Abstract
Alignments of homologous genes typically reveal a great diversity of intron locations, far more than could fit comfortably in a single gene. Thus, a minority of these intron positions could be inherited from a single ancestral gene, but the larger share must be attributed to subsequent events of intron gain or intron “sliding” (movement from one position to another within a gene). Intron sliding has been argued from cases of discordant introns and from putative spatial clustering of intron positions. A list of 32 cases of discordant introns is presented here. Most of these cases are found to be artefactual. The spatial and phylogenetic distributions of intron positions from five published compilations of gene data, comprising 205 intron positions, have been examined systematically for evidence of intron sliding. The results suggest that sliding, if it occurs at all, has contributed little to the diversity of intron positions.
The Problem of Intron Position Diversity
The locations of introns in homologous genes do not always coincide, the proportion of shared intron positions decreasing with increased evolutionary distance. In early comparisons of eukaryotic protein-coding genes (e.g., ref. 1), it seemed possible to attribute all such differences to loss of introns inherited from an intron-rich ancestral gene. Such a view has become problematic, due to the greatly increased numbers of intron positions now known, and to the increasing recognition that individual intron positions typically show a restricted phylogenetic distribution indicative of a recent origin (2–5). If all 205 different intron positions documented in published compilations of gene data for actins (6), glyceraldehyde-3-phosphate dehydrogenase (GAPDH; ref. 7), small G proteins (3), triose-phosphate isomerase (TPI; ref. 5), and tubulins (8) are packed into hypothetical ancestral genes (with a combined length of 1,603 codons), they would break up the genes into exons with a mean length of only 23 bp and a median length of only 14 bp, with many minuscule exons (e.g., 26% would be 1–6 bp in length). If only half of these 205 introns occupy ancestral positions, this still would imply a mean ancestral exon size of just 15 codons, three times smaller than the mean exon size observed for the most intron-rich extant genomes known (9, 10).
Thus, the vast diversity of (typically phylogenetically restricted) intron positions suggests that the majority of intron locations in extant eukaryotic genes do not represent divisions present in genes of a eukaryotic common ancestor—much less the spacers between mini-genes in an even more ancient hypothetical progenitor (11). A modest proportion of intron positions could represent ancient features, as required minimally in an introns-early view (12), but most extant divisions in split genes are more recent in origin, owing to one or more processes that have operated during the divergence of eukaryotes. Two candidate processes, sliding of old introns to new positions (13, 14), and addition of introns to genes [by insertion (15) or by duplication of splice signals (16)], have been proposed and discussed in relation to several sets of intron data (2, 4, 5, 8, 17–20).
Two Hypotheses of Intron Sliding
The term “sliding” and its apparent synonyms (“migration,” “frameshifting,” “shifting,” “slippage,” “displacement,” and “drift”) appear frequently in the literature on intron evolution (7, 8, 13, 21–26), but the nature of this process, the evidence supporting it, its underlying molecular mechanism, and its significance for gene evolution, are often unclear. These issues can be clarified briefly as follows. The term “junctional sliding” originally was used to refer to the reassignment of a single upstream or downstream splice junction so as to produce an indel (insertion or deletion) in the encoded mRNA and protein (27). Currently, sliding and its various synonyms are used ambiguously to refer to this process of junctional sliding, as well as to the distinct phenomenon of apparent shifts of an entire intron (which do not produce an indel), referred to here as “intron sliding.” Junctional sliding is relevant here only in that it is invoked as a component process in some models of intron sliding.
In spite of suggestive evidence, intron sliding has not been demonstrated to occur. A diagnosis of intron sliding would be nearly unavoidable for a reliable case in which demonstrably homologous introns occur at slightly different positions in closely related genes. To our knowledge, no such case has yet been found. A claim of homology has been made for introns in two histone genes of Volvox carteri (28), but the introns are different in length and the sequence similarity is largely due to biased nucleotide composition: the alignment of the native sequences is not significantly better than that of the randomly scrambled sequences (R. F. Doolittle, personal communication). The difficulty is not in finding closely spaced introns, which are common, but instead may lie in detecting their homology: spliceosomal introns diverge so rapidly that sequence similarity indicative of homology quickly vanishes (e.g., ref. 3). In the absence of sequence evidence, close spacing itself has been interpreted as evidence of homology of introns. This argument takes two forms (described in more detail below), the discordant introns argument and the clustering argument, neither of which has been evaluated systematically.
