Abstract
Productive infection by herpes simplex virus type 1 (HSV-1), which occurs in the host cell nucleus, is accompanied by dramatic modifications of the nuclear architecture, including profound alterations of nucleolar morphology. Here, we show that the three most abundant nucleolar proteins—nucleolin, B23, and fibrillarin—are redistributed out of the nucleoli as a consequence of HSV-1 infection. We show that the amount of nucleolin increases progressively during the course of infection. We demonstrate for the first time that a nucleolar protein, i.e., nucleolin, colocalizes with ICP8 in the viral replication compartments, at the time when viral replication is effective, suggesting an involvement of nucleolin in the HSV-1 DNA replication process. At later times of infection, a granular form of nucleolin localizes to the cytoplasm, in structures that display the characteristic features of aggresomes, indicating that this form of nucleolin is very probably destined for degradation. The delocalization of nucleolin from the nucleoli requires the viral ICP4 protein or a factor(s) whose expression involves ICP4. Using small interfering RNA technology, we show that viral replication requires a high level of nucleolin expression, demonstrating for the first time a direct role for a nucleolar protein in herpes simplex virus biology.
Herpes simplex virus type 1 (HSV-1) is a human herpesvirus consisting of an outer envelope, a tegument, a capsid, and a linear double-stranded DNA. Productive infection consists of a highly ordered program of viral gene expression, DNA replication, and virion assembly that leads to the formation of infectious viral progeny and cell death (48). The viral DNA contains at least 80 genes whose expression is sequentially and temporally regulated by complex regulatory mechanisms. Viral genes can be divided into immediate-early, early, and late genes, according to their kinetics of expression. Proteins encoded by immediate-early genes are involved in the regulation of the synthesis of early and late proteins. Proteins encoded by early genes participate in viral DNA replication, and proteins encoded by late genes are mainly the structural components of the viral particles.
Viral transcription, DNA replication, assembly of new capsids, and packaging of HSV-1 DNA occur in the host cell nucleus. As a consequence, HSV-1-infected cells undergo a variety of changes including dramatic modifications of the nuclear architecture. The formation of viral replication compartments (VRC), which are the sites of replication, transcription, and encapsidation of HSV-1 genomes, is accompanied by the marginalization of chromatin and the disruption of the nuclear lamina and of PML bodies, as well as by a profound modification of nucleolar morphology (2, 15, 33, 39). Soon after infection, nucleoli increase in size, localize close to the nuclear membrane, and finally become fragmented in small pieces (39, 49). In addition, several viral proteins, including ICP0, ICP4, ICP27, US11, and gamma 34.5, localize at least transiently to nucleoli during infection (8, 10, 31, 37, 40, 50).
The nucleolus is the most prominent compartment of the cell nucleus and the most extensively studied nuclear domain. It is well known to be the site of rRNA transcription, processing, and assembly into the ribosomal subunits. Recent studies, however, have highlighted the fact that nucleoli are dynamic structures involved in additional nonclassical roles, including cell cycle regulation, viral replication, and cellular stress responses (3, 11). Moreover, it is becoming more and more clear that different types of viruses induce important alterations of nucleoli and that these alterations may participate directly in specific processes that are crucial for the outcome of infection, like viral DNA replication, virus assembly, and control of intracellular trafficking (for reviews see references 22 and 23).
The nucleolar structure integrity depends on RNA polymerase (pol) I activity and on the correct expression of the nucleolar protein nucleolin (65). Nucleolin is one of the most abundant nonribosomal proteins of the nucleolus (6). It represents about 5 to 10% of the nucleolar protein content of exponentially growing eukaryotic cells. Nucleolin is a ubiquitous, highly phosphorylated, multifunctional, and mobile protein that shuttles between nucleoli, nucleoplasm, cytoplasm, and cell surface. It has been described as being a part of many pathways, from interactions with viruses at the cellular membrane (42) to essential roles in the regulation of gene expression, like chromatin remodeling (1), DNA recombination and replication, RNA transcription by RNA pol I and II, rRNA processing, mRNA stabilization, cytokinesis, and apoptosis (for reviews see references 38 and 61). Nucleolin is also a major actor in promoting cell proliferation. The nucleolin level is higher in tumors and actively dividing cells (13, 52, 59) whereas nucleolin depletion by RNA interference promotes cell cycle arrest and apoptosis (65). B23 and fibrillarin are two other abundant nucleolar proteins involved in ribosome biogenesis and in other functions. B23 constantly shuttles between nucleoli and the cytoplasm (5); it is also redistributed from nucleoli in response to cytotoxic drugs and genotoxic stress (68) and plays a role in the cell cycle (13, 59). B23 binds different cellular and viral proteins (for reviews see references 22 and 23).
At present, the roles that may be played by the HSV-1-induced nucleolar perturbations in viral replication and host cell functions are not elucidated, although two reports describe the delocalization of nucleolar proteins at the late stage of infection (34, 47). Nucleolin and fibrillarin are delocalized from nucleoli in HSV-1-infected cells, and the leaky late UL24 viral protein is necessary for the late dispersal of nucleoplasmic nucleolin. However, nothing is known about the early delocalization of nucleolar proteins, and the fine kinetics of this process is unknown.
Therefore, because of the importance of nucleolar proteins like nucleolin for nucleolar structure integrity and function and because some of the nucleolar functions accomplished by these proteins could be required at some stage of the viral cycle, we have hypothesized that HSV-1-induced nucleolar perturbations might occur through the deregulation of these nucleolar proteins. Furthermore, if a nucleolar “experience” is required for HSV-1 infection, we might expect that some nucleolar proteins, at least, are involved in this process.
Here, we have analyzed the behavior of nucleolin and also that of two other abundant nucleolar proteins, B23 and fibrillarin, during the course of infection. We have investigated the expression and distribution of these proteins using extensive time course analyses spanning from 3 h postinfection (p.i.) to 24 h p.i. We have tracked the intracellular distribution of these proteins by fine cell fractionation procedures and immunofluorescence experiments. We show that these proteins are redistributed out of the nucleoli after infection through an immediate-early viral gene expression-dependent mechanism. Using small interfering RNA (siRNA) technology, we show that viral replication requires a high level of nucleolin expression, demonstrating for the first time a direct role for a nucleolar protein in HSV biology.
MATERIALS AND METHODS
Cell lines, virus strain, and infection of cells.
HeLa monolayer cells were grown in Eagle's minimum essential medium (E-MEM; Sigma-Aldrich, St. Louis, MO) containing 100 U/ml penicillin and 100 μg/ml streptomycin and with or without 5% heat-inactivated fetal calf serum (FCS; Eurobio, Les Ulis, France) at 37°C under 5% CO2. HSV-1 strains used in this study included the wild-type (wt) 17+ strain (HSV-1-17) and dl110 (HSV-1ΔICP-0; ICP0 gene deleted) obtained from P. Lomonte (Université Lyon 1, Villeurbanne, France); HP66 (HSV-1ΔDNAPol; UL30 gene deleted) was provided by D. Coen (Harvard University, Boston, MA) (36) and grown on polB3 cells expressing the HSV UL30 gene (24) (provided by C. Hwang, SUNY Health Science Center, Syracuse, NY). HSV-1-KOS 5dl1.2 (HSV-1ΔICP-27; ICP27 gene deleted) and HSV-1-17 Cgal delIE3 (HSV-1ΔICP-4; ICP4 gene deleted) were produced, respectively, on Vero cells or on the 7b complementing cell line (34, 35) (provided by P. Marconi, University of Ferrara, Ferrara, Italy) expressing both the ICP27 and the ICP4 proteins.
Cells were infected just before confluence with the HSV-1 strain at a multiplicity of infection (MOI) of 10 PFU per cell, or at the MOI specified in the figure legend, in E-MEM in the presence of 1% FCS. After 1 h of adsorption of the virus at 33°C under 5% CO2, the medium was removed and the cells were washed and then incubated in E-MEM at 37°C until harvesting. Times postinfection were calculated from the time of addition of the virus.
Plaque assays.
Vero cells in six-well plates were inoculated with serial 10-fold dilutions of progeny virus. After a 1-h adsorption period at 33°C, the inoculum was removed and the cells were thoroughly washed with phosphate-buffered saline (PBS) and then overlaid with E-MEM. Plaques were counted and visualized at 72 h p.i. by fixing and staining the cells with 5% methylene blue in 70% methanol.
To measure virus production after inhibition of nucleolin expression, cells were treated with nucleolin siRNAs for 5 days. The cell monolayers were then infected with HSV-1 at an MOI of 0.4 to 10 PFU per cell. Cells and cell culture supernatants were collected at the indicated times postinfection. Virus titers were determined by plaque assay.
Pharmacological agents.
The following agents (all from Sigma-Aldrich except where noted) were used at the indicated concentration: cycloheximide (20 μg/ml), nocodazole (5 μg/ml), N-acetyl-leucyl-leucyl-norleucinal (ALLN) (5 μg/ml), and actinomycin D (10 ng/ml). For drug treatments, nocodazole and ALLN were added to the culture medium 12 h before infection.
