Abstract
The cellular ESCRT pathway functions in membrane remodeling events that accompany endosomal protein sorting, cytokinesis, and enveloped RNA virus budding. In the last case, short sequence motifs (termed late domains) within human immunodeficiency virus type 1 (HIV-1) p6Gag bind and recruit two ESCRT pathway proteins, TSG101 and ALIX, to facilitate virus budding. We now report that overexpression of the HECT ubiquitin E3 ligase, NEDD4L/NEDD4-2, stimulated the release of HIV-1 constructs that lacked TSG101- and ALIX-binding late domains, increasing infectious titers >20-fold. Furthermore, depletion of endogenous NEDD4L inhibited the release of these crippled viruses and led to cytokinesis defects. Stimulation of virus budding was dependent upon the ubiquitin ligase activity of NEDD4L and required only the minimal HIV-1 Gag assembly regions, demonstrating that Gag has ubiquitin-dependent, cis-acting late domain activities located outside of the p6 region. NEDD4L stimulation also required TSG101 and resulted in ubiquitylation of several ESCRT-I subunits, including TSG101. Finally, we found that TSG101/ESCRT-I was required for efficient release of Mason-Pfizer monkey virus, which buds primarily by using a PPXY late domain to recruit NEDD4-like proteins. These observations suggest that NEDD4L and possibly other NEDD4-like proteins can ubiquitylate and activate ESCRT-I to function in virus budding.
The structural proteins of many enveloped RNA viruses, including human immunodeficiency virus type 1 (HIV-1) Gag, contain short cis-acting sequence motifs, termed “late domains,” that recruit host factors to facilitate virus budding (5, 29, 35, 59). To date, three well-characterized late-domain sequences and their cellular binding partners have been described: PT(S)AP late domains bind TSG101 (6, 12, 31, 57), YPXL late domains bind ALIX/AIP1 (51) (as well as the AP-2 adaptor complex [2]), and PPXY late domains bind various members of the large family of mammalian NEDD4 ubiquitin E3 ligases (hereafter called NEDD4-like proteins; reviewed in references 22, 29, and 46). Each of the late domain binding partners can be linked to the cellular ESCRT (endosomal sorting complex required for transport) pathway, indicating that this pathway functions in virus release.
Within the cell, the ESCRT pathway also plays important roles in helping to sort membrane proteins into vesicles that bud into late endosomes/multivesicular bodies (MVB) (21, 38, 61). These vesicles carry membrane proteins from the limiting endosomal membrane into the endosomal lumen, where their contents can ultimately be degraded when MVBs fuse with lysosomes. The ESCRT pathway can also be recruited to function in the abscission stage of cytokinesis (1), a process in which the thin membranous midbody that connects two dividing cells is severed to complete the process of cell division. In addition to their shared dependence on the ESCRT pathway, the processes of MVB vesicle formation, abscission, and virus budding are similar in that the membrane fission events required for vesicle/cell/virus release must be catalyzed from the cytoplasmic face of the bud neck. Thus, these membrane remodeling events share important mechanistic requirements that may be provided by the ESCRT machinery.
In both humans and yeast, most ESCRT pathway proteins function as subunits of one of four different multiprotein complexes, termed ESCRT-0, -I, -II, and -III. In humans, TSG101 functions as the unique central subunit of the heterotetrameric ESCRT-I complex. The N-terminal UEV domain of TSG101 binds directly to viral PTAP late domains (40), and this interaction thereby recruits ESCRT-I to function in virus release. Similarly, ALIX, though not a constitutive member of any ESCRT complex, binds directly to viral YPXL late domains (51, 64) and thereby links viral proteins to the CHMP4 subset of cellular ESCRT-III proteins (10, 55). Thus, PTAP and YPXL late domains connect directly to known ESCRT complexes.
Connections between PPXY viral late domains and the ESCRT pathway are less firmly established, although several lines of evidence link NEDD4-like proteins to the ESCRT pathway, including the following. (i) The sole NEDD4-like protein in yeast, Rsp5p, is required for ESCRT pathway function (25) and is responsible for ubiquitylation, internalization, and vacuolar degradation of various membrane proteins, including the α-factor receptor Ste2p, the amino acid permease Gap1p, and the uracil permease Fur4p (8, 11, 19, 48). (ii) Mammalian membrane proteins can also be bound, ubiquitylated, and targeted for lysosomal degradation by NEDD4-like proteins. For example, the mammalian NEDD4 and NEDD4L E3 ligases bind directly to PPXY sequences on the epithelial Na+ channel, ubiquitylate the protein, and thereby direct its lysosomal degradation (4, 17, 49). (iii) Several human NEDD4-like proteins colocalize together with other ESCRT pathway components at endosomal compartments, and this colocalization requires their C-terminal catalytic E3 HECT (homologous to E6-associated protein C-terminus) domains, indicating that there must be physical interaction(s) between this domain and another ESCRT pathway factor(s) (30). These interactions have not yet been characterized, however, and PPXY viral late domains therefore cannot yet be tied directly to the ESCRT pathway. Furthermore, these analyses are complicated by the complexity (and possible redundancy) of the mammalian NEDD4-like proteins and also by uncertainties that surround the role(s) and target(s) of their ubiquitin E3 ligase activities.
Many enveloped RNA viruses contain multiple copies of different late domains (5, 29, 35, 59). For example, the Gag protein of the murine leukemia virus contains binding sites for TSG101, ALIX, and NEDD4-like E3 ubiquitin ligases, all of which can stimulate virus budding, albeit to different extents (45). Similarly, the HIV-1 Gag p6 region contains functional binding sites for both TSG101 and ALIX. Although the different late domains typically differ in functional importance, their retention and conservation implies that even “auxiliary” late domains must provide viruses with an evolutionary advantage. In principle, such advantages could include synergistic interactions between the different late-domain binding partners and/or the capacity to replicate in multiple cell types that express different levels of late-domain partners. It is therefore of interest to screen for additional viral late-domain activities, even in viruses, such as HIV-1, where a dominant late domain (PTAP) has already been identified.
A number of lines of evidence indicate that efficient virus budding through the PPXY and PTAP (but not YPXL) late domains is also dependent upon ubiquitin transfer (15, 36, 37, 42, 44, 47, 50, 52; for reviews, see references 58 and 29). For PPXY late domains, this dependence can apparently be explained, at least in part, by the ubiquitin E3 ligase activity of their NEDD4-like binding partners (30, 56). In contrast, it is not clear why viruses like HIV-1 that bud primarily through PTAP/ESCRT-I interactions also require ubiquitin transfer. We therefore sought to identify and characterize ubiquitin E3 ligases that can function in HIV-1 budding, with the long-term goal of understanding the ubiquitin dependence of HIV-1 release.
MATERIALS AND METHODS
Cell cultures.
293T and HeLa cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum.
Expression constructs for HIV-1 and NEDD4-like proteins.
NEDD4-like proteins were expressed from pCIneo vectors (Promega) as N-terminally FLAG-tagged proteins, and these constructs are summarized in Table S1 in the supplemental material. The parental HIV-1NL4-3 R9 proviral vector and HIV-1ΔPTAP and HIV-1ΔPTAP/ΔYP derivatives have been described (10). HIV-1 Gag expression plasmids used for virus-like particle (VLP) production used the pcDNA3.1.myc.His.B(−) backbone (see Table S1 in the supplemental material). Briefly, HIV-1HXB2 GagΔPTAP and GagΔp6 were made by amplifying/mutating the appropriate coding sequences from the codon-optimized HIV-1 Gag expression construct pGag-GFP (20) (obtained as a generous gift from Marilyn Resh, Sloan-Kettering) and subcloned into the EcoRI and BamHI sites of the pcDNA3.1.myc.His.B(−) vector (Invitrogen). Constructs were confirmed by DNA sequencing. Constructs expressing HIV-1HXB2 Gag with the CREB leucine zipper in place of NC were obtained by PCR amplification of the CREB leucine zipper coding sequence from HIVgptWTZIP (65) (obtained as a generous gift from Eric Barklis, Oregon Health Sciences Center) and subcloning back into pcDNA.Gag. The resulting junction sequence (between HIV-1 Gag and the CREB leucine zipper) is GCT ACC ATA ATG GCT CGA GAG TGT CGT (HIV-1 sequences are in plain font, the leucine zipper sequences are in italics, and an XhoI site is in bold). The construct for minimal HIV-1HXB2 GagΔ-ZIP expressed a fusion protein composed of the N-terminal 7 amino acids of MA, Gag residues 278 to 377, and the CREB leucine zipper. Full cloning details are available upon request. A vector expressing a mutant MVB12B protein with a 172PPQY175AAAA mutation was created using the megaprimer method, using the parental MVB12B expression vector as a template (33).
Assays for virion release and infectivity.
To assay the effects of different NEDD4-like E3 ligases on virion release and infectivity, ∼8 × 105 293T cells/well (6-well plates) were cotransfected with 1 μg of HIV-1 or Gag expression plasmids and 0.5 to 4 μg of each NEDD4-like E3 ligase expression plasmid using Lipofectamine 2000 (Invitrogen). To equalize protein expression levels, the experiment shown in Fig. 1B used 4 μg AIP4 and AIP5 expression vectors and 2 μg of all other NEDD4-like expression vectors. Virions were harvested 24 h (HIV-1ΔPTAP and GagΔPTAP) or 48 h (other Gag constructs) posttransfection, centrifuged through a 20% sucrose cushion, and analyzed by Western blotting using a polyclonal rabbit anti-CA antibody (1:15,000). Note that 3× more sample was loaded for the GagΔ-ZIP construct (see Fig. 5D), because anti-CA antibodies detected this sample less efficiently. Band intensities were quantified using an Odyssey scanner (Li-Cor Biosciences).