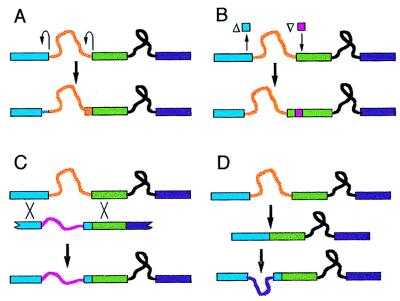
Several intron sliding mechanisms have been proposed. The two most commonly invoked mechanisms, shown in Fig. 1 A and B, are here referred to collectively as “DNA-based sliding.” The mechanism in Fig. 1A calls for a double event of junctional sliding in which nucleotide changes alter splicing signals so as to effect balanced reassignments of the upstream and downstream splice junctions (13, 29). The mechanism in Fig. 1B invokes balanced indels (26, 28). Martinez et al. (14) propose an RNA-mediated mechanism, in which a spliced intron is inserted by the splicing machinery (reverse-spliced) into a nearby site, reverse-transcribed, and incorporated into DNA by recombination. More generally, retropositional movement of introns can be expected to create the appearance of sliding to the extent that the RNA substrate is a spliced (rather than unspliced) mRNA, because the recombination event that incorporates the intron will tend to convert flanking sites to their intron-lacking states (Fig. 1C). Given this, in the comments below, we do not distinguish between the model of Martinez et al. (14) and the coupled insertion/deletion model (Fig. 1C), because the only relevant difference in their implications is that the former implies homology of closely spaced introns, a phenomenon for which no evidence currently exists.
Figure 1.
Mechanisms to account for closely spaced introns. (A) Intron sliding by balanced junction reassignments. Upstream and downstream splice junctions each are reassigned by junctional sliding (curved arrows) to new positions offset in the same direction, by the same distance. In the example shown, both splice junctions are offset in the 5′ direction. (B) Intron sliding by balanced indels. In the example shown, a segment from the upstream exon is deleted, and a segment of equal length is inserted in the downstream exon. As with the balanced junction reassignments model, the exonic sequences (of the ancestral and derived genes) between the two intron locations are not homologous. (C) Coupled insertion/deletion. The fragment shown below the ancestral gene represents a cDNA copy of a hybrid mRNA created by reverse-splicing of an intron into a novel site. Homologous recombination (at points marked by X) incorporates a segment of this intron-bearing cDNA into the chromosome, resulting in the addition of one intron and the accompanying loss of an intron at a nearby site. (D) Separate loss and gain. The ancestral intron is lost, and nothing hinders the gain of a new intron, which may occur at a closely spaced or distantly spaced site.
Alternatively, if introns do not slide, instances of apparent sliding would be due to separate events of loss and gain of introns, or to separate events of gain. As an explanation for a pair of closely spaced intron positions, the hypothesis of separate gain is usually a reasonable alternative to intron sliding, because usually no evidence exists that either intron position was present in a common ancestor. Even when one of two closely spaced introns appears to be ancestral (based on an outgroup comparison), an apparent slide could be due to loss of the ancestral intron followed by a separate event of gain, the separate loss and gain model (Fig. 1D).
What is the relevance of intron sliding for the origin and evolution of intron-containing genes? Based on the assumption that intron sliding is widespread, some authors advocate an introns-early view in which all (or nearly all) differences in intron positions are attributable to sliding and loss of primordial introns, with no need to invoke widespread gain of introns (8, 14, 20, 24). Others advocate an introns-late view in which sliding is insignificant, and all (or nearly all) differences in intron positions are attributable to recent gain and loss (2, 5). One of these contrasting interpretations may be correct, but the dichotomy is rhetorical in origin and does not reflect an underlying logical necessity: though an introns-late view clearly requires extensive intron gain, it is not incompatible with sliding subsequent to gain; nor is intron sliding the only available means to address the problem of intron diversity from an introns-early perspective.
The issue of intron sliding may be separated from polemics by considering two hypotheses independently of any view on the ultimate origin of introns: the strong intron sliding hypothesis would be that a substantial proportion of observed introns are shifted from their original locations (regardless of how and when those original locations were established), whereas the weak intron sliding hypothesis is merely that intron sliding has occurred, if but rarely. These proposals are addressed below, using case studies of discordant introns and an extensive set of data on the phylogenetic and spatial distribution of introns.
