Abstract
The human asialoglycoprotein receptor H2a subunit contains a charged pentapeptide, EGHRG, in its ectodomain that is the only sequence absent from the H2b alternatively spliced variant. H2b exits the endoplasmic reticulum (ER) even when singly expressed, whereas H2a gives rise to a cleaved soluble secreted ectodomain fragment; uncleaved membrane-bound H2a molecules are completely retained and degraded in the ER. We have inserted the H2a pentapeptide into the sequence of the H1 subunit (H1i5), which caused complete ER retention but, unexpectedly, no degradation. This suggests that the pentapeptide is a determinant for ER retention not colocalizing in H2a with the determinant for degradation. The state of sugar chain processing and the ER localization of H1i5, which was unchanged at 15°C or after treatment with nocodazole, indicate ER retention and not retrieval from the cis-Golgi or the intermediate compartment. H1i5 folded similarly to H1, and both associated to calnexin. However, whereas H1 dissociated with a half time of 45 min, H1i5 remained bound to the chaperone for prolonged periods. The correct global folding of H2a and H1i5 and of other normal precursors and unassembled proteins and the true ER retention, and not exit and retrieval, suggest a difference in their quality control mechanism compared with that of misfolded proteins, which does involve retrieval. However, both pathways may involve calnexin.
Keywords: endoplasmic reticulum degradation, retention signal, signal peptidase, protein folding, T cell antigen receptor
Retention and degradation of proteins in the ER, also called “pre-Golgi quality control,” prevents the traffic through the secretory pathway of malfolded polypeptides and unassembled subunits of oligomeric membrane proteins (1, 2). It takes place in a compartment before the Golgi in the secretory pathway without the involvement of lysosomes (3, 4). Recently it was shown for several proteins that after retention in the ER, the ubiquitin pathway is involved in the proteins’ degradation (5). In several cases it is the transmembrane domain and, more specifically, charged residues in that region that are responsible for the ER retention and degradation [e.g., unassembled α and β subunits of the T cell antigen receptor (TCR) (6, 7)]. This could point to a mechanism of retention in quality control similar to the one involved in the retention by the transmembrane domain of some ER resident proteins like HMGCoA reductase (8), ER Ca2+ pump (9), and cytochrome P450 (10, 11). The ER chaperone calnexin is involved in the quality control of several proteins, and its role in ER retention was directly shown for some of them (12, 13). When naturally expressed in HepG2 cells, the human asialoglycoprotein receptor (ASGPR) H2b subunit forms the cell surface heterooligomeric complex with the H1 subunit, whereas the alternatively spliced variant H2a is cleaved next to the membrane span to give a soluble secreted form of the receptor (14). The only difference between H2a and H2b is an additional charged pentapeptide, EGHRG, which is present in the ectodomain of H2a next to its transmembrane segment. Even when singly expressed, H2b can escape ER degradation by leaving the ER [with an efficiency of about 30% (15)] as does the soluble H2a ectodomain fragment. On the other hand membrane-bound H2a cannot escape the ER quality control machinery. It serves only as a precursor of the soluble secreted form. Uncleaved precursor molecules are subject to ER retention and degradation. As we show here this is not caused by a misfolding of H2a compared with H2b but is due to a direct action of the extra lumenal juxtamembrane pentapeptide, EGHRG, as a determinant for ER retention.
MATERIALS AND METHODS
Materials.
Rainbow 14C-labeled methylated protein standards were obtained from Amersham. Pro-mix cell-labeling mix ([35S]methionine plus [35S]cysteine) was from Amersham (>1,000 Ci/mmol; 1 Ci = 37 GBq). Protein A-Sepharose was from Repligen. Endo-N-acetyl glucosaminidase H (endo H) was from New England Biolabs. N-glycanase, endo-N-acetyl glucosaminidase D (endo D), and N-acetyl-leucyl-leucyl-norleucinal (ALLN) were from Boehringer Mannheim. Swainsonine was from Genzyme. Jack bean α-mannosidase, inhibitors, and other common reagents were from Sigma.
DNA Constructs.
The 15 bp from H2a cDNA that differentiate it from H2b and code for the juxtamembrane pentapeptide EGHRG (15) were inserted into H1 cDNA (16) by overlap extension PCR (17). The sequence of the mutated H1 (H1i5) was verified by double-stranded dideoxy sequencing using the Sequenase 2 kit (United States Biochemical). It was then subcloned using BamHI and EcoRI sites into the pMEX-neo mammalian expression vector containing a Moloney murine sarcoma virus long terminal repeat (LTR) as promoter (18).
Cell Lines and Culture.
NIH 3T3 cells were transfected with pMEX-neo containing H1i5 using a calcium phosphate transfection protocol (19). After selection with G418 a representative clone of these cells (5-D) or 3T3 cell lines expressing H1 (1–7 cells), H2a (2–18 cells), or H2b (2C cells) (14) were grown in DMEM plus 10% calf serum under 5% CO2.
Antibodies.
Polyclonal antibodies specific for peptides corresponding to the carboxy termini of H1 (anti-H1) or H2 or the H2a-specific pentapeptide (anti-H2a) were the ones used in earlier studies (14, 15). For immunofluorescence IgGs were purified from the antisera. A mouse monoclonal anti-rat PDI and a rabbit polyclonal against the carboxy terminus of calnexin were purchased from Stressgen Biotechnologies (Victoria, Canada). Rabbit polyclonal antibodies against purified Golgi membranes (20) were a gift of Daniel Louvard, and anti-p58 antibodies (21) were a gift of Jakko Saraste.
Metabolic Labeling, Treatment with Inhibitors, and Immunoprecipitation.