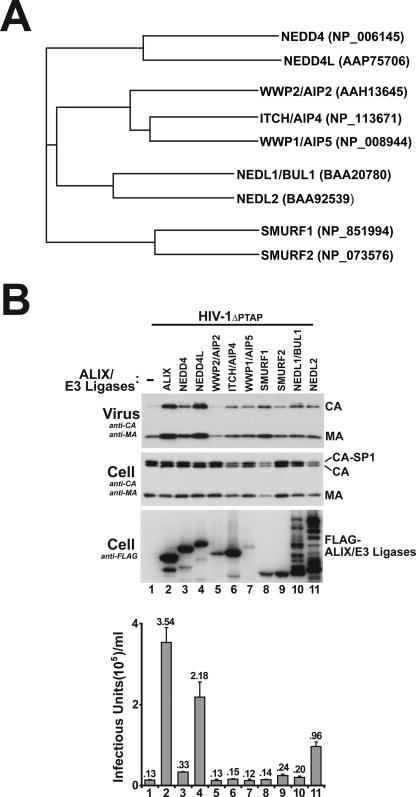
FIG. 1.
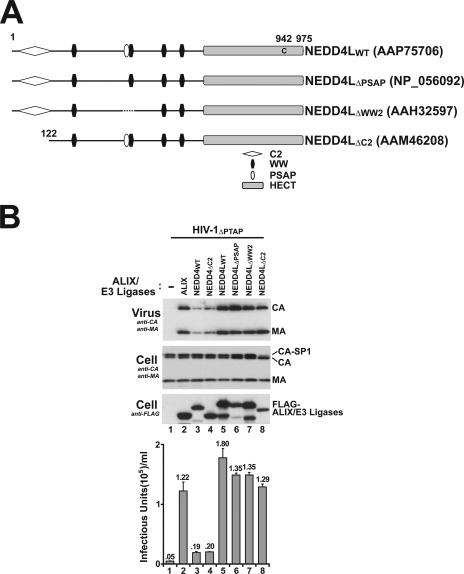
NEDD4L overexpression stimulates HIV-1ΔPTAP release and infectious titers. (A) Phylogenetic tree of nine different NEDD4-like E3 ubiquitin ligases showing the four different subclasses and their members. NCBI accession numbers for the different proteins used in the alignment are given at right (in parentheses). (B) Levels of HIV-1ΔPTAP release and titers upon coexpression with an empty expression vector (−; lanes 1) or with vectors expressing FLAG-tagged ALIX (lanes 2) or FLAG-tagged NEDD-like E3 ubiquitin ligases (lanes 3 to 11). Western blots show virion-associated levels of CAGag and MAGag proteins released into the supernatant (panel 1), cellular Gag protein levels (panel 2, anti-CA and -MA), and levels of exogenous, overexpressed ALIX or NEDD4-like E3 protein (panel 3, anti-FLAG). Viral titers (panel 4) were measured in single-cycle MAGIC assays (n = 3 ± standard deviation). Note that cellular levels of CAGag and MAGag were similar in all cases (panel 2) and that the expression levels of exogenous ALIX and NEDD4-like proteins (panel 3) were also similar in all cases except that of WWP1 (lanes 7, low expression levels) and NEDL1 and -2 (lanes 10 and 11, evidence of protein degradation).
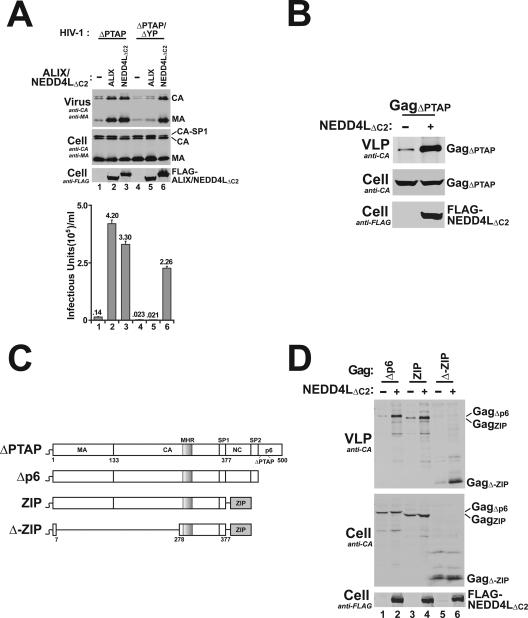
FIG. 5.
Gag278-377 is sufficient for NEDD4L-dependent HIV-1ΔPTAP release. (A) The ALIX binding site in p6Gag is not required for NEDD4L-dependent HIV-1ΔPTAP release. Lanes 1 to 3 show levels of HIV-1ΔPTAP release and titers upon cotransfection with an empty expression vector (−) or with an expression vector for ALIX or NEDD4LΔC2. Lanes 4 to 6 show levels of HIV-1ΔPTAP/ΔYP release and titers upon coexpression with an empty expression vector (−) or with an expression vector for ALIX or NEDD4LΔC2. Note that the Gag protein expressed by the HIV-1ΔPTAP/ΔYP proviral construct cannot bind ALIX (10) and that NEDD4LΔC2 overexpression stimulated HIV-1ΔPTAP/ΔYP release and infectivity whereas ALIX overexpression did not (compare lanes 6 to lanes 4 and 5). (B) NEDD4LΔC2 overexpression stimulates release of VLPs formed by the HIV-1 GagΔPTAP protein alone. Western blotting analyses show an expression vector for HIV-1HXB2 Gag (GagΔPTAP) cotransfected with an empty expression vector (lane 1) or with the NEDD4LΔC2 expression vector. Panel 1 shows VLP-associated Gag protein release (anti-CA), panel 2 shows cellular Gag protein levels (anti-CA), and panel 3 shows levels of exogenously expressed NEDD4LΔC2 (anti-FLAG). Note that NEDD4LΔC2 overexpression also stimulates GagΔPTAP VLP release (compare lane 2 to 1 in panel 1). (C) Schematic illustration of the Gag constructs used to define the minimal NEDD4L-responsive region. GagΔPTAP has a 7LIRL10 mutation in place of the 7PTAP10 late domain of p6Gag. GagΔp6 lacks the entire p6 region. GagZIP lacks the SP2 and p6 regions and has the CREB leucine zipper in place of the NC region. GagΔ-ZIP contains only an N-terminal membrane binding/myristoylation signal, the C-terminal domain of the CA domain (including the major homology region), and the SP1 spacer (Gag residues 278 to 377) fused to the CREB leucine zipper. (D) Gag278-377 is sufficient for NEDD4L-dependent VLP release. The indicated Gag expression constructs were cotransfected either with an empty expression vector (−) (lanes 1, 3, and 5) or with the NEDD4LΔC2 expression vector (lanes 2, 4, and 6). VLP production, cellular Gag protein levels, and exogenous NEDD4LΔC2 levels were analyzed as described above for part B. Note that NEDD4LΔC2 overexpression increased the levels of VLP production for all three Gag constructs (compare lanes 1 and 2, 3 and 4, and 5 and 6).
NEDD4L depletion.
Twenty-one-nucleotide RNA duplexes with symmetric 2-nucleotide 3′ (2′-deoxy)T overhangs corresponding to NEDD4LWT coding nucleotides 1888 to 2006 (GAGUCCUAUCGGAGAAUUA) were synthesized and annealed. Fifty nanomolar small interfering RNA (siRNA) was transfected twice into 293T or HeLa cells (24-h intervals) using Lipofectamine 2000. For the experiments shown in Fig. 4, 1 μg of the proviral HIV-1ΔPTAP expression plasmid was cotransfected with the second siRNA sample and virus was harvested 24 h later. NEDD4L levels were analyzed by Western blotting using affinity-purified rabbit anti-NEDD4L antibody raised by Covance Inc. against a pure recombinant polypeptide corresponding to NEDD4LΔWW2 residues 227 to 409 (UT517; 1:500 dilution). The major endogenous NEDD4L isoform shown in Western blots (see Fig. 4 and 7B; molecular mass = ∼125 kDa) was larger than the NEDD4LWT isoform (molecular mass = 112 kDa). Several smaller endogenous NEDD4L isoforms/degradation products were also observed and were depleted as efficiently as the largest NEDD4L isoform (see Fig. S1 in the supplemental material). HIV-1ΔPTAP release and titers were assayed as described above.
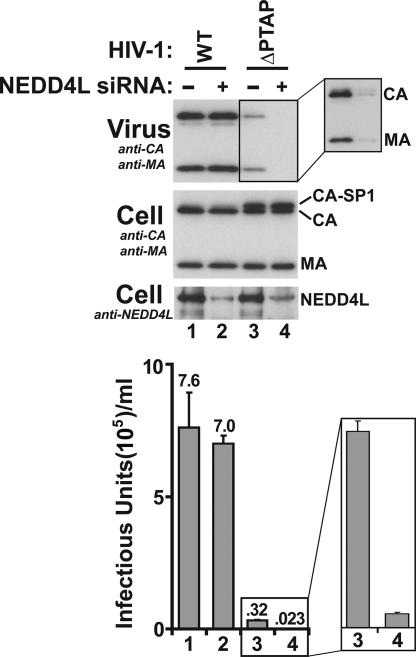
FIG. 4.
Requirement for endogenous NEDD4L in HIV-1 release. Expression vectors for wild-type (WT) HIV-1 (lanes 1 and 2) or HIV-1ΔPTAP (ΔPTAP) (lanes 3 and 4) were cotransfected with an irrelevant control siRNA (lanes 1 and 3) or with an siRNA targeting endogenous NEDD4L (lanes 2 and 4, respectively). Levels of the endogenous NEDD4L protein were analyzed by Western blotting (panel 3; anti-NEDD4L). HIV-1 release, cellular Gag protein levels, and viral titers were analyzed as described in the legend to Fig. 1 (n = 3 ± standard deviation). Quantitative analyses revealed than >90% of the endogenous NEDD4L was depleted by the siRNA treatment. Note that smaller NEDD4L isoforms/degradation were also comparably reduced by siRNA treatment (compare lanes 2 and 4 to lanes 1 and 3 in panel 3; also see Fig. S1 in the supplemental material). Note also that siRNA depletion of endogenous NEDD4L reduced HIV-1ΔPTAP release and infectivity 14-fold (compare lanes 4 to lanes 3). Expansions show enhanced exposure of the Western blot (upper panel) and a vertical expansion of the infectivity data from lanes 3 and 4.
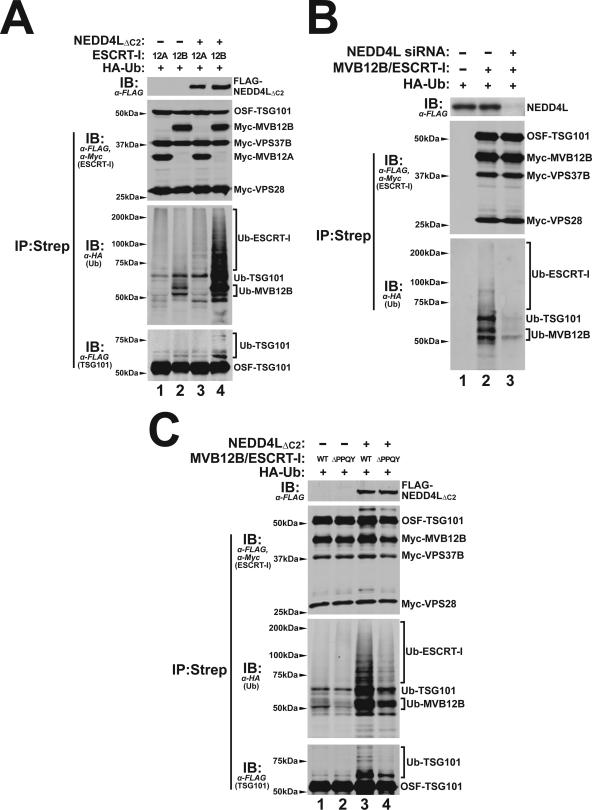
FIG. 7.
NEDD4L stimulation of ESCRT-I ubiquitylation. (A) NEDD4LΔC2 overexpression stimulates ubiquitylation of ESCRT-I complexes that contain MVB12B subunits. HA-tagged ubiquitin (HA-Ub) was co-overexpressed together with ESCRT-I complexes containing MVB12A (lanes 1 and 3) or MVB12B (lanes 2 and 4) subunits with empty vector controls (lanes 1 and 2) or with NEDD4LΔC2 (lanes 3 and 4). Panel 1 shows a Western blot of the input extract, and panels 2 to 4 show Western blots of ESCRT-I complexes that were purified by incubating with Strep-Tactin Sepharose (which bound the OSF-TSG101 subunit). Panel 2 shows the four purified ESCRT-I subunits, with anti-FLAG (α-FLAG; detecting OSF-TSG101) and anti-Myc (α-Myc; detecting other ESCRT-I subunits); panel 3 shows ubiquitylated proteins that copurified with the ESCRT-I complex (α-HA [anti-HA]). Panel 4 shows an overexpressed blot probed for OSF-TSG101 (anti-FLAG). Note that NEDD4LΔC2 overexpression dramatically increased ubiquitylation levels of ESCRT-I complexes that contained MVB12B subunits (compare lanes 2 and 4, lower two panels) but not of ESCRT-I complexes that contained MVB12A subunits (lanes 1 and 3). Note also that that the ESCRT-I complexes containing MVB12B had higher basal ubiquitylation levels than ESCRT-I complexes that contained MVB12A (compare lanes 1 and 2) and that ubiquitin modifications could be detected on TSG101 (panel 4, compare lanes 3 and 4) and on MVB12B (not shown). (B) Depletion of endogenous NEDD4L reduces the basal levels of MVB12B/ESCRT-I ubiquitylation. HA-tagged ubiquitin was cotransfected with empty control vectors (lanes 1) or with expression vectors for the four subunits of ESCRT-I (OSF-TSG101, Myc-MVB12B, and Myc-VPS37B; lanes 2 and 3). 293T cells used in the experiments shown in lanes 1 and 2 were expressing wild-type levels of endogenous NEDD4L, whereas the 293T cells used in the experiment shown in lanes 3 were depleted of NEDD4L by siRNA treatment. Panels 1 to 3 correspond to the analogous three panels in part A, except that an anti-NEDD4L antibody was used to detect endogenous NEDD4L in panel 1. Note that NEDD4L depletion decreased MVB12B/ESCRT-I ubiquitylation to nearly undetectable levels (compare lane 2 to lane 3 in panel 3). (C) The MVB12B PPQY motif is required for efficient NEDD4L-dependent ESCRT-I ubiquitylation. This experiment is analogous to the experiments shown in part A for the MVB12B/ESCRT-I complexes except that the ESCRT-I complexes expressed in lanes 2 and 4 contained MVB12B proteins with 172PPQY175-to-172AAAA175 mutations (MVB12BΔPPQY). Note that NEDD4L-dependent ESCRT-I ubiquitylation was decreased by the MVB12BΔPPQY mutation (compare lanes 3 and 4) and basal ESCRT-I ubiquitylation levels were also decreased by the MVB12B mutation (compare lanes 1 and 2).
TSG101 depletion and reconstitution.
Methods for depleting endogenous TSG101 and reexpressing an siRNA-resistant TSG101 construct (termed TSG101*) were described previously (12). Where noted, 1 μg of the HIV-1ΔPTAP expression vector and 1 μg of expression vectors for ALIX (10) or 0.5 μg NEDD4LΔC2 were cotransfected with the second anti-TSG101 siRNA. Virus was harvested 24 h after the second siRNA transfection.
ESCRT-I ubiquitylation assays.
For the experiments shown in Fig. 7A and C, 293T cells (6 × 106 cells/55-cm2 dish, 50 to 70% confluent) were cotransfected with 2.5 μg (each) of ESCRT-I expression plasmids for TSG101, VPS28, VPS37B, and MVB12A/B (33) and hemagglutinin (HA)-tagged ubiquitin (a generous gift from Heinrich Göttlinger, University of Massachusetts Medical Center) using polyethylenimine (25,000 kDa; Polysciences, Warrington, PA) as described previously (9). Briefly, 45 μl polyethylenimine (1 mg/ml in water) was added to plasmid DNA (in 1 ml Opti-MEM (Invitrogen)), incubated at room temperature for 10 min, and added dropwise onto the culture. The medium was changed 12 h posttransfection, and cells were harvested 48 h posttransfection by incubation in 300 μl lysis buffer (1% Nonidet P-40, 100 mM NaCl, 0.05% sodium dodecyl sulfate, 0.5% sodium deoxycholate in phosphate-buffered saline [PBS]) supplemented with proteinase inhibitor cocktail (1:100 dilution; Sigma) and 20 mM N-ethylmaleimide. Lysates were clarified by centrifugation (18,000 × g, 10 min, 4°C) and incubated with Strep-Tactin Sepharose (40 μl slurry; IBA GmbH, Gottingen, Germany) (2 h, 4°C). The matrix was washed five times in lysis buffer and eluted using 40 μl 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer. ESCRT-I complexes were analyzed by Western blotting with rabbit polyclonal anti-FLAG (Sigma, 1:3,000), mouse monoclonal anti-HA (Covance, 1:500), or mouse monoclonal anti-Myc (Covance, 1:3,000) antibody. In addition to the data shown in Fig. 7, Western blots were also performed in which the anti-HA antibodies (detecting ubiquitin) were mixed with anti-Myc and/or anti-FLAG antibodies (detecting ESCRT-I subunits) in order to confirm that ubiquitin signals comigrated with the assigned protein band (not shown).
Assays for M-PMV release.
Proviral expression constructs for Mason-Pfizer monkey virus (M-PMV) expressing wild-type Gag (pMPMVwt) or Gag proteins with mutations in the PPXY (pMPMVΔPPPY) or PSAP (pMPMVΔPSAP) late domain or both (pMPMVΔPPPY/ΔPSAP) have been described (14). To assay the effects of TSG101 depletion on M-PMV VLP release, TSG101-specific or control siRNAs were transfected twice into 293T cells (50 nM siRNA, six-well plate) at 24-h intervals using Lipofectamine 2000 (12). The second (co)transfection included 1 μg of M-PMV proviral plasmid, and M-PMV virions were harvested 48 h later by centrifugation through a 20% sucrose cushion. Particle production was analyzed by Western blotting using anti-M-PMV mouse monoclonal antibody 10.10 (15 μg/ml), which binds the p12 domain of M-PMV Gag (43) (obtained as a generous gift from Eric Hunter, Emory University).
Immunofluorescence imaging.
HeLa cells transfected with 50 nM siRNA against NEDD4L or 10 nM siRNA against TSG101 (12) were fixed 24 h posttransfection with 3% paraformaldehyde in PBS. Confocal immunofluorescence images were acquired using the Fluoview software program on a FV300 IX81 Olympus microscope. Images are single confocal slices. Microtubules and nuclei were stained using anti-α-tubulin (red, 1:1,000; Sigma) and Sytox green, respectively (34).
FACS analyses of cellular DNA levels.
HeLa cells transfected with 50 nM siRNA against NEDD4L were collected 24 h posttransfection and resuspended in propidium iodide (PI) solution (50 μg of PI per ml PBS, 0.1% Triton X-100, 0.25 mg RNase/ml, 30 min, 4°C). PI-positive cells were counted with a FACScan (BD Bioscience) fluorescence-activated cell sorter (FACS), and peak volumes associated with each DNA content were analyzed using CellQuest software (34).
RESULTS
NEDD4L overexpression rescues HIV-1ΔPTAP release.
The high efficiency of virus release from 293T cells via the p6Gag PTAP late domain makes it difficult to use wild-type HIV-1 to analyze alternative virus release mechanisms. In contrast, HIV-1 constructs that lack PTAP late domains are released much less efficiently and therefore serve as sensitive reporter systems for detecting trans-acting cellular factors that stimulate virus budding. This approach was recently used to show that ALIX strongly stimulates the release of infectious HIV-1ΔPTAP from 293T cells (10, 55), and we have applied the same approach to test whether any of the different human NEDD4 family ubiquitin E3 ligases can stimulate HIV-1 release.
As shown in Fig. 1A, four subfamilies comprising a total of nine different human NEDD4-like ubiquitin E3 ligases were identified using NCBI BLAST searches for homologs of human NEDD4 (and see reference 22). Expression plasmids for N-terminally FLAG-tagged versions of the nine E3 ligases were cotransfected into 293T cells together with a proviral HIV-1 construct that lacked the p6Gag PTAP motif (HIV-1ΔPTAP) and analyzed for the ability to stimulate virus release and infectivity (Fig. 1B, lanes 3 to 11). For controls, the HIV-1ΔPTAP construct was also cotransfected with an empty expression vector (lanes 1, negative control) and with a FLAG-ALIX expression construct (lanes 2, positive control). As shown in panel 3, FLAG-tagged proteins that corresponded to full-length versions of all nine E3 ligases were detected in Western blots. Note, however, that WWP1 was expressed at low levels (lanes 7), and the two largest E3 ligases, NEDL1 and NEDL2, exhibited significant degradation (lanes 10 and 11).
The effects of overexpressing the different NEDD4-like proteins on HIV-1ΔPTAP particle release and infectious titers were analyzed using Western blotting and single-cycle infectivity assays. As expected, ALIX overexpression stimulated HIV-1ΔPTAP release very substantially, increasing both particle release (panel 1, compare lanes 1 and 2) and viral titers by 27-fold (lower graph), to a level that was ∼20% of that for wild-type HIV-1 (not shown). Importantly, overexpression of NEDD4L (also known as NEDD4-2) also substantially increased both HIV-1ΔPTAP release (compare lanes 1 and 4) and infectivity (17-fold in this experiment and 15- to 40-fold in multiple repetitions). Importantly, NEDD4L overexpression did not alter cellular Gag protein levels, indicating that the titer increases were not simply due to elevated Gag expression (panel 2; also data not shown). We therefore conclude that increasing cellular NEDD4L levels can partially overcome the budding defects of an HIV-1 construct that lacks a PTAP late domain.
While no other NEDD4-like protein increased HIV-1ΔPTAP release and titers as dramatically as NEDD4L, the negative results for WWP1, NEDL1, and NEDL2 must be viewed with caution owing to expression problems. Indeed, despite substantial protein degradation, NEDL2 overexpression increased HIV-1ΔPTAP titers significantly (sevenfold in this experiment and five- to sevenfold in other repetitions), indicating that this E3 ligase could also stimulate virus release, albeit to a lesser extent than NEDD4L. Several other NEDD4-like proteins also increased HIV-1ΔPTAP release and titers slightly, but in all cases the titers were within threefold of the levels for the negative control. To control for possible inhibitory effects at high protein overexpression levels (see below), this experiment was also performed using fourfold-lesser quantities of each NEDD4-like expression plasmid. Once again, NEDD4L and NEDL2 were the best of the NEDD4-like proteins in stimulating HIV-1ΔPTAP release (not shown). Thus, NEDD4L (and to a lesser extent NEDL2) can potently rescue the release of an HIV-1 construct that cannot bind TSG101/ESCRT-I.
Activities of different NEDD4L variants.
A number of different NEDD4L isoforms are produced via alternate transcriptional start sites and alternative splicing, and these isoforms can differentially regulate epithelial Na+ channel activity (3, 7, 23). We therefore tested the activities of the four representative, naturally occurring NEDD4L isoforms summarized in Fig. 2A (3, 17, 28, 53). As shown in Fig. 2B, all four NEDD4L isoforms strongly stimulated HIV-1ΔPTAP release and infectivity (26- to 36-fold; lanes 5 to 8). ALIX overexpression again stimulated HIV-1ΔPTAP release and infectivity (25-fold; positive control, lanes 2), whereas control NEDD4WT and NEDD4ΔC2 constructs stimulated release and infectious titers only modestly (∼4-fold; lanes 2 and 3). These observations indicate that neither the C2 domain, the PSAP motif, nor the second WW domain is absolutely required for NEDD4L stimulation of HIV-1ΔPTAP release.
FIG. 2.
Overexpression of four different NEDD4L isoforms rescues HIV-1ΔPTAP release and titer. (A) Four representative, naturally occurring NEDD4L isoforms, with abbreviations and domain structures. Note that the wild type (NEDD4LWT) was used in the experiments shown in Fig. 1. (B) Levels of HIV-1ΔPTAP release and infectivity upon coexpression with an empty expression vector (−; lanes 1), with a vector expressing FLAG-tagged ALIX (lanes 2), with wild-type or NEDD4ΔC2 expression constructs (specificity controls; lanes 3 and 4), or with vectors expressing the different NEDD4L isoforms (lanes 5 to 8). The four panels here are analogous to those in Fig. 1B. Note that cellular CAGag and MAGag levels are similar in all cases but that accumulation of the CA-SP1-processing intermediate, which is characteristic of delayed virus budding, is dramatically reduced upon NEDD4LΔC2 overexpression (panel 2, lane 8).
One notable difference between the different NEDD4L isoforms was that the NEDD4LΔC2 isoform uniquely reduced the accumulation of cell-associated Gag processing intermediates (Fig. 2B; also data not shown). This effect can be seen by comparing the high steady-state levels of the CA-SP1Gag intermediate that accumulated for budding-defective HIV-1ΔPTAP constructs under most conditions (Fig. 2B, second panel, lanes 1 to 7) to the reduced level of this intermediate observed upon overexpression of NEDD4LΔC2 (lane 8). Defects in processing at the CA-SP1 junction typically accompany defects or delays in HIV-1 budding (13), and we therefore infer that the NEDD4LΔC2 isoform is particularly effective at rescuing release of HIV-1ΔPTAP. Hence, the naturally occurring NEDD4LΔC2 isoform was used in all subsequent experiments.
Optimizing NEDD4L stimulation of HIV-1ΔPTAP release.
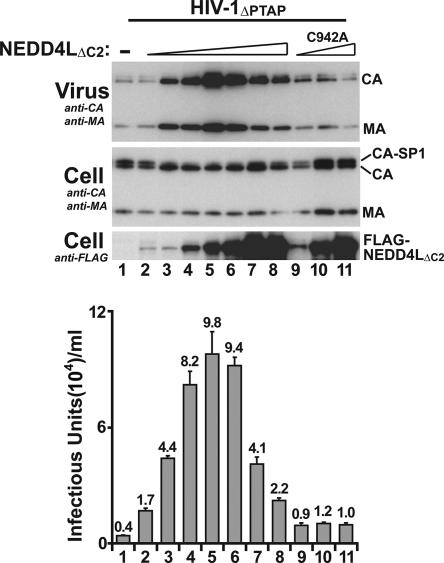
NEDD4LΔC2 was titrated to identify protein levels that optimally rescued HIV-1ΔPTAP release and titers. As shown in Fig. 3, viral titers initially increased with increasing levels of exogenous NEDD4LΔC2 (compare lanes 1 to 5), with maximal increases in virus release and titer (25-fold) observed upon transfection of 0.5 μg of the NEDD4LΔC2 expression construct (lanes 5). This NEDD4LΔC2 level also maximally reduced accumulation of the CA-SP1 processing intermediate. Higher NEDD4LΔC2 levels actually increased CA-SP1 accumulation and decreased virus release and infectivity (lanes 6 to 8), indicating that NEDD4L can dominantly inhibit virus release when highly overexpressed. A NEDD4LΔC2 expression plasmid concentration of 0.5 μg was therefore used in all subsequent experiments.
FIG. 3.
Optimized expression and requirement for NEDD4LΔC2 catalytic activity in the stimulation of HIV-1ΔPTAP release and titer. Levels of HIV-1ΔPTAP release and infectivity upon cotransfection of 1 μg of HIV-1ΔPTAP proviral expression plasmid with an empty expression vector (−; lanes 1), or with 0.05, 0.1, 0.3, 0.5, 1, 2, or 4 μg of the NEDD4LΔC2 expression vector (lanes 2 to 8, respectively), or with 0.1, 0.5, or 2 μg of an expression vector for the catalytically inactive NEDD4LΔC2,C942A protein (lanes 9 to 11, respectively). Note that optimal HIV-1ΔPTAP release and titer were obtained when 0.5 μg of the NEDD4LΔC2 vector was used (lanes 5) and that NEDD4LΔC2,C942A overexpression did not stimulate HIV-1ΔPTAP release or titer at any dose (lanes 9 to 11).
NEDD4LΔC2 catalytic activity is required to stimulate HIV-1ΔPTAP release.
Ubiquitin ligase enzymatic activities are typically required for NEDD4-like protein functions (8, 16, 18, 25, 30), including murine leukemia virus release through interactions between the PPXY late domains and the WWP1/WWP2/ITCH subset of NEDD4-like proteins (30). We therefore tested the function of an enzymatically inactive NEDD4L protein that had a mutation in the active-site cysteine (Cys942Ala). As shown in Fig. 3, the mutant NEDD4LΔC2,C942A protein failed to increase HIV-1ΔPTAP release or infectious titers at any of the three different protein expression levels tested (lanes 9 to 11), implying that NEDD4L enzymatic activity is required to stimulate virus release.
Endogenous NEDD4L also stimulates HIV-1ΔPTAP release.
While the experiments described above demonstrated that high levels of exogenous NEDD4L proteins can stimulate HIV-1ΔPTAP release, it was also important to test whether endogenous NEDD4L was functionally required for virus release. The effect of depleting endogenous NEDD4L on the release of both wild-type and ΔPTAP HIV-1 constructs was therefore tested (Fig. 4). The siRNA used in these studies targeted a conserved sequence present in all known NEDD4L isoforms and reduced endogenous NEDD4L protein levels more than 10-fold (panel 3, compare lanes 1 and 3 to 2 and 4). As shown in panels 1 and 4, NEDD4L depletion did not significantly reduce the release of wild-type HIV-1 (compare lanes 1 and 2) but did substantially reduce HIV-1ΔPTAP release, resulting in a further 14-fold decrease in the residual titer of this crippled virus (compare lanes 3 and 4 and expansions). Thus, although HIV-1ΔPTAP is released poorly from 293T cells, the release that does occur is strongly dependent upon the presence of endogenous NEDD4L.
NEDD4L stimulation does not require the ALIX binding site on p6Gag.
A series of experiments were performed to test the viral requirements for NEDD4L-stimulated release. The first set of experiments showed that the other well-characterized late domain of HIV-1, the ALIX-binding YPXL site, was not required for NEDD4L activity, because NEDD4LΔC2 stimulated release of HIV-1ΔPTAP constructs to nearly the same extent whether or not the ALIX binding site was present (Fig. 5A, compare lanes 3 and 6). Control experiments behaved as expected in that ALIX overexpression rescued release and infectivity of the HIV-1ΔPTAP construct (compare lanes 1 and 2) but did not rescue the HIV-1ΔPTAP/ΔYP construct (compare lanes 2 and 5). Hence, NEDD4L stimulation does not require ALIX binding to its p6Gag late domain.
