Abstract
To determine the extent of homologous recombination in human influenza A virus, we assembled a data set of 13,852 sequences representing all eight segments and both major circulating subtypes, H3N2 and H1N1. Using an exhaustive search and a nonparametric test for mosaic structure, we identified 315 sequences (∼2%) in five different RNA segments that, after a multiple-comparison correction, had statistically significant mosaic signals compatible with homologous recombination. Of these, only two contained recombinant regions of sufficient length (>100 nucleotides [nt]) that the occurrence of homologous recombination could be verified using phylogenetic methods, with the rest involving very short sequence regions (15 to 30 nt). Although this secondary analysis revealed patterns of phylogenetic incongruence compatible with the action of recombination, neither candidate recombinant was strongly supported. Given our inability to exclude the occurrence of mixed infection and template switching during amplification, laboratory artifacts provide an alternative and likely explanation for the occurrence of phylogenetic incongruence in these two cases. We therefore conclude that, if it occurs at all, homologous recombination plays only a very minor role in the evolution of human influenza A virus.
Influenza A viruses are a major cause of respiratory disease in humans, responsible for 36,000 annual deaths in the United States alone (7, 28) and occasional widespread pandemics associated with much higher levels of mortality and morbidity (27). The viral genome is comprised of eight negative-strand RNA segments, with a combined length of ∼13.6 kb, that can evolve through a variety of mechanisms. Most notably, the lack of a proofreading mechanism during RNA replication results in a high frequency of point mutations which, when combined with large population sizes and short generation times, gives influenza A virus the ability to generate quickly both antigenic variants that can escape host immunity—a process termed antigenic drift (5, 29)—and genotypes that provide resistance to antiviral agents, such as the adamantanes (9) and neuraminidase (NA) inhibitors (2). In addition to generating genetic diversity by rapid mutation, when multiple viruses coinfect a single cell, the eight segments of the influenza virus genome can reassort and yield progeny virions with a novel combination of segments, a process termed antigenic shift. Such reassortment is well documented among those viral strains that differ in their host species, such as humans and birds. Reassortment of this type, involving the acquisition from avian hosts of new polymerase PB1, hemagglutinin (HA), and/or NA segments to which there was no prior human immunity, played a major role in the genesis of the human influenza pandemics of 1957 and 1968 (15, 22). More recently, intrasubtype reassortment has also been shown to occur frequently among cocirculating human H3N2 influenza A viruses (14, 18), which may also impact ongoing antigenic evolution (14). In addition to reassortment among RNA segments, intragenic recombination between different RNA segments, commonly referred to as nonhomologous recombination (3, 20, 25), as well as intragenic recombination between viral RNA and exogenous RNA (16), has been observed and may possibly play a role in determining pathogenicity (25).
More controversial, however, is the occurrence of homologous recombination in influenza viruses, most likely involving copy choice (template-switching) replication of RNA molecules that coinfect a single cell. Although bioinformatic evidence for homologous recombination has been suggested (13, 19), these results remain unsubstantiated, with extensive lineage-specific rate variation a likely source of a false-positive signal for at least some putative recombination events (24, 31). Indeed, because the genomic RNA generated during replication is rapidly packaged with ribonucleoprotein, which acts to prevent the occurrence of template-switching that is central to copy choice replication, homologous RNA recombination is thought to occur rarely, if at all, in both influenza viruses (17) and negative-strand RNA viruses in general (8). In particular, a comprehensive phylogenetic analysis of recombination in negative-sense RNA viruses found only sporadic evidence for recombination, and not among influenza viruses (8), although the process was recently demonstrated in Zaire Ebola virus, an unsegmented negative-sense single-stranded RNA virus (30). If proven to occur, homologous recombination would facilitate two evolutionary processes in influenza virus: the purging of deleterious mutations and the rapid generation of novel genotypes, potentially including new antigenic and drug-resistant variants.
To assess whether homologous recombination has played a role in shaping the genetic diversity of human influenza A virus, we compiled a data set of 13,852 sequences representing all eight RNA segments of isolates of the A/H1N1 and A/H3N2 subtypes. Using an exhaustive search method (4), we statistically assessed the possibility of every potential two-breakpoint homologous-recombination event, considering each sequence as a possible recombinant and searching over all possible parents and all possible breakpoints. In our data set, this translated into considering over 7 billion sequence triplets, where two of the sequences in each triplet are posited to have recombined to form the third sequence in the triplet. For those sequences identified by this method to contain putative recombinant sections longer than 100 nucleotides (nt), we used more stringent phylogenetic methods to further verify that they contained an evolutionary signal (i.e., phylogenetic incongruence) compatible with the action of homologous recombination.
MATERIALS AND METHODS
Sequence data.
Nucleotide sequences of human influenza A virus were obtained from the Influenza Virus Resource (http://www.ncbi.nlm.nih.gov/genomes/FLU/Database/select.cgi) and aligned using MUSCLE (10). Sixteen sets of sequences, two for each RNA segment, were obtained by downloading all of the full-length subtype A/H3N2 and subtype A/H1N1 sequences generated through the NIH/NIAID Influenza Genome Sequencing Project. In addition, two previously published data sets comprising 413 HA and NA segments of human A/H3N2 viruses were also included in the analysis (18). After removing duplicate sequences that were identical at the nucleotide level, a final subset of 10,492 sequences was analyzed.
Recombination analysis.
As an initial screen for possible recombination, each of the 18 data sets was first analyzed using the 3SEQ program (4). 3SEQ tests all possible two-breakpoint recombination events for each triplet of sequences in the data set, assigns a P value (rejecting clonality) to each sequence triplet, and infers breakpoints. Breakpoint pairs are found using a parsimony criterion, with the most likely breakpoint positions being those that minimize the number of mutations between the putative recombinant sequence and a two-breakpoint mosaic of the parental sequences. Breakpoint pairs are reported as ranges of nucleotide sites, since there are multiple pairs of breakpoints that can satisfy this parsimony criterion. 3SEQ reports a P value by calculating the exact probability that this type of recombination signal would be observed under the null hypothesis of clonal (nonrecombinant) evolution. Finally, all P values are corrected with a Dunn-Šidák correction for the large number of triplets tested. If a particular sequence triplet had a corrected P of <0.05, and if the inferred breakpoints guaranteed that the shortest possible recombinant segment was longer than 100 nt (which we deemed suitable for phylogenetic analysis), a secondary phylogenetic analysis of the data was used as an independent verification of putative homologous recombination identified within these sequence triplets. Given that 3SEQ is one of the most powerful methods for detecting recombination (4) and is the only method available that can scan hundreds of sequences at a time and identify the candidate recombinants with breakpoints and P values, it is an appropriate method for detecting recombination in large data sets of influenza A virus. However, although simulations show that 3SEQ is generally robust to false-positive results (4), lineage-specific rate variation can generate apparent recombinants that triplet methods (like 3SEQ) detect as real recombinants.
To minimize the possibility of false-positive results, we performed a secondary phylogenetic analysis of recombination in our data sets of influenza A virus. For each putative recombinant (or set of recombinants with the same breakpoints), the entire data set alignment was divided at the breakpoint positions established by 3SEQ. If two recombination breakpoints were found in a single sequence, the sequence region between the breakpoints was denoted the “minor” region, generated by the minor parent, and the remainder was referred to as the “major” region, generated by the major parent. Because of the very large size of the data sets in this study, initial neighbor-joining phylogenetic trees were inferred using the PAUP* package (26) on either side of the putative breakpoints. If evidence for phylogenetic incongruence was apparent due to a change in the topological positions of specific sequences, a more detailed analysis using maximum likelihood (ML) phylogenetic trees was undertaken. In this case, phylogenetically representative sequences, along with those closely related to the putative recombinants, were selected from the data sets to comprise a final data set of 30 to 40 sequences on which rigorous phylogenetic analyses could be undertaken using the breakpoints determined with 3SEQ. For these analyses, the best-fit model of nucleotide substitution was determined using MODELTEST (21) (details available from the authors on request), and phylogenetic trees were inferred under this model using the ML method available in PAUP* (26), employing tree bisection-reconnection branch swapping in each case. Finally, to assess the degree of support for the differing phylogenetic positions of each putative recombinant, a bootstrap-resampling analysis was undertaken using 1,000 replicate neighbor-joining trees inferred under the best-fit substitution model.
