Abstract
The translation of laboratory innovations into clinical tools is dependent upon the development of regulatory arrangements designed to ensure that the new technology will be used reliably and consistently. A case study of a key post-genomic technology, gene chips or microarrays, exemplifies this claim. The number of microarray publications and patents has increased exponentially during the last decade and diagnostic microarray tests already are making their way into the clinic. Yet starting in the mid-1990s, scientific journals were overrun with criticism concerning the ambiguities involved in interpreting most of the assumptions of a microarray experiment. Questions concerning platform comparability and statistical calculations were and continue to be raised, in spite of the emergence by 2001 of an initial set of standards concerning several components of a microarray experiment. This article probes the history and ongoing efforts aimed at turning microarray experimentation into a viable, meaningful, and consensual technology by focusing on two related elements:
1) The history of the development of the Microarray Gene Expression Data Society (MGED), a remarkable bottom-up initiative that brings together different kinds of specialists from academic, commercial, and hybrid settings to produce, maintain, and update microarray standards; and
2) The unusual mix of skills and expertise involved in the development and use of microarrays. The production, accumulation, storage, and mining of microarray data remain multi-skilled endeavors bridging together different types of scientists who embody a diversity of scientific traditions. Beyond standardization, the interfacing of these different skills has become a key issue for further development of the field.
Introduction
If asked what the term “science” evokes, most people likely will reply “invention” and “discovery,” certainly not “standard-setting activities,” the latter being most often associated with industry or even boring, repetitive tasks. And yet, as many contributors to the field of Science & Technology Studies have shown, standards are a sine qua non of modern science. For one thing, without scientific instruments, there would be no experimental practice, and, most importantly, the nature of knowledge itself would be affected [1]. A machine as an isolated entity does not produce meaningful results, and debates about the factual status of a given result hinge upon successful claims about the reproducibility of the experiment that produces it, which in turn depends on establishing equivalences between the instruments and research materials used in different settings [2]. Much of modern biology and medicine, for instance, is predicated upon the existence of standardized mice [3,4].
In the last 10 to 15 years, standards and other forms of regulation have ranked high on the research agenda of sociologists and historians of science. O’Connell, for instance, convincingly has shown that labor-intensive metrological practices underlie the universality of scientific and technical claims, and Mallard has further explored the diversity and heterogeneity of metrological networks [5,6,7]. In both cases, the authors focused on physical entities for which “absolute standards” can, at least in principle, be defined. The situation is more complex in biomedicine, where no absolute or pre-set standards exist for, say, blood cells: In this domain, metrology and standardization are based on the degree of consensus regarding samples analyzed in different laboratories [8]. In other words, relative standards and consistency of measurements, rather than the search for a “true value,” are the rule of the game. As forcefully argued by French philosopher Georges Canguilhem, physiological norms are ultimately dependent on the environment and, thus, cannot be taken as stand-alone, external standards. As a result, normalization does not presuppose a norm; instead, norms are the result of normalization processes [9]. As we will discuss in this article, a relatively novel situation arises from the fact that the technologies of contemporary biomedicine are increasingly defined by a combination of computerized instruments and molecular entities such as genes and enzymes. Ideally, the standard-setting activities that target, using different assumptions and approaches, the engineering and biological components of a given technology should be articulated or at least aligned, but this is not necessarily the case.
Standards come in different forms and are enforced and utilized for different purposes. According to a typology proposed by two medical sociologists, design standards define the elements of complex systems of action and allow them to function; terminological standards fix the terms used to classify and describe phenomena in order to achieve comparability among them; performance standards define outcomes or results acceptable in particular situations; and finally, procedural standards that depend on the existence of the former three types of standards specify the actions or protocols that must be followed in given situations [10]. While this typology of biomedical standards is certainly useful, we should contextualize it by asking whether and how the transformation of biomedical research over the last 50 years, in particular the development of molecular biology and, most recently, genomics, has affected processes of standardization and regulation. One of us has argued elsewhere that this is indeed the case, insofar as the direct interaction of biology and medicine that has increasingly characterized the evolution of Western biomedicine since World War II has been accompanied by a systematic recourse to the collective production of evidence involving the establishment of conventions, sometimes tacit and unintentional, but most often arrived at through concerted programs of action [11]. These programs involve the entities and protocols produced and maintained by extended networks of researchers, clinicians, and industrial actors. Examples of these range from multi-center cancer clinical trials carried out by cooperative groups to the production of a standardized nomenclature of human leukocyte reagents and the establishment of predictive cancer genetics services [12,13,14].
In this paper, we would like to pursue this line of research by investigating the standardization and regulation of microarrays, a tool that has become emblematic of post-genomic medicine. After briefly situating the role of this new technology, we will examine how its development was predicated upon the establishment of hybrid regulatory networks that have become increasingly extended as microarrays reach a stage where they are now ready to “take on the clinic.”