Subcellular fractionation.
At the indicated times after infection, the cell medium was removed and cells were washed with cold PBS, pH 7.4, and then scraped into PBS. The whole-cell lysates or cytoplasmic, nuclear, nucleolar, and nucleoplasmic extracts were prepared as described previously (54). Briefly, nucleoli were purified using a procedure adapted from previously published protocols (41, 44). HeLa cells were incubated on ice in a hypotonic buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, and 1 mM MgCl2) and then lysed by addition of 0.3% Nonidet P-40. After centrifugation, the supernatant and the pellet containing the cytoplasmic and nuclear fractions, respectively, were harvested. Nuclei were then purified by centrifugation through an 0.88 M sucrose cushion containing 0.05 mM MgCl2. Purified nuclei were resuspended in 0.34 M sucrose-0.05 mM MgCl2 and disrupted by ultrasonication. Nucleoli were then purified by centrifugation of the resulting homogenate through an 0.88 M sucrose cushion containing 0.05 mM MgCl2. The supernatant containing the nucleoplasmic fraction devoid of nucleoli and the pellet containing the nucleoli were harvested.
siRNA transfection.
Two siRNA duplexes, siRNA#2 (5′-UCCAAGGUAACUUUAUUUCUU-3′) and siRNA#4 (5′-UUCUUUGACAGGCUCUUCCUU-3′), obtained from Dharmacon (Perbio Science, France) and targeting sequences in exon 13 and exons 4 and 5, respectively, of human nucleolin, were used to deplete nucleolin as previously described (65). Briefly, 1 day before transfection, HeLa cells were trypsinized, resuspended in medium without antibiotics, and transferred to six-well plates at a density of 5 × 105 cells per well. Transfection was performed with lipid transfection reagent DharmaFECT 1 from Dharmacon (Perbio Science, France) according to the manufacturer's instructions, with a final concentration of 100 nM of mixtures of the two functional siRNA duplexes specific for human nucleolin. A nonspecific siRNA duplex (RISC-free siRNA Dharmacon D-001600-01-05) was used for control transfections. Mock-transfected cells were treated with transfection reagent only. One day after, cells were submitted to a second transfection under the same conditions. Five days after the first transfection, cells were subjected to HSV-1 infection for different periods of time and then processed for appropriate analysis.
Western blot analysis.
Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred onto a polyvinylidene difluoride membrane (Immobilon-P; Millipore). The membranes were incubated with a 500× dilution of either a rabbit polyclonal antinucleolin antibody (19); antifibrillarin (N-15; Santa Cruz), anti-ICP27, or anti-US11 antibodies (14); or mouse monoclonal anti-B23 (H-106; Sigma), anti-β-actin (AC-15; Sigma), anti-histone H3 (Abcam), or anti-UL42 antibodies. Anti-ICP27 and anti-UL42 antibodies were kindly provided by H. Marsden (antibodies 42 and Z1F11, respectively) (53, 58). Proteins were revealed by chemiluminescence (ECL from Amersham Biosciences) using an anti-rabbit or an anti-mouse peroxidase-conjugated antibody (Sigma) or an anti-goat peroxidase-conjugated antibody (Santa Cruz), diluted 1:10,000.
Indirect immunofluorescence assay and confocal microscopy.
For immunostaining, HeLa cells grown on glass slides were mock infected or infected for various times with HSV-1, as indicated in the figure legends. Then, cells were washed with PBS, fixed with formaldehyde (3% in PBS), and then permeabilized with 1% Triton in PBS. Glass slides were incubated at room temperature for 1 h with the blocking buffer (1% bovine serum albumin and 0.5% Tween 20 in PBS) containing 3% FCS and then for 2 h with the primary antibodies diluted in the blocking buffer plus 1% FCS. Cells were then washed several times before treatment for 45 min with the secondary antibodies in the same manner. Subsequently, glass slides were washed three times with PBS containing 10 mM glycine and mounted in Fluoromount G mounting medium (Southern Biotechnology) containing DAPI (4′,6′-diamidino-2-phenylindole; Fluka) to stain the DNA. Cells were analyzed by confocal laser scanning microscopy (Leica TCS SP2). Appropriate emission filters were used for double- and triple-labeling experiments. Images were merged by computer. To control for cross-reactivity, samples were stained with one primary antibody and appropriate secondary antibodies. No overlap between the channels was observed for any of the samples at the settings used.
The primary antibodies were either rabbit polyclonal antibodies—antinucleolin (1:200 dilution) (19), anti-US11 (1:200 dilution) (14), and anti-ICP27 (1:100 dilution)—or mouse monoclonal antibodies—anti-B23 (H-106; Sigma), anti-ICP8 (39-S; ATCC) (1:5 dilution), anti-lamin A/C (636; Santa Cruz), anti-γ-tubulin (GTU-88; Sigma), and anti-vimentin (V9; Dako). The conjugated secondary antibodies used were donkey anti-rabbit or anti-mouse Fluoprobes 488 (green)- or 546 (red) (Molecular Probes)-conjugated goat anti-rabbit antibody or goat anti-rabbit or anti-mouse Alexa Fluor 633 (long red) (Invitrogen)-conjugated antibody.
The immunofluorescence figures in this report show representative data. Each experiment was reproduced multiple times, and the cells are representative of the overall effects observed under each set of conditions.
RESULTS
(i) Increase in the amount of nucleolar proteins during HSV-1 infection.
We investigated whether the steady state, profiles, and intracellular distribution of nucleolin, B23, and fibrillarin were modified after HSV-1 infection. HeLa cells were infected for different periods of time. Light microscopy observations revealed that, as expected, morphology of nucleoli were modified during the course of infection, although the nucleoli remained visible even at late times of infection (data not shown).
The amounts of nucleolar proteins were tracked by Western blot analyses. We first focused on the total amount of nucleolin in infected cells. As seen in Fig. 1A, during the first hours of infection the global amount of nucleolin was not dramatically modified. Then, from 6 h p.i., this amount was progressively increased, whereas that of β-actin, a control housekeeping protein, was not affected and that of the late viral protein US11, used as a control for viral infection, increased progressively.
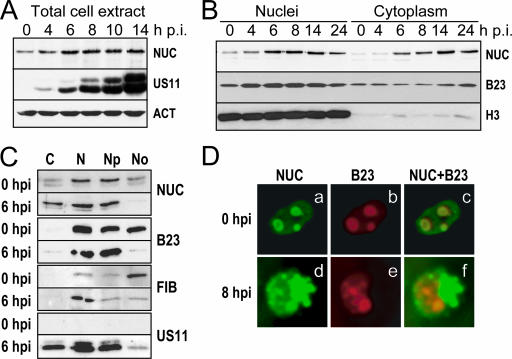
FIG. 1.
Analysis of nucleolar protein behavior during HSV-1 infection. HeLa cells were either mock infected (0 h p.i.) or infected for different periods of time (4 to 24 h) with 10 PFU of the 17+ wt strain of HSV-1 per cell. Quantitation of the amounts of nucleolar proteins was performed by Western blot analysis (A to C) using polyclonal anti-human nucleolin (NUC) or fibrillarin (FIB) antibodies or monoclonal anti-human B23 antibody (B23). (A) Nucleolin analysis from 20 μg total cell protein extracts. (B) Nucleolin and B23 analysis in protein extracts from 20-μg cytoplasmic and 20-μg nuclear subfractions. (C) Nucleolin, B23, and fibrillarin analysis in protein extracts from 10-μg cytoplasmic (lane C), 10-μg nuclear (lane N), 10-μg nucleoplasmic (lane Np, nuclei devoid of nucleoli), and 2.5-μg nucleolar (lane No) subfractions. Antibodies to the viral late protein US11 (A and C), to the housekeeping β-actin protein (ACT) (A), or to the nuclear protein histone H3 (H3) (B) were used as a control for infection, protein loading, or the quality of the fractionation procedure, respectively. (D) Fluorescence microscopy views (magnification, ×40) of intracellular location of nucleolin and B23. Mock-infected and 8-h-infected HeLa cells grown on glass slides were fixed and stained with a rabbit polyclonal antibody to human nucleolin (a and d) and with a mouse monoclonal antibody to B23 (b and e). Merged images of the two channel signals are given (c and f).
Western blot analyses were then performed with proteins purified from cytoplasmic and nuclear fractions of mock-infected cells and cells infected for 4 to 24 h. Data presented in Fig. 1B show that, as expected, nucleolin was barely detectable in the cytoplasm of mock-infected cells. This was the case also in cells infected for 4 h. At 6 h p.i. the amount of nucleolin increased in the nuclei as well as in the cytoplasm. Histone H3 used as a control for specific nuclear protein was present in large amount in the nuclei and almost undetectable in the cytoplasm, indicating that the presence of cytoplasmic nucleolin in infected cells was not due to a contamination of the cytoplasmic fraction with nuclei during the fractionation procedure. These results suggest that the intracellular distribution of nucleolin is modified during the course of infection.