Gag278-377 is sufficient for NEDD4L-dependent release of HIV-1ΔPTAP.
In principle, NEDD4L could act through any viral protein to stimulate HIV-1ΔPTAP release. We therefore tested whether NEDD4LΔC2 overexpression stimulated release of VLPs formed by HIV-1 GagΔPTAP in the absence of any other viral proteins. As shown in Fig. 5B, NEDD4LΔC2 overexpression strongly stimulated GagΔPTAP VLP release (40-fold in this experiment), implying that Gag is the target of NEDD4L (either directly or indirectly) and that no other viral proteins are required for NEDD4L stimulation.
Truncated and chimeric Gag constructs were used to map the minimal region of Gag required for NEDD4L stimulation (Fig. 5C). As shown in Fig. 5D, NEDD4LΔC2 overexpression stimulated VLP production by a Gag construct that lacked the entire p6 region (GagΔp6; compare lanes 1 and 2), indicating that neither of the known HIV-1 late domains nor their flanking sequences were required for NEDD4L stimulation. To test for the involvement of NCGag, we took advantage of the previous observation that NCGag/RNA “tethering” interactions essential for virion assembly can be functionally replaced by a heterologous leucine zipper dimerization sequence from the human CREB protein (27, 65). NEDD4LΔC2 overexpression also stimulated the release of a chimeric Gag construct (GagZIP) that lacked the SP2 and p6 regions and had the CREB leucine zipper in place of the NC region (compare lanes 3 and 4). Thus, the NC region, which also contains an ALIX binding site (39), is dispensable for NEDD4L stimulation. Finally, Göttlinger and colleagues have shown that particle assembly can be mediated by a minimal Gag construct (GagΔ-ZIP) that contains only an N-terminal membrane binding/myristoylation signal, the C-terminal domain of the CA domain (including the major homology region), and the SP1 spacer (Gag residues 278 to 377) fused to a leucine zipper. NEDD4LΔC2 overexpression also stimulated the release of this minimal assembly construct, and we therefore conclude that the element(s) that determine NEDD4L responsiveness are located within the minimal assembly region(s) of the HIV-1 Gag protein.
TSG101 is required for NEDD4L-dependent HIV-1ΔPTAP release.
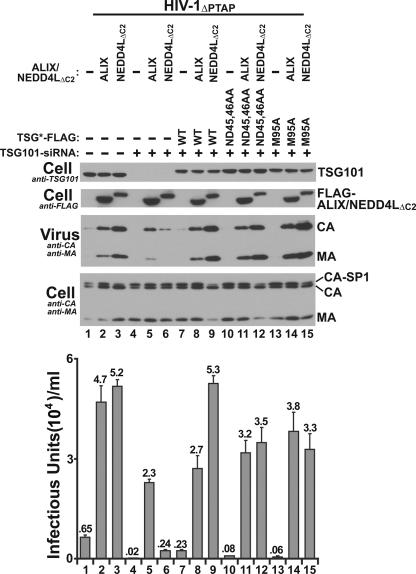
In principle, NEDD4L could act upstream, downstream, or independently of the TSG101/ESCRT-I complex. In the former case, NEDD4L stimulation of HIV-1ΔPTAP release should require TSG101, whereas in the latter two cases, it should not. As shown in Fig. 6, siRNA depletion of TSG101 blocked NEDD4L stimulation of HIV-1ΔPTAP release. TSG101 was efficiently depleted using siRNA (panel 1, compare lanes 4 to 6 to lanes 1 to 3), and the absence of TSG101 potently inhibited NEDD4LΔC2-mediated release of HIV-1ΔPTAP (compare lanes 3 and 6). Importantly, reexpression of an exogenous siRNA-resistant form of TSG101 (termed TSG101-FLAG*) fully restored the ability of NEDD4LΔC2 to stimulate HIV-1ΔPTAP release (compare lanes 9 to lanes 6 and 3), demonstrating that the siRNA effects observed in lane 6 were specifically due to TSG101 depletion and not off-target effects. Unlike results with NEDD4LΔC2, the stimulation of HIV-1ΔPTAP release observed upon ALIX overexpression was largely independent of TSG101 (compare lanes 2 to lanes 8, 12, and 15), and ALIX overexpression therefore served as a control case in which virus release did not require TSG101/ESCRT-I. These experiments indicate that NEDD4L acts upstream of (or together with) TSG101 in promoting HIV-1ΔPTAP release.
FIG. 6.
TSG101 requirements for NEDD4L-dependent HIV-1ΔPTAP release: HIV-1ΔPTAP release and titers upon cotransfection with an empty expression vector (−) (lanes 1, 4, 7, 10, and 13) or with vectors expressing ALIX (lanes 2, 5, 8, 11, and 14) or NEDD4LΔC2 (lanes 3, 6, 9, 12, and 15). Analogous experiments were performed in the presence of endogenous TSG101 (control siRNA; lanes 1 to 3) or in the absence of endogenous TSG101 (siRNA depletion; lanes 4 to 15). Cells used for the experiments in lanes 7 to 15 also expressed exogenous, siRNA-resistant TSG101 (TSG101*), corresponding to the wild-type protein (lanes 7 to 9), a TSG101 protein that cannot bind ubiquitin (ND45,46AA; lanes 10 to 12), or a TSG101 mutant that cannot bind PTAP motifs (M95A; lanes 13 to 15). Note that TSG101 depletion inhibits NEDD4L-dependent HIV-1ΔPTAP release and reduces titers (compare lanes 3 and 6) but does not inhibit ALIX-dependent HIV-1ΔPTAP release (compare lanes 2 and 5). Note also that TSG101 mutants that lack ubiquitin or PTAP binding activities can support NEDD4L-dependent HIV-1ΔPTAP release nearly as well as wild-type TSG101 (compare lanes 12 and 15 to lanes 9). TSG101 depletion and rescue were analyzed by Western blotting (panel 1, anti-TSG101). HIV-1ΔPTAP release, cellular Gag protein levels, exogenous ALIX and NEDD4LΔC2 expression levels, and infectious titers were analyzed as described in the legend to Fig. 1.
Conceivably, NEDD4L might function by transferring ubiquitin to a target protein, such as HIV-1 Gag, which could then be recognized by the ubiquitin binding activity of the TSG101 UEV domain. This was not the case, however, because a mutant TSG101 protein (TSG101*ND45,46AA) that lacked the ability to bind ubiquitin (41, 54) still supported NEDD4L-mediated HIV-1ΔPTAP release nearly as well as the wild-type protein (compare lanes 12 to lanes 9). Similarly, a mutant TSG101 protein (TSG101*M95A) that lacked the ability to bind PTAP sequences (40, 41) also supported NEDD4L-mediated release (compare lanes 15 to lanes 9). Thus, although TSG101 is required for NEDD4L-mediated HIV-1ΔPTAP release, the ubiquitin and PTAP binding activities of the TSG101 UEV domain are not.
NEDD4L induces ESCRT-I ubiquitylation.
We next tested whether NEDD4LΔC2 overexpression altered ESCRT-I ubiquitylation. Human ESCRT-I is a family of related complexes, each of which contains a TSG101 subunit, a VPS28 subunit, one of two MVB12 subunits (A and B), and one of four VPS37 subunits (A to D) (33). As shown in Fig. 7A, HA-tagged ubiquitin was overexpressed with ESCRT-I complexes that contained either MVB12A (lanes 1 and 3) or MVB12B subunits (lanes 2 and 4). The overexpressed ESCRT-I complexes were then affinity purified (panel 2) and tested for ubiquitin modifications by immunoblotting against HA-Ub (panel 3). In both cases, basal ESCRT-I ubiquitylation was detected even in the absence of NEDD4L overexpression, with MVB12B/ESCRT-I showing higher levels of modification (compare lanes 1 and 2). Although we have not identified all of the ESCRT-I-associated proteins that are ubiquitylated, the banding patterns indicate that both MVB12B and TSG101 are ubiquitylated (panel 4; also data not shown).
Importantly, NEDD4LΔC2 overexpression dramatically increased ubiquitylation of MVB12B/ESCRT-I (Fig. 7A. compare lanes 2 and 4) and also increased MVB12A/ESCRT-I ubiquitylation (compares lanes 1 and 3). Conversely, siRNA depletion of NEDD4L reduced basal levels of MVB12B/ ESCRT-I ubiquitylation to nearly undetectable levels (Fig. 7B, panel 3, compare lanes 2 and 3). Hence, endogenous NEDD4L proteins are responsible for ubiquitylation of the MVB12B/ESCRT-I complex, and higher levels of NEDD4L increase ESCRT-I ubiquitylation.
The simplest explanation for our data is that NEDD4L proteins are recruited through direct interactions with the MVB12B/ESCRT-I complex. NEDD4-like proteins are commonly recruited through interactions between their WW domains and PPXY-like binding sites on their substrates (22, 24, 49). MVB12B has a single PPQY sequence motif (residues 172 to 175), and we tested the effect of mutating this sequence on ubiquitylation of the MVB12B/ESCRT-I complex. As shown in Fig. 7C, the mutant MVB12B protein was expressed well and formed stable complexes with the other ESCRT-I subunits, and the mutation reduced but did not eliminate MVB12B/ESCRT-I ubiquitylation, both when NEDD4LCΔ2 was overexpressed (compare lanes 3 and 4) and when only endogenous NEDD4L was present (compare lanes 1 and 2). Thus, the MVB12B PPQY sequence increases ubiquitylation of the MVB12B/ESCRT-I complex but cannot be the only element responsible for NEDD4L recruitment.
TSG101 is also required for M-MPV Gag release.