RESULTS AND DISCUSSION
Two of the 10 human influenza A/H3N2 virus data sets (PB2 and NP) analyzed here contained sequences with statistically significant mosaic structure, as determined by 3SEQ, and with putative recombinant sections that were each sufficiently long (>100 nt) that they could be reanalyzed by phylogenetic recombination detection methods. Three of the remaining eight A/H3N2 data sets (PA, NA, and MP) and one of the A/H1N1 data sets (NA) also resulted in 3SEQ P values that revealed a strong signal of mosaicism, but in all these cases, the inferred breakpoints were either close to the gene segment's endpoints or very close to each other, making it impossible to infer a credible phylogeny. The remaining five A/H3N2 data sets (PB1, HA, HA413, NS, and NA413 [the 413 suffix means that it is the HA or NA data set containing 413 sequences]) and seven of the A/H1N1 data sets (PB2, PB1, PA, HA, NP, MP, and NS) did not contain any statistically significant mosaic signals that survived a Dunn-Šidák correction in 3SEQ. The recombination analysis results are summarized in Table 1 for A/H3N2 and Table 2 for A/H1N1. The two putative recombinant data sets are discussed in more detail below.
TABLE 1.
Results of recombination analysis of 10 A/H3N2 influenza virus data sets
| Segment | No. of sequences (no. distinct) | Alignment length (nt) | No. of variable sites | 3SEQ P value | No. “recombinant” | No. “recombinant” >100 nt | Phylogenetic signal for recombination |
|---|---|---|---|---|---|---|---|
| PB2 | 1,086 (912) | 2,347 | 898 | 10−3 | 24 | 1 | Weak |
| PB1 | 878 (715) | 2,341 | 849 | 0.49 | 0 | 0 | No |
| PA | 1,365 (1,156) | 2,242 | 947 | 2.6 × 10−6 | 28 | 0 | No |
| HA | 1,365 (1,154) | 1,772 | 918 | 1 | 0 | 0 | No |
| 413 (336) | 1,711 | 518 | 1 | 0 | 0 | No | |
| NP | 1,256 (938) | 1,570 | 620 | 2.9 × 10−6 | 6 | 1 | Weak |
| NA | 1,365 (1,059) | 1,475 | 754 | 1.2 × 10−10 | 240 | 0 | No |
| 413 (274) | 1,407 | 438 | 1 | 0 | 0 | No | |
| MP | 1,250 (682) | 1,028 | 366 | 0.04 | 1 | 0 | No |
| NS | 630 (344) | 906 | 318 | 1 | 0 | 0 | No |
TABLE 2.
Results of the recombination analysis of eight A/H1N1 influenza virus data sets
| Segment | No. of sequences (no. distinct) | Alignment length (nt) | No. of variable sites | 3SEQ P value | No. “recombinant” | No. “recombinant” >100 nt | Phylogenetic signal for recombination |