Subconfluent cell monolayers were metabolically labeled as described (14) but with the following modifications: 0.5 mCi/ml of Pro-Mix mixture of [35S]cysteine and [35S]methionine were used per 60-mm dish with addition of 5 mM unlabeled methionine. Treatments with swainsonine were with 4 μg/ml (from a 2-mg/ml stock in water) during the starvation, pulse, and chase periods, and brefeldin A (BFA) was added at 5 μg/ml (5-mg/ml stock in methanol) only during the pulse and chase. Cell lysis and immunoprecipitations from cell lysates or supernatants and endo H and N-glycanase treatments were performed as described before (14). For treatment with endo D immunoprecipitates were washed and then boiled in 10 μl of 0.5% SDS in 50 mM sodium citrate, pH 6.0. Then, 10 μl of 50 mM sodium citrate (pH 5.5), 40 mM EDTA (pH 7.5), and 3% Triton X-100 plus 0.4 mU of endo D were added and incubated overnight at 37°C. Samples were boiled with sample buffer before SDS/PAGE. Treatment with α-mannosidase was performed as described (22). For the experiments of association with calnexin, cells were lysed as in ref. 23. Sequential immunoprecipitation was done first with anti-calnexin antibody. The pellets were then washed and boiled with 1% SDS and 2 mM DTT. The supernatants were diluted with 10 volumes of 1% Triton X-100, 0.5% sodium deoxycholate, and 2 mM oxidized glutathione and reimmunoprecipitated with anti-H1 antibody.
Gel Electrophoresis, Fluorography, and Quantitation.
Reducing SDS/PAGE was performed on 10% Laemmli gels except where stated otherwise. For nonreducing conditions sample buffer without 2-mercaptoethanol was used. The gels were analyzed by fluorography using 20% 2,5-diphenyloxazole and exposing to BioMax MR film from Eastman Kodak. For more sensitive detection of the coprecipitation of H1 with calnexin, the gels were blotted onto nitrocellulose and the blots were exposed to a Fuji Bas 1000 phosphorimager screen. Quantitations were also performed in the phosphorimager.
Immunofluorescence Microscopy.
The procedures employed were essentially as described previously (15) with the following modifications. Fixation and permeabilization of the cells grown on coverslips were done by sequential incubation with methanol and methanol:acetone 1:1 for 5 min each at −20°C. As secondary antibodies we used indocarbocyanine (Cy3)- or fluoresceine isothiocyanate (FITC)-conjugated goat anti-rabbit IgG or FITC-conjugated goat anti-mouse IgG, all from The Jackson Laboratory. For treatment with nocodazole (Nz), cells on coverslips were incubated with medium containing 20 μM Nz (5 mg/ml stock in DMSO) at 37°C in a CO2 incubator. Incubations of cells on coverslips at 15°C were done in a water bath with medium containing 20 mM Hepes buffer, pH 7.4.
Photography on an Axioskop fluorescence microscope (Zeiss) was done at identical exposure times between samples to be able to compare signal intensities.
RESULTS
The Additional Pentapeptide in H2a Does Not Affect Folding of the Polypeptide.
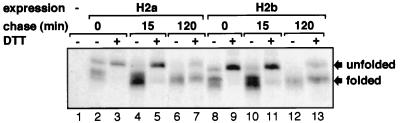
Protein malfolding is a known cause for ER retention and degradation (2). One possible explanation for the different behavior of H2a and H2b could be malfolding triggered by the extra pentapeptide. In nonreducing SDS/PAGE, proteins with several disulfide bonds migrate after synthesis as a group of bands that are converted into one higher mobility band upon folding (24). The S—S bonds also become progressively buried into the structure and thus resistant to reduction by incubation of the cells with a low concentration of DTT. This was shown to occur with the ASGPR H1 subunit (25). We analyzed the bands obtained on nonreducing SDS/PAGE after a short pulse of NIH 3T3 cells expressing H2a or H2b with [35S]cysteine followed by chase for different times and immunoprecipitation from cell lysates. Before lysis, cells were treated with 0.1 M iodoacetamide to block free sulfhydryl groups and prevent disulfide bonding during or after the lysis. The immunoprecipitates were treated with N-glycanase to eliminate heterogeneity in the run due to the sugar chains. H2a and H2b migrated after a pulse in a heterogeneous pattern (Fig. 1, lanes 2 and 8), with all bands being completely sensitive to reduction and “unfolding” by DTT (Fig. 1, lanes 3 and 9). After 15-min chase the pattern became less heterogeneous (Fig. 1, lanes 4 and 10), and after 2-h chase only a faster-running band could be seen, which was mostly resistant to the effect of DTT (Fig. 1, lanes 6, 7, 12, and 13). No difference can be seen in the pattern of migration or in the kinetics of conversion to the folded species between H2a and H2b. Although we cannot discard the possibility of a small change in conformation in the region of the pentapeptide that might not be detected in this experiment, we can conclude that at least there is no gross misfolding of H2a. This hinted to a specific role of the pentapeptide in the ER retention and degradation of H2a, prompting us to transfer it to the sequence of another protein to determine whether it is an autonomous signal.
Figure 1.
H2a and H2b fold at similar rates. NIH 3T3 cells (lane 1) or the same cells stably transfected with expression vectors encoding H2a (2–18 cell line, lanes 2–7) or H2b (2C cell line, lanes 8–13) were metabolically labeled with [35S]cysteine for 10 min (lanes 1, 2, and 8) or for 5 min plus another 5 min in the presence of 5 mM DTT (lanes 3 and 9) and chased with complete medium for different times as indicated. For some dishes the last 5 min of chase were in the presence of 5 mM DTT as indicated. Cells were then incubated with 0.1 M iodoacetamide for 5 min at 4°C. Cell lysates were immunoprecipitated with anti-H2 carboxyl-terminal antibodies. Immunoprecipitates were treated with N-glycanase and analyzed on nonreducing SDS/PAGE followed by fluorography. Note that all H2a bands migrate slightly slower than the corresponding H2b bands due to the presence of the extra pentapeptide.