Case Studies of Discordant Introns
For some gene comparisons, it has been proposed that the numbers of introns per gene, or the general locations of introns, are conserved in spite of differences in the exact positions of one or more introns, which are called “discordant” or “quasi-conserved” (13, 14, 19, 20, 26, 29). For example, Brenner and Corrochano (29) report that the histidyl-tRNA synthetase genes of pufferfish and hamster each have 12 introns, exactly matching in position except for the eighth introns, which differ in position by a mere 5 bp. Similarly, Jellie et al. (26) present the intron/exon structure of a gene for a nine-domain globin in Artemia salina, noting that three of the nine domain-encoding regions lack an ancestral intron position, yet each such region has an intron at a different position. For several years, we have been cataloguing reports of discordant introns in concordant contexts as they appear in the literature and attempting to verify the physical evidence for them, resulting in a database of information on 32 cases (available from the authors).
Most of these discordances are artefacts. Table 1 lists the 12 of 32 cases of apparently discordant introns in concordant contexts that are now known (on the basis of information described in Table 1) to arise from errors in a published sequence. This list includes the discordant introns reported recently by Brenner and Corrochano (29). In an additional eight cases, apparently discordant introns with the same phase (location relative to the triplet reading frame) occur in regions of alignment ambiguity, such that alternative alignments place the introns at exactly the same position (data not shown). For any isolated case, it is impossible to judge conclusively that the putatively discordant introns are actually concordant (i.e., that one alignment is true and the other false). However, because the vast majority (95%) of other introns in these same genes are concordant, the same is likely to be true of the majority of introns in poorly aligned regions, the appearance of discordance arising from alignment errors.
Table 1.
Apparently discordant introns attributable to errors
| Gene | Source of discordant intron
|
Dist, bp | Flanking introns
|
|||
|---|---|---|---|---|---|---|
| Erroneous position | Other position | Matches
|
Diffs
|
|||
| 5′ | 3′ | 5′ + 3′ | ||||
| Ca-ATPase | Oryctolagusa | Homo | 1 | 11 | 10 | 0 |
| Ca II | Musb | Vert. CA II | 14 | 3 | 2 | 0 |
| GAPDH | Phanerochaetec | Other fungi | 1 | 4 | 0 | 4 |
| GAPDH | Gallusd | Homo, Mus | 1 | 2 | 4 | 3 |
| His-TRS | Mesocricetuse | Fugu | 1 | 4 | 7 | 0 |
| His-TRS | Mesocricetuse | Fugu | 5 | 7 | 4 | 0 |
| ITF | Mus, Rattusf | Homo | 3 | 1 | 0 | 0 |
| Lamin b | Xenopusg | Vert., paralogs | 1 | 5 | 4 | 0 |
| Laminin β2 | Mush | Homo | 1 | 12 | 18 | 0 |
| Ryr | Susi | Homo | 1 | ≥4 | ≥23 | ≥0 |
| TPI | Lactucaj | 3 other plants | 1 | ≥0 | ≥0 | ≥0 |
| Tubulin β4 | D. aurariak | D. melanogaster | 1 | ≥1 | ≥1 | ≥0 |
The first column gives a gene family. The second and third columns give taxonomic sources (and isotypes, as appropriate) for each intron position (references for all gene sequences are available from the authors or at http://is.dal.ca/~arlin/slpd). The fourth (Dist) column gives the apparent (uncorrected) distance between the intron positions. The last three columns provide information on the concordance of other introns in the same genes (matching pairs of introns on the 5′ side of the discordant pair, matching pairs on the 3′ side, and total count of differing introns on both sides; minimum estimates are due to incompletely sequenced genes). Errors were identified on the basis of the following information:
D. H. MacLennan and A. Odermatt, personal communication;
personal communication cited by Hewett-Emmett and Tashian (30);
F. Schuren, personal communication;
internal discrepancy in GenBank accession M11213;
F. Tsui, personal communication;
subsequent publication (31);
R. Stick, personal communication;
M. Durkin, M. Gautam, and U. Wewer, personal communication;
published correction (32);
R. Michelmore, personal communication;
H. Domdey, personal communication. Ca-ATPase, calcium-dependent sarcomeric ATPase; CA II, carbonic anhydrase type II; ITF, intestinal trefoil factor; His-TRS, histidyl-tRNA synthetase; Ryr, ryanodine receptor; Vert., vertebrate.
The remaining 12 cases of discordant introns, shown in Table 2, do not arise from alignment ambiguities and are not known to be attributable to errors. In the absence of confirmatory sequencing and crucial evidence for the homology of introns, these cases remain open to multiple interpretations.
Table 2.