To verify whether these modifications are common to other abundant nucleolar proteins, we performed the same analysis with B23 and found that the amount of B23 protein was less modified in the nuclei and cytoplasm of infected cells (Fig. 1B).
To gain insight into the intracellular distribution of nucleolar proteins in infected cells, we compared the amounts of nucleolin, B23, and fibrillarin in different subcellular compartments, including cytoplasm, nuclei, nucleoli, and nucleoplasm (nuclei devoid of nucleoli), in mock-infected cells, and in cells infected for 6 h. The results of Western blot analysis in Fig. 1C confirm the localization of nucleolin in the cytoplasm of infected cells and reveal that the amount of nucleolin in the nucleolar fraction was strongly decreased since it was not detectable at 6 h p.i. In contrast, at 6 h p.i., B23 and fibrillarin were barely or not at all detectable in the cytoplasm of infected cells; these two proteins remained essentially nuclear. However, the amount of B23 and fibrillarin in nucleoli decreased, and the decrease of nucleolar B23 was more drastic than that of nucleolar fibrillarin.
Immunofluorescence analyses performed on mock-infected and 8-h-infected cells confirmed that nucleolin and B23 are visible essentially in nucleoli of mock-infected cells where they colocalize (Fig. 1D, panels a to c). Conversely, in infected cells, the localization of the two proteins was very different from that in mock-infected cells, and at 8 h p.i. only a part of nucleolin and B23 colocalized (Fig. 1D, panels d to f).
These results indicate that upon HSV-1 infection the amount and localization of nucleolar proteins are differentially affected.
(ii) Nucleolin localizes in different intracellular compartments during HSV-1 infection.
To investigate the behavior of nucleolin during HSV-1 infection, we carried out a time course of nucleolin delocalization in HeLa cells infected for 4 to 24 h with a wt strain of HSV-1. The intracellular distribution of nucleolin was examined by multiple-immunofluorescence detection of proteins, followed by confocal microscope observations (Fig. 2). The VRC were visualized with specific antibody directed against the ICP8 viral protein. ICP8 is an essential DNA replication protein with single-stranded DNA (ssDNA)-binding activity that is found exclusively in the VRC in the nucleus where viral transcription and DNA replication occur (7). Cells were simultaneously counterstained with the DNA-binding fluorochrome DAPI to reveal the intranuclear distribution of the proteins. In these and all subsequent colocalization experiments, representative cells are shown, and in all instances, the majority of the cells displayed the colocalization pattern shown and results have been verified in at least two independent experiments.
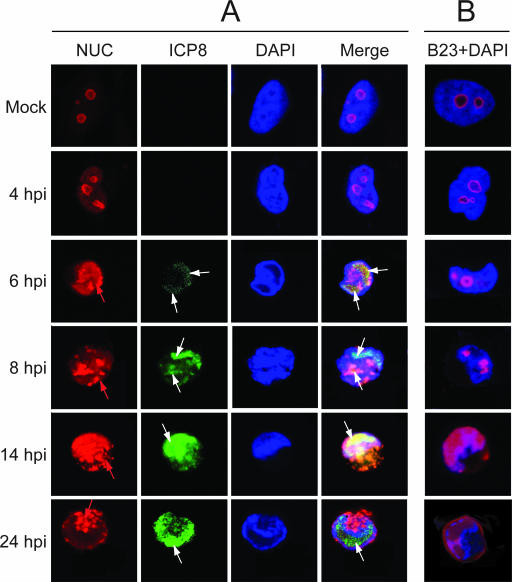
FIG. 2.
Dynamic localization of nucleolin and B23 during the course of HSV-1 infection. HeLa cells grown in a coverslip chamber were infected with the HSV-1 17+ strain at a multiplicity of 10 PFU per cell. The cells were examined at different times postinfection (4 to 24 h), and mock-infected HeLa cells were used as a control. (A) Immunofluorescence and subsequent confocal laser scanning microscopy analysis were carried out with antibodies to either nucleolin (NUC; red) or ICP8 (green) as a VRC-specific protein and with DAPI (blue) to stain the DNA. The positions of the VRC are indicated by white arrows, and those of aggregated nucleolin are indicated by red arrows. The merge of nucleolin, ICP8, and DAPI signals is given. Representative pictures of three independent experiments. (B) Colabeling of B23 nucleolar protein (red) and DAPI (blue) in HeLa cells from the same experiment as that presented in panel A. The confocal images were collected with a focal depth of 0.5 μm.
In mock-infected cells and at 4 h p.i., nucleolin was found exclusively in nucleoli (Fig. 2A). Later, infection of cells leads to rapid changes in nucleolin localization. From 6 to 24 h p.i., nucleolin exhibited different patterns and was present in two different forms, a diffuse one and a granular one indicated by a red arrow. From 6 h p.i., nucleolin increased in amount, and up to 6 to 8 h p.i., it was mainly present in the nucleus. As early as ICP8 was detected (6 h p.i.) and while nuclear membrane invagination occurred, part of nucleolin colocalized with sites of ICP8 staining. From 6 to14 h p.i., the signals of ICP8 and of part of nucleolin clearly overlapped, indicating that at least a fraction of nucleolin colocalizes with ICP8 in the VRC in the nucleus during the process of infection.
As infection progressed, the presence of nucleolin was not restricted to the VRC. Indeed, nucleolin was found also in structures devoid of ICP8 (Fig. 2A, 14 to 24 h p.i.); granular dots of nucleolin appeared as early as 6 to 8 h after infection, and they progressively increased in size and amount. At 14 h p.i. a part of nucleolin was still present in the VRC; however, the granular form of nucleolin became predominant and accumulated in structures that did not contain ICP8. This granular form of nucleolin was located in the part of the cell that was not stained by DAPI, suggesting an extranuclear localization. At 24 h p.i. nucleolin did not colocalize anymore with ICP8 and was present mainly in structures located at the periphery of the nucleus and in the invagination of the nucleus. These structures that contain granular nucleolin resemble aggresome in their morphology and localization (see below).
The pattern of delocalization of nucleolin seems to be specific since the nucleolar protein B23 behaves in a different way, as seen in Fig. 2B. Until 6 h p.i., B23 remained in structures that displayed a nucleolar morphology. Then, from 8 to 24 h p.i., it localized in a structure that was devoid of the characteristic blue DAPI staining, indicating that this could be VRC. At 24 h p.i. part of B23 was present at the periphery of the nucleus. In contrast to nucleolin, B23 was never seen in a granular form.
Therefore, during HSV-1 infection, the two abundant nucleolar proteins, nucleolin and B23, are delocalized from nucleoli. Nucleolin was detected in VRC as early as 6 h p.i., whereas B23 localized from 8 h p.i. in a structure that could be VRC and continued to localize in this structure at very late times of infection (24 h p.i.) when nucleolin seemed to be excluded from the VRC and was instead present in granular structures and at the periphery of the nucleus. At 24 h p.i. parts of both nucleolin and B23 were present at the periphery of the nucleus. In addition, the DAPI labeling confirmed that, from 4 h p.i., the nucleus was morphologically modified and the labeling also revealed that granular nucleolin was proximal to the invagination of the nucleus. This was particularly obvious at late times of infection.
(iii) Immediate-early viral functions are necessary for nucleolin delocalization.
Infection of cells with HSV-1 results in the selective and progressive inhibition of cellular protein synthesis (reviewed in reference 49). The delocalization of nucleolin upon infection could be the result of the inhibition of cellular protein synthesis. To test this hypothesis, mock-infected and infected cells were treated with cycloheximide, which inhibits the elongation of polypeptides during the translation process. In infected cells, cycloheximide was added either simultaneously with the viral particles or from 2 or 4 h p.i. until the end of the experiment. Its effect on nucleolin was observed by immunofluorescence analysis (Fig. 3A). These observations show that when cycloheximide was added together with the infectious viral particles, nucleolin remained exclusively nucleolar (Fig. 3A, fourth panel from left), as was the case for mock-infected control cells (compare Fig. 3A, two leftmost panels and panel fourth from left). In contrast, addition of cycloheximide to cells from 2 or 4 h p.i. led to delocalization of nucleolin out of the nucleolus (Fig. 3A, two rightmost panels, respectively) in a manner similar to that of infected control cells that were not submitted to treatment (Fig. 3A, third panel from left). At these times of infection at least some of the immediate-early and early proteins, respectively, are already synthesized. Thus, the delocalization of nucleolin in infected cells requires ongoing viral gene expression and is not simply a consequence of the inhibition of host cell translation.
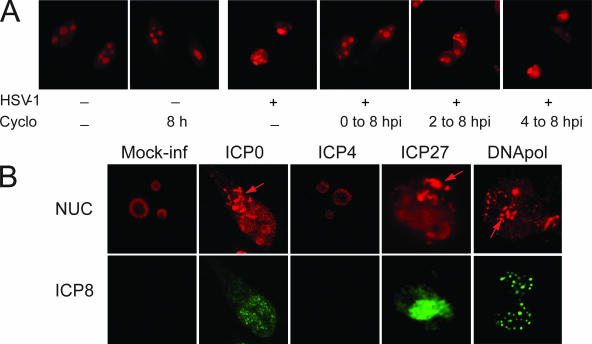
FIG. 3.
Viral functions necessary for nucleolin delocalization. (A) HeLa cells infected for 8 h with 10 PFU per cell of HSV-1 (four rightmost panels) were either untreated (third panel from left) or treated with 20 μg/ml of cycloheximide (Cyclo, three rightmost panels) from the beginning of infection (fourth panel from left) or from 2 h p.i. (fifth panel from left) or 4 h p.i. (rightmost panel) until the end of the experiment. Mock-infected HeLa cells were used as a control; they were either untreated (leftmost panel) or treated for 8 h with cycloheximide (second panel from left). After harvesting, cells were fixed and stained with antinucleolin antibody and analyzed by fluorescence microscopy (magnification, ×40). (B) HeLa cells were either mock infected (leftmost panels) or infected for 16 h (four rightmost panels on top and bottom) with 10 PFU per cell of the viral strains dl110 (second panels from left), HSV-1-17 Cgal delIE3 (third panels from left), HSV-1-KOS 5dl1.2 (fourth panels from left), and HP66 (rightmost panels) which are defective in the specific viral genes ICP0, ICP4, ICP27, and DNA pol as indicated on the top of the corresponding panels. Immunofluorescence and subsequent confocal laser scanning microscopy analysis were carried out with antibodies to either nucleolin (in red; top panels) or ICP8 (in green; bottom panels) as a control for viral infection and for the presence of VRC or pre-VRC. The positions of the aggregated form of nucleolin are indicated by red arrows.
In order to better understand the viral requirements for the delocalization of nucleolin and for the formation of nucleolin granular dots, we analyzed the nucleolin localization by immunofluorescence in cells infected with HSV-1 strains defective in specific viral genes. The mutant viruses used derived from wt strains and correspond to viruses in which the ICP0 (dl110), ICP4 (HSV-1-17 Cgal delIE3), and ICP27 (HSV-1-KOS 5dl1.2) immediate-early genes or the DNA pol (HP66) early viral gene has been deleted. Results of immunofluorescence analysis are presented in Fig. 3B. Redistribution of nucleolin was seen in cells infected with HSV-1 mutants with deletions of ICP0 and ICP27 immediate-early genes (Fig. 3B, top row, second and fourth panels from left) and for DNA pol early gene (Fig. 3B, top row, rightmost panel). In these cells, nucleolin was present in both diffused and granular forms, and there was a concordance of ICP8 staining and of the diffused form of nucleolin (Fig. 3B, bottom row, second panel from left and two rightmost panels), although in the case of DNA pol mutant, there was formation of punctate prereplication compartments as expected. In contrast, in cells infected with a viral mutant with a deletion of the ICP4 immediate-early gene, nucleolin remained exclusively in the nucleoli, and as expected ICP8-containing replication compartments were not formed (Fig. 3B, third panels from left). These data suggest that ICP4 bears functions that are absolutely required for the delocalization of nucleolin upon infection and that the viral processes that occur before the beginning of ICP4 synthesis are not sufficient. Furthermore, the delocalization of nucleolin seen in cells infected with ΔDNA pol mutant indicates that late viral functions are not required for the process.
(iv) Inhibition of nucleolin expression results in reduction of viral proteins and of infectious progeny virus yields.
Our present results show that nucleolin undergoes important modifications in HSV-1-infected cells, suggesting that nucleolin could be one of the cellular protein that plays a role in viral replication. We predicted that inhibition of its expression would result in changes in viral replication. To test this possibility, we carried out experiments using siRNA strategy to knock down nucleolin expression. HeLa cells were transfected with either nucleolin-specific siRNAs that have already been shown to down-regulate nucleolin expression or control scrambled siRNAs (65). Five days posttransfection, the level of nucleolin was assayed by immunofluorescence using antibody to nucleolin (Fig. 4A). Nucleolin staining was very low or undetectable in most of the cells transfected with nucleolin siRNAs, compared to cells transfected with control scrambled siRNAs, which have normal levels of nucleolin (compare Fig. 4A, panels a and d). This confirms that the nucleolin siRNAs used induce a considerable reduction in nucleolin levels upon transfection. Nucleoli from silenced and nonsilenced cells were visualized by the use of an antibody to B23, the other abundant standard nucleolar protein. All the cells showed an intense B23 staining as demonstrated by the merge of B23 and nucleolin immunostaining images (Fig. 4A, panels b and c and panels e and f), indicating that the level of B23 and the integrity of the nucleoli were not affected in cells where nucleolin expression was knocked down.
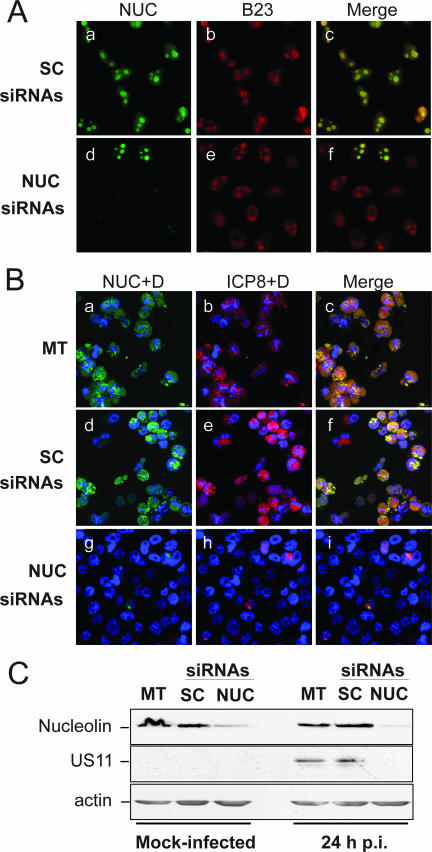
FIG. 4.
Down-regulation of nucleolin by siRNA induces inhibition of HSV-1 infection. HeLa cells were transfected with either nucleolin siRNAs (NUC siRNAs) or control scrambled siRNAs (SC siRNAs) or were mock transfected (MT). After 24 h, cells were transfected again with the same siRNAs. Five days after the first transfection, cells were infected for 24 h with 10 PFU of wt HSV-1 per cell. Six days after the first transfection, cells were submitted either to immunofluorescence or to Western blot analysis. Nucleolin was visualized by the use of a specific polyclonal antibody (in green). (A) Detection of nucleolin (NUC) in green (a, c, d, and f) of mock-infected cells 6 days after the first transfection, showing effective depletion of nucleolin. Detection of B23 in red (b, c, e, and f) was used as a control for nucleolar integrity. (B) Immunofluorescence images of transfected cells infected for 24 h showing inhibition of viral infection in nucleolin knockdown cells. Detection of nucleolin (a, c, d, f, g, and i). Detection of ICP8 (b, c, e, f, h, and i) was used as a control for viral infection and for the VRC. DAPI staining (D) is in blue. (C) Western blot analysis of experiments presented in panels A and B, using antibodies to nucleolin and the viral late protein US11 or to the housekeeping β-actin protein as a control for infection or for protein loading, respectively.
To determine if nucleolin depletion can affect the efficiency of viral infection, cells were submitted to HSV-1 infection 5 days posttransfection with the siRNAs. The effect on viral infection was determined 24 h p.i. by immunofluorescence using an anti-ICP8 antibody (Fig. 4B). It appears very clearly that in cells transfected with nucleolin siRNAs, the ICP8 staining was restricted to the small numbers of cells that have not been transfected by the siRNA showing a detectable amount of nucleolin (Fig. 4B, panels g to i), while it was detected in all control cells, both in mock-transfected cells (Fig. 4B, panels a to c) and in cells transfected with scrambled siRNAs (Fig. 4B, panels d to f). Western blot analysis presented in Fig. 4C confirmed that, in mock-infected cells and in cells infected for 24 h, the amount of nucleolin was drastically decreased in cells transfected with nucleolin siRNAs (NUC), compared to mock-transfected cells (MT) or cells transfected with scrambled siRNAs (SC). This indicates that the efficient down-regulation of nucleolin was specific and was not due to the transfection procedure. In addition, by using antibodies directed against the late viral protein US11 and against the housekeeping protein β-actin we observed that in infected cells, the levels of US11 protein were strongly decreased only in cells transfected with nucleolin siRNAs while the level of the control β-actin was not significantly modified. These data confirmed the results of the immunofluorescence experiment and demonstrate that viral expression was inhibited in cells where nucleolin expression was down regulated. Altogether, these results suggest that HSV-1 expression requires the presence of nucleolin, since both the expression of the early ICP8 (Fig. 4B, panels h and i) and that of the late US11 (Fig. 4C) viral protein is inhibited in its absence.
It has been shown that nucleolin could interfere with viral penetration. Indeed, in the case of human immunodeficiency virus, nucleolin is a receptor for viral penetration, and functional anchorage of virus particles on target cells requires nucleolin (42). Therefore, the effects of changes in nucleolin expression on virus adsorption and production were estimated by plaque titration assays (Fig. 5).
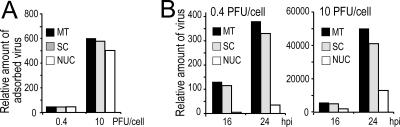
FIG. 5.
Knockdown of endogenous nucleolin by siRNA results in decrease in virus titers. HeLa cells were transfected as indicated in the legend to Fig. 4. Five days after the first transfection cells were infected at an MOI of 0.4 or 10 PFU of wt HSV-1 per cell, as indicated in each panel. After harvesting, virus titers were determined by plaque assay. (A) Estimation of the amount of viral particles adsorbed on the cell surface. Cells were harvested after the first 2 h of contact with the virus at 4°C. Viral titers were determined from cells after removal of the medium and thorough washing of the cells. (B) Estimation of the amounts of progeny viruses from cells and cell culture supernatants harvested after 16 or 24 h of infection at 37°C as indicated.
Five days after transfection with siRNAs, cells were infected for 2 h at 4°C using various amounts of HSV-1 particles (MOI = 0.4 to 10 PFU per cell) and then harvested (Fig. 5A). These experimental conditions allowed viral adsorption and not viral replication. There was no significant difference in the amounts of infectious viral particles adsorbed on the surface of cells where nucleolin was knocked down compared to cells having a normal nucleolin level, even when infection was performed with high amounts of virus (MOI = 10 PFU per cell). This suggests that adsorption of the virus is not repressed in cells where nucleolin expression is inhibited.
We then determined the effect of repression of nucleolin expression on the amounts of progeny virus produced in cells infected for 16 h or 24 h (Fig. 5B). We observed a modest reduction in the titers of virus produced from cells transfected with the control scrambled siRNAs compared to mock-transfected control cells. This indicates that transfection of cells with nonspecific siRNAs has no significant consequence on viral production. In contrast, virus yields were reduced 23- and 10-fold in cells transfected with nucleolin siRNA and infected for 16 or 24 h, respectively, at an MOI of 0.4 PFU/cell, compared to cells transfected with control scrambled siRNAs. In cells infected at a very high MOI (10 PFU/cell), virus yields were reduced more than threefold in nucleolin-knockdown cells. These data clearly confirm that viral infection requires nucleolin for efficient infection.
(v) Granular nucleolin mainly localized in extranuclear aggresome structures in infected cells.
Our results show that nucleolin is required for the outcome of HSV-1 infection and that, soon after infection, nucleolin increases in amount and is first delocalized in the VRC and then accumulates in granular dots located in juxtanuclear structures that display morphological homology with aggresomes. It has been established that aggresomes are formed by the deposition of misfolded proteins in a large structure surrounding the microtubule-organizing center (MTOC) and is enclosed in a characteristic vimentin cage. This process, which is accompanied by a reorganization of the intermediate filaments of the cytoskeleton, requires an intact microtubule cytoskeleton and does not require proteasome inhibition (16, 26, 27).
We therefore undertook analyses in order to verify whether these structures containing granular dots of nucleolin satisfy the definition of aggresomes. For this, mock-infected cells and cells infected for 8 h were submitted to indirect coimmunofluorescence experiments using specific antibodies to nucleolin and to lamin, which is a component of the lamina that delimitate the inner face of the nuclear envelope (55). In mock-infected cells, nucleolin staining was exclusively in the nucleus (Fig. 6A, panels a to c). At 8 h p.i., the nongranular form of nucleolin was located inside the lamin layer, while the granular dots of nucleolin were located outside the lamin layer (Fig. 6A, panels d to f). This result led us to conclude that the structures containing the granular dots of nucleolin were in the cytoplasm of infected cells and raises the question of whether they represent some type of aggresome.
FIG. 6.
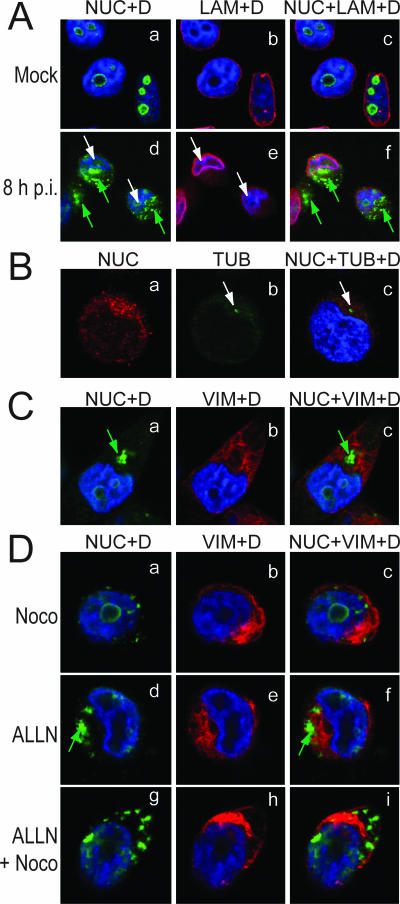
Aggregated nucleolin has the characteristics of aggresomes. (A) Aggregated nucleolin has a cytoplasmic localization. The subcellular localization of aggregated nucleolin was determined by double immunolabeling of HeLa cells either mock infected (a to c) or infected for 8 h with 10 PFU per cell of the 17+ wt strain of HSV-1 (d to f). Confocal images were obtained from immunofluorescence analysis carried out with antibodies to nucleolin (NUC in green, panels a and d) and to lamin (LAM in red, panels b and e), a component of the lamina that delimitates the inner face of the nuclear envelope. DAPI staining (D) is in blue. The white and green arrowheads indicate VRC and aggregated nucleolin, respectively. The merge of nucleolin, lamin, and DAPI signals is given in panels c and f. (B) Aggregated nucleolin localizes around the centrosome. HeLa cells infected for 8 h were labeled with antibodies to nucleolin (NUC in red) and to γ-tubulin (TUB in green) and with DAPI (D) as indicated at the top of the confocal images. The position of the centrosome is indicated by a white arrow. (C) Aggregated nucleolin is enclosed in a vimentin cage. HeLa cells infected for 8 h were stained with antibodies to nucleolin (NUC in green) and to vimentin (VIM in red). DAPI staining (D) is in blue. (D) Formation of large aggregates of nucleolin depends on microtubule integrity and not on proteasome inhibition. HeLa cells were incubated for 12 h with 5 μg/ml nocodazole (Noco) (a to c), 5 μg/ml ALLN (d to f), or both drugs (g to i) and then infected for 8 h as indicated in panel A, in the presence of the same concentrations of drugs. Formation of the large perinuclear aggregated nucleolin is prevented by nocodazole treatment (a to c and g to i) and not by ALLN treatment (d to f). DAPI staining (D) is indicated. In panels C and D, green arrows indicate the position of the caged nucleolin. Digital images were obtained by confocal laser scanning. Representative data from several independent experiments.
To answer this question, we first performed colocalization studies using antibodies specific to nucleolin and to γ-tubulin, a marker of MTOC. We showed that in infected cells granular dots of nucleolin localized around γ-tubulin at the MTOC (indicated by an arrow in Fig. 6B, panels b and c), near the characteristic distortion region of the nucleus, indicating that they localize around the MTOC. Then using antibodies to vimentin (Fig. 6C), we show that at 8 h p.i. vimentin was distributed throughout the cell and in concentrated foci within the cytoplasm near the distortion of the nucleus, and vimentin formed a ring surrounding the core of the intense perinuclear signal of granular nucleolin, indicating that this large structure containing nucleolin was enclosed inside a typical vimentin cage.
Since granular dots of nucleolin have typical features of proteins present in aggresomes, we further tested whether the formation of these nucleolin-containing structures depends on microtubule integrity. Cells were treated prior to infection with 5 μM nocodazole, which is an antimitotic agent that causes the depolymerization of microtubules (67). In cells infected for 8 h, treatment with nocodazole alone resulted in an increase in the number of cytoplasmic structures containing nucleolin (compare panel a of Fig. 6D to panel a of Fig. 6C). These structures were smaller and were located throughout the nucleus and the cytoplasm of infected cells as evidenced by nucleolin-vimentin and DAPI staining (Fig. 6D, panels a to c). These smaller nucleolin structures were not surrounded by a vimentin cage, indicating that the formation of large perinuclear structures containing nucleolin requires intact microtubules.
To assess the role of the proteasome in the formation of granular dots of nucleolin, HeLa cells were treated with 5 μg/ml of the specific proteasome inhibitor ALLN prior to infection. At this concentration there was no clear evidence of cell toxicity, as reported by other studies using ALLN (4, 18). In the presence of ALLN alone, there was accumulation of large nucleolin-containing structures surrounded by vimentin (Fig. 6D, panels d to f), indicating that inhibition of proteasome has no effect on the formation and localization of granular nucleolin dots.