Microarrays: a post-genomic tool of choice
Microarrays are an emergent technology of contemporary biomedicine. A single DNA microarray contains thousands of short DNA sequences arrayed on a solid surface. Compared to previous molecular genetic approaches, a microarray experiment involves the simultaneous analysis of many hundreds or thousands of genes as opposed to single genes, thus making them a key tool of the post-genomic era. According to a PubMed search, the number of articles based on microarray technology has grown exponentially during the last decade, from fewer than 200 in 1999 to more than 6,000 in 2005, for a total of more than 27,000 articles in six years. These articles are not confined to experimental reports, as an increasing number of publications describe clinical applications of microarrays [15-19]. A search in the Computer Retrieval of Information on Scientific Projects (CRISP) database of the projects funded by the National Institutes of Health yielded 13,954 awards by the end of 2006. A search in the international patent database Derwent Innovation Index found approximately 6,500 patents (see Figure 1) [20]. On the industrial side, world DNA microarray revenue was $596 million in 2003 and revenue for the total U.S. DNA microarray market was approximately $446.8 million in 2005 [21,22]. The 2006 revenues of two leading companies in the field, Affymetrix and Illumina, were, respectively, $355 million and $184 million [23]. In short, microarray technology is not only growing rapidly, but it also is being translated simultaneously into the scientific, clinical, and commercial domains.
Figure 1.
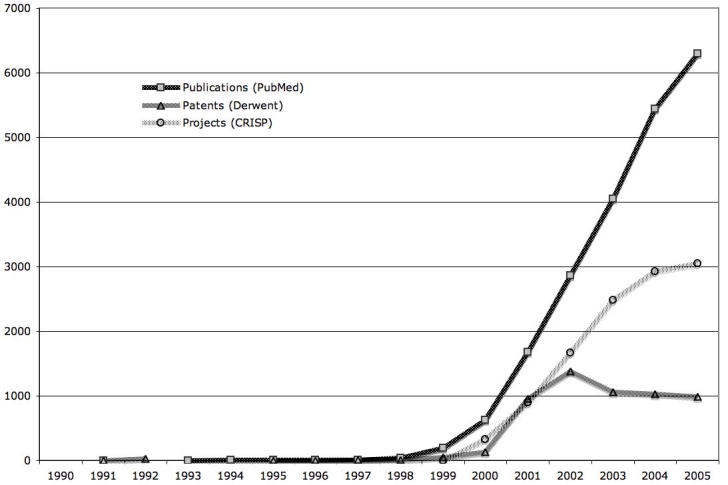
Growth in the number of microarray publications, patents, and projects. Since the late 1990’s there has been a steep increase in productivity in all three domains.
In their “Strategic Analysis of World DNA Microarray Markets,” the authors of a Frost & Sullivan report predict that the acceptance of microarrays as diagnostic tools will be contingent on the manufacturers’ ability to overcome “laboratory-to-laboratory and platform-to-platform inconsistencies” [22]. This clearly understates the issue, and we argue here that standardization of the many different elements that partake in a microarray experiment do not simply relate to the diagnostic use of the technology but more broadly to the shared acceptance of the technology as a reliable tool for producing genomic findings.
The beginnings of microarrays
As is most often the case with scientific instruments and technologies, especially those that resort in part to pre-existing techniques, it is difficult to establish an exact and uncontroversial origin for microarray technology. Several groups and people claim to have played a role in its development, and we can follow the somewhat meandering, technical, and scientific lineage of microarrays by examining publication routes and patent applications. The microarray is an example of a scientific instrument that has been produced through networks of collaboration, and as we will see, microarray technology cannot be reduced to its instrumental parts. The tool only acquires its full meaning once it is related to specific bodies of knowledge (in the present case, molecular genetics) and connected to specific sets of issues. As noted by Baird, the question of knowing whether a scientific instrument works is intimately related to the question of knowing how it works [1]. The point is that in order to produce meaningful scientific results, a tool must be entrenched in an evidential context, i.e., a network of scientific questions, entities, and issues as well as laboratory and clinical routines for which its use is deemed relevant.