|---|---|---|---|---|---|---|---|
| PB2 | 478 (421) | 2,346 | 871 | 1 | 0 | 0 | No |
| PB1 | 478 (399) | 2,341 | 870 | 0.95 | 0 | 0 | No |
| PA | 478 (380) | 2,238 | 776 | 0.96 | 0 | 0 | No |
| HA | 482 (440) | 1,781 | 804 | 0.82 | 0 | 0 | No |
| NP | 478 (332) | 1,566 | 544 | 1 | 0 | 0 | No |
| NA | 481 (405) | 1,464 | 657 | 10−5 | 16 | 0 | No |
| MP | 478 (248) | 1,027 | 296 | 1 | 0 | 0 | No |
| NS | 478 (297) | 890 | 336 | 1 | 0 | 0 | No |
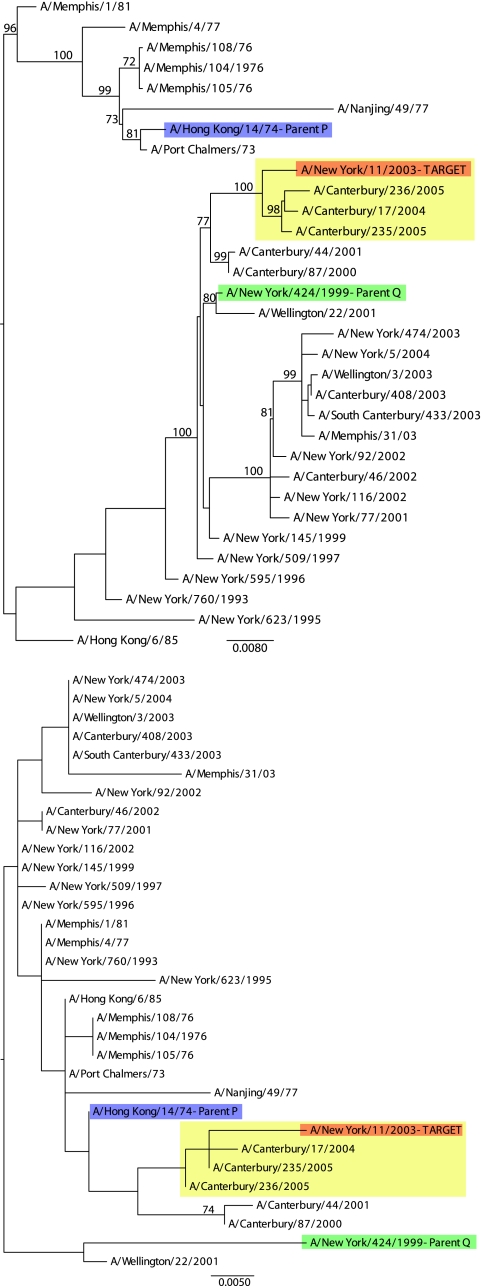
The H3N2 PB2 data set assembled here contained 912 distinct sequences, one of which, A/New York/11/2003, statistically supported a mosaic structure with both mosaic regions longer than 100 nt. The two most likely parental sequences, identified as A/Hong Kong/14/1974 (major parent) and A/New York/424/1999 (minor parent), revealed a strong mosaic signal (corrected P = 0.013) in relation to A/New York/11/2003. However, while the phylogenies inferred for the minor (positions 202 to 2189) (Fig. 1, top) and major (positions 1 to 201 and 2190 to 2347) (Fig. 1, bottom) segments revealed topological movement of the putative recombinant sequence relative to the parental sequences, a general lack of phylogenetic resolution, reflected in low levels of bootstrap support (particularly in the major segment), meant that there was insufficient signal to infer phylogenetic incongruence. Since support for phylogenetic incongruence is necessarily made up of two components, the phylogenetic relationships among the parents and recombinant on the major and minor trees, we called the signal “weak” if one of the components received only low bootstrap support.
FIG. 1.
ML phylogenetic trees for nucleotide positions 202 to 2189 (minor segment) (top) and 1 to 201 and 2190 to 2347 (major segment) (bottom) of the PB2 data set of 1086 A/H3N2 sequences. All bootstrap values greater than 50% are shown. The tree is midpoint rooted for purposes of clarity only, and all branch lengths are drawn to a scale of nucleotide substitutions per site. The red sequence is the candidate recombinant sequence, with the yellow box identifying three additional sequences that cluster with A/New York/11/2003 in both phylogenies. The blue sequence is the candidate major parent (“Parent P”), and the green sequence is the candidate minor parent (“Parent Q”).
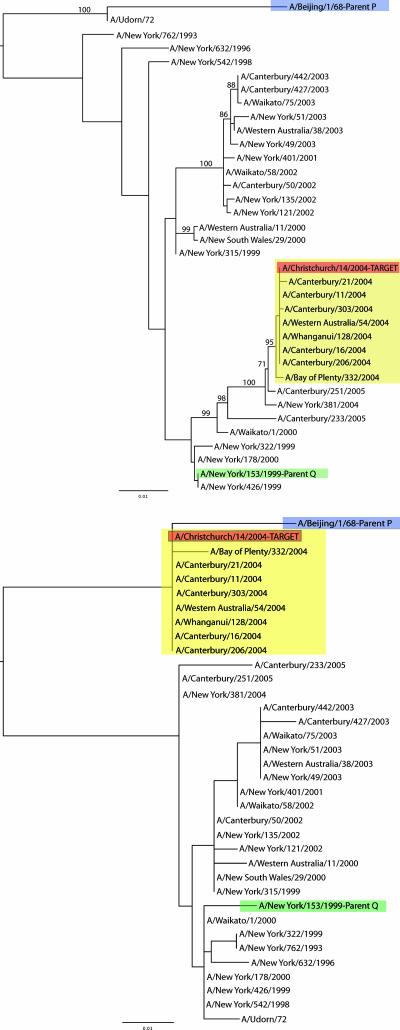
For the NP data set of A/H3N2 viruses, a single sequence, A/Christchurch/14/2004, supported a mosaic structure with both candidate recombinant regions longer than 100 nt. The candidate parental sequences identified by 3SEQ were A/Beijing/1/1968 as the major parent and A/New York/153/1999 as the minor parent (clonality among these three isolates was rejected at a corrected P of 0.032). The ML tree for the region 98 to 1454 is presented in Fig. 2, top, while that for regions 1 to 97 and 1455 to 1570 is shown in Fig. 2, bottom. In these phylogenies, the putative recombinant sequence was clearly more closely related to a different parent in each sequence region. The phylogenies also revealed sequence A/New York/381/2004 as a better candidate for the minor parent than A/New York/153/1999; the mosaic signal when assuming A/New York/381/2004 as the minor parent in the recombination event was still strong (corrected P = 0.052). However, as in the PB2 data set, the lack of bootstrap support in the phylogeny inferred for the major segment indicates that there is in reality an insufficiently strong signal for phylogenetic incongruence to conclude that homologous recombination has occurred.
FIG. 2.
ML phylogenetic trees for nucleotide positions 98 to 1454 (minor segment) (top) and 1 to 97 and 1455 to 1570 (major segment) (bottom) of the NP data set of 1,256 A/H3N2 sequences. All bootstrap values greater than 50% are shown. The tree is midpoint rooted for purposes of clarity only, and all branch lengths are drawn to a scale of nucleotide substitutions per site. The red sequence is the candidate recombinant sequence, with the yellow box identifying eight additional sequences that cluster with A/Christchurch/14/2004 in both phylogenies. The blue sequence is the candidate major parent (“Parent P”), and the green sequence is the candidate minor parent (“Parent Q”).
For the two candidate recombinants, A/New York/11/2003 (PB2) and A/Christchurch/14/2004 (NP), it is also puzzling that the parental sequences were sampled 25 and 31 years apart, respectively. Hence, for one of these recombination events to have occurred, a lineage of viruses closely related to an “archaic” virus (either A/Hong Kong/14/1974 or A/Beijing/1/1968) must have circulated until at least 1999 and recombined with A/New York/424/1999 or A/New York/153/1999. Given the rapid rate of influenza A virus mutation through frequent RNA polymerase errors, as well as the rapid lineage turnover driven by positive selection on the major antigenic proteins (6, 11, 12, 23), this scenario seems extremely unlikely. Thus, laboratory error, such as template switching during amplification in a mixed or contaminated sample, is a likely explanation of these apparent homologous recombination events.
In sum, our study has revealed that no sequence of human influenza A virus contains a clear signature of phylogenetic incongruence indicative of the action of homologous RNA recombination. Given that more than 10,000 distinct sequences were analyzed, this constitutes strong evidence that homologous recombination plays only a very minor role, if any, in the evolution of human influenza A virus. More generally, the occurrence of phylogenetic incongruence does not in itself constitute conclusive evidence for this process. Specifically, because our analysis is necessarily based on viral consensus sequences rather than the myriad individual viral molecules that characterize any infection, it is equally plausible that the “recombinants” detected here in fact represent cases of mixed infection in individual hosts, followed by the amplification and sequencing of different viral molecules, thereby producing laboratory-generated artificial recombinants. Hence, to demonstrate conclusively the occurrence of homologous recombination in influenza A virus, it will be necessary either to clone (or plaque purify) and sequence multiple viral genomes from an individual host and demonstrate the presence of the recombinant and both parental genotypes within the sample (1) or to show that recombinant sequences form a distinct circulating lineage, with readily identifiable parents, that is transmitted among multiple individuals in a population (30).
Finally, although there were 315 sequences in the data analyzed here that carried a strong mosaic signal as identified by 3SEQ, it was impossible to verify the vast majority of these as recombinants, since the putative recombinant regions were too short to infer a credible phylogenetic history. It is therefore possible that homologous recombination, should it occur in influenza A virus, more commonly involves the transfer of very short sections of RNA, a process that would be undetectable by the majority of other methods devised to detect recombination. If homologous recombination of short segments is determined to be a relevant process in influenza A virus evolution, the basis of our more frequent observation of mosaicism in A/H3N2 viruses compared to A/H1N1 viruses will need to be investigated further. However, by far the strongest signal in the influenza A virus sequence data analyzed here is that of strict clonality, supporting most models of influenza virus evolution proposed to date.
Acknowledgments
The research undertaken in this study was funded in part by Resources for the Future (M.F.B.), NIH/NIGMS grant P50GM071508 (M.F.B.), National Institutes of Health grant GM28016 (M.F.B.), NIH grant number GM080533-01 (E.C.H.), and the Intramural Research Program of the NIH and the NIAID (J.K.T.).
We thank John Zollweg and Linda Woodard at the Cornell University Center for Advanced Computing for suggesting algorithmic improvements to 3SEQ, as well as two anonymous reviewers for helpful suggestions.
Footnotes
Published ahead of print on 19 March 2008.
REFERENCES
- 1.Aaskov, J., K. Buzacott, E. Field, K. Lowry, A. Berlioz-Arthaud, and E. C. Holmes. 2007. Multiple recombinant dengue type 1 viruses in an isolate from a dengue patient. J. Gen. Virol. 883334-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoki, F. Y., G. Boivin, and N. Roberts. 2007. Influenza virus susceptibility and resistance to oseltamivir. Antivir. Ther. 12603-616. [PubMed] [Google Scholar]
- 3.Bergmann, M., A. Garcia-Sastre, and P. Palese. 1992. Transfection-mediated recombination of influenza A virus. J. Virol. 667576-7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boni, M. F., D. Posada, and M. W. Feldman. 2007. An exact nonparametric method for inferring mosaic structure in sequence triplets. Genetics 1761035-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Both, G. W., M. J. Sleigh, N. J. Cox, and A. P. Kendal. 1983. Antigenic drift in influenza virus H3 hemagglutinin from 1968 to 1980: multiple evolutionary pathways and sequential amino acid changes at key antigenic sites. J. Virol. 4852-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bush, R. M., W. M. Fitch, C. A. Bender, and N. J. Cox. 1999. Positive selection on the H3 hemagglutinin gene of human influenza virus A. Mol. Biol. Evol. 161457-1465. [DOI] [PubMed] [Google Scholar]
- 7.CDC. 2007. Deaths: final data for 2004. National Vital Statistics Reports 55. National Center for Health Statistics, Hyattsville, MD. [PubMed]
- 8.Chare, E. R., E. A. Gould, and E. C. Holmes. 2003. Phylogenetic analysis reveals a low rate of homologous recombination in negative-sense RNA viruses. J. Gen. Virol. 842691-2703. [DOI] [PubMed] [Google Scholar]
- 9.Deyde, V. M., X. Xu, R. A. Bright, M. Shaw, C. B. Smith, Y. Zhang, Y. Shu, L. V. Gubareva, N. J. Cox, and A. I. Klimov. 2007. Surveillance of resistance to adamantanes among influenza A(H3N2) and A(H1N1) viruses isolated worldwide. J. Infect. Dis. 196249-257. [DOI] [PubMed] [Google Scholar]
- 10.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 321792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferguson, N. M., A. P. Galvani, and R. M. Bush. 2003. Ecological and immunological determinants of influenza evolution. Nature 422428-433. [DOI] [PubMed] [Google Scholar]
- 12.Fitch, W. M., R. M. Bush, C. A. Bender, and N. J. Cox. 1997. Long term trends in the evolution of H(3) HA1 human influenza type A. Proc. Natl. Acad. Sci. USA 947712-7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibbs, M. J., J. S. Armstrong, and A. J. Gibbs. 2001. Recombination in the hemagglutinin gene of the 1918 “Spanish Flu”. Science 2931842. [DOI] [PubMed] [Google Scholar]
- 14.Holmes, E. C., E. Ghedin, N. Miller, J. Taylor, Y. M. Bao, K. St George, B. T. Grenfell, S. L. Salzberg, C. M. Fraser, D. J. Lipman, and J. K. Taubenberger. 2005. Whole-genome analysis of human influenza A virus reveals multiple persistent lineages and reassortment among recent H3N2 viruses. PLoS Biol. 3e300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawaoka, Y., S. Krauss, and R. G. Webster. 1989. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J. Virol. 634603-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khatchikian, D., M. Orlich, and R. Rott. 1989. Increased viral pathogenicity after insertion of a 28 S ribosomal RNA sequence into the haemagglutinin gene of an influenza virus. Nature 340156-157. [DOI] [PubMed] [Google Scholar]
- 17.Kilbourne, E. D. 1978. Molecular epidemiology—influenza as archetype. Harvey Lectures 73225-258. [PubMed] [Google Scholar]
- 18.Nelson, M. I., L. Simonsen, C. Viboud, M. A. Miller, J. Taylor, K. S. George, S. B. Griesemer, E. Ghedin, N. A. Sengamalay, D. J. Spiro, I. Volkov, B. T. Grenfell, D. J. Lipman, J. K. Taubenberger, and E. C. Holmes. 2006. Stochastic processes are key determinants of short-term evolution in influenza A virus. PLoS Pathog. 2e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niman, H. 2007. Swine influenza A evolution via recombination—genetic drift reservoir. Nat. Precedings http://hdl.handle.net/10101/npre.2007.385.1.
- 20.Orlich, M., H. Gottwald, and R. Rott. 1994. Nonhomologous recombination between the hemagglutinin gene and the nucleoprotein gene of an influenza virus. Virology 204462-465. [DOI] [PubMed] [Google Scholar]
- 21.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14817-818. [DOI] [PubMed] [Google Scholar]
- 22.Scholtissek, C., W. Rohde, V. Von Hoyningen, and R. Rott. 1978. On the origin of the human influenza virus subtypes H2N2 and H3N2. Virology 8713-20. [DOI] [PubMed] [Google Scholar]
- 23.Smith, D. J., A. S. Lapedes, J. C. de Jong, T. M. Bestebroer, G. F. Rimmelzwaan, A. D. M. E. Osterhaus, and R. A. M. Fouchier. 2004. Mapping the antigenic and genetic evolution of influenza virus. Science 305371-376. [DOI] [PubMed] [Google Scholar]
- 24.Strimmer, K., K. Forslund, B. Holland, and V. Moulton. 2003. A novel exploratory method for visual recombination detection. Genome Biol. 4R33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suarez, D. L., D. A. Senne, J. Banks, I. H. Brown, S. C. Essen, C. W. Lee, R. J. Manvell, C. Mathieu-Benson, V. Moreno, and J. C. Pedersen. 2004. Recombination resulting in virulence shift in avian influenza outbreak, Chile. Emerg. Infect. Dis. 10693-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swofford, D. L. 2003. PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, MA.
- 27.Taubenberger, J. K., and D. M. Morens. 2006. 1918 influenza: the mother of all pandemics. Emerg. Infect. Dis. 1215-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson, W. W., D. K. Shay, E. Weintraub, L. Brammer, N. Cox, L. J. Anderson, and K. Fukuda. 2003. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 289179-186. [DOI] [PubMed] [Google Scholar]
- 29.Webster, R. G., W. J. Bean, O. T. Gorman, T. M. Chambers, and Y. Kawaoka. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56152-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wittmann, T. J., R. Biek, A. Hassanin, P. Rouquet, P. Reed, P. Yaba, X. Pourrut, L. A. Real, J.-P. Gonzalez, and E. M. Leroy. 2007. Isolates of Zaire ebolavirus from wild apes reveal genetic lineage of recombinants. Proc. Natl. Acad. Sci. USA 10417123-17127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Worobey, M., A. Rambaut, O. G. Pybus, and D. L. Robertson. 2002. Questioning the evidence for genetic recombination in the 1918 “Spanish Flu” virus. Science 296211. [DOI] [PubMed] [Google Scholar]