In cells treated with both nocodazole and ALLN, the amount and the size of nucleolin aggregates were increased compared to cells treated with nocodazole alone (compare panels a and g of Fig. 6D). However, these aggregates were smaller than those present in untreated cells or in cells treated with ALLN alone (compare panel a in Fig. 6C and panel d in Fig. 6D to panels a and g in Fig. 6D) and were not caged by vimentin.
Altogether, these results indicate that in infected cells, the perinuclear structures containing granular nucleolin have the characteristics of aggresomes.
DISCUSSION
In this paper we present evidence that the behavior of nucleolar proteins is rapidly modified after infection with HSV-1. The analysis of the intracellular distribution of the three most abundant nucleolar proteins—nucleolin, B23, and fibrillarin—revealed that after infection, these proteins are delocalized from the nucleoli. This is in agreement with previous reports (34, 47). In addition, we demonstrate that at early times of infection, while nucleolin is translocated to both the nucleoplasm and the cytoplasm, the two other proteins remain in large part nuclear.
In addition, the amount of nucleolin increases progressively during the course of infection. This is obvious from 6 h p.i. in the nuclei of infected cells, and the increase is more pronounced in the cytoplasmic fraction, given that cytoplasmic nucleolin is almost undetectable in mock-infected cells and becomes very abundant as early as 6 h p.i. However, the levels of B23 and fibrillarin are less significantly affected in HSV-1-infected cells. Therefore, the expression and localization of these three nucleolar proteins are differentially regulated during the course of infection.
It is well documented that in HSV-1-infected cells there is a repression of the synthesis of most of the host proteins. In previous reports we have speculated that the small number of cellular proteins that continue to be efficiently synthesized upon infection could play a major role in determining the outcome of infection (20, 21, 57). We have already shown that this is the case for the S-adenosylmethionine decarboxylase, whose synthesis is increased after infection, and the inhibition of its activity prevents HSV-1 infection. Our present observation demonstrating that knockdown of nucleolin expression by the siRNA strategy results in a drastic decrease in the production of infectious virus strongly supports the notion that nucleolin is also a member of this class of cellular proteins whose function is required for an efficient HSV-1 outcome. This result is reinforced by the observation that infection occurs exclusively in the small number of cells in which nucleolin remains detectable after siRNA treatment.
Several publications have reported that nucleolin is involved in different steps during infection with various viruses (for reviews see references 22 and 23). At this stage of the study we cannot firmly conclude by which molecular mechanism nucleolin promotes HSV-1 outcome, although, based upon our results, it can be proposed that nucleolin very probably plays a role during the early phase of the viral cycle and could affect viral DNA replication. Obviously the initial step of infection is not inhibited, since viral adsorption is as efficient in cells transfected with nucleolin siRNA as in cells transfected with scrambled siRNAs or in mock-transfected cells. This clearly shows that nucleolin is not involved in the virus penetration step, as is the case for human immunodeficiency virus (42). One cannot exclude as well the possibility that HSV-1-induced alteration of nucleolin and B23 is related to the blockage of cell cycle progression at the S phase that is observed after infection, since it has been shown recently that inhibition of nucleolin expression results in a cell growth arrest, accumulation in G2, and an increase of apoptosis (65).
At early times of infection, nucleolin first delocalizes from the nucleolus to the nucleoplasm and colocalizes in part with ICP8 in the VRC where viral DNA replication takes place. ICP8 is one of the seven early viral proteins essential for viral genome replication that interacts with ssDNA (9, 29, 30). Therefore, because nucleolin is a multifunctional protein, involved in chromatin structure and dynamics (1, 38, 60, 61), and because it localizes in the VRC at the time when viral replication is effective, it can be speculated that nucleolin is involved in some aspects of viral DNA replication. Indeed, several reports suggest that nucleolin could play a role in DNA metabolism including DNA replication, recombination, and repair. In vitro, nucleolin forms a ternary complex with the simian virus 40 helicase T-antigen and topoisomerase (56). Nucleolin mediates the cohesion of the T-antigen helicase holoenzyme during plasmid unwinding, enabling the formation of a functional complex at the replication fork.
It has been also shown that nucleolin interacts in vitro with replication protein A (RPA) (12, 28). RPA is an ssDNA binding protein of eukaryotic cells which is necessary for both initiation and elongation steps of chromosomal DNA replication (23, 25), and it has been shown that RPA is associated with ICP8 within VRC (64), although it is not known if RPA is involved in HSV replication. Nucleolin binding to RPA inhibits DNA replication initiation (12). B23 is also translocated in a structure that could be VRC, and this protein was also shown to play a role in DNA replication (45, 62, 63). However, the presence of B23 in this structure is delayed compared to localization of nucleolin in VRC, suggesting that the two proteins very probably display different functions.
Later on, nucleolin delocalizes from the nucleus to the cytoplasm of infected cells. However, while nuclear nucleolin has a diffused pattern, the cytoplasmic nucleolin is granular and is concentrated at the periphery of the nucleus. These nucleolin-containing aggregates have the characteristic localization and morphology of aggresomes. Our data clearly demonstrate that this is the case. Indeed, the aggregate structures containing nucleolin are localized in the cytoplasm of infected cells. Like aggresomes, they are located close to centrosomes and are enclosed in a characteristic vimentin cage. In further agreement with aggresomes, our data suggest that small aggregates of nucleolin are transported by intact microtubules to form larger aggregates, since inhibition of microtubule dynamics by the antimitotic agent nocodazole abrogates the formation of large nucleolin aggregates. Furthermore, as is the case for aggresomes, proteasome inhibition lead to an increase in aggregated nucleolin formation. Therefore, the results of our immunocytochemical studies indicate that the structures containing aggregated nucleolin are almost identical to what have been referred to as aggresomes (16, 27, 51). The aggregated structures observed in HSV-1- and HSV-2-infected cells are different since HSV-2 aggresome-like structures are not enclosed in a vimentin cage surrounding the MTOC, in contrast to the HSV-1 aggresomal structures containing nucleolin described here, suggesting different functions (43).
Nucleolin is delocalized from the nucleolus to the VRC of HSV-1-infected cells at early times of infection until at least 14 h p.i. and then to juxtanuclear bodies containing aggregated nucleolin. Therefore, one can imagine that HSV-1 induces a stabilization of nucleolin which is delocalized from nucleoli to reach the VCR to fulfill a transitory function and that the excess amount of nucleolin, either newly synthesized or preexisting, is sequestered within the aggresome due to the saturation of proteasome activity that can be anticipated after HSV-1 infection. The aggresome accumulates misfolded proteins destined for degradation by the ubiquitin-proteasome pathway, and it was proposed that the formation of an aggresome is a general cellular response to the presence of aggregated, nondegraded proteins (17). In the case of HSV-1 and HSV-2, the composition, the function, and the dynamics of cytoplasmic aggresomes or aggresome-like structures are far from being elucidated (43, 66).
The results of our experiments using cycloheximide and HSV-1 mutants demonstrate that the early delocalization of nucleolin from the nucleolus is not the consequence of the release of viral proteins from the infecting particles, or of cellular modifications induced by these proteins, but requires viral gene expression. We show that nucleolin does not delocalize from nucleolus in cells infected with a mutant lacking ICP4, while it delocalizes in cells infected with mutants lacking the ICP0 or ICP27 gene. This shows that the immediate-early protein ICP4 is required for the early delocalization of nucleolin from the nucleolus. This strongly suggests that the potential function of nucleolin delocalization on HSV-1 metabolism, very probably induced by ICP4 or by a factor depending on ICP4, occurs after the immediate-early phase and plays a major role, since ICP4 is an essential viral gene without which HSV-1 infection is precluded.
It has been recently shown that the leaky-late UL24 viral protein is necessary for the late dispersal of nucleolin occurring from 9 h p.i. to 18 h p.i. (34). It is known that the expression of UL24 requires ICP27 protein (46). However, our results show that the presence of ICP27 and the replication of the viral DNA are not required for the early delocalization of nucleolin out of the nucleoli and for the formation of aggregates of nucleolin observed as soon as 6 h p.i., when UL24 protein is almost undetectable. Altogether, these results show that nucleolin is delocalized from the nucleolus very early after the beginning of HSV-1 infection. The factor(s) responsible for the early delocalization of nucleolin from the nucleolus remains to be identified. However, our results suggest that the expression of these factors does not require the presence of ICP0 and ICP27 but requires that of ICP4, since the early delocalization of nucleolin does not occur in cells infected with an ICP4-defective HSV-1 mutant.
Acknowledgments
This work was supported by CNRS, INSERM, and Université Claude Bernard Lyon-1. A.C., A.G., and J.-J.D. are members of INSERM.
We are grateful to H. Marsden (MRC, Glasgow, United Kingdom) for the generous gift of anti-ICP27 and anti-UL42 antibodies; to P. Schaffer (Harvard Medical School, Boston, MA), D. Parris (Ohio State University, Columbus, OH), and P. Marconi (University of Ferrara, Ferrara, Italy) for generous gifts of HSV-1-KOS 5dl1.2 (HSVΔICP27) and HSV-1-17 Cgal delIE3 (HSVΔICP4) HSV-1 deletion mutants and the 7b complementing cell line, respectively (35); to P. Lomonte (University of Lyon, Lyon, France) for the generous gift of dl110 (HSV-1ΔICP-0) (32); and to D. Coen (Harvard University, Boston, MA) (36) and C. Hwang (24) (SUNY Health Science Center, Syracuse, NY) for the generous gifts of HP66 (HSV-1ΔPol) and polB3 cells expressing the HSV UL30 gene, respectively. We are grateful to Denis Ressnikoff (Centre Commun de Quantimétrie, Faculté de Médecine Rockefeller-Lyon, France) for generous advice in confocal microscopy experiments and to Linda Betroune and Marie Chapoton for technical assistance.
Footnotes
Published ahead of print on 5 March 2008.
REFERENCES
- 1.Angelov, D., V. A. Bondarenko, S. Almagro, H. Menoni, F. Mongelard, F. Hans, F. Mietton, V. M. Studitsky, A. Hamiche, S. Dimitrov, and P. Bouvet. 2006. Nucleolin is a histone chaperone with FACT-like activity and assists remodeling of nucleosomes. EMBO J. 251669-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Besse, S., and F. Puvion-Dutilleul. 1996. Distribution of ribosomal genes in nucleoli of herpes simplex virus type 1 infected cells. Eur. J. Cell Biol. 7133-44. [PubMed] [Google Scholar]
- 3.Boisvert, F. M., S. van Koningsbruggen, J. Navascues, and A. I. Lamond. 2007. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 8574-585. [DOI] [PubMed] [Google Scholar]
- 4.Boncoeur, E., O. Tabary, E. Bonvin, C. Muselet, A. Fritah, E. Lefait, G. Redeuilh, A. Clement, J. Jacquot, and A. Henrion-Caude. 2006. Oxidative stress response results in increased p21WAF1/CIP1 degradation in cystic fibrosis lung epithelial cells. Free Radic. Biol. Med. 4075-86. [DOI] [PubMed] [Google Scholar]
- 5.Borer, R. A., C. F. Lehner, H. M. Eppenberger, and E. A. Nigg. 1989. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell 56379-390. [DOI] [PubMed] [Google Scholar]
- 6.Bugler, B., M. Caizergues-Ferrer, G. Bouche, H. Bourbon, and F. Amalric. 1982. Detection and localization of a class of proteins immunologically related to a 100-kDa nucleolar protein. Eur. J. Biochem. 128475-480. [DOI] [PubMed] [Google Scholar]
- 7.Bush, M., D. R. Yager, M. Gao, K. Weisshart, A. I. Marcy, D. M. Coen, and D. M. Knipe. 1991. Correct intranuclear localization of herpes simplex virus DNA polymerase requires the viral ICP8 DNA-binding protein. J. Virol. 651082-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Catez, F., M. Erard, N. Schaerer-Uthurralt, K. Kindbeiter, J.-J. Madjar, and J.-J. Diaz. 2002. Unique motif for nucleolar retention and nuclear export regulated by phosphorylation. Mol. Cell. Biol. 221126-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Challberg, M. D. 1986. A method for identifying the viral genes required for herpesvirus DNA replication. Proc. Natl. Acad. Sci. USA 839094-9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng, G., M. E. Brett, and B. He. 2002. Signals that dictate nuclear, nucleolar, and cytoplasmic shuttling of the γ134.5 protein of herpes simplex virus type 1. J. Virol. 769434-9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coute, Y., J. A. Burgess, J. J. Diaz, C. Chichester, F. Lisacek, A. Greco, and J. C. Sanchez. 2006. Deciphering the human nucleolar proteome. Mass Spectrom. Rev. 25215-234. [DOI] [PubMed] [Google Scholar]
- 12.Daniely, Y., and J. A. Borowiec. 2000. Formation of a complex between nucleolin and replication protein A after cell stress prevents initiation of DNA replication. J. Cell Biol. 149799-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derenzini, M., V. Sirri, D. Trere, and R. L. Ochs. 1995. The quantity of nucleolar proteins nucleolin and protein B23 is related to cell doubling time in human cancer cells. Lab. Investig. 73497-502. [PubMed] [Google Scholar]
- 14.Diaz, J.-J., D. Simonin, T. Massé, P. Deviller, K. Kindbeiter, L. Denoroy, and J.-J. Madjar. 1993. The herpes simplex virus type 1 Us11 gene product is a phosphorylated protein found to be non-specifically associated with both ribosomal subunits. J. Gen. Virol. 74397-406. [DOI] [PubMed] [Google Scholar]
- 15.Everett, R. D., P. Freemont, H. Saitoh, M. Dasso, A. Orr, M. Kathoria, and J. Parkinson. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 726581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Mata, R., Z. Bebok, E. J. Sorscher, and E. S. Sztul. 1999. Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J. Cell Biol. 1461239-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Mata, R., Y. S. Gao, and E. Sztul. 2002. Hassles with taking out the garbage: aggravating aggresomes. Traffic 3388-396. [DOI] [PubMed] [Google Scholar]
- 18.Gerber, A., A. Heimburg, A. Reisenauer, A. Wille, T. Welte, and F. Bühling. 2004. Proteasome inhibitors modulate chemokine production in lung epithelial and monocytic cells. Eur. Respir. J. 2440-48. [DOI] [PubMed] [Google Scholar]
- 19.Ghisolfi-Nieto, L., G. Joseph, F. Puvion-Dutilleul, F. Amalric, and P. Bouvet. 1996. Nucleolin is a sequence-specific RNA-binding protein: characterization of targets on pre-ribosomal RNA. J. Mol. Biol. 26034-53. [DOI] [PubMed] [Google Scholar]
- 20.Greco, A., N. Bausch, Y. Coute, and J. J. Diaz. 2000. Characterization by two-dimensional gel electrophoresis of host proteins whose synthesis is sustained or stimulated during the course of herpes simplex virus type 1 infection. Electrophoresis 212522-2530. [DOI] [PubMed] [Google Scholar]
- 21.Greco, A., A. Calle, F. Morfin, D. Thouvenot, M. Cayre, K. Kindbeiter, L. Martin, O. Levillain, and J. J. Diaz. 2005. S-Adenosyl methionine decarboxylase activity is required for the outcome of herpes simplex virus type 1 infection and represents a new potential therapeutic target. FASEB J. 191128-1130. [DOI] [PubMed] [Google Scholar]
- 22.Hiscox, J. 2007. RNA viruses: hijacking the dynamic nucleolus. Nat. Rev. Microbiol. 5119-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hiscox, J. 2002. The nucleolus—a gateway to viral infection? Arch. Virol. 1471077-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hwang, Y. T., B. Y. Liu, D. M. Coen, and C. B. Hwang. 1997. Effects of mutations in the Exo III motif of the herpes simplex virus DNA polymerase gene on enzyme activities, viral replication, and replication fidelity. J. Virol. 717791-7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iftode, C., Y. Daniely, and J. A. Borowiec. 1999. Replication protein A (RPA): the eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 34141-180. [DOI] [PubMed] [Google Scholar]
- 26.Johnston, J. A., M. E. Illing, and R. R. Kopito. 2002. Cytoplasmic dynein/dynactin mediates the assembly of aggresomes. Cell Motil. Cytoskeleton 5326-38. [DOI] [PubMed] [Google Scholar]
- 27.Johnston, J. A., C. L. Ward, and R. R. Kopito. 1998. Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 1431883-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, K., D. D. Dimitrova, K. M. Carta, A. Saxena, M. Daras, and J. A. Borowiec. 2005. Novel checkpoint response to genotoxic stress mediated by nucleolin-replication protein A complex formation. Mol. Cell. Biol. 252463-2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, C. K., and D. M. Knipe. 1985. An immunoassay for the study of DNA-binding activities of herpes simplex virus protein ICP8. J. Virol. 54731-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lehman, I. R., and P. E. Boehmer. 1999. Replication of herpes simplex virus DNA. J. Biol. Chem. 27428059-28062. [DOI] [PubMed] [Google Scholar]
- 31.Leopardi, R., and B. Roizman. 1996. Functional interaction and colocalization of the herpes simplex virus 1 major regulatory protein ICP4 with EAP, a nucleolar-ribosomal protein. Proc. Natl. Acad. Sci. USA 934572-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lomonte, P., and R. D. Everett. 1999. Herpes simplex virus type 1 immediate-early protein Vmw110 inhibits progression of cells through mitosis and from G1 into S phase of the cell cycle. J. Virol. 739456-9467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez-Iglesias, C., F. Puvion-Dutilleul, J. Cebrian, and M. E. Christensen. 1988. Herpes simplex virus type 1-induced modifications in the distribution of nucleolar B-36 protein. Eur. J. Cell Biol. 46259-269. [PubMed] [Google Scholar]
- 34.Lymberopoulos, M. H., and A. Pearson. 2007. Involvement of UL24 in herpes-simplex-virus-1-induced dispersal of nucleolin. Virology 363397-409. [DOI] [PubMed] [Google Scholar]
- 35.Marconi, P., D. Krisky, T. Oligino, P. L. Poliani, R. Ramakrishnan, W. F. Goins, D. J. Fink, and J. C. Glorioso. 1996. Replication-defective herpes simplex virus vectors for gene transfer in vivo. Proc. Natl. Acad. Sci. USA 9311319-11320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marcy, A. I., D. R. Yager, and D. M. Coen. 1990. Isolation and characterization of herpes simplex virus mutants containing engineered mutations at the DNA polymerase locus. J. Virol. 642208-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mears, W. E., and S. A. Rice. 1998. The herpes simplex virus immediate-early protein ICP27 shuttles between nucleus and cytoplasm. Virology 242128-137. [DOI] [PubMed] [Google Scholar]
- 38.Mongelard, F., and P. Bouvet. 2007. Nucleolin: a multiFACeTed protein. Trends Cell Biol. 1780-86. [DOI] [PubMed] [Google Scholar]
- 39.Monier, K., J. C. Armas, S. Etteldorf, P. Ghazal, and K. F. Sullivan. 2000. Annexation of the interchromosomal space during viral infection. Nat. Cell Biol. 2661-665. [DOI] [PubMed] [Google Scholar]
- 40.Morency, E., Y. Coute, J. Thomas, P. Texier, and P. Lomonte. 2005. The protein ICP0 of herpes simplex virus type 1 is targeted to nucleoli of infected cells. Brief report. Arch. Virol. 1502387-2395. [DOI] [PubMed] [Google Scholar]
- 41.Muramatsu, M., and T. Onishi. 1978. Isolation and purification of nucleoli and nucleolar chromatin from mammalian cells. Methods Cell Biol. 17141-161. [DOI] [PubMed] [Google Scholar]
- 42.Nisole, S., B. Krust, and A. Hovanessian. 2002. Anchorage of HIV on permissive cells leads to coaggregation of viral particles with surface nucleolin at membrane raft microdomains. Exp. Cell Res. 276155-173. [DOI] [PubMed] [Google Scholar]
- 43.Nozawa, N., Y. Yamauchi, K. Ohtsuka, Y. Kawaguchi, and Y. Nishiyama. 2004. Formation of aggresome-like structures in herpes simplex virus type 2-infected cells and a potential role in virus assembly. Exp. Cell Res. 299486-497. [DOI] [PubMed] [Google Scholar]
- 44.Ochs, R. L. 1998. Methods used to study structure and function of the nucleolus. Methods Cell Biol. 53303-321. [DOI] [PubMed] [Google Scholar]
- 45.Okuwaki, M., A. Iwamatsu, M. Tsujimoto, and K. Nagata. 2001. Identification of nucleophosmin/B23, an acidic nucleolar protein, as a stimulatory factor for in vitro replication of adenovirus DNA complexed with viral basic core proteins. J. Mol. Biol. 31141-55. [DOI] [PubMed] [Google Scholar]
- 46.Pearson, A., D. M. Knipe, and D. M. Coen. 2004. ICP27 selectively regulates the cytoplasmic localization of a subset of viral transcripts in herpes simplex virus type 1-infected cells. J. Virol. 7823-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Puvion-Dutilleul, F., E. Pichard, P. Sheldrick, F. Amalric, and E. Puvion. 1986. Appearance of host-specific nucleolar proteins in intranuclear “dense bodies” following herpes simplex infection. Eur. J. Cell Biol. 39458-468. [PubMed] [Google Scholar]
- 48.Roizman, B., and A. Sears. 1990. Herpes simplex viruses and their replication, p. 1795-1842. In B. N. Fields, D. M. Knipe, R. M. Chanock, M. S. Hirsch, J. L. Melnick, T. P. Monath, and B. Roizman (ed.), Fundamental virology. Raven Press, New York, NY.
- 49.Roizman, B., and A. E. Sears. 1993. Herpes simplex viruses and their replication, p. 11-68. In B. Roizman, R. J. Whitley, and C. Lopez (ed.), The human herpes viruses, 3rd ed. Raven Press, New York, NY.
- 50.Roller, R. J., L. L. Monk, D. Stuart, and B. Roizman. 1996. Structure and function in the herpes simplex virus 1 RNA-binding protein US11: mapping of the domain required for ribosomal and nucleolar association and RNA binding in vitro. J. Virol. 702842-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosevear, E. R., M. McReynolds, and R. D. Goldman. 1990. Dynamic properties of intermediate filaments: disassembly and reassembly during mitosis in baby hamster kidney cells. Cell Motil. Cytoskeleton 17150-166. [DOI] [PubMed] [Google Scholar]
- 52.Roussel, P., and D. Hernandez-Verdun. 1994. Identification of Ag-NOR proteins, markers of proliferation related to ribosomal gene activity. Exp. Cell Res. 214465-472. [DOI] [PubMed] [Google Scholar]
- 53.Schenk, P., S. Pietschmann, H. Gelderblom, G. Pauli, and H. Ludwig. 1988. Monoclonal antibodies against herpes simplex virus type 1-infected nuclei defining and localizing the ICP8 protein, 65K DNA-binding protein and polypeptides of the ICP35 family. J. Gen. Virol. 6999-111. [DOI] [PubMed] [Google Scholar]
- 54.Scherl, A., Y. Couté, C. Déon, A. Callé, K. Kindbeiter, J.-C. Sanchez, A. Greco, D. Hochstrasser, and J.-J. Diaz. 2002. Functional proteomic analysis of the human nucleolus. Mol. Biol. Cell 134100-4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schirmer, E. C., and L. Gerace. 2005. The nuclear membrane proteome: extending the envelope. Trends Biochem. Sci. 30551-558. [DOI] [PubMed] [Google Scholar]
- 56.Seinsoth, S., H. Uhlmann-Schiffler, and H. Stahl. 2003. Bidirectional DNA unwinding by a ternary complex of T antigen, nucleolin and topoisomerase I. EMBO Rep. 4263-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simonin, D., J. J. Diaz, T. Masse, and J. J. Madjar. 1997. Persistence of ribosomal protein synthesis after infection of HeLa cells by herpes simplex virus type 1. J. Gen. Virol. 78435-443. [DOI] [PubMed] [Google Scholar]
- 58.Sinclair, M., J. McLauchlan, H. Marsden, and S. Brown. 1994. Characterization of a herpes simplex virus type 1 deletion variant (1703) which under-produces Vmw63 during immediate early conditions of infection. J. Gen. Virol. 751083-1089. [DOI] [PubMed] [Google Scholar]
- 59.Sirri, V., P. Roussel, M. C. Gendron, and D. Hernandez-Verdun. 1997. Amount of the two major Ag-NOR proteins, nucleolin, and protein B23 is cell-cycle dependent. Cytometry 28147-156. [PubMed] [Google Scholar]
- 60.Srivastava, M., and H. B. Pollard. 1999. Molecular dissection of nucleolin's role in growth and cell proliferation: new insights. FASEB J. 131911-1922. [PubMed] [Google Scholar]
- 61.Storck, S., M. Shukla, S. Dimitrov, and P. Bouvet. 2007. Functions of the histone chaperone nucleolin in diseases. Subcell. Biochem. 41125-144. [DOI] [PubMed] [Google Scholar]
- 62.Takemura, M., K. Sato, M. Nishio, T. Akiyama, H. Umekawa, and S. Yoshida. 1999. Nucleolar protein B23.1 binds to retinoblastoma protein and synergistically stimulates DNA polymerase alpha activity. J. Biochem. (Tokyo) 125904-909. [DOI] [PubMed] [Google Scholar]
- 63.Tarapore, P., M. Okuda, and K. Fukasawa. 2002. A mammalian in vitro centriole duplication system: evidence for involvement of CDK2/cyclin E and nucleophosmin/B23 in centrosome duplication. Cell Cycle 175-81. [PubMed] [Google Scholar]
- 64.Taylor, T., and D. Knipe. 2004. Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J. Virol. 785856-5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ugrinova, I., K. Monier, C. Ivaldi, M. Thiry, S. Storck, F. Mongelard, and P. Bouvet. 2007. Inactivation of nucleolin leads to nucleolar disruption, cell cycle arrest and defects in centrosome duplication. BMC Mol. Biol. 866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wileman, T. 2007. Aggresomes and pericentriolar sites of virus assembly: cellular defense or viral design? Annu. Rev. Microbiol. 61149-167. [DOI] [PubMed] [Google Scholar]
- 67.Wojcik, C., D. Schroeter, S. Wilk, J. Lamprecht, and N. Paweletz. 1996. Ubiquitin-mediated proteolysis centers in HeLa cells: indication from studies of an inhibitor of the chymotrypsin-like activity of the proteasome. Eur. J. Cell Biol. 71311-331. [PubMed] [Google Scholar]
- 68.Wu, M. H., J. H. Chang, C. C. Chou, and B. Y. Yung. 2002. Involvement of nucleophosmin/B23 in the response of HeLa cells to UV irradiation. Int. J. Cancer 97297-305. [DOI] [PubMed] [Google Scholar]