Additional apparently discordant introns
| Gene | Source of discordant intron
|
Dist, bp | Flanking introns
|
|||
|---|---|---|---|---|---|---|
| Unique position | Other position | Matches
|
Diffs
|
|||
| 5′ | 3′ | 5′ + 3′ | ||||
| ADH | Oryza ADH-2 | Monocot, dicot | 1 | 3 | 5 | 0 |
| ADH | Rattus | Mus, Homo | 1 | 2 | 5 | 0 |
| ARF | Drosophila | Homo | 1 | 0 | 1 | 1 |
| Asp AT | Mus mito. | Gallus mito. | 1 | 1 | 7 | 0 |
| CA | Homo CA 4,9 | Vert. CA 1,2,3,5,7 | 1 | 3 | 1 | ≥2 |
| Chol. est. | Rattus | Homo, Mus | 1 | 6 | 3 | ≤2 |
| Chol. est. | Rattus | Home, Mus | 4 | 6 | 3 | ≤2 |
| Globin | Artemia dom. 3 | Artemia dom. 6 | 1 | 1 | 0 | 0 |
| Globin | Artemia dom. 4 | Many animals | 1 | 0 | 0 | 1 |
| Histone H3 | Volvox H3-I | Volvox H3-II | 1 | 0 | 0 | 0 |
| β-Tubulin | Erysiphe | Many other fungi | 1 | 2 | 3 | 0 |
| β-Tubulin | Plasmodium falc. | Plasmodium sp. | 2 | 0 | 1 | 0 |
Conventions are as given for Table 1. Maxima appearing in the final (Diffs) column for the cholesterol esterase case reflect the fact that the only potential differences among flanking introns are subject to doubt. The intron position in the leftward column is unique (not found in any related gene) and therefore more subject to doubt than that in the rightward column, which has been identified multiple times, with the following exceptions: Histone H3 (both positions are unique) and carbonic anhydrase (neither position is unique). ADH, alcohol dehydrogenase; ARF, ADP-ribosylation factor; Asp AT, aspartate aminotransferase; CA, carbonic anhydrase; Chol. est., cholesterol esterase; cp., chloroplastic isozyme; dom., domain (of a multiple-domain-encoding gene); mito., mitochondrial isozyme.
Intron Sliding and Phylogeny
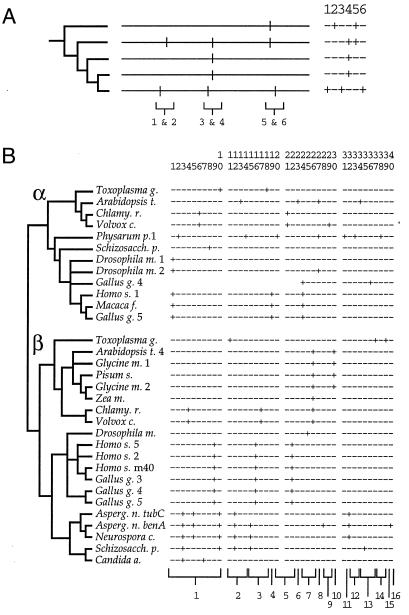
The phylogenetic hallmark of the mechanisms shown in Fig. 1 would be a distribution in which one intron position is nested within the distribution of another, as illustrated in Fig. 2A. This pattern of nesting is evident in some isolated instances in which intron sliding has been suggested (e.g., ref. 26), but its occurrence in more extensive sets of data has not been considered. For example, Liaud et al. (8) have argued that intron positions found among diverse tubulin genes fall into 16 regularly spaced clusters, as though each cluster represented the descendants of an ancestral intron that has slid, in localized fashion, to neighboring positions. Yet, the phylogenetic distribution of tubulin intron positions compiled by Liaud et al. (8) shows little sign of the nesting expected from such extensive intron sliding (Fig. 2B): nested distributions are absent from 15 of the 16 putative clusters.
Figure 2.
Intron sliding and phylogeny. (A) Hypothetical examples of nested and nonnested distributions. (Left) A phylogenetic tree. (Center) Maps of intron positions. (Right) A table of occurrences for the six different intron positions. Brackets indicate pairs of closely spaced positions. Introns 1 and 2 do not show a nested distribution. Intron 3 is nested within the distribution of intron 4 in a manner suggestive of sliding. Intron 6 is nested within the distribution of intron 5, in a manner suggestive of separate loss and gain, because both introns are absent in ingroups. (B) Intron positions in tubulin genes. (Left) A phylogenetic tree relating the genes (based largely on ref. 33). (Right) A table of occurrences of 40 intron positions (slightly modified from ref. 8). The 16 numbered sets of introns indicated by brackets at the bottom denote the putative clusters proposed by Liaud et al. (8). Four of the introns, numbered 10, 12, 27, and 30, have patchy distributions, suggesting either loss or sliding to a different position. In the case of intron 10, other introns are closely spaced and phylogenetically nested: if intron 10 is ancestral to α- and β-tubulins, then introns 6–9 could have arisen from it by one of the mechanisms shown in Fig. 1.