Insertion of the H2a Pentapeptide into the Sequence of H1 Causes Complete ER Retention but not Degradation.
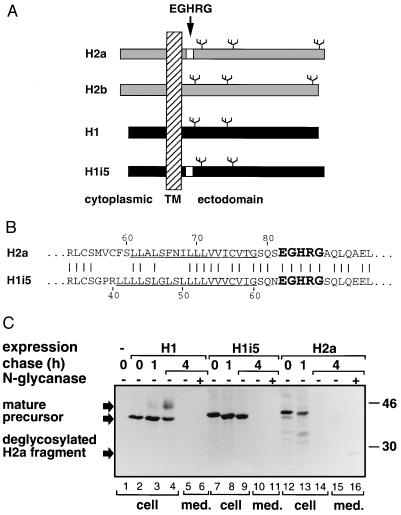
To prevent a deleterious effect on folding and to transfer the pentapeptide to a similar context, we chose the H1 subunit as a recipient protein. Like H2b, H1 can exit the ER when singly expressed (26). H1 and H2 share about 60% identity, so the 15-bp coding for the pentapeptide could be inserted into H1 cDNA by PCR mutagenesis into the same position as it exists in H2a, near the transmembrane domain (Fig. 2 A and B). The construct (H1i5) was inserted into an expression vector (pMEX-neo) and stably transfected into NIH 3T3 cells. Several selected clones were analyzed by metabolic labeling with [35S]cysteine followed by chase for different periods, immunoprecipitation from cell lysates or cell supernatants, and SDS/PAGE. Analysis of a representative cell line (5-D) is shown in Fig. 2C, compared with cells expressing wild-type H1 or H2a. H1 matures after chase to a 46-kDa Golgi-processed form (Fig. 2C, lanes 2–4). H2a produces a 35-kDa fragment and is almost completely degraded after 4-h chase (Fig. 2C, lanes 12–14). A small portion of the H2a fragment is Golgi processed; it is seen secreted after 4-h chase and observed as a sharper band (28 kDa) after deglycosylation with N-glycanase (Fig. 2C, lanes 15–16). As for H1i5 after chase there is no conversion to a Golgi-processed species, which suggests ER retention. Unexpectedly no degradation could be seen either (Fig. 2C, lanes 8 and 9). Neither H1 nor H1i5 form an intracellular or a secreted fragment (Fig. 2C, lanes 5, 6, 10, and 11). Cells were metabolically labeled and chased for longer periods (data not shown), and we could extrapolate a half life of about 14 h for H1i5, which is similar to that of H1 or H2b in HepG2 cells (12 h) (27).
Figure 2.
(A) Schematic representation of human ASGPR subunits and the H1i5 mutant. Human ASGPR subunit H1 is in black as well as H1 with insertion of the EGHRG sequence (H1i5). The EGHRG insert is represented by a white box in H1i5 and H2a. H2a and H2b are in gray. H1 and H2 sequences can be aligned except for an extra 18-aa insert in the cytoplasmic tail of H2. The three N-linked sugar chains in H2 and two in H1 are depicted. (B) Sequence of the transmembrane domain plus flanking residues. The transmembrane domains of H2a and H1i5 are underlined. The EGHRG insert is in bold. H2b and H1 sequences in this region are identical to those of H2a and H1i5, respectively, but without EGHRG. Vertical lines indicate identical residues. (C) H1i5 is not processed through the Golgi and is not degraded. NIH 3T3 cells or the same cells stably transfected with expression vectors encoding H1 (1–7 cell line), H1i5 (5-D cell line), or H2a (2–18 cell line) were metabolically labeled with [35S]cysteine for 20 min, and some were then chased with complete medium for several times as indicated. Cell lysates (cell) or cell supernatants (med.) were immunoprecipitated with anti-H1 antibody (lanes 1–11) or anti-H2a antibody (lanes 12–16), followed in some cases (lanes 6, 11, and 16) by treatment with N-glycanase. The products were then analyzed on 12% SDS/polyacrylamide gel followed by fluorography. On the right are indicated the migrations of molecular mass standards in kilodaltons. On the left the migration of precursors, Golgi processed (mature) proteins, and secreted deglycosylated H2a fragment.
Therefore, the insertion of the H2a pentapeptide causes ER retention of H1 but does not trigger its ER degradation or cleavage.
H1i5 is Localized in the ER.
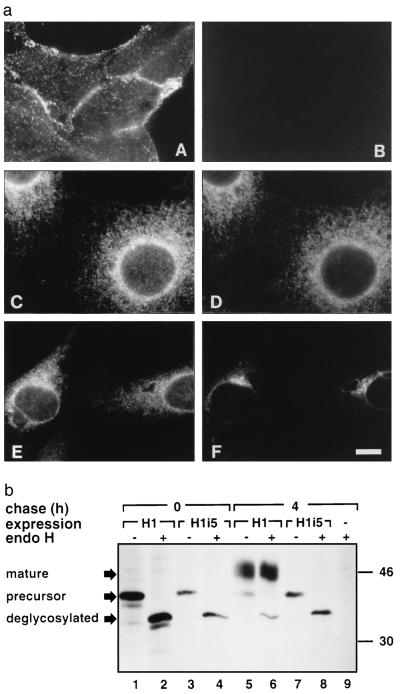
Using anti-H1 carboxyl-terminal antibodies, no fluorescence was seen on the surface of nonpermeabilized cells expressing H1i5 compared with those expressing H1 (Fig. 3a, A and B). When the cells were permeabilized H1i5 appeared in a pattern similar to that revealed by an antibody against PDI (an ER resident protein) and different from that shown by an antibody directed against a Golgi subcellular fraction (Fig. 3a, C–F). This suggests an ER localization for H1i5 as had been seen for singly expressed H2a (4, 15).
Figure 3.
H1i5 remains in the ER with high mannose glycosylation. (a) A and B show immunofluorescence staining of nonpermeabilized cells expressing H1 (1–7 cell line, A) or H1i5 (5-D cell line, B) reacted with anti-H1 antibody and a secondary goat anti-rabbit IgG antibody conjugated to Cy3. C and D show a double-labeling experiment on permeabilized cells expressing H1i5 with anti-H1 antibody (secondary goat anti-rabbit IgG-Cy3, C) and an anti-PDI mouse monoclonal antibody (as ER marker) (secondary goat anti-mouse IgG FITC, D). E and F show double labeling with anti-H1/cy3 (E) and anti-Golgi subfraction antibody/FITC (F). (Bar = 10 μm.) (b) NIH 3T3 cells or the cell lines expressing H1 or H1i5 were metabolically labeled with [35S]cysteine for 20 min (lanes 1–4) or for 20 min and then chased with complete medium for 4 h (lanes 5–9). Cell lysates were immunoprecipitated with anti-H1 antibody followed in the indicated cases by treatment with endo H and analyzed on 12% SDS/polyacrylamide gel followed by fluorography. On the right are indicated the migrations of molecular mass standards in kilodaltons. On the left are the migration of precursors, Golgi processed (mature), and deglycosylated proteins.
H1 and H1i5 precursors were completely sensitive to endo H (Fig. 3b, lanes 1–4). After 4-h chase most of H1 became resistant to endo H (Fig. 3b, lanes 5 and 6), whereas H1i5 was still completely sensitive (Fig. 3b, lanes 7 and 8), indicating no complex type N-glycosylation and a premedial Golgi location. Even after 8-h chase all H1i5 is sensitive to endo H (data not shown). To rule out the possibility that N-linked sugar chains in H1i5 are not able to be processed to a complex type, cells expressing H1i5 were metabolically labeled and chased in the presence of BFA. By relocalizing Golgi modification enzymes to the ER, BFA caused H1i5 to become resistant to endo H treatment, indicating its competence for complex-type modification (data not shown).
H1i5 is Retained in the ER and Does Not Recycle Through the Golgi or the ER–Golgi Intermediate Compartment.
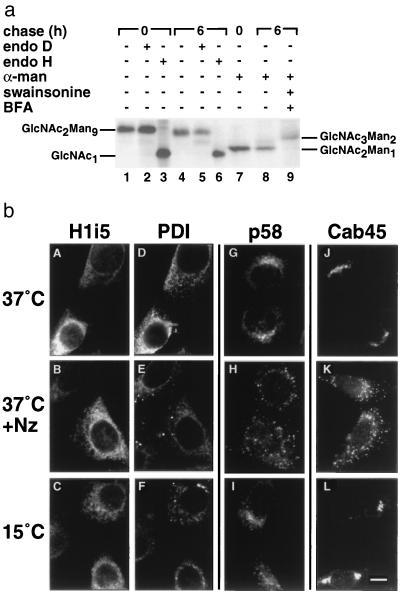
The immunofluorescence pattern of H1i5 (Fig. 3a) indicates a predominant ER localization but does not rule out the possibility of its exit to the Golgi or the ER-to-Golgi intermediate compartment (ERGIC) and quick retrieval to the ER as occurs with many ER resident proteins (28). The lack of complex-type glycosylation (Fig. 3b) would exclude transit through the medial- but not the cis-Golgi. In the cis-Golgi H1i5 would be exposed to Golgi α-mannosidase I, which would process its sugar chains to GlcNAc2Man5 (29). Molecules containing GlcNAc2Man5 should accumulate after several cycles through the cis-Golgi and back to the ER, and they should be sensitive to endo D (30). The N-linked sugar chains of H2a and its cleaved fragment are trimmed to GlcNAc2Man6 and not to GlcNAc2Man5 (31). H2a was also shown to be resistant to endo D in CHO15B cells, which lack GlcNAc-transferase I and as a result should accumulate GlcNAc2Man5 if H2a would reach the cis-Golgi. Nevertheless, the rapid degradation of H2a does not allow long chase periods. These were possible with H1i5. Metabolically labeled H1i5 was treated with endo D after immunoprecipitation, showing total resistance after pulse (Fig. 4a, lane 2) and only a very slight sensitivity after 6-h chase (Fig. 4a, lane 5). This suggests two possibilities: (i) Most H1i5 is retained in the ER and there is “leakage” of some H1i5 molecules to the cis-Golgi, or (ii) all H1i5 is retained in the ER and there is some trimming in the ER to GlcNAc2Man5. Trimming of mannose residues occurs after prolonged residence in the ER (22), and both Man9-mannosidase and soluble ER mannosidase have some activity that trims the last α1,2-mannosidic linkage in GlcNAc2Man6 (32, 33).
Figure 4.
H1i5 does not recycle through the cis-Golgi or the ERGIC. (a) Resistance of H1i5 to endo D and sensitivity to α-mannosidase after metabolic labeling and long chases. Cells expressing H1i5 were metabolically labeled with [35S]cysteine for 20 min and chased with complete medium for the indicated times. One of the samples was treated with 4 μg/ml swainsonine and 5 μg/ml BFA (lane 9). Cell lysates were immunoprecipitated with anti-H1 antibody followed in the indicated cases by treatment with endo D, endo H, or jack bean α-mannosidase and analyzed on SDS/polyacrylamide gel followed by fluorography. On the sides are indicated the migrations of H1i5 molecules with the deduced composition of their N-linked sugar chains. (b) Unchanged pattern of H1i5 after treatment with nocodazole or incubation at 15°C. A–F show a double-labeling experiment on permeabilized cells expressing H1i5 (5-D cell line) with anti-H1 antibody (secondary goat anti-rabbit IgG-Cy3; A–C) and an anti-PDI mouse monoclonal antibody (secondary goat anti-mouse IgG-FITC; D–F). G–I show labeling with an antibody against an ERGIC marker, p58, and J–L show labeling with a lumenal Golgi marker, Cab45, both with a secondary goat anti-rabbit IgG–Cy3. Cells were untreated (37°C, A, D, G, and J), treated for 5 h with 20 μM Nz (37°C+Nz, B, E, H, and K), or incubated for 5 h at 15°C (C, F, I, and L). Note the changes in the staining pattern of PDI after treatment of the cells, while the ER-like pattern of H1i5 remains unchanged. (Bar = 10 μm.)
One last intermediate in sugar chain processing in the Golgi that would be sensitive to endo H and resistant to endo D could be GlcNAc3Man5 formed by the action of GlcNAc-transferase I in the medial Golgi. Nevertheless, GlcNAc3Man5 would be partially resistant to jack bean α-mannosidase, giving as a product GlcNAc3Man2. In fact, this is the product carried by H1i5 molecules obtained by treating cells with BFA (to allow processing by Golgi enzymes) plus swainsonine (to inhibit Golgi α-mannosidase II) and then treating the immunoprecipitated H1i5 with α-mannosidase (Fig. 4a, lane 9). Without treatment, H1i5 is totally sensitive to α-mannosidase before or after chase, giving molecules that carry GlcNAc2Man1 (Fig. 4a, lanes 7 and 8). Taken together the resistance to endo D and sensitivity to α-mannosidase indicate that H1i5 does not reach the Golgi after long chases.
To corroborate this conclusion with another approach, cells expressing H1i5 were treated with nocodazole (Nz). This agent disrupts microtubules, leading to a disintegration of the Golgi and interruption of traffic between the Golgi, the ERGIC, and the ER (21, 34). The fate of H1i5 was compared with that of an ER resident protein, PDI, that contains a KDEL signal for retrieval. Both H1i5 and PDI showed an ER pattern by double label immunofluorescence on cells without treatment (Fig. 4b, A and D). After treatment for 5 h with 20 μM Nz the ER staining of PDI faded, whereas much of the protein concentrated in large punctate structures (Fig. 4b, E). Following the same treatment H1i5 remained unchanged in an ER pattern (Fig. 4b, B). The same result was obtained after treatment for shorter or longer times, up to 16 h, including cycloheximide in some samples to block de novo synthesis (data not shown). An ERGIC marker, p58 (21), showed the expected perinuclear staining (more diffuse than Golgi) without treatment (Fig. 4b, G), which changed to more punctate staining after treatment with Nz (Fig. 4b, H). A soluble lumenal Golgi resident, Cab45 (35), showed a typical Golgi-concentrated perinuclear staining before treatment (Fig. 4b, J), which disintegrated into punctate structures after treatment (Fig. 4b, K). The punctate staining of p58 and that of Cab45 after treatment showed only a minor colocalization with that of PDI (data not shown). PDI may be concentrating in these conditions at ER exit sites, whereas the other proteins may localize in structures further ahead in the ER–Golgi pathway.
Anterograde movement of proteins from the ER can be blocked at 15°C, at which temperature they accumulate in the ERGIC. After incubation for 5 h at 15°C the ER pattern of PDI faded and the protein accumulated in perinuclear patches and punctate structures (Fig. 4b, F). Under the same conditions the ER pattern of H1i5 was unchanged (Fig. 4b, C).
Taken together the results show that H1i5 is retained in the ER and does not exit or recycle through the Golgi or the ERGIC.
Prolonged Association of H1i5 to Calnexin.
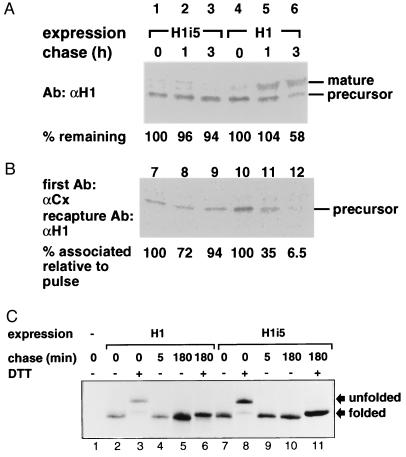
Because calnexin was found to be involved in the quality control of other proteins, we analyzed its association to H1 and H1i5. Cells expressing H1 or H1i5 were metabolically labeled, and immunoprecipitation of cell lysates was done first with anti-calnexin antibody followed by elution and reimmunoprecipitation (recapture) with anti-H1 antibody. As shown in Fig. 5B, both H1 and H1i5 interact with calnexin after the pulse. However, after chase, calnexin dissociates from H1 with a half time of about 45 min whereas it remains associated to H1i5 for very long periods (Fig. 5B, compare lanes 7–9). The chase was done in the presence of the protease inhibitors ALLN [to preclude any ER degradation because ALLN was shown to inhibit degradation of H2a (36)] and leupeptin (to inhibit degradation of H1 in its transit through endosomes/lysosomes). The inhibition of the degradation of H1 was partial as seen in Fig. 5A. Nevertheless, the rate of degradation of H1 is much slower than the dissociation from calnexin (compare Fig. 5A, lanes 4–6 and Fig. 5B, lanes 10–12).