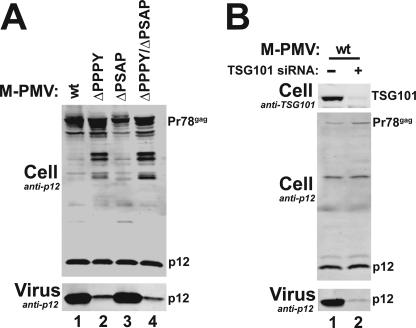
The data described above are consistent with a model in which NEDD4L acts upstream to recruit, ubiquitylate, and/or otherwise activate ESCRT-I. These observations suggested the possibility that other retroviruses, like M-PMV, that use PPXY late domains to recruit NEDD4-like proteins might also act through ESCRT-I. M-PMV Gag contains both PPXY and PSAP late domains, but the PPXY late domain appears dominant in most contexts (14, 62). This is illustrated in Fig. 8A, where mutation of the PPPY late domain inhibited Gag processing and diminished virion release from 293T cells (compare lanes 1 to lanes 2 and 4) whereas the effects of mutating the PSAP late domain were much less severe (compare lanes 1 and 3). In contrast, M-PMV release was almost entirely blocked when TSG101 was depleted from the producer cells (Fig. 8B, compare lanes 1 and 2). Thus, although the PPXY motif is the dominant late domain, TSG101/ESCRT-I is still required for release of M-PMV from 293T cells.
FIG. 8.
Endogenous TSG101 is required for efficient release of M-PMV. (A) The PPPY late domain is essential for M-PMV release from 293T cells. pM-PMVwt, pMPMVΔPPPY, pMPMVΔPSAP, and pMPMVΔPPXY/ΔPSAP (lanes 1 to 4, respectively) were expressed, and Western blots measured levels of M-PMV Gag/p12 (upper panel) and virion-associated M-PMV Gag/p12 (lower panel). (B) TSG101 depletion strongly inhibits release of wild-type M-PMV from 293T cells. Control siRNA (−; lane 1) or TSG101 siRNA (lane 2) were cotransfected with wild-type M-PMV proviral expression vectors. Western blots measured levels of cellular TSG101 (upper panel), cellular M-PMV Gag/p12 (middle panel), and virion-associated M-PMV Gag/p12 (lower panel).
NEDD4L is required for efficient HeLa cell cytokinesis.
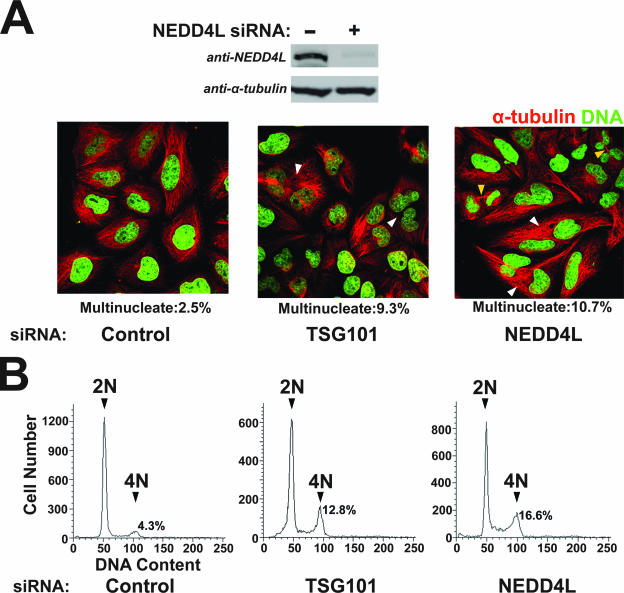
TSG101/ESCRT-I is recruited to the HeLa cell midbody, where it also functions in the final abscission step of cytokinesis (1). We therefore tested whether NEDD4L, like TSG101/ ESCRT-I, was also required for cytokinesis. As reported previously, siRNA depletion of TSG101 induced accumulation of multinuclear cells, as assayed both by direct counting of multinuclear cells (Fig. 9A, middle panel; 9.3% multinuclear cells) and by FACS analyses that measured the ratios of cells with 2N versus 4N DNA contents (Fig. 9B, middle panel, 12.8% of cells with 4N DNA content) (1, 34). Hence, as expected, TSG101 depletion increased the fraction of multinuclear cells compared to an irrelevant siRNA control in both the direct counting assay (2.5%) and the FACS assay (4.2%). siRNA depletion of NEDD4L proteins from HeLa cells similarly induced accumulation of multinuclear cells in both assays (right panels; 10.7% in the direct counting assay and 16.6% in the FACS assay) and also led to an increase in the number of cells with visible midbodies (Fig. 9A). Thus, NEDD4L depletion induces at least as strong a cytokinesis defect as does TSG101 depletion.
FIG. 9.
NEDD4L is required for efficient HeLa cell cytokinesis. (A) Immunofluorescent images of HeLa cells treated with a control siRNA (left, negative control) or with siRNAs targeting TSG101 (middle, positive control) or NEDD4L (right). Microtubules (red; anti-α-tubulin) and nuclei (Sytox green) were stained for reference. White arrowheads highlight multinuclear cells, and yellow arrowheads indicate cells with visible midbodies. Mono- and multinucleated cells were quantified by microscopic counting, and the percentages of multinuclear cells are reported below each panel. (B) FACS analyses of the DNA content of cells treated with a control siRNA (left; negative control) or with siRNAs targeting TSG101 (middle; positive control) or NEDD4L (right). Peaks corresponding to 2N and 4N DNA contents are labeled, and the integrated peak volumes are provided.
DISCUSSION
Viral requirements for NEDD4L stimulation.
Our data, together with those in the accompanying article by Göttlinger and colleagues (55a), demonstrate that the NEDD4-like ubiquitin HECT E3 ligase, NEDD4L, can stimulate the release and infectivity of HIV-1 viruses that lack the PTAP and YPXL late domains. This is the first indication that NEDD4-like proteins can also function in the release of enveloped RNA viruses like HIV-1 that lack PPXY late domains. Mapping studies and deletion analyses revealed that NEDD4L exerts its effects through the viral Gag protein but that most Gag regions are dispensable, including the globular domain of MA, the CA N-terminal domain, NC, SP2, and p6. Thus, the minimal regions of Gag required for NEDD4L stimulation match the minimal regions required for particle assembly. This implies that the CACTD-SP1Gag region must contain cis-acting late-domain activities that can support efficient virion budding, provided cellular NEDD4L levels are sufficiently high. We do not yet understand how NEDD4L activity is physically connected to the HIV-1 Gag assembly region, however, since we were unable to demonstrate a stable interaction between HIV-1 Gag and NEDD4L. We did observe that overexpression of the catalytically active NEDD4LΔC2 protein resulted in a substantial enhancement in the overall levels of ubiquitylated proteins, including Gag, in purified VLPs (data not shown; also see the accompanying paper by Göttlinger and colleagues [55a]). This observation raises the possibility that NEDD4L may exert its effects by binding and ubiquitylating Gag. However, we also observed that NEDD4LΔC2 overexpression substantially increased overall protein ubiquitylation levels in cell lysates, and therefore, we cannot be certain that increases in virion-associated Gag ubiquitylation levels seen upon NEDD4L overexpression do not simply mirror global increases in cellular protein ubiquitylation levels.
NEDD4L requirements.
NEDD4L was significantly more active than other human NEDD4-like proteins in stimulating HIV-1ΔPTAP release, and this effect was specific, because another subfamily member, NEDD4, was far less active despite sharing 63% amino acid identity. Cells express a number of different NEDD4L isoforms (3, 7, 23), and all four of the NEDD4L isoforms that we tested stimulated HIV-1ΔPTAP release. Thus, stimulation did not require the C2, PSAP, or WW2 elements, each of which was missing in a subset of the different NEDD4L isoforms. The dispensability of the PSAP motif is somewhat surprising, since this element is a consensus binding site for the TSG101 UEV domain, and we speculate that although this element is not absolutely required, it is likely to bind TSG101 in some contexts.
The activity of the NEDD4LΔC2 construct indicates that regions outside the C2 core must dictate substrate selection, as has been seen for other NEDD4-like proteins (30, 46). Indeed, the C2 domain actually appeared to inhibit NEDD4L-stimulated HIV-1ΔPTAP release slightly, since the NEDD4LΔC2 isoform uniquely rescued the Gag processing defect that typically accompanies viral budding defects (13). The C2 domain of a related NEDD4-like protein, SMURF2, was recently shown to bind and autoinhibit the catalytic activity of its HECT domain (60), and it is reasonable to expect that the C2 domain of NEDD4L inhibits virus release in a similar fashion.
Most importantly, the stimulation of HIV-1ΔPTAP release required the ubiquitin E3 ligase enzymatic activity, implying that NEDD4L functions in virus release by transferring ubiquitin onto target protein(s). Thus, NEDD4L can provide a connection between HIV-1 budding and ubiquitin. We note, however, that NEDD4L alone cannot fully explain why efficient release of wild-type HIV-1 requires ubiquitin transfer, because NEDD4L depletion did not significantly inhibit the release of the wild-type virus (Fig. 4). Nevertheless, depletion of endogenous NEDD4L did substantially inhibit the already low release of the crippled HIV-1ΔPTAP virus, implying that endogenous NEDD4L plays a significant role in HIV-1 release when TSG101 recruitment is compromised.
NEDD4L acts through ESCRT-I.
siRNA depletion of TSG101 blocked the ability of NEDD4LΔC2 to stimulate HIV-1ΔPTAP release, demonstrating that NEDD4L must exert its effects upstream of (or in concert with) TSG101/ESCRT-I. NEDD4L and ALIX differ in this respect, since ALIX strongly stimulated HIV-1ΔPTAP release whether or not TSG101 was present. Although the role of TSG101/ESCRT-I in NEDD4L stimulation remains to be characterized fully, the PTAP and ubiquitin binding activities of the TSG101 UEV domain are not required. These observations argue against the otherwise attractive possibility that NEDD4L proteins might simultaneously bind TSG101 (e.g., through TSG101 UEV-PSAP interactions) and ubiquitylate Gag, thereby creating a mechanism for recruiting TSG101/ESCRT-I to sites of virus budding (e.g., through TSG101 UEV-Ub interactions). We have also found that TSG101/ESCRT-I complexes that lack ubiquitin binding activity can support the budding of wild-type HIV-1 (not shown). Hence, there is currently no evidence that ubiquitin binding by TSG101/ESCRT-I is functionally important for the release of HIV-1 via either TSG101/ESCRT-I or NEDD4L.
NEDD4L interactions with ESCRT-I.
Our studies raise the question of how NEDD4L interacts with TSG101/ESCRT-I. The best evidence for such an interaction comes from the studies of Medina et al., who showed that Rous sarcoma virus release can be mediated by an interaction between a PPXY sequence in Rous sarcoma virus Gag and the chicken NEDD4-like protein, LDI-1 (late domain interacting protein 1) (32). Importantly, they also showed that human TSG101 could immunoprecipitate HA-tagged LDI-1, providing a physical link between TSG101 and a NEDD4-like protein. Although those authors emphasized the similarity between chicken LDI-1 and human NEDD4 (64% identity), LDI-1 is actually a much better match with human NEDD4L (93% identity). Thus, their experiments indicate that under some conditions, NEDD4L proteins can physically associate with TSG101/ESCRT-I (either directly or indirectly). Unfortunately, we have been unable to detect reproducible interactions between human TSG101/ESCRT-I and human NEDD4L, either in coprecipitation experiments or in biochemical experiments with pure recombinant complexes (not shown). We were also unable to observe colocalization of ESCRT-I with either endogenous or exogenous NEDD4L in the midbody during cytokinesis (not shown). Hence, we cannot confirm that human NEDD4L and ESCRT-I can form stable complexes, either in vitro or in vivo.
Nevertheless, our experiments support the idea that NEDD4L and TSG101/ESCRT-I can interact, at least transiently, because we find that NEDD4L overexpression increases ubiquitylation of ESCRT-I subunits, including TSG101 and MVB12B, and that depletion of endogenous NEDD4L reduces their ubiquitylation (Fig. 7). Although we cannot rule out the possibility of indirect interactions, NEDD4L-induced ubiquitylation is greater for ESCRT-I complexes that contain MVB12B subunits, and mutation of a potential NEDD4L WW domain docking site within MVB12 reduced (but did not eliminate) ESCRT-I ubiquitylation. Multiple ESCRT-I isoforms with different subunit compositions form stable complexes in cells (33), and this is the first example of an isoform-specific phenotype, raising the possibility that different effectors like NEDD4L may exert differential effects through specific subsets of ESCRT-I complexes.
Ubiquitylation may activate ESCRT-I.
NEDD4L overexpression results in ubiquitylation of TSG101/ESCRT-I, suggesting that ubiquitylation may activate ESCRT-I to function in HIV-1 release. Similarly, depletion of endogenous NEDD4L reduced the efficiency of HeLa cell cytokinesis, indicating that TSG101 and NEDD4L may also work together in this cellular process. Furthermore, another ubiquitin E3 ligase, Mahogunin, was recently shown to bind TSG101 and catalyze its monoubiquitylation in vitro and in vivo (26). In this case, monoubiquitylation of TSG101 appeared to enhance ESCRT activity, as reflected in the requirement for Mahogunin activity in the lysosomal degradation of the epidermal growth factor receptor. Thus, there are growing indications that ubiquitylation can positively regulate TSG101/ESCRT-I. Interestingly, recent studies indicate that retroviral release through PTAP late domains can require ubiquitylation of a cellular factor(s) (63), and our experiments suggest that TSG101/ESCRT-I may be one such factor.
Role for TSG101/ESCRT-I in release of PPXY-containing viruses.
The roles of PPXY late domains in virus budding are not yet fully understood and appear to be quite complicated, possibly because different PPXY late domains can utilize distinct or redundant NEDD4-like binding partners. There are, however, several indications that PPXY motifs and their NEDD4-like binding partners can act in concert with TSG101/ESCRT-I to facilitate virus release. For example, the Marburg, lymphocytic choriomeningitis, and lassa fever viruses use PPXY late domains, yet are still dependent upon TSG101 (37a, 54a). We found that this can also be true for retroviruses, since TSG101 depletion strongly inhibited M-PMV release, even though M-PMV relies more heavily on its PPXY motif than on its PSAP motif (Fig. 8). Thus, these all appear to be cases in which NEDD4-like proteins bind PPXY late domains and then function via ESCRT-I, perhaps in analogy to the NEDD4L-stimulated release of HIV-1ΔPTAP described here.
Summary and implications.
In summary, we find that NEDD4L exhibits a remarkable ability to stimulate HIV-1ΔPTAP release and infectivity and that the NEDD4L ubiquitin E3 ligase activity can link the assembling virion, either directly or indirectly, to ESCRT-I. We speculate that NEDD4L may also function in the release of wild-type HIV-1 in contexts where the virus cannot recruit TSG101 as efficiently as it does in 293T cells. Thus, in cell types where Gag or TSG101/ ESCRT-I levels are lower (or where NEDD4L levels are higher), NEDD4L may cooperate with other late domain binding proteins to enhance the efficiency of virus release. Indeed, we envision the possibility that ESCRT-I and NEDD4L (and possibly even ALIX) can contact Gag and one another at multiple sites to form fully functional HIV-1 budding complexes that optimize the efficiency of virus release.
Supplementary Material
Acknowledgments
We thank Bob Weis and Jean-Marc Lalouel for helpful discussions regarding NEDD4L variants.
This research was supported by NIH grant AI51174 (to W.I.S.).
Footnotes
Published ahead of print on 5 March 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Carlton, J. G., and J. Martin-Serrano. 2007. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science 3161908-1912. [DOI] [PubMed] [Google Scholar]
- 2.Chen, C., O. Vincent, J. Jin, O. A. Weisz, and R. C. Montelaro. 2005. Functions of early (AP-2) and late (AIP1/ALIX) endocytic proteins in equine infectious anemia virus budding. J. Biol. Chem. 28040474-40480. [DOI] [PubMed] [Google Scholar]
- 3.Chen, H., C. A. Ross, N. Wang, Y. Huo, D. F. MacKinnon, J. B. Potash, S. G. Simpson, F. J. McMahon, J. R. DePaulo, Jr., and M. G. McInnis. 2001. NEDD4L on human chromosome 18q21 has multiple forms of transcripts and is a homologue of the mouse Nedd4-2 gene. Eur. J. Hum. Genet. 9922-930. [DOI] [PubMed] [Google Scholar]
- 4.Debonneville, C., S. Y. Flores, E. Kamynina, P. J. Plant, C. Tauxe, M. A. Thomas, C. Munster, A. Chraibi, J. H. Pratt, J. D. Horisberger, D. Pearce, J. Loffing, and O. Staub. 2001. Phosphorylation of Nedd4-2 by Sgk1 regulates epithelial Na(+) channel cell surface expression. EMBO J. 207052-7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Demirov, D. G., and E. O. Freed. 2004. Retrovirus budding. Virus Res. 10687-102. [DOI] [PubMed] [Google Scholar]
- 6.Demirov, D. G., A. Ono, J. M. Orenstein, and E. O. Freed. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. USA 99955-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunn, D. M., T. Ishigami, J. Pankow, A. von Niederhausern, J. Alder, S. C. Hunt, M. F. Leppert, J. M. Lalouel, and R. B. Weiss. 2002. Common variant of human NEDD4L activates a cryptic splice site to form a frameshifted transcript. J. Hum. Genet. 47665-676. [DOI] [PubMed] [Google Scholar]
- 8.Dunn, R., and L. Hicke. 2001. Domains of the rsp5 ubiquitin-protein ligase required for receptor-mediated and fluid-phase endocytosis. Mol. Biol. Cell 12421-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durocher, Y., S. Perret, and A. Kamen. 2002. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 30E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fisher, R. D., H. Y. Chung, Q. Zhai, H. Robinson, W. I. Sundquist, and C. P. Hill. 2007. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell 128841-852. [DOI] [PubMed] [Google Scholar]
- 11.Galan, J. M., V. Moreau, B. Andre, C. Volland, and R. Haguenauer-Tsapis. 1996. Ubiquitination mediated by the Npi1p/Rsp5p ubiquitin-protein ligase is required for endocytosis of the yeast uracil permease. J. Biol. Chem. 27110946-10952. [DOI] [PubMed] [Google Scholar]
- 12.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 10755-65. [DOI] [PubMed] [Google Scholar]
- 13.Gottlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 883195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gottwein, E., J. Bodem, B. Muller, A. Schmechel, H. Zentgraf, and H. G. Krausslich. 2003. The Mason-Pfizer monkey virus PPPY and PSAP motifs both contribute to virus release. J. Virol. 779474-9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottwein, E., S. Jager, A. Habermann, and H. G. Krausslich. 2006. Cumulative mutations of ubiquitin acceptor sites in human immunodeficiency virus type 1 Gag cause a late budding defect. J. Virol. 806267-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goulet, C. C., K. A. Volk, C. M. Adams, L. S. Prince, J. B. Stokes, and P. M. Snyder. 1998. Inhibition of the epithelial Na+ channel by interaction of Nedd4 with a PY motif deleted in Liddle's syndrome. J. Biol. Chem. 27330012-30017. [DOI] [PubMed] [Google Scholar]
- 17.Harvey, K. F., A. Dinudom, D. I. Cook, and S. Kumar. 2001. The Nedd4-like protein KIAA0439 is a potential regulator of the epithelial sodium channel. J. Biol. Chem. 2768597-8601. [DOI] [PubMed] [Google Scholar]
- 18.Heidecker, G., P. A. Lloyd, F. Soheilian, K. Nagashima, and D. Derse. 2007. The role of WWP1-Gag interaction and Gag ubiquitination in assembly and release of human T-cell leukemia virus type 1. J. Virol. 819769-9777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hein, C., J. Y. Springael, C. Volland, R. Haguenauer-Tsapis, and B. Andre. 1995. NPl1, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol. Microbiol. 1877-87. [DOI] [PubMed] [Google Scholar]
- 20.Hermida-Matsumoto, L., and M. D. Resh. 2000. Localization of human immunodeficiency virus type 1 Gag and Env at the plasma membrane by confocal imaging. J. Virol. 748670-8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hurley, J. H., and S. D. Emr. 2006. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 35277-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ingham, R. J., G. Gish, and T. Pawson. 2004. The Nedd4 family of E3 ubiquitin ligases: functional diversity within a common modular architecture. Oncogene 231972-1984. [DOI] [PubMed] [Google Scholar]
- 23.Itani, O. A., J. B. Stokes, and C. P. Thomas. 2005. Nedd4-2 isoforms differentially associate with ENaC and regulate its activity. Am. J. Physiol. Renal Physiol. 289F334-F346. [DOI] [PubMed] [Google Scholar]
- 24.Kanelis, V., D. Rotin, and J. D. Forman-Kay. 2001. Solution structure of a Nedd4 WW domain-ENaC peptide complex. Nat. Struct. Biol. 8407-412. [DOI] [PubMed] [Google Scholar]
- 25.Katzmann, D. J., S. Sarkar, T. Chu, A. Audhya, and S. D. Emr. 2004. Multivesicular body sorting: ubiquitin ligase Rsp5 is required for the modification and sorting of carboxypeptidase S. Mol. Biol. Cell 15468-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, B. Y., J. A. Olzmann, G. S. Barsh, L. S. Chin, and L. Li. 2007. Spongiform neurodegeneration-associated E3 ligase Mahogunin ubiquitylates TSG101 and regulates endosomal trafficking. Mol. Biol. Cell 181129-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loriaux, M. M., R. P. Rehfuss, R. G. Brennan, and R. H. Goodman. 1993. Engineered leucine zippers show that hemiphosphorylated CREB complexes are transcriptionally active. Proc. Natl. Acad. Sci. USA 909046-9050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malbert-Colas, L., G. Nicolas, C. Galand, M. C. Lecomte, and D. Dhermy. 2003. Identification of new partners of the epithelial sodium channel alpha subunit. C. R. Biol. 326615-624. [DOI] [PubMed] [Google Scholar]
- 29.Martin-Serrano, J. 2007. The role of ubiquitin in retroviral egress. Traffic 81297-1303. [DOI] [PubMed] [Google Scholar]
- 30.Martin-Serrano, J., S. W. Eastman, W. Chung, and P. D. Bieniasz. 2005. HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J. Cell Biol. 16889-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 71313-1319. [DOI] [PubMed] [Google Scholar]
- 32.Medina, G., Y. Zhang, Y. Tang, E. Gottwein, M. L. Vana, F. Bouamr, J. Leis, and C. A. Carter. 2005. The functionally exchangeable L domains in RSV and HIV-1 Gag direct particle release through pathways linked by Tsg101. Traffic 6880-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morita, E., V. Sandrin, S. Alam, D. Eckert, S. P. Gygi, and W. I. Sundquist. 2007. Identification of the human MVB12 proteins as ESCRT-I subunits that function in HIV budding. Cell Host Microbe 241-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morita, E., V. Sandrin, H. Y. Chung, S. G. Morham, S. P. Gygi, C. K. Rodesch, and W. I. Sundquist. 2007. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 264215-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morita, E., and W. I. Sundquist. 2004. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20395-425. [DOI] [PubMed] [Google Scholar]
- 36.Ott, D. E., L. V. Coren, E. N. Chertova, T. D. Gagliardi, and U. Schubert. 2000. Ubiquitination of HIV-1 and MuLV Gag. Virology 278111-121. [DOI] [PubMed] [Google Scholar]
- 37.Ott, D. E., L. V. Coren, T. D. Copeland, B. P. Kane, D. G. Johnson, R. C. Sowder II, Y. Yoshinaka, S. Oroszlan, L. O. Arthur, and L. E. Henderson. 1998. Ubiquitin is covalently attached to the p6Gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and to the p12Gag protein of Moloney murine leukemia virus. J. Virol. 722962-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37a.Perez, M., R. C. Craven, and J. C. de la Torre. 2003. The small RING finger protein Z drives arenavirus budding: implications for antiviral strategies. Proc. Natl. Acad. Sci. USA 10012978-12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piper, R. C., and D. J. Katzmann. 2007. Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Dev. Biol. 23519-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Popov, S., E. Popova, M. Inoue, and H. G. Gottlinger. 2008. Human immunodeficiency virus type 1 Gag engages the Bro1 domain of ALIX/AIP1 through the nucleocapsid. J. Virol. 821389-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pornillos, O., S. L. Alam, D. R. Davis, and W. I. Sundquist. 2002. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat. Struct. Biol. 9812-817. [DOI] [PubMed] [Google Scholar]
- 41.Pornillos, O., S. L. Alam, R. L. Rich, D. G. Myszka, D. R. Davis, and W. I. Sundquist. 2002. Structure and functional interactions of the Tsg101 UEV domain. EMBO J. 212397-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Putterman, D., R. B. Pepinsky, and V. M. Vogt. 1990. Ubiquitin in avian leukosis virus particles. Virology 176633-637. [DOI] [PubMed] [Google Scholar]
- 43.Sakalian, M., S. D. Parker, R. A. Weldon, Jr., and E. Hunter. 1996. Synthesis and assembly of retrovirus Gag precursors into immature capsids in vitro. J. Virol. 703706-3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schubert, U., D. E. Ott, E. N. Chertova, R. Welker, U. Tessmer, M. F. Princiotta, J. R. Bennink, H. G. Krausslich, and J. W. Yewdell. 2000. Proteasome inhibition interferes with Gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. USA 9713057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Segura-Morales, C., C. Pescia, C. Chatellard-Causse, R. Sadoul, E. Bertrand, and E. Basyuk. 2005. Tsg101 and Alix interact with murine leukemia virus Gag and cooperate with Nedd4 ubiquitin ligases during budding. J. Biol. Chem. 28027004-27012. [DOI] [PubMed] [Google Scholar]
- 46.Shearwin-Whyatt, L., H. E. Dalton, N. Foot, and S. Kumar. 2006. Regulation of functional diversity within the Nedd4 family by accessory and adaptor proteins. Bioessays 28617-628. [DOI] [PubMed] [Google Scholar]
- 47.Shehu-Xhilaga, M., S. Ablan, D. G. Demirov, C. Chen, R. C. Montelaro, and E. O. Freed. 2004. Late domain-dependent inhibition of equine infectious anemia virus budding. J. Virol. 78724-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Springael, J. Y., J. O. De Craene, and B. Andre. 1999. The yeast Npi1/Rsp5 ubiquitin ligase lacking its N-terminal C2 domain is competent for ubiquitination but not for subsequent endocytosis of the gap1 permease. Biochem. Biophys. Res. Commun. 257561-566. [DOI] [PubMed] [Google Scholar]
- 49.Staub, O., S. Dho, P. Henry, J. Correa, T. Ishikawa, J. McGlade, and D. Rotin. 1996. WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle's syndrome. EMBO J. 152371-2380. [PMC free article] [PubMed] [Google Scholar]
- 50.Strack, B., A. Calistri, M. A. Accola, G. Palu, and H. G. Gottlinger. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 9713063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strack, B., A. Calistri, S. Craig, E. Popova, and H. G. Gottlinger. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114689-699. [DOI] [PubMed] [Google Scholar]
- 52.Strack, B., A. Calistri, and H. G. Gottlinger. 2002. Late assembly domain function can exhibit context dependence and involves ubiquitin residues implicated in endocytosis. J. Virol. 765472-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strausberg, R. L., et al. 2002. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. USA 9916899-16903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sundquist, W. I., H. L. Schubert, B. N. Kelly, G. C. Hill, J. M. Holton, and C. P. Hill. 2004. Ubiquitin recognition by the human TSG101 protein. Mol. Cell 13783-789. [DOI] [PubMed] [Google Scholar]
- 54a.Urata, S., T. Noda, Y. Kawaoka, S. Morikawa, H. Yokosawa, and J. Yasuda. 2007. Interaction of Tsg101 with Marburg virus VP40 depends on the PPPY motif, but not the PT/SAP motif as in the case of Ebola virus, and Tsg101 plays a critical role in the budding of Marburg virus-like particles induced by VP40, NP, and GP. J. Virol. 814895-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Usami, Y., S. Popov, and H. G. Gottlinger. 2007. Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. J. Virol. 816614-6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55a.Usami, Y., S. Popov, E. Popova, and H. G. Göttlinger. 2008. Efficient and specific rescue of human immunodeficiency virus type 1 budding defects by a Nedd4-like ubiquitin ligase. J. Virol. 824898-4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vana, M. L., Y. Tang, A. Chen, G. Medina, C. Carter, and J. Leis. 2004. Role of Nedd4 and ubiquitination of Rous sarcoma virus Gag in budding of virus-like particles from cells. J. Virol. 7813943-13953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55Gag. Proc. Natl. Acad. Sci. USA 987724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vogt, V. M. 2000. Ubiquitin in retrovirus assembly: actor or bystander? Proc. Natl. Acad. Sci. USA 9712945-12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Welsch, S., B. Muller, and H. G. Krausslich. 2007. More than one door—budding of enveloped viruses through cellular membranes. FEBS Lett. 5812089-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wiesner, S., A. A. Ogunjimi, H. R. Wang, D. Rotin, F. Sicheri, J. L. Wrana, and J. D. Forman-Kay. 2007. Autoinhibition of the HECT-type ubiquitin ligase Smurf2 through its C2 domain. Cell 130651-662. [DOI] [PubMed] [Google Scholar]
- 61.Williams, R. L., and S. Urbe. 2007. The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell Biol. 8355-368. [DOI] [PubMed] [Google Scholar]
- 62.Yasuda, J., and E. Hunter. 1998. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J. Virol. 724095-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhadina, M., M. O. McClure, M. C. Johnson, and P. D. Bieniasz. 2007. Ubiquitin-dependent virus particle budding without viral protein ubiquitination. Proc. Natl. Acad. Sci. USA 10420031-20036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhai, Q., R. D. Fisher, H. Y. Chung, D. G. Myszka, W. I. Sundquist, and C. P. Hill. 2008. Structural and functional studies of ALIX interactions with YPX(n) L late domains of HIV-1 and EIAV. Nat. Struct. Mol. Biol. 1543-49. [DOI] [PubMed] [Google Scholar]
- 65.Zhang, Y., H. Qian, Z. Love, and E. Barklis. 1998. Analysis of the assembly function of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J. Virol. 721782-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.