To evaluate phylogenetic evidence for intron sliding more systematically, the phylogenetic distributions of introns for the five sets of intron data discussed above were examined. [Nexus files combining intron data and phylogenetic trees taken from analyses of the corresponding protein sequences (3, 5, 33–35) are available from the authors.]
Of the total set of 205 intron positions, 157 (76.6%) show a distribution that is consistent with a single origin followed by faithful inheritance (i.e., no events of loss); 24 positions (11.7%) show a distribution consistent with a single origin followed by 1–3 apparent events of loss; and 24 positions (11.7%) exhibit various complex patterns suggesting multiple (≥2) origins or many (≥4) losses. Note that an event of origin may be an event of either sliding or gain, and that an event of loss may represent either actual loss or sliding to a different position. Thus, the intron positions with patchy distributions are candidates for ancestral introns that may have slid elsewhere, to closely spaced positions that can be identified readily by a pattern of phylogenetic nesting.
In the five sets of intron data, 40 pairs of intron positions are closely spaced (1–30 bp apart) and show a nested phylogenetic distribution (a database describing the 40 cases of closely spaced nested pairs is available from the authors). For 25 of the 40 nested pairs, both introns are absent in an ingroup so as to suggest (as explained in Fig. 2A) the model of separate loss and gain (Fig. 1D). Nevertheless, because such patterns also might arise by intron sliding (or coupled insertion/deletion; Fig. 1 A–C) followed by separate loss events, it is not possible to draw a conclusion from these numbers in the absence of a reference standard or an exact quantitative model.
A standard of comparison can be established by considering that events of sliding (and, to a lesser degree, coupled insertion/deletion) are spatially localized, whereas separate events of loss and gain are not. Closely spaced, phylogenetically nested pairs of introns may be due to either cause, whereas distantly spaced pairs must be attributed to whatever nonsliding processes are operative (inheritance of ancestral introns, and separate events of gain and loss). If nested distributions are mainly due to localized intron sliding, then with increasing distance between introns, nested phylogenetic distributions will be rare; but if sliding never occurs, one expects similar phylogenetic patterns regardless of the distance between the introns. To test these implications, nested pairs of introns 31–60 bp apart and 61–90 bp apart were identified by the same criteria used to identify the closely spaced (1–30 bp) cases.
The comparison between these three subsets yields a simple result. The numbers of cases (40, 44, and 27 for the short, medium, and long distances, respectively) are not significantly different (χ2 = 4.3, two degrees of freedom, P > 0.05), indicating that separate loss and gain is sufficient to explain the observed number of closely spaced nested pairs. Furthermore, the proportions of cases in which the phylogenetic distribution of the introns suggests an intron-lacking intermediate (25 of 40, 28 of 44, and 13 of 27, for the short, medium, and long classes, respectively) are not significantly different (χ2 = 1.9 for the 3 × 2 contingency test, two degrees of freedom, P > 0.05). Closely spaced nested pairs of introns, which represent the best candidates for intron sliding, are neither more frequent nor more suggestive of sliding (as opposed to separate loss and gain) than more distantly spaced pairs, which are not candidates for intron sliding. Thus, a phylogenetic signal that might justify invoking a special process to explain closely spaced introns is not detected.
Intron Sliding and the Spatial Distribution of Intron Positions
Apparent clustering and excess closeness have been mentioned in regard to gene data for tubulins (8), as mentioned above, and also for TPI (5, 25, 36), GAPDH (4, 7, 19, 20), and globins (17, 18, 24, 37). However, a nonrandom degree of closeness or clustering has not been demonstrated.
The spatial distributions of intron positions for the five sets of data discussed previously, along with sample random distributions, are shown in Fig. 3. The observed distributions do not seem to exhibit clusters more prominent or more regularly spaced than those generated at random. The possibility of excess closeness can be evaluated more rigorously by comparing the nearest-neighbor distances to an exponential distribution (5), and the possibility of clustering (under-dispersion) can be evaluated by applying the covariance test of Goss and Lewontin (38).
Figure 3.