When DNA microarrays emerged on the public scientific scene in the early 1990s, only a very limited number of actual biological experiments had been carried out. The technology was still a tool looking for a problem to which it could be applied. Work on early prototypes of microarray instrumentation was under way by the late 1980s, namely within a team led by Stephen Fodor of the Affymax Research Institute and future president of Affymetrix, currently the leading manufacturer of microarrays and related equipment. A paper by Fodor and colleagues entitled “Light-directed, spatially addressable parallel chemical synthesis” appeared in the February 15, 1991, issue of Science outlining a new technique that combined elements from the semiconductor industry and combinatorial chemistry [24]. The authors mentioned that the new technique could be of use in the discovery of principles behind molecular interactions, but the article was primarily one introducing a new technique, rather than a scientific article reporting the results of a biological experiment. Moreover, observers have argued that rather than inventing a new technique from scratch, the authors succeeded in miniaturizing pre-existing technologies. Some noted that macroarrays were a precursor to microarrays, which have now in turn led to protein arrays [25]. Although these are distinct technologies in themselves, they move on the same trajectory to scientific discovery [26]. Between 1992 and 1995, Fodor and colleagues published several more articles focusing on methods for sequencing DNA. In September 1995, members of Affymetrix published yet another article that presented the company’s products as standardized tools, in contrast to the “homemade” arrays produced by individual laboratories:
The collection of data forms an “image” of the array. Image processing software, GeneChip™ software (Affymetrix, Santa Clara, CA, USA), is used to segment the image into synthesis sites and integrate the data over each synthesis site. A robust benchtop version of this detection instrument, the GeneChip scanner (Affymetrix) has been developed. [27]
Affymetrix, however, was not the only player in the field of microarray technology. The company’s “engineering” approach to microarray production involved the use of photolithographic methods to synthesize hundreds of thousand of oligonucleotides on a small glass surface, but an alternative, less “industrial” approach was available. Developed in the early 1990s by Patrick Brown’s team at Stanford University, this more distinctively biological technique used a robot to deposit cDNA spots onto a coated glass slide [28].
A month after the Affymetrix article praising its “GeneChip” technology, the Stanford University team published a widely cited Science article (3,327 citations by September 2007, according to the Web of Science) often mentioned as the first published paper to report actual biomedical results based on parallel microarray analysis. Entitled “Quantitative Monitoring of Gene Expression Patterns with a Complementary DNA Microarray” [29], and using the “spotted microarray” approach, the paper compared gene expression in the root and leaf tissue of a weed called Arabidopsis thaliana (a model organism for molecular genetics). The weed itself was used more as a “proof of concept” than as an end in itself, for the article ended on a “promissory note” mentioning the advent of personalized medicine:
A wide variety of acute and chronic physiological and pathological conditions might lead to characteristic changes in the patterns of gene expression in peripheral blood cells or other easily sampled tissues. In concert with cDNA microarrays for monitoring complex expression patterns, these tissues might therefore serve as sensitive in vivo sensors for clinical diagnosis. Microarrays of cDNAs could thus provide a useful link between human gene sequences and clinical medicine. [29]
Speculations concerning the use-value of a technology contribute to the shaping of its future directions [30]. While it is now widely accepted that DNA microarrays are useful for detecting genes expressed in diseases and this kind of information eventually may inform medical treatment [31], this was not necessarily the case at the time. The Schena et al. article was strategically placed in a Science “Genome Issue” that contained a report proudly announcing that we were “Entering a Postgenome Era” and hailing microarray experimentation as a breakthrough [32]. That same report, however, simultaneously warned of the mess ahead in sorting out gene expressions and cited a Johns Hopkins scientist as saying “we are now going to have expression maps of 100,000 different genes. Good luck figuring that out!” [32] This was one of the first public mentions of an impending need to tame the avalanche of unstandardized data expected to emerge from an increasing number of laboratories as the new technique became widely adopted.
A threatening flood of data
In the early 1990s, when the first technical articles on microarrays appeared, no major concerns were expressed about the possibility of analyzing, validating, and comparing data emerging from the use of the novel technique. The situation changed after 1995, as soon as the scientific meaning of the new tool began to take shape. Following the publication of the Schena et al. article and the subsequent publication of a 1996 article resulting from the collaboration of the Stanford team with a NIH team, focusing this time on human cancer genes [33], scientific journals increasingly began to wonder about the many parameters involved in interpreting a microarray experiment, as well as the lack of global comparability of results [34,35,36]. In 1998, one of the leaders of this rapidly evolving field, David Botstein, commented in an editorial that “[w]hat began as a trickle of such data now threatens to be a flood; yet for even the most important results, all that most of us have ever seen are sets of stills or collapsed flat representations.” [37]
In 2002, the NIH had begun funding the Toxicogenomics Research Consortium, a group of seven research centers with the goal of assessing the causes of gene expression variation across labs [35]. Many questions were being raised: How was anyone to know whether the different platforms being used were comparable or even how they were implemented? How were statistics calculated? How could anyone assure that the many uncertainties within each experiment were leading to reliable results? [38,39] These concerns did not dampen enthusiasm for potential microarray applications, but instead incited supporters of the new technology to realize the need for standard-building initiatives in order to solve the problem of comparability.
At this stage, readers unfamiliar with microarrays may wonder why it is so difficult to assess the results obtained with this technology. Microarray instrumentation does not produce, strictly speaking, raw data — as with other recent computerized apparatuses, the readings made by the instrument are generated and transmitted via several hardware components, such as photomultipliers, and software programs that implement statistical calculations and produce visual representations of the results [40]. Observers of the field routinely note that a lot of the messiness involved in getting reliable, reproducible, and comparable experimental results pertains to the extremely large data sets produced from a single experiment. The enormous matrices produced by each array inevitably contain noise and uncertainty, casting doubts upon the interpretation of significant results.
Each experiment uses an array similar to a momentary snapshot or photograph of gene expression, meaning each photograph must be interpreted by the observer to a certain extent. At a recent bioinformatics workshop, one of the participants noted that the first level of quality control is as blunt as “look at the image!”¹ A preliminary glance at the computerized image — that, once again, is the result of statistical treatments — will reveal clusters and spot intensities. Data are then normalized in order to identify the possible role of spots, at which time meaning is extracted from the data structure in order to explain biological behavior. A plenitude of statistical devices are employed throughout the process such as t-tests, K-means, ANOVA tests, and Euclidean distance. Workshop participants were told to use multiple hypothesis testing in order to lower the number of predicted false positives, since false positives are a given in microarray research and lowering their emergence early on will improve results. In terms of statistical measures, the choice of a given statistical device can produce an interestingly random variation in results. The procedure for generating K-means, for example, starts by randomly guessing K-centers between spots. This process defines clusters that may or may not have been there to begin with but provides a good estimate of where they might be if they are lurking in the data matrix. The (unpublished) workbook distributed during the workshop reminded participants that “the outcome depends on the initial guesses,” thus making the point that they would most likely not encounter “actual clusters” but instead statistically produced estimates of clustering.
A top NCI statistician recently objected to the claim that the main problem for microarray research lies in the massive datasets generated by each experiment: “[Myth]: That the greatest challenge is managing the mass of microarray data. … [Truth]: Biologists need both good analysis tools and good statistical collaborators. Both are in short supply.” [41] Although he also extended his criticism to the systematic resort to complex classification algorithms such as neural networks and to techniques such as data mining and pattern recognition, the fact remains that the emergence of the new specialty known as bioinformatics — whose practitioners are skilled users of these techniques — has accompanied the growth of microarrays. While a discussion of the development of this specialty goes beyond the limits of this article, it should be noted that, as maintained by sociologists of science and technology, in order to follow the trajectory of an instrument or a technique one has to analyze it as part of a growing network of related tools, artifacts, skills, institutions, and people that in the present case include bioinformatics as a new domain of activity [42]. In turn, by becoming part of existing networks, such as cancer research, a novel technology such as microarrays contributes to the reconfiguration of biomedical activities. In particular, problems concerning the results produced by microarrays do not only concern the hapless technicians working with them and do not stop at the laboratory’s doors, as data variability poses a further challenge to genomic research initiatives built on the premise that data produced in different settings are comparable. To achieve comparability, a multi-tiered process involving industrial, academic, and government actors had to be put in place.
MGED: Grassroots practices for standards
As shown by Figure 1, by the late 1990s the number of publications reporting microarray experiments was on the rise, yet this growth also was marked by fears the technology was developing more quickly than the community could understand it. The field of genomics already provided multiple examples of “bottom-up” initiatives, such as research networks and consortia designed to tackle specific problems (e.g., sequencing a given chromosome), as well as more formal organizations such as the Human Genome Organization (HUGO). Moreover, problems raised by the uncontrolled growth of new biomedical technologies, such as that of monoclonal antibodies, had been solved in the past by resorting to distributed regulatory networks [43]. Given these departures, when a group at the European Bioinformatics Institute (EBI) decided in the late 1990s to take it upon themselves to launch an initiative that would bring scientists closer to understanding and comparing microarray data, the move was not completely unexpected.
The EBI group ideally was situated for heading this movement, as it already constituted a leading bioinformatics cluster well positioned within the genomic discovery network. EBI is a satellite station of the European Molecular Biology Laboratory (EMBL) and closely affiliated with the Wellcome Trust Sanger Institute, a central institution in the Human Genome Project. The head of the microarray group, Alvis Brazma, worked to assemble a task force for the creation and dissemination of microarray standards. His task certainly was facilitated by the fact that the field itself was characterized by the presence of a fairly limited set of highly visible industrial and academic actors. On the industrial side, we already mentioned the leading role played by Affymetrix. On the academic side, a recent search in the Web of Science showed that two researchers — Patrick Brown and David Botstein, both from Stanford — dominate the field in terms of the number of publications (116 and 77, respectively) and the number of citations received (31,793 and 21,524, respectively; one of their joint papers has received 4,231 citations). Moreover, 17 of Brown’s papers and 15 of Botstein’s figure among the top 0.01 percent most cited papers in their domain. Authors further down the list also demonstrate a strong performance in terms of these same parameters. A co-authorship network of the most cited authors in the field (not shown here) [20] indicates these authors occupy a strategic “bridge” position between the main clusters of co-authors.
Before long, the EBI group joined forces with Affymetrix and Stanford University, the dominant players in microarray experimentation, to establish a self-styled grassroots movement called the Microarray Gene Data Expression Society (MGED) with a basic aim to standardize the field. From the very outset, MGED was a “hybrid” organization, as it included industrial and university members. EBI, while officially a public institution, is itself a “hybrid” organization in the sense that, as convincingly argued by McMeekin and Harvey, its operations are predicated upon the existence of knowledge flows between interdependent public/private databases [44]. In spite of its “grassroots” status, MGED’s legitimacy was grounded in a solid infrastructure, allowing it to move forward swiftly with plans for standard-setting.
MGED officially was founded in November 1999 at an EBI meeting, now known as MGED1, a brief, two-day event, with the stated goal of putting an end to the messiness of microarray experimentation by establishing data communication standards ensuring the reproducibility and, thus, the reliability of results. A minimum of information, it contended, was absolutely necessary in order to effectively communicate the meaning of results. Fittingly, the standards sought by the organization were called “Minimum Information About a Microarray Experiment” (MIAME) [45]. In April 2001, with MIAME nearly eight months from its official completion, an article in Science further discussed the problem of comparability between experiments and proudly announced that data standards were on the horizon [46]. In addition to enthusiastically welcoming the coming of standards, the article mentioned that the “putting aside of egos” in the quest for a common standard was essential to the process. The implication here was that one needed to address the differing needs and concerns of the several categories of people involved in microarray experiments, pointing once again to the fact that a coordination (rather than a division) of labor underlies this line of work.
In February 2000, MGED-related EBI scientists called attention to the problem of the existence of many public and private data repositories that lacked compatibility [47]. As a possible solution to this quandary, the article announced that “[m]eanwhile, the EBI, in collaboration with the German Cancer Research Centre, is developing ArrayExpress, a gene-expression database compliant with the current recommendations.” [47] By interlinking standards and the repository, assurance could be given that minimum standards would be met by all of the datasets deposited. One of the outcomes of MGED1 had been the establishment of five working groups on such issues as dataset compatibility². As planned, the five groups assembled at the European Molecular Biology Laboratory (EMBL) in Heidelberg, Germany in May 2000 for MGED2. The goal of that second meeting was to finalize recommendations for MIAME through a synthesis of results received from the five task groups³. The number of participants was restricted to 250, and they were able to agree upon standards for annotation, storage, and communication of microarray data. MGED3 took place in March 2001 at Stanford, by which time a paper describing MIAME was almost ready. It was published in the December 2001 issue of Nature Genetics [45] and called for the deposition of experiment information into public data repositories but did not insist on specific ones.
In June 2002, the MGED Society became a not-for-profit organization, and by October 2002, it had managed to convince a number of key scientific journals such as Nature, Cell, and The Lancet to require MIAME for publication of microarray results. As stated in an open letter to scientific journals requesting formal compliance with the MIAME standards:
For microarray experiments, simply defining the appropriate data has been a challenge, because the large quantity of data generated in each experiment and the typical complexity of the ancillary information needed to interpret the results are unlike anything that has yet faced the biological research community. [48]
In addition to enforcing publication standards, MGED requested that primary experimental data be deposited into a permanent public data repository⁴. Their request was largely successful, mostly because many journal committees already were searching for experiment standards to aid them in the selection of “good scientific work” buried within the growing amount of microarray submissions. A Nature editorial expressed relief that they could “at last” enforce microarray standards [49]. The same editorial went on to explain how frustrating it had been to review microarray papers in the absence of standardized experimental details that would allow referees to assess the reliability of reported results. Interim, homemade solutions to this issue had proved problematic; for instance: “[m]any journals allowed authors to put the huge data files on their own websites for the review process, until it became clear that unscrupulous authors compromised the anonymity of referees by tracking who had visited their website.” [49] By enforcing the MIAME requirements, journals could rest assured they were overcoming these problems: “[h]arried editors can rejoice that, at last, the community is taming the unruly beast that is microarray information.”[49]
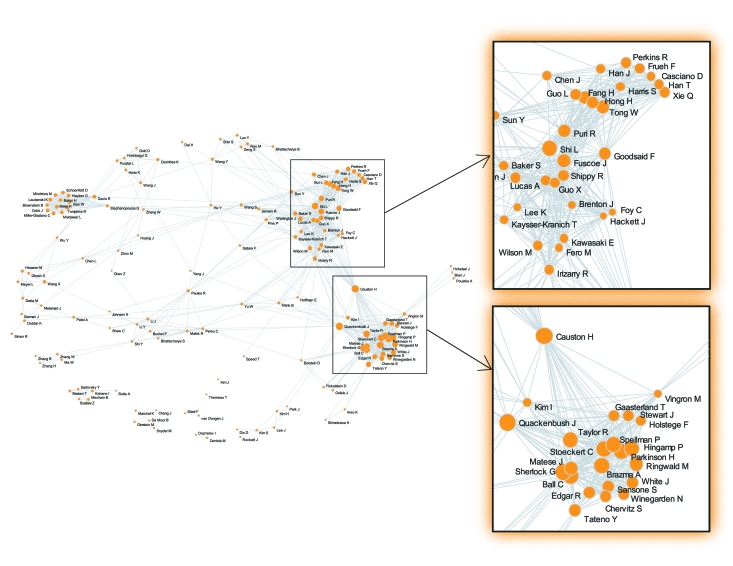
From MGED3 onward, the meetings drew hundreds of participants as its international scope widened. MGED4 took place again in the United States, but MGED5-8 took place in Japan, France, Canada, and Norway. In 2006, MGED9 returned to the United States (Seattle) and drew more than 200 participants. Figure 2 shows a co-authorship map of publications concerning microarray standards. The cluster at the bottom right of the map consists mostly of MGED authors with the previously mentioned Alvis Brazma at its center, while the cluster above it predominantly includes representatives from industry and the FDA, with notable academic representation from Stanford and Johns Hopkins. The map shows the existence of a close international collaboration between academic scientists in biostatistics and computer science that in turn entwines government/industrial actors. The role of a “bridge” between the two highlighted clusters is played mainly by Helen Causton (MRC Clinical Sciences Centre, Imperial College, London) and, to a minor extent, by John Quackenbush (Dana-Farber Cancer Institute, Boston) and Irene Kim (Johns Hopkins School of Medicine). Thus, as is generally the case within the field, microarray standardization processes result from the concerted action of a relatively small number of actors whose cooperative endeavor cuts across government, academic, and industrial boundaries.
Figure 2.
Microarray standards co-authorship network. The map was produced using ReseauLu, a software package for heterogeneous network analysis (www.aguidel.com). We queried PubMed for microarray articles, using the subheading “standards” (1991-present). The size of the nodes is proportional to the number of articles co-authored. Only the top 150 authors are represented on the map, and each author is represented by a single node.
Criticisms of standards (can they please everyone?)
Microarray standards must simultaneously appeal to biologists, computer scientists (bioinformatics specialists), and statisticians (and, of course, industrial actors). However, these different groups do not necessarily share a consensus concerning the definition of what should count as “reliable” or “reproducible.” Biologists generally insist on observable facts, while statisticians produce a different type of fact through statistical manipulation; as for bioinformaticians and computer science specialists, their activities focus on database inputs and outputs. While, in principle, these different elements should be aligned to achieve a successful microarray experiment, tensions remain, turning the task of establishing mutually acceptable data standards into a difficult challenge. The standardization movement, as we have seen, began at EBI and thus can be said to have been propelled primarily by bioinformaticians who worry about reliable databases. Resistance to these standards often comes from laboratory biologists who are hesitant to invest financial resources in order to follow stricter experimental standards. Undoubtedly, keeping the cost of a single microarray experiment down means that one can perform more experiments. This issue is acknowledged in the first description of MIAME that evoked potential financial commitments from its users:
Storing the primary image files would require a significant quantity of disk space, and there is no community consensus as to whether this would be cost-effective or whether this should be the task of public repositories or the primary authors [45].
The creation of public repositories soon provided a solution, but in addition to being expensive, standard enforcement could be time-consuming. MIAME authors were aware they would have to sell their initiative to a community that was likely to receive it with mixed feelings. On the one hand, standards were needed in order to produce scientifically meaningful results. On the other hand, they could potentially become a hindrance for this fast-developing field. Aware of these criticisms, MIAME attempted to negotiate a compromise between finely detailed annotation and a lack of information:
Too much detail may be too taxing for data producers and may complicate data recording and database submission, whereas too little detail may limit the usefulness of the data. MIAME is an informal specification, the goal of which is to guide cooperative data providers. It is not designed to close all possible loopholes in data submission requirements [45].
While some biologists continued to allege that MIAME was a resource-consuming endeavor with little payback for them in the end, others made a more conceptual criticism, arguing it was time to go beyond standards because replication and consistency do not say enough about experimental outcomes: “[c]onsistent standards between laboratories would help improve the consistency of results — but consistency is not enough — after all the results within a laboratory were all consistent but the results can be consistently wrong.” [50] Yet other biologists contend that despite MGED’s assertion it is a grassroots organization, the standards are being dictated from the top down, by bioinformaticians who primarily deal with experimental end points and want to impose their vision upon the biologists who do all of the experimental work from its inception to lab bench conclusion⁵.
MIAME is not enough: the proliferation of standards
Today, MGED oversees five workgroups in addition to MIAME: MAGE (Microarray and Gene Expression), MISFISHIE (Minimum Information Specification For In Situ Hybridization and Immunohistochemistry Experiments), OWG (Ontology workgroup), RSBI WGs (Reporting Structure for Biological Investigations Working Groups), and Transformations. MIAME, in other words, was only the starting point of a motley of standardization initiatives that now include several additional aspects of microarray experimental work and also extend to other types of genomic and proteomic experimentation [51,52,53,54]. For instance, MAGE, one of a variety of open-source projects, contains different extensions for standardizing microarray expression data and simplify their translation across different data systems. Ontology is another key domain, since standardization now targets, in addition to statistical aspects, the annotations used to report results. Once again, the purpose behind this initiative is to tame massive amounts of data by using a classificatory vocabulary that lacks the vagaries of natural language. Several related initiatives transit directly and indirectly through MGED members. They include groups such as the Gene Ontology Consortium, which optimistically hopes to develop universal terminology for the global genomics community:
Ontologies have long been used in an attempt to describe all entities within an area of reality and all relationships between those entities. An ontology comprises a set of well-defined terms with well-defined relationships. The structure itself reflects the current representation of biological knowledge as well as serving as a guide for organizing new data [55].
Ontologies contain controlled vocabularies that are expected to permit the translation of microarray data across and within scientific domains. Since, however, ontologies are never complete, they are ongoing projects that require continual revisions and additions from those working closely with the technology and its results.
All of the standardization initiatives mentioned in the previous paragraph bear the imprint of bioinformatics. As a new discipline, bioinformatics is still riding the wave of major initiatives such as the Human Genome Project and the HapMap Project and is negotiating its own scientific identity by defining work practices and training programs. At the same time, it must contend with the other communities of practice that have a stake in the development of microarrays, including biology, statistics, genetics, toxicology, pharmacogenomics, and cancer research. The introduction of microarrays in the clinic has further transformed the field, mobilizing new actors and adding targets and layers of regulatory intervention. The FDA, for instance, recently sponsored a MicroArray Quality Control (MAQC) project in order to examine the contentious issue of inter-platform reproducibility of microarray experiments [56]. MAQC, in other words, differs from MGED insofar as it tackles the potentially more thorny issues of data generation and validity, thus directly confronting the messiness of experimental work. The project involved 137 participants from 51 organizations who were asked to compare seven different microarray platforms. Results were qualified as promising, but there was still more comparability among certain platforms than others, and as with all microarray experiments, no two experiments were entirely identical. Irrespective of these results, the interesting issue from a social science point of view lies in the fact that in order to confront this new technology, standard-setting initiatives must not only proliferate, but also have to experiment with novel regulatory approaches that mobilize extended networks of research and industrial organizations. In other words, as science studies scholars, we witness the co-production of a technology and the arrangements (social, technical, institutional, and material) that shape its trajectory [57,58]. While this is not the place to examine the complex issue of the burgeoning standardization of microarrays as clinical tools, we should note that a similar conclusion applies. A single example will suffice: By examining, at once, thousands of genes, one can in principle tremendously reduce the time previously allotted to the task of molecular investigations. One of the predicted results is that this will allow the conflation of diagnosis and therapy (arguably, on an individualized basis), a process for which a name has already been found: theranostics [59,60]. Diagnosis and therapy are distinct medical and regulatory categories. If theranostics succeeds, regulators and the standard-setting specialists on whom they rely will have to go back to the drawing board.
Conclusion
Despite debate and controversy, the MGED Society has consistently maintained that the application of MIAME, MAGE, and related products can, and should, at least in theory, lead to increased comparability and reproducibility among reported experiments. As social scientists, we are interested in standard-setting initiatives such as the ones propelled by MGED because they exemplify new forms of collaborative work among biomedical researchers, computer scientists, and statisticians from public, industrial, and, increasingly, hybrid research organizations. The goal of these endeavors is to produce the conventions that increasingly regulate biomedical work. By using the term “conventions,” we do not mean they are entirely arbitrary, for they are grounded in the technical and experimental infrastructure of the field, but they are to a large extent the result of negotiations and compromise. Indeed, as we have seen, collaborative work does not exclude controversy, and in this article we discussed some of the fault lines that have appeared in this respect within the microarray community.
MIAME and its fellow standards are certainly an important achievement. The original version of MIAME has been updated several times, allowing for more detailed specifications of the software and tools which support it as well as for more precise experimental descriptions . Many practitioners, however, still feel as though there is a long way to go in the way of comparability of results between labs. As prices of individual chips are expected to continue to fall [61], microarray experiments are likely to proliferate, thus making the task of finding a solution to the comparability issue even more urgent. Moreover, increasing resort to microarrays for human diagnostic purposes [62] adds a distinctively clinical dimension to the regulatory conundrum [15,63,64]. In short, microarray technology provides a fascinating case study of how social institutions (research organizations, biotech companies, regulatory agencies) choose standards and how, in turn, standards produce new social and scientific institutions [65].
Acknowledgments
The authors would like to thank Bertrand Jordan for his comments on the initial draft of this paper.
Abbreviations
- CRISP
Computer Retrieval of Information on Scientific Projects
- HUGO
Human Genome Organization
- EBI
European Bioinformatics Institute
- EMBL
European Molecular Biology Laboratory
- MGED
Microarray Gene Data Expression Society
- MIAME
Minimum Information About a Microarray Experiment
- MAGE
Microarray and Gene Expression
- MISFISHIE
Minimum Information Specification For In Situ Hybridization and Immunohistochemistry Experiments
- OWG
Ontology Workgroup
- RSBI WGs
Reporting Structure for Biological Investigations Working Groups
- MAQC
MicroArray Quality Control
Footnotes
¹This paragraph is based on field-work observations made on April 8-9, 2006, at an “Exploratory transcriptomics workshop” hosted by the Réseau québécois de bio-informatique (BioneQ) and sponsored by Genome Québec, Université du Québec à Montréal, and Université de Montréal.
²The five working groups were: a) Experiment description and data representation standards; b) Microarray data XML exchange format; c) Ontologies for sample description; d) Normalization, quality control and cross-platform comparison; and e) Future user group: queries, query language, data mining.
³This event was extended to both academic and industry participants, and was sponsored by Incyte Genomics, GeneLogic, and GlaxoWellcome.
⁴At the time, the two main repositories were GEO, overseen by the National Center for Biotechnology Information (NCBI) (www.ncbi.nlm.gov/geo/), and ArrayExpress, overseen by the European Bioinformatics Institute (EBI) (www.ebi.ac.uk/arrayexpress).
⁵These statements are taken from an interview with two biologists at EMBL, August 2006.
References
- Baird D. Thing knowledge. A philosophy of scientific instruments. Berkeley: University of California Press; 2004. [Google Scholar]
- Collins HM. Changing order: replication and induction in scientific practice. 2nd ed. Chicago: University of Chicago Press; 1992. [Google Scholar]
- Rader K. Making mice: Standardizing animals for American biomedical research, 1900-1955. Princeton: Princeton University Press; 2004. [Google Scholar]
- Löwy I, Gaudillière JP. Disciplining cancer: Mice and the practice of genetic purity. In: Gaudillière JP, Löwy I, editors. The Invisible industrialist: manufacturers and the construction of scientific knowledge. London: Macmillan; 1998. pp. 209–249. [Google Scholar]
- O'Connell J. Metrology: The creation of universality by the circulation of particulars. Social Studies of Science. 1993;23:129–173. [Google Scholar]
- Mallard A. Compare, standardize and settle agreement. On some usual metrological problems. Social Studies of Science. 1998;28:571–601. [Google Scholar]
- Schaffer S. Late Victorian metrology and its instrumentation: A manufactory of Ohms. In: Bud R, Cozzens SE, editors. Invisible connections: instruments, institutions, and science. Bellingham, Washington: SPIE Optical Engineering Press; 1992. pp. 23–56. [Google Scholar]
- Keating P, Cambrosio A. Interlaboratory life: regulating flow cytometry. In: Gaudillière JP, Löwy I, editors. The Invisible industrialist: manufacturers and the construction of scientific knowledge. London: Macmillan; 1998. pp. 250–295. [Google Scholar]
- Canguilhem G. The normal and the pathological. New York: Zone Books; 1989. [Google Scholar]
- Timmermans S, Berg M. The gold standard: the challenge of evidence-based medicine and standardization in health care. Philadelphia: Temple University Press; 2003. [Google Scholar]
- Cambrosio A, Keating P, Schlich T, Weisz G. Regulatory objectivity and the generation and management of evidence in medicine. Soc Sci Med. 2006;63:189–199. doi: 10.1016/j.socscimed.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Keating P, Cambrosio A. Cancer clinical trials: The emergence and development of a new style of practice. Bull Hist Med. 2007;81:197–223. doi: 10.1353/bhm.2007.0003. [DOI] [PubMed] [Google Scholar]
- Cambrosio A, Keating P, Mogoutov A. Mapping collaborative work and innovation in biomedicine: A computer-assisted analysis of antibody reagent workshops. Social Studies of Science. 2004;34:325–364. [Google Scholar]
- Bourret P, Mogoutov A, Julian-Reynier C, Cambrosio A. A new clinical collective for French cancer genetics: A heterogeneous mapping analysis. Science, Technology, & Human Values. 2006;31:431–464. [Google Scholar]
- Harris NL, Horning SJ. Burkitt’s lymphoma: The message from microarrays. N Engl J Med. 2006;354:2495–2498. doi: 10.1056/NEJMe068075. [DOI] [PubMed] [Google Scholar]
- Quackenbush J. Microarray analysis and tumour classification. N Engl J Med. 2006;354:2463–2472. doi: 10.1056/NEJMra042342. [DOI] [PubMed] [Google Scholar]
- Perkel JM. Medicine gets personal. With new regulations and new diagnostics, pharmacogenetics comes to the clinic. The Scientist. 2005;19(8):34. [Google Scholar]
- Simon R, Wang SJ. Use of genomic signatures in therapeutics development in oncology and other diseases. Pharmacogenomics J. 2006;6:166–173. doi: 10.1038/sj.tpj.6500349. [DOI] [PubMