Abstract
Sixty-six haplotypes at a locus containing a simple dinucleotide (CA)n microsatellite repeat were isolated by PCR–single-strand conformational polymorphism from populations of the horseshoe crab Limulus polyphemus. These haplotypes were sequenced to assess nucleotide variation directly. Thirty-four distinct sequences (alleles) were identified in a region 570 bp long that included the microsatellite motif. In the repeat region itself, CA-number varied in integer values from 5 to 11 across alleles, except that a (CA)8 class was not observed. Differences among alleles were due also to polymorphisms at 22 sites in regions immediately flanking the microsatellite repeats. Nucleotide substitutions in these regions were used to estimate phylogenetic relationships among alleles, and the gene phylogeny was used to trace the evolution of length variation and CA repeat numbers. A low correlation between size variation and genealogical relationships among alleles suggests that absolute fragment size (as normally scored in microsatellite assays) is an unreliable indicator of historical affinities among alleles. This finding on the molecular fine structure of microsatellite variation suggests the need for caution in the use of repeat counts at microsatellite loci as secure indicators of allelic relationships.
Interspersed throughout the genomes of most eukaryotic organisms are simple sequence repeats (SSRs) or microsatellite loci, each consisting of a tandem-repeat array of short (2–5 bp) DNA sequence units. Because of the great variability in the number of repeat units at most SSR loci, microsatellites provide an important source of molecular markers for many areas of genetic research (1–3). Slippage during DNA replication is thought to be an important causal factor generating length mutations at microsatellite loci (4–6), but knowledge of the precise mode of evolution of repeat numbers in populations is far from complete. To account for the distribution of microsatellite alleles in populations, a “stepwise mutation” model has been proposed that assumes the loss or gain of single repeat units, one at a time. Although computer simulations (7, 8) have shown that this model is consistent with observed population distributions of microsatellite alleles, Di Rienzo et al. (9) provided evidence that a modified (two-step) stepwise mutation model may better predict patterns of variation at these loci. Regardless of the exact mutational model generating allelic variation, the range of repeat numbers found at microsatellite loci appears to be constrained, implying that allelic size alone is unlikely to be an unambiguous indicator of phylogenetic affinities among alleles (10, 11).
Traditional genetic surveys of microsatellite loci capitalize upon SSR variation because it is easy to score DNA fragments by size. Length variation is the conspicuous and usually the sole criterion employed to characterize allelic diversity at loci displaying variable numbers of tandem repeats. However, recent reports of DNA sequence variation at microsatellite loci have noted size homoplasy within (12–14) and between species (11, 13, 15). In other words, an allelic size-class can include alleles identical by descent (homology) and alleles that have achieved the same length via convergent evolutionary events, parallelisms, or reversions (length homoplasy). The extent to which homoplasy at microsatellite loci occurs in population genetic studies, and how flanking regions may contribute to its magnitude, remain open issues (16).
Fine structure analyses of variation among microsatellite alleles have shown unexpected complexities in the mutational process. Allelic differences can involve not only the number of SSRs, but also different kinds of “interruptions” within a tandem-repeat array (11–13, 17, 18) as well as nucleotide substitutions and insertions/deletions (indels) in regions flanking the repeat motif (11, 14, 15). These findings are relevant to the design of statistical methods for estimating genetic distances between microsatellite alleles distinguished by fragment size (19–21). Clearly, the propriety of any such statistic is critically dependent upon the nature of molecular interconversion among microsatellite allelic classes.
Variation in flanking regions provides a potential source of information on genealogical relationships among length-defined microsatellite alleles. Gene genealogies are used increasingly to illuminate biological processes at many levels of the biological hierarchy (22). Inferences based on phylogenetic trees are drawn either from the structure of a tree or from the way character states map onto the tree. For example, DNA sequences from microsatellite flanking regions were used to infer phylogenetic relationships among the principal lineages of cichlid fishes and two other families of the suborder Labroidei that diverged more than 80–100 million years ago (23). Analyses of the phylogenetic histories of flanking sequences also should be informative with regard to the molecular basis of microsatellite mutations (24) and to the development of appropriate conceptual and statistical models of microsatellite evolution.
However, almost no data are available for the assessment of genealogical relationships among microsatellite alleles. The DNA sequences obtained from flanking regions usually are short and contain few polymorphic sites because (i) size-fractionated genomic libraries with fragment size <400 bp conventionally are used to screen for microsatellite loci, and (ii) the PCR primers employed to screen for population-level variation typically are designed expressly to amplify only minimal flanking regions. Even when microsatellite loci of adequate length are obtained, the physical isolation of alleles before sequencing typically involves labor-intensive cloning procedures. The only published intraspecific phylogeny for a microsatellite locus was inferred indirectly from that of a linked HLA gene in humans (24). Here we initiate genealogical appraisals of microsatellite variation within species using single-strand conformational polymorphism (SSCP) to gel-isolate alleles for direct sequence analysis (25). DNA sequence data at flanking regions can be determined directly from the isolated haplotypes and used for phylogenetic analysis.
MATERIALS AND METHODS
A dinucleotide (CA)n microsatellite locus was isolated and screened for variation from the genomic DNA of horseshoe crabs (Limulus polyphemus) originally collected by Saunders et al. (26). Isolation of the microsatellite locus followed a procedure similar to that described by Sültmann et al. (27). A primer 5′-CCTTATATAAAGCAAACCAGACGGCAGCA-3′ was used to generate randomly amplified PCR fragments using DNA samples from several individuals. Conditions for the PCR were as follows: 1.5 mM MgCl2, 0.2 mM of each dNTP, 0.8 μM of primer, 1× buffer, and 0.2 unit of Taq polymerase (Promega) in 25 μl final volume. Thermocycling conditions were 1 min at 94°C, 1 min at 53°C, and 1 min at 72°C for 27 cycles, plus a final extension step at 72°C for 2 min. PCR products were selected by size (between 0.5 and 1.5 kbp) and isolated using a 2% agarose gel. DNA fragments purified from the gel were cloned with the pGem-T vector system (Promega), and cloned inserts were sequenced from various individuals following an automated sequencing protocol (Applied Biosystems model PE 373A).
DNA sequences from several individuals were aligned for each of three different fragments, and searched for polymorphisms. A PCR fragment 1.5 kbp long was found to contain a simple dinucleotide (CA) repeat. Based on the alignment, specific PCR primers for this locus were designed: LPF3s, 5′-TTAAAGTGCGAGGAAGATTTTG; and LPF3a, 5′-AGCCTTACCGCTCAAATATCG. These primers amplify a fragment ≈640 bp long containing the simple repeat region.
DNA sequence variation at this locus was screened among 37 horseshoe crabs from widespread coastal locations between New Hampshire and the Gulf coast of Florida. Genomic DNA samples were used as templates for amplification by the PCR, with primers LPF3s and LPF3a applied under the following conditions: 1.8 mM MgCl2, 0.2 mM of each dNTP, 0.6 μM of each primer, 1× buffer, and 0.2 unit of Taq polymerase (Promega) in 50 μl final volume. Cycling conditions were 1 min at 94°C, 1 min at 52°C, and 1 min at 72°C for 28 cycles. The presence of a single clear band was verified in 2% agarose gels.
Separation and isolation of alleles prior to sequencing was performed by nonisotopic SSCP (28–30) following the protocol detailed by Ortí et al. (25). Conditions for SSCP were as follows: 15 μl of unpurified PCR product (roughly 0.5–1.6 μg of DNA) were mixed with 2.5 μl of denaturing loading buffer containing 0.4 μl of 1 M methylmercury hydroxide (Matthey Electronics, Ward Hill, MA), 1.8 μl of 15% Ficoll loading buffer (with 0.25% bromophenol blue and 0.25% xylene cyanol), and 0.3 μl of 1× TBE buffer (90 mM Tris/92 mM boric acid/2.5 mM EDTA). This mixture was heated for 4 min at 85°C to denature the DNA and immediately chilled on ice before loading into 10% polyacrylamide (39:1 acrylamide to bisacrylamide) gels. SSCP gels were run with 1× TBE buffer on a vertical electrophoresis system (Fisher Biotech model VE16-1) at constant power (15 W) and temperature (5°C) for 21 h.
Gels were stained for 20 min with SYBR Green II (Molecular Probes) for single-stranded nucleic acids, in a stock solution diluted 1:5000 in 1× TBE (pH 8.0). Bands were visualized and photographed under UV light. In most cases three to four bands were present, as expected for heterozygous individuals (only two bands are present in homozygotes or when allelic differences are not big enough to allow separation by SSCP). A small fraction of each band was excised from the gel with the tip of a 200-μl glass micropipette. These acrylamide plugs were placed individually in tubes with 50 μl of distilled water and stored at −20°C. The gel samples were heated to 80°C for 10 min and then used as template to generate double-stranded PCR products using the same LPF3 primers. PCR products (50 μl) were purified (High Pure PCR Product Purification Kit, Boehringer Mannheim) and 3 μl of the purified product was used in cycle-sequencing reactions with each of the LPF3 primers (SequiTherm Excel DNA Sequencing Kit, Epicentre Technologies, Madison, WI).
DNA sequences were aligned by eye and analyzed phylogenetically with paup version 3.1.1 (31). To assess the phylogenetic distribution of length variation and its relationship to the number of CA repeats, a genealogy for this locus was inferred using only base substitutions in the regions flanking the microsatellite.
RESULTS
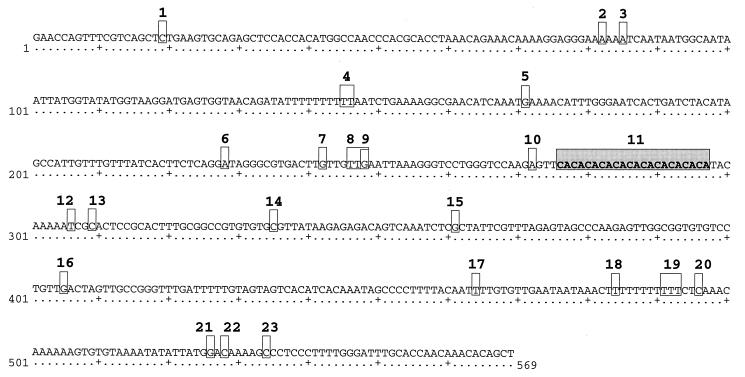
DNA sequences ≈570 bp long were obtained for each of 66 haplotypes isolated by SSCP. A full-length consensus sequence is shown in Fig. 1. Twenty-two polymorphic sites were detected in the flanking sequences of the CA repeat, 4 of which involved length variation (indels of one or more bases). The number of CA repeats at the microsatellite region itself (Fig. 1, character 11) occurred in all integer values between 5 and 11, with the exception of repeat count 8 which was not observed in our samples. Two regions with homopolymeric stretches (poly-Ts; Fig. 1, characters 4 and 19) each displayed 1–3 bp of length variation among alleles. The fourth site involving length variation (Fig. 1, character 8) is a 3-bp indel in a region with either two or three copies of TTR (R = A or G). All other polymorphic sites involved base substitutions.
Figure 1.
Full-length consensus sequence of the LPF3 locus in horseshoe crabs (GenBank accession AF020906). The shaded box is the microsatellite repeat region; numbers identify polymorphic sites in the sample of 66 alleles sequenced.
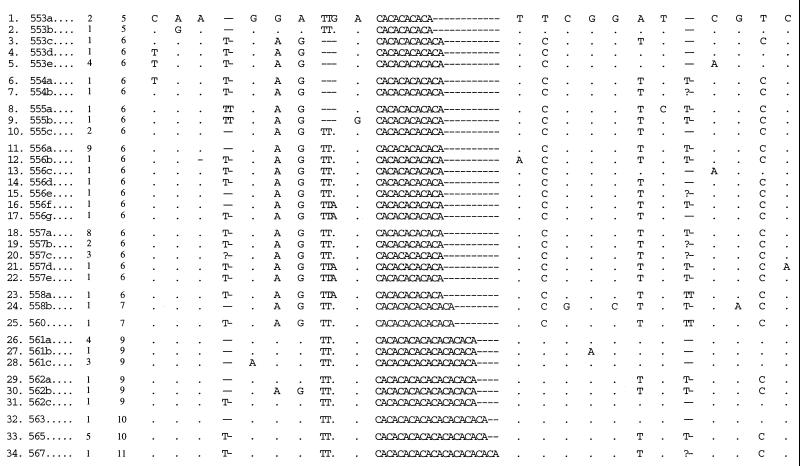
The combination of size variation and base substitutions at the 23 polymorphic sites (Fig. 1) defined a total of 34 different alleles among the samples (Table 1). These grouped into 12 distinct size classes. The total size of the sequenced fragments varied from 553 to 567 bp (Table 1).
Table 1.
Variation at 23 polymorphic sites among 34 alleles from the horseshoe crab locus LPF3
| Allele† | n‡ | (CA) | Polymorphic site number*
|
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 + 9§ | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | ||||
Note that dots indicate identity with the allelic state of allele 1. Characters 3, 4, 8, 11, and 19 are indels.
Polymorphic sites are as labeled in Fig. 1.
Alleles are labeled and sorted by size class.
Number of alleles found among 66 assayed.
Character 8 is the presence/absence of the TTR repeat; character 9 is either G or A when TTR is present.
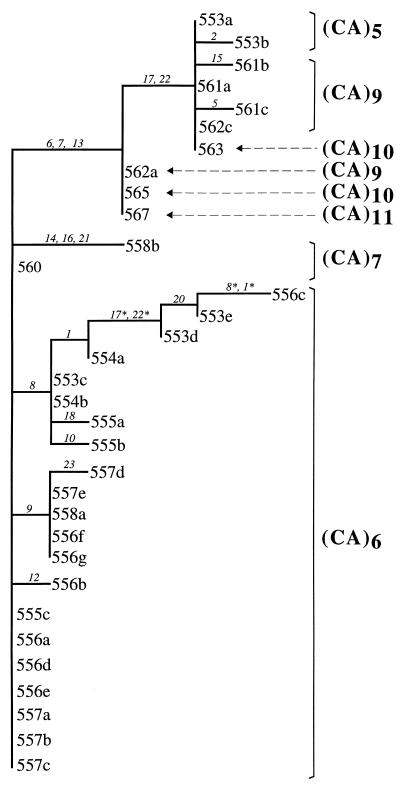
Phylogenetic analysis of the nucleotide substitutions in the microsatellite flanking regions resulted in a single most-parsimonious tree (Fig. 2) of length = 23 steps and Rescaled Consistency Index = 0.77 (32). A neighbor-joining analysis (33) supports the same topology (data not shown). The two main branches on this tree are defined unambiguously by changes at characters 6, 7, and 13 (Table 1 and Fig. 2). CA repeat number and characters involving size variation in the flanking region (characters 3, 4, and 19) were excluded from the analysis. The only exception involved characters 8 and 9 (presence/absence of TTG or TTA), which were included in the analysis because the indel was associated with an informative nucleotide substitution. Character 11, the number of CA repeats itself, subsequently was mapped onto the tree based on the flanking sequences (Fig. 2).
Figure 2.
Allele genealogy for the horseshoe crab LPF3 locus. Shown is the most-parsimonious tree (length = 23 steps, Rescaled Consistency Index = 0.77) reconstructed using polymorphic sites in the microsatellite flanking regions (excluding characters 3, 4, 11, and 19; see Fig. 1). Branch lengths are proportional to numbers of changes; branches of length = zero are collapsed. The number of CA repeats for each allele is indicated on the right. Numbers on branches are particular sequence characters (labeled by number as in Table 1) inferred by the parsimony algorithm to have changed along each branch of the tree. Asterisks indicate character state reversions or parallel changes.
Allele 30 (562b) also was excluded from the phylogenetic analysis because it appeared to be a recombinant between other surveyed alleles that differ in their 5′ and 3′ ends. The postulated recombination event may have involved, for example, the common alleles 11 and 29 (Table 1): the 5′ region of allele 30 shares nucleotide states at characters 1–10 with allele 11, whereas the 3′ end shares nucleotide states at characters 11–26 with allele 29 (Table 1). The apparent recombination event that produced allele 30 probably occurred in nature because the individual carrying it was scored as a 558a/562b heterozygote. An in vitro recombination event in the PCR tube would have produced a third allele detectable in the sequencing assay (34). Furthermore, SSCP separation of PCR products, rather than cloning, reduces the chance of detecting such in vitro artifacts (25).
A phylogenetic approach was used to test for correlations between genealogical relationship and fragment size. Alternative hypotheses involving the evolution of CA repeat number and absolute fragment size (as traditionally scored in microsatellite assays) were tested. Searches were conducted with topological constraints such that either all alleles of the same size or all alleles harboring the same number of CA repeats were forced into monophyletic groups. The resulting topologies were 11 and 3 steps longer, respectively, than the most parsimonious tree (length = 23) shown in Fig. 2. Forcing alleles with same fragment size into monophyletic groups resulted in a significantly worse (P < 0.05) phylogenetic hypothesis (35), suggesting that overall fragment size is a poor indicator of allelic phylogeny for this locus. Forcing alleles with the same number of repeat units into monophyletic groups resulted in a three-step increase in tree-length that was not statistically significant.
DISCUSSION
In agreement with observations from other species (11–13, 17, 18), genetic diversity in this microsatellite region of L. polyphemus is not solely the result of number variation in the primary tandem repeat unit (CA in this case). Two indels and two variable-length homopolymeric (poly-T) runs in sequences immediately flanking the CA repeat motif also contributed to the electrophoretic size variation detected on conventional microsatellite gels. Variation in the number of CA repeats alone would have produced only six “electromorphs” in our sample at this locus. All of the remaining 28 “true alleles” were distinguished by nucleotide substitutions or by the above-mentioned length variants in flanking regions. Among the total of 12 size-class alleles detected, 8 consisted of multiple DNA sequence variants. Such findings extend to the population level conclusions about allelic diversity at microsatellite loci previously drawn from flanking-sequence differences among species or higher taxa (13, 15).
The phylogenetic distribution and high representation of the (CA)6 class of alleles (Fig. 2) suggest that the mutation rate at this microsatellite motif may be lower than the rate of substitutions in flanking regions and/or that evolutionary convergence to this size class has been common. Among the 66 haplotypes assayed in total, the (CA)6 motif was present in 43 copies that displayed 22 different variations in flanking sequence. The possibility of a low mutation rate in repeat count (and an old age) of the (CA)6 lineage gained support from the observation that alleles with (CA)7–11 were less frequent among the samples and harbored less flanking region variation. Similar observations and lines of reasoning were advanced for a (CA)9 class of alleles at a human microsatellite locus (24).
The genealogical placements of identical and similar-length haplotypes in the allelic phylogeny (Fig. 2) further demonstrate that (i) considerable molecular heterogeneity often underlies a given CA repeat class [for example, 21 alleles masqueraded within the (CA)6 class, 5 within (CA)9, and 2 each within (CA)5 and (CA)10]; and (ii) haplotypes in identical and adjacent size classes of CA repeats are not invariably closer to one another phylogenetically than are those in nonadjacent size classes. For example, alleles with (CA)5 are related more closely to alleles with (CA)9–11 than to alleles with (CA)6. Also, the two (CA)10 alleles differed from one another by the same pair of nucleotide sequence characters (17 and 22) that distinguished several non-(CA)10 alleles. Thus, microsatellite alleles of identical or similar size can be members of different genealogical clades as defined by flanking nucleotide sequences. Variation at the poly-T regions (Fig. 1 and Table 1, characters 4 and 19) was highly homoplasious. These characters had a rescaled consistency index = 0.2 when mapped onto the most parsimonious tree. The phylogenetic placement of SSR number calls into question an implicit assumption often underlying estimates of genetic distance based on the stepwise mutation model—that allelic size is a reliable indicator of phylogenetic affinity.
If the current findings should prove common or typical for SSR loci, they will have implications for interpretations of variable numbers of tandem repeat data in population genetic studies. First, even at known “hypervariable” loci, many more alleles can be present than are detectable by DNA length variation alone. Thus, individual heterozygosities may be seriously underestimated in conventional microsatellite assays. Second, homoplasy with respect to size class for conventionally assayed microsatellite alleles could compromise appraisals of genetic relatedness between individuals, as well as determinations of allelic identities and genetic distances between populations or species.
Most previous empirical studies of microsatellite evolution have not addressed these issues directly because (i) size variation was the sole criterion for genotyping, (ii) only one representative of each allelic size class was sequenced, or (iii) the flanking regions sequenced were too short for informative genealogical comparisons (11, 14). Lehmann et al. (36) used the existence of null alleles to examine a microsatellite locus in Anopheles mosquitoes. They found two parallel series of alleles with similar size distributions but that differed by a point mutation in the flanking region at one of the priming sites. Considerable homoplasy also was inferred, with alleles from both the primary and the null series represented in all allelic size classes. The general implications are similar to those of the present study—microsatellite alleles of identical size are not necessarily identical by descent, even within a species. As shown here, SSCP assays are well suited to distinguish same-size alleles that differ in flanking regions by one or more base substitutions. Thus, these assays constitute a promising alternative for scoring microsatellite loci and minimizing size homoplasy.
Compound or imperfect microsatellites (those with variations on a simple repeat motif theme) have more complex evolutionary patterns than do perfect microsatellites. Conventional wisdom and some empirical evidence suggest that compound microsatellites may conform more closely to an “infinite alleles” model, which in turn should reduce the opportunity for “homoplasic noise” (12) because of the larger number of potentially achievable allelic states (refs. 19 and 37; but see ref. 13). Additional considerations for both compound and simple microsatellites include mutational biases or selective constraints on allelic size that may truncate and converge allelic distributions even in otherwise divergent populations or species (11, 21, 38–40).
The microsatellite locus analyzed in this study was simple in the sense that only perfect CA repeats were involved in the SSR region itself, but imperfect in the sense that extensive additional variation (including size variation) was present in flanking regions. Genealogical analyses of nucleotide sequence variation in these flanking regions have given some hint of the mutational complexity and evolutionary diversity that can underlie conventionally detected size differences among microsatellite alleles.
Acknowledgments
We thank other members of the Avise lab and G. Kochert and C. Schlötterer for constructive comments on the manuscript. This work was supported by the National Science Foundation and by funds made available from the University of Georgia.
ABBREVIATIONS
- SSR
simple sequence repeat
- SSCP
single-strand conformational polymorphism
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF020906).
References
- 1.Tautz D. Nucleic Acids Res. 1989;17:6463–6471. doi: 10.1093/nar/17.16.6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weber L, May P E. Am J Human Genet. 1989;44:388–396. [PMC free article] [PubMed] [Google Scholar]
- 3.Kashi Y, Tikochinski Y, Genislav E, Iragi F, Nave A, Beckmann J S, Gruenbaum Y, Soller M. Nucleic Acids Res. 1990;18:1129–1132. doi: 10.1093/nar/18.5.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levinson G, Gutman G A. Mol Biol Evol. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- 5.Schlötterer C, Tautz D. Nucleic Acids Res. 1992;20:211–215. doi: 10.1093/nar/20.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strand M, Prolla T A, Liskay R M, Petes T D. Nature (London) 1993;365:274–276. doi: 10.1038/365274a0. [DOI] [PubMed] [Google Scholar]
- 7.Shriver M D, Jin L, Chakraborty R, Bowerwinkle E. Genetics. 1993;134:983–993. doi: 10.1093/genetics/134.3.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valdes A M, Slatkin M, Freimer N B. Genetics. 1993;133:737–749. doi: 10.1093/genetics/133.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Rienzo A, Peterson A C, Garza J C, Valdes A M, Slatkin M, Freimer N. Proc Natl Acad Sci USA. 1994;91:3166–3170. doi: 10.1073/pnas.91.8.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowcock A M, Ruiz-Linares A, Tomfohrde J, Minch E, Kidd J R, Cavalli-Sforza L L. Nature (London) 1994;368:455–457. doi: 10.1038/368455a0. [DOI] [PubMed] [Google Scholar]
- 11.Garza J C, Slatkin M, Freimer N B. Mol Biol Evol. 1995;12:594–603. doi: 10.1093/oxfordjournals.molbev.a040239. [DOI] [PubMed] [Google Scholar]
- 12.Estoup A, Tailliez C, Cornuet J- M, Solignac M. Mol Biol Evol. 1995;12:1074–1084. doi: 10.1093/oxfordjournals.molbev.a040282. [DOI] [PubMed] [Google Scholar]
- 13.Angers B, Bernatchez L. Mol Biol Evol. 1997;14:230–238. doi: 10.1093/oxfordjournals.molbev.a025759. [DOI] [PubMed] [Google Scholar]
- 14.Grimaldi M-C, Crouau-Roy B. J Mol Evol. 1997;44:336–340. doi: 10.1007/pl00006151. [DOI] [PubMed] [Google Scholar]
- 15.FitzSimmons N N, Moritz C, Moore S S. Mol Biol Evol. 1995;12:432–440. doi: 10.1093/oxfordjournals.molbev.a040218. [DOI] [PubMed] [Google Scholar]
- 16.Jarne P, Lagoda P J L. Trends Ecol Evol. 1996;11:424–429. doi: 10.1016/0169-5347(96)10049-5. [DOI] [PubMed] [Google Scholar]
- 17.Adams M, Urquhart A, Kimpton C, Gill P. Hum Mol Genet. 1993;2:1373–1376. doi: 10.1093/hmg/2.9.1373. [DOI] [PubMed] [Google Scholar]
- 18.Urquhart A, Kimpton C P, Gill P. Hum Genet. 1993;92:637–638. doi: 10.1007/BF00420957. [DOI] [PubMed] [Google Scholar]
- 19.Goldstein D B, Linares A R, Cavalli-Sforza L L, Feldman M W. Genetics. 1995;139:463–471. doi: 10.1093/genetics/139.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slatkin M. Genetics. 1995;139:457–462. doi: 10.1093/genetics/139.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nauta M J, Weissing F J. Genetics. 1996;143:1021–1032. doi: 10.1093/genetics/143.2.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harvey P H, Leigh Brown A J, Maynard Smith J, Nee S. New Uses for New Phylogenies. Oxford: Oxford Univ. Press; 1996. [Google Scholar]
- 23.Zardoya R, Vollmer D M, Craddock C, Streelman J T, Karl S, Meyer A. Proc R Soc London B. 1996;263:1589–1598. doi: 10.1098/rspb.1996.0233. [DOI] [PubMed] [Google Scholar]
- 24.Jin L, Macaubas C, Hallmayer J, Kimura A, Mignot E. Proc Natl Acad Sci USA. 1996;93:15285–15288. doi: 10.1073/pnas.93.26.15285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ortí G, Hare M P, Avise J C. Mol Ecol. 1997;6:575–580. doi: 10.1046/j.1365-294x.1997.00212.x. [DOI] [PubMed] [Google Scholar]
- 26.Saunders N C, Kessler L G, Avise J C. Genetics. 1986;112:613–627. doi: 10.1093/genetics/112.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sültmann H, Mayer W E, Figueroa F, Tichy H, Klein J. Mol Biol Evol. 1995;12:1033–1047. doi: 10.1093/oxfordjournals.molbev.a040279. [DOI] [PubMed] [Google Scholar]
- 28.Orita M, Iwahana H, Kanazawa H, Hayashi K, Sekiya T. Proc Natl Acad Sci USA. 1989;86:2766–2770. doi: 10.1073/pnas.86.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orita M, Suzuki Y, Sekiya T, Hayashi K. Genomics. 1989;5:874–879. doi: 10.1016/0888-7543(89)90129-8. [DOI] [PubMed] [Google Scholar]
- 30.Hongyo T, Buzard G S, Calvert R J, Weghorst C M. Nucleic Acids Res. 1993;21:3637–3642. doi: 10.1093/nar/21.16.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swofford D L. paup: Phylogenetic Analysis Using Parsimony. Champaign: Illinois Natural History Survey; 1993. , Version 3.1.1. [Google Scholar]
- 32.Farris J S. Cladistics. 1989;5:417–419. doi: 10.1111/j.1096-0031.1989.tb00573.x. [DOI] [PubMed] [Google Scholar]
- 33.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 34.Bradley R D, Hillis D M. Mol Biol Evol. 1997;14:592–593. doi: 10.1093/oxfordjournals.molbev.a025797. [DOI] [PubMed] [Google Scholar]
- 35.Templeton A R. Evolution. 1983;37:221–244. doi: 10.1111/j.1558-5646.1983.tb05533.x. [DOI] [PubMed] [Google Scholar]
- 36.Lehmann T, Hawley W A, Collins F H. Genetics. 1996;144:1155–1163. doi: 10.1093/genetics/144.3.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feldman M W, Bergman A, Pollock D D, Goldstein D B. Genetics. 1997;145:207–216. doi: 10.1093/genetics/145.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Slatkin M. Mol Biol Evol. 1995;12:373–480. doi: 10.1093/oxfordjournals.molbev.a040222. [DOI] [PubMed] [Google Scholar]
- 39.Amos W, Sawcer S J, Feakes R W, Rubinsztein D C. Nat Genet. 1996;13:390–391. doi: 10.1038/ng0896-390. [DOI] [PubMed] [Google Scholar]
- 40.Primmer C R, Ellegren H, Saino N, Møller A P. Nat Genet. 1996;13:391–393. doi: 10.1038/ng0896-391. [DOI] [PubMed] [Google Scholar]