Spatial distribution of intron positions for five genes. Positions of introns are shown as vertical hatches on a horizontal bar (the scale at the bottom is in bp). Beneath each observed distribution are two random distributions (drawn at random, without replacement, from all possible inter-nucleotide sites in the coding region). Intron positions within one codon of each other may appear as a thickened line, rather than as separate lines. (A) Actin, 40 distinct intron positions in 375 codons, mapped relative to human α-actin (6). (B) GAPDH, 46 positions, 337 codons, relative to Zea Gap4 (modified slightly from ref. 7). (C) Small G proteins, 58 positions, 191 codons, relative to human H-Ras (modified slightly from ref. 3). (D) TPI, 21 positions, 248 codons, relative to chicken TPI (5). (E) Tubulin, 40 positions, 452 codons, relative to Pisum Tub1 (8).
Using the exponential test and the covariance test, we find no significant deviation from randomness for four of the five data sets, and a significant deviation for both tests for the case of tubulin (P < 0.05 for the Kolmogorov–Smirnov exponential test; P < 0.005 for the Goss–Lewontin covariance test). This deviation appears to be due, not to regularly spaced clusters, but to a bias in intron density: half of the tubulin introns map to the first 20% of the gene (Fig. 3E). Such a bias in intron density (strongest for the tubulin data, but also seen for actin and GAPDH, Fig. 3) is a corollary of the well known tendency for exons to be shorter toward the 5′ end of a gene (9). If the tubulin data are split into 5′ and 3′ partitions (the region containing the first 20 intron positions and the region containing the last 20) to compensate crudely for the observed bias in intron density, no significant deviation from randomness (P > 0.1) is found for either partition, for either statistical test.
A final test can be made with specific reference to DNA-based sliding, for which it is possible to predict something of the distribution of sliding distances based on (i) the implication of these models that the exonic sequences between the two slid introns are not homologous (Fig. 1 A and B); and (ii) the apparently contradictory empirical result that the aligned exonic sequences between closely spaced introns are typically so similar as to give the appearance of homology (14), as illustrated by the example shown in Fig. 4A. If closely spaced intron positions result from DNA-based sliding, it must be the case that successful slides are limited to the rare instances in which, by chance, high sequence identify is achieved. Longer slides will be less likely to succeed, thus will be increasingly rare, the steepness of the drop in frequency being a function of the required sequence identity (Fig. 4B). The sequence identity in the exonic interval for the set of 40 closely spaced nested pairs of introns is 71%, thus, based on Fig. 4B, the distribution of sliding distances is expected to drop precipitously, with nearly all slides being 1–5 bp.
Figure 4.
Distance test for DNA-based sliding. (A) An example of the high sequence identity (in this case, 6/7 or 86%) observed for the aligned exonic sequences between closely spaced intron positions. The locations of introns are marked by ∗; the aligned exonic sequences between the intron positions are shown in bold; the amino acid sequence, which is identical for the two genes compared, is given above (in single-letter code; the example is a nested pair, introns 10 and 8 from the tubulin data set, represented by gene sequences for Toxoplasma gondii α-tubulin and Schizosaccharomyces pombe α-tubulin). (B) Dependence of DNA-based intron sliding distances on sequence identity. In general, the chance that a sequence of length N will match (to a given degree of identity) an unrelated sequence decreases as N increases, thus the distance over which introns slide by a DNA-based mechanism will show a decreasing frequency distribution, the sharpness of the decrease being dependent on the sequence identity required (data from computer simulations). (C) Observed distribution of short pairwise distances between intron positions. The complete set refers to all 323 pairs of intron positions that are within 30 bp of each other and that never occur together in the same gene. The nested subset refers to 40 of these pairs that are phylogenetically nested.
However, the observed distribution of pairwise inter-intron-position distances (Fig. 4C) is flat, not sharply decreasing. In particular, distances for the nested subset of cases (the best candidates for sliding) are no less flat than for the complete set. The conclusion that DNA-based intron sliding is negligible would seem difficult to avoid. Indeed, Martinez et al. (14) previously made what is essentially a nonquantitative version of this same argument, concluding that DNA-based intron sliding cannot account for intron positions separated by more than a few codons of highly conserved sequence.
More generally, the lack of evidence for excess closeness or for spatial clustering suggests that localized intron sliding—of any type—must be either so infrequent as to be negligible, or so rampant as to disperse clusters beyond recognition. The lack of a significant phylogenetic signal indicative of sliding favors the former alternative.
Summary
Because the introns observed in extant genes occur at far too many positions to have been present all together in a common ancestral gene, most intron positions must have arisen more recently, by intron sliding, intron gain, or both. Case studies of discordant intron positions do not resolve the question of whether intron sliding occurs, because most such cases are artefactual and the remaining cases are ambiguous in the absence of crucial evidence for the homology of the discordant introns. The weak intron sliding hypothesis (i.e., that intron sliding occurs) remains viable in the absence of clear evidence for or against it.
The strong intron sliding hypothesis has been evaluated on the basis of implications with respect to the spatial and phylogenetic distribution of intron positions, using data from five sets of genes comprising 205 distinct intron positions. The phylogenetic distributions of introns suggest that closely spaced nested pairs of introns, which are consistent with intron sliding, are no more common than expected from a comparison with distantly spaced nested pairs, which are not. The spatial distribution of intron positions reveals no sign of the excess closeness or clustering expected from sliding. These results suggest that the influence of intron sliding is negligible, intron position diversity arising primarily by the addition of introns to genes during eukaryotic evolution.
Acknowledgments
We thank B. Diaz and E. Raff for providing data on the tubulin β4 gene of Drosophila melanogaster, and L. Corrochano, H. Domdey, M. Durkin, H. Edenberg, M. Gautam, D. Hui, D. MacLennan, R. Michelmore, A. Odermatt, F. Schuren, J. Sherwood, R. Stick, U. Wewer, R. Wu, Y. Xie, and especially F. Tsui for providing corrections or confirmations of published sequence information. This work was supported by the Program in Evolutionary Biology of the Canadian Institute for Advanced Research (A.S., J.M.L., and W.F.D.), Medical Research Council Grant MT4467 (to W.F.D.), National Science Foundation Grant MCB-9318858 (to J.D.P.), and an Alfred P. Sloan Foundation/National Science Foundation Fellowship in Molecular Evolution (to J.M.L.).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: GAPDH, glyceraldehyde-3-phosphate dehydrogenase; TPI, triose-phosphate isomerase.
References
- 1.Crabtree G R, Comeau C M, Fowlkes D M, Fornace A J, Malley J D, Kant J A. J Mol Biol. 1985;185:1–19. doi: 10.1016/0022-2836(85)90179-2. [DOI] [PubMed] [Google Scholar]
- 2.Dibb N J, Newman A J. EMBO J. 1989;8:2015–2021. doi: 10.1002/j.1460-2075.1989.tb03609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dietmaier W, Fabry S. Curr Genet. 1994;26:497–505. doi: 10.1007/BF00309940. [DOI] [PubMed] [Google Scholar]
- 4.Logsdon J M, Jr, Palmer J D. Nature (London) 1994;369:526. doi: 10.1038/369526a0. [DOI] [PubMed] [Google Scholar]
- 5.Logsdon J M, Jr, Tyshenko M G, Dixon C, D-, Jafari J, Walker V K, Palmer J D. Proc Natl Acad Sci USA. 1995;92:8507–8511. doi: 10.1073/pnas.92.18.8507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weber K, Kabsch W. EMBO J. 1994;13:1280–1286. doi: 10.1002/j.1460-2075.1994.tb06380.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kersanach R, Brinkmann H, Liaud M, Zhang D, Martin W, Cerff R. Nature (London) 1994;367:387–389. doi: 10.1038/367387a0. [DOI] [PubMed] [Google Scholar]
- 8.Liaud M-F, Brinkmann H, Cerff R. Plant Mol Biol. 1992;18:639–651. doi: 10.1007/BF00020007. [DOI] [PubMed] [Google Scholar]
- 9.Smith M W. J Mol Evol. 1988;27:45–55. doi: 10.1007/BF02099729. [DOI] [PubMed] [Google Scholar]
- 10.Palmer J D, Logsdon J M., Jr Curr Opin Genet Dev. 1991;1:470–477. doi: 10.1016/s0959-437x(05)80194-7. [DOI] [PubMed] [Google Scholar]
- 11.Gilbert W. Cold Spring Harbor Symp Quant Biol. 1987;52:901–905. doi: 10.1101/sqb.1987.052.01.098. [DOI] [PubMed] [Google Scholar]
- 12.Doolittle W F. Am Nat. 1987;130:915–928. [Google Scholar]
- 13.Rogers J. Trends Genet. 1986;12:223. [Google Scholar]
- 14.Martinez P, Martin W, Cerff R. J Mol Biol. 1989;208:551–565. doi: 10.1016/0022-2836(89)90147-2. [DOI] [PubMed] [Google Scholar]
- 15.Cavalier-Smith T. Trends Genet. 1991;7:145–148. [PubMed] [Google Scholar]
- 16.Rogers J H. Trends Genet. 1989;5:213–216. doi: 10.1016/0168-9525(89)90084-x. [DOI] [PubMed] [Google Scholar]
- 17.Moens L, Vanfleteren J, De Baere I, Jellie A M, Tate W, Trotman C N. FEBS Lett. 1992;312:105–109. doi: 10.1016/0014-5793(92)80915-4. [DOI] [PubMed] [Google Scholar]
- 18.Pohajdak B, Dixon B. FEBS Lett. 1993;320:281–283. doi: 10.1016/0014-5793(93)80603-r. [DOI] [PubMed] [Google Scholar]
- 19.Cerff R, Martin W, Brinkmann H. Nature (London) 1994;369:527–528. [Google Scholar]
- 20.Cerff R. In: Tracing Biological Evolution in Protein and Gene Structures. Go M, Schimmel P, editors. New York: Elsevier; 1995. pp. 205–227. [Google Scholar]
- 21.Holland S K, Blake C C F. BioSystems. 1987;20:181–206. doi: 10.1016/0303-2647(87)90044-x. [DOI] [PubMed] [Google Scholar]
- 22.Yoshihara C M, Lee J D, Dodgson J B. Nucleic Acids Res. 1987;15:753–770. doi: 10.1093/nar/15.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nata K, Sugimoto T, Kohri K, Hidaka H, Hattori E, Yamamoto H, Yonekura H, Okamoto H. Gene. 1993;130:183–189. doi: 10.1016/0378-1119(93)90418-3. [DOI] [PubMed] [Google Scholar]
- 24.Moens L, Vanfleteren J, De Baere I, Jellie A M, Tate W, Trotman C N A. FEBS Lett. 1993;320:284–287. doi: 10.1016/0014-5793(92)80915-4. [DOI] [PubMed] [Google Scholar]
- 25.Gilbert W, Glynias M. Gene. 1993;135:137–144. doi: 10.1016/0378-1119(93)90058-b. [DOI] [PubMed] [Google Scholar]
- 26.Jellie A M, Tate W P, Trotman C N A. J Mol Evol. 1996;42:641–647. doi: 10.1007/BF02338797. [DOI] [PubMed] [Google Scholar]
- 27.Craik C S, Rutter W J, Fletterick R. Science. 1983;220:1125–1129. doi: 10.1126/science.6344214. [DOI] [PubMed] [Google Scholar]
- 28.Muller K, Schmitt R. Nucleic Acids Res. 1988;16:4121–4136. doi: 10.1093/nar/16.9.4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brenner S, Corrochano L M. Proc Natl Acad Sci USA. 1996;93:8485–8489. doi: 10.1073/pnas.93.16.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hewett-Emmett D, Tashian R E. In: The Carbonic Anhydrases: Cellular Physiology and Molecular Genetics. Dodgson S J, Tashian R E, Gros G, Carter N D, editors. New York: Plenum; 1991. pp. 15–32. [Google Scholar]
- 31.Chinery R, Poulsom R, Cox H M. Gene. 1996;171:249–253. doi: 10.1016/0378-1119(96)00074-1. [DOI] [PubMed] [Google Scholar]
- 32.Wen G, Leeb T, Reinhart B, Schmoelzl S, Brenig B. Anim Genet. 1996;27:297–304. doi: 10.1111/j.1365-2052.1996.tb00973.x. [DOI] [PubMed] [Google Scholar]
- 33.Baldauf S L, Palmer J D. Proc Natl Acad Sci USA. 1993;90:11558. doi: 10.1073/pnas.90.24.11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drouin G, de Sa M M, Zuker M. J Mol Evol. 1995;41:841–849. doi: 10.1007/BF00173163. [DOI] [PubMed] [Google Scholar]
- 35.Roger A. Ph.D. thesis. Halifax, Nova Scotia, Canada: Dalhousie University; 1996. [Google Scholar]
- 36.Stoltzfus A, Spencer D, Doolittle W F. Comput Appl Biosci. 1995;11:509–515. doi: 10.1093/bioinformatics/11.5.509. [DOI] [PubMed] [Google Scholar]
- 37.Stoltzfus A, Doolittle W F. Curr Biol. 1993;3:215–217. doi: 10.1016/0960-9822(93)90336-m. [DOI] [PubMed] [Google Scholar]
- 38.Goss P J E, Lewontin R C. Genetics. 1996;143:589–602. doi: 10.1093/genetics/143.1.589. [DOI] [PMC free article] [PubMed] [Google Scholar]