Figure 5.
H1i5 shows prolonged association to calnexin, though its folding is similar to that of H1. (A) The cell lines expressing H1 or H1i5 were metabolically labeled for 40 min with [35S]cysteine and chased for the indicated times with complete medium in the presence of 150 μM ALLN and 200 μg/ml leupeptin. Cell lysates were immunoprecipitated with anti-H1 antibody and analyzed by SDS/polyacrylamide gel followed by phosphorimager detection. At the bottom of the panel are phosphorimager quantitations of H1 remaining after the chases relative to pulse. (B) The same as in A except that cell lysates were immunoprecipitated with anti-calnexin antibody and immunoprecipitates were boiled in SDS. Supernatants were then diluted with excess of Triton X-100 and reimmunoprecipitated with anti-H1 antibody, as described in Materials and Methods. The coprecipitated proteins were analyzed by SDS/PAGE and fluorography. On the right are indicated the migrations of precursor and Golgi processed (mature) H1. At the bottom of the panel are phosphorimager quantitations of H1 associated to calnexin after the chases relative to that associated after the pulse. (C) NIH 3T3 cells or the cells expressing H1 or H1i5 were metabolically labeled and processed in the same manner as in Fig. 1. Cell lysates were immunoprecipitated with anti-H1 antibodies. Immunoprecipitates were treated with N-glycanase and analyzed on nonreducing SDS/polyacrylamide gel followed by fluorography.
Therefore, the ER retention of H1i5 correlates with a prolonged association to calnexin.
Folding of H1i5 is Normal.
As stated above, H1 had been chosen as a recipient for the H2a pentapeptide because of its similarity with H2, allowing a sequence alignment and insertion into the same position. This position had the further advantage of being both in H1 and H2 at a junction between exons, which could preclude the disruption of folding of the two independent domains. Nevertheless, a possibility existed that the pentapeptide caused misfolding in H1i5, which in turn could cause tight binding to calnexin and ER retention. To analyze this the same assay was used as in Fig. 1. As shown in Fig. 5C the proteins seen on nonreducing SDS/PAGE after pulse labeling are almost completely sensitive to in vivo reduction by DTT (Fig. 5C, lanes 2, 3, 7, and 8). After chase they migrate slightly faster and become resistant to DTT (Fig. 5C, lanes 5, 6, 10, and 11). H1 and H1i5 yield a very similar pattern of bands and exhibit similar kinetics of folding, indicating normal folding of the mutant protein. Hence, the ER retention of H1i5 is not due to misfolding but to a direct action of the EGHRG lumenal juxtamembrane peptide as a determinant for retention.
DISCUSSION
Although there seem to be no primary sequence signals shared by different proteins that undergo ER retention and degradation, there are some common structural features. Charged residues in the transmembrane domain and flanking regions (as for the H2a EGHRG pentapeptide) are generally involved in membrane protein ER quality control [e.g., in TCR-α and -β subunits (6, 37)], which may hint to a common mechanism involving a secondary structure determinant. Proline substitution of alanine-82 in H2b (the same position where EGHRG is in H2a; Fig. 2B) had the same effect of causing ER retention and degradation (38). Proline is predicted to induce a β turn, the same conformational change predicted for the pentapeptide. Therefore, we suggest that the ER retention is caused by a lax “signal peptide” type of determinant in the transmembrane and juxtamembrane regions and not by a primary KDEL-type sequence. In the case of H2a this determinant is the EGHRG sequence. It must be located on a membrane-bound protein because the H2a soluble ectodomain fragment contains the pentapeptide and is nonetheless secreted (14).
Transfer of charged transmembrane residues from the TCR-α subunit to interleukin 2 receptor α chain caused ER retention and degradation of the latter, suggesting a unified process or a colocalization of both determinants (37). This was also true in the case of a soluble protein. When a carboxy-terminal cysteine was transferred from IgM to cathepsin D, the latter was retained and degraded in the ER, whereas addition of a carboxyl-terminal KDEL caused ER retention but no degradation (39). In the case of ASGPR H2a we have been able to separate the two events and their structural determinants because the insertion of the pentapeptide into H1 caused its ER retention but not degradation. The degradation of H2a must be caused by another determinant. The effect of EGHRG on H1 and of KDEL on cathepsin D suggest that a determinant for ER retention is not enough to cause degradation for which another signal is needed. For H2a this degradation signal remains to be found, but it might well be in the transmembrane region based on the colocalized signals for retention and degradation in the transmembrane domains of the other proteins mentioned above.
The presence of the pentapeptide does not cause general misfolding of H2a (Fig. 1) or of H1i5 (Fig. 5C). Similarly, the determinant for ER retention and degradation of TCR-α chain also did not cause global misfolding (37). Unassembled α subunit of the nicotinic acetylcholine receptor also folds to a native-like conformation (40) and so do proteins containing an uncleaved glycosylphosphatidylinositol signal (41). Taken together these findings suggest that although misfolding can cause ER retention and degradation, it may usually not be involved in the quality control of normal unassembled chains and protein precursors. Prolonged association of these proteins to calnexin, as we saw for H1i5 and not for H1 (Fig. 5B), may involve interaction not with unfolded domains but with specific determinants for retention, as, in our case, the EGHRG sequence or this sequence in combination with the transmembrane domain. Although the quality control of malfolded proteins may also involve calnexin, as was seen for a mutant α1-antitrypsin (42), it might take place through a different pathway than that of normal proteins. This is suggested by a recent report in which mutant α1-antitrypsin Z was inefficiently degraded in the ER of cells derived from patients with mutant α1-antitrypsin-related liver disease while the degradation of ASGPR H2a in those cells was not impaired (43).
That H1i5 is completely sensitive to endo H and α-mannosidase and resistant to endo D after metabolic labeling followed by long chases (Fig. 3b and 4a) indicates that the EGHRG sequence is a determinant for true ER retention and not for retrieval. Consistent with this conclusion, the ER-like immunofluorescent pattern of H1i5 was unchanged by incubation of the cells at 15°C, at which temperature proteins trafficking to the Golgi are arrested in the ERGIC (44). No change was observed as well by incubation of the cells with Nz, which by depolymerizing microtubules inhibits traffic between the cis-Golgi, the ERGIC, and the ER (21, 34) (Fig. 4b). In contrast to our results with H1i5, the quality control of several misfolded proteins involves exit from the ER and retrieval, e.g., tsO45 vesicular stomatitis virus (VSV) G protein (45) and misfolded unassembled MHC class I molecules (46, 47). On the other hand, true ER retention is also responsible for the quality control of unassembled IgM intermediates (48). We could speculate that two, distinct mechanisms may exist for protein quality control, one for retrieval to the ER of misfolded proteins and the other for true ER retention of unassembled proteins or protein precursors.
The prolonged association of H1i5 with calnexin suggests a possible role for this transmembrane chaperone in ER retention. Calnexin was shown to play a role in the retention of unassembled MHC class I and TCR subunits (12, 13). Although calnexin has a KKXX-type signal for its retrieval to the ER, it might have an additional determinant for true ER retention, as is the case for calreticulin (49). Consistent with this hypothesis, in the case of VSV G ts045, although it interacts both with calnexin and heavy chain binding protein (BiP), it was shown to exit and recycle back to the ER only together with the latter (45).
Acknowledgments
We thank Malka Okon for technical assistance, Jakko Saraste and Daniel Louvard for antibodies, Shoshana Bar-Nun for helpful advice, and Rachel Ehrlich and Yoav Henis for critically reading the manuscript. This work was supported by Grant 91-00-218 from the United States–Israel Binational Science Foundation and Grant 425/94-2 from the Israel Academy of Sciences and Humanities.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: ASGPR, asialoglycoprotein receptor; endo H, endo-N-acetylglucosaminidase H; endo D, endo-N-acetylglucosaminidase D; TCR, T cell antigen receptor; PDI, protein disulfide isomerase; BFA, brefeldin A; Nz, nocodazole; ALLN, N-acetyl-leucyl-leucyl-norleucinal; ERGIC, ER–Golgi intermediate compartment.
References
- 1.Klausner R D, Sitia R. Cell. 1990;62:611–614. doi: 10.1016/0092-8674(90)90104-m. [DOI] [PubMed] [Google Scholar]
- 2.Hammond C, Helenius A. Curr Opin Cell Biol. 1995;7:523–529. doi: 10.1016/0955-0674(95)80009-3. [DOI] [PubMed] [Google Scholar]
- 3.Lippincott S J, Bonifacino J S, Yuan L C, Klausner R D. Cell. 1988;54:209–220. doi: 10.1016/0092-8674(88)90553-3. [DOI] [PubMed] [Google Scholar]
- 4.Amara J F, Lederkremer G, Lodish H F. J Cell Biol. 1989;109:3315–3324. doi: 10.1083/jcb.109.6.3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kopito R R. Cell. 1997;88:427–430. doi: 10.1016/s0092-8674(00)81881-4. [DOI] [PubMed] [Google Scholar]
- 6.Wileman T, Carson G R, Shih F F, Concino M F, Terhorst C. Cell Regul. 1990;1:907–919. doi: 10.1091/mbc.1.12.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonifacino J S, Suzuki C K, Klausner R D. Science. 1990;247:79–82. doi: 10.1126/science.2294595. [DOI] [PubMed] [Google Scholar]
- 8.Kumagai H, Chun K T, Simoni R D. J Biol Chem. 1995;270:19107–19113. doi: 10.1074/jbc.270.32.19107. [DOI] [PubMed] [Google Scholar]
- 9.Foletti D, Guerini D, Carafoli E. FASEB J. 1995;9:670–680. doi: 10.1096/fasebj.9.8.7768360. [DOI] [PubMed] [Google Scholar]
- 10.Murakami K, Mihara K, Omura T. J Biochem (Tokyo) 1994;116:164–175. doi: 10.1093/oxfordjournals.jbchem.a124489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szczesnaskorupa E, Ahn K S, Chen C D, Doray B, Kemper B. J Biol Chem. 1995;270:24327–24333. doi: 10.1074/jbc.270.41.24327. [DOI] [PubMed] [Google Scholar]
- 12.Jackson M R, Cohendoyle M F, Peterson P A, Williams D B. Science. 1994;263:384–387. doi: 10.1126/science.8278813. [DOI] [PubMed] [Google Scholar]
- 13.Rajagopalan S, Xu Y H, Brenner M B. Science. 1994;263:387–390. doi: 10.1126/science.8278814. [DOI] [PubMed] [Google Scholar]
- 14.Tolchinsky S, Yuk M H, Ayalon M, Lodish H F, Lederkremer G Z. J Biol Chem. 1996;271:14496–14503. doi: 10.1074/jbc.271.24.14496. [DOI] [PubMed] [Google Scholar]
- 15.Lederkremer G Z, Lodish H F. J Biol Chem. 1991;266:1237–1244. [PubMed] [Google Scholar]
- 16.Spiess M, Schwartz A L, Lodish H F. J Biol Chem. 1985;260:1979–1982. [PubMed] [Google Scholar]
- 17.Landt O, Grunert H P, Hahn U. Gene. 1990;96:125–128. doi: 10.1016/0378-1119(90)90351-q. [DOI] [PubMed] [Google Scholar]
- 18.Martin-Zanca D, Oskam R, Mitra G, Copeland T, Barbacid M. Mol Cell Biol. 1989;9:24–33. doi: 10.1128/mcb.9.1.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen C A, Okayama H. BioTechniques. 1988;6:632–638. [PubMed] [Google Scholar]
- 20.Louvard D, Reggio H, Warren G. J Cell Biol. 1982;92:92–107. doi: 10.1083/jcb.92.1.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saraste J, Svensson K. J Cell Sci. 1991;100:415–430. doi: 10.1242/jcs.100.3.415. [DOI] [PubMed] [Google Scholar]
- 22.Hebert D N, Foellmer B, Helenius A. Cell. 1995;81:425–433. doi: 10.1016/0092-8674(95)90395-x. [DOI] [PubMed] [Google Scholar]
- 23.Ou W J, Cameron P H, Thomas D Y, Bergeron J J M. Nature (London) 1993;364:771–776. doi: 10.1038/364771a0. [DOI] [PubMed] [Google Scholar]
- 24.Braakman I, Helenius J, Helenius A. Nature (London) 1992;356:260–262. doi: 10.1038/356260a0. [DOI] [PubMed] [Google Scholar]
- 25.Lodish H F, Kong N, Wikstrom L. J Biol Chem. 1992;267:12753–12760. [PubMed] [Google Scholar]
- 26.Spiess M. Biochemistry. 1990;29:1009–1018. doi: 10.1021/bi00495a001. [DOI] [PubMed] [Google Scholar]
- 27.Bischoff J, Lodish H F. J Biol Chem. 1987;262:11825–11832. [PubMed] [Google Scholar]
- 28.Pelham H R B. Curr Opin Cell Biol. 1995;7:530–535. doi: 10.1016/0955-0674(95)80010-7. [DOI] [PubMed] [Google Scholar]
- 29.Kornfeld R, Kornfeld S. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 30.Tai T, Yamashita K, Ogata A M, Koide N, Muramatsu T. J Biol Chem. 1975;250:8569–8575. [PubMed] [Google Scholar]
- 31.Wikstrom L, Lodish H F. J Cell Biol. 1991;113:997–1007. doi: 10.1083/jcb.113.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bause E, Breuer W, Schweden J, Roeser R, Geyer R. Eur J Biochem. 1992;208:451–457. doi: 10.1111/j.1432-1033.1992.tb17207.x. [DOI] [PubMed] [Google Scholar]
- 33.Bischoff J, Kornfeld R. J Biol Chem. 1986;261:4758–4765. [PubMed] [Google Scholar]
- 34.Cole N B, Sciaky N, Marotta A, Song J, Lippincottschwartz J. Mol Biol Cell. 1996;7:631–650. doi: 10.1091/mbc.7.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scherer P E, Lederkremer G Z, Williams S, Fogliano M, Baldini G, Lodish H F. J Cell Biol. 1996;133:257–268. doi: 10.1083/jcb.133.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neumann D, Yuk M H, Lodish H F, Lederkremer Z. Biochem J. 1996;313:391–399. doi: 10.1042/bj3130391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bonifacino J S, Cosson P, Shah N, Klausner R D. EMBO J. 1991;10:2783–2793. doi: 10.1002/j.1460-2075.1991.tb07827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuk M H, Lodish H F. J Cell Biol. 1993;123:1735–1749. doi: 10.1083/jcb.123.6.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alberini C M, Bet P, Milstein C, Sitia R. Nature (London) 1990;347:485–487. doi: 10.1038/347485a0. [DOI] [PubMed] [Google Scholar]
- 40.Blount P, Merlie J P. J Biol Chem. 1988;263:1072–1080. [PubMed] [Google Scholar]
- 41.Field M C, Moran P, Li W, Keller G A, Caras I W. J Biol Chem. 1994;269:10830–10837. [PubMed] [Google Scholar]
- 42.Liu Y, Choudhury P, Cabral C M, Sifers R N. J Biol Chem. 1997;272:7946–7951. doi: 10.1074/jbc.272.12.7946. [DOI] [PubMed] [Google Scholar]
- 43.Teckman J H, Perlmutter D H. J Biol Chem. 1996;271:13215–13220. doi: 10.1074/jbc.271.22.13215. [DOI] [PubMed] [Google Scholar]
- 44.Hauri H P, Schweizer A. Curr Opin Cell Biol. 1992;4:600–608. doi: 10.1016/0955-0674(92)90078-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hammond C, Helenius A. J Cell Biol. 1994;126:41–52. doi: 10.1083/jcb.126.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raposo G, Vansanten H M, Leijendekker R, Geuze H J, Ploegh H L. J Cell Biol. 1995;131:1403–1419. doi: 10.1083/jcb.131.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hsu V W, Yuan L C, Nuchtern J G, Lippincott S J, Hammerling G J, Klausner R D. Nature (London) 1991;352:441–444. doi: 10.1038/352441a0. [DOI] [PubMed] [Google Scholar]
- 48.Isidoro C, Maggioni C, Demoz M, Fra A M, Sitia R. J Biol Chem. 1996;271:26138–26142. doi: 10.1074/jbc.271.42.26138. [DOI] [PubMed] [Google Scholar]
- 49.Sonnichsen B, Fullekrug J, Van P N, Diekmann W, Robinson D G, Mieskes G. J Cell Sci. 1994;107:2705–2717. doi: 10.1242/jcs.107.10.2705. [DOI] [PubMed] [Google Scholar]