J. Virol. 805733-5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang, K., F. Homa, and J. D. Baines. 2007. Putative terminase subunits of herpes simplex virus 1 form a complex in the cytoplasm and interact with portal protein in the nucleus. J. Virol. 816419-6433. [DOI] [PMC free article] [PubMed] [Google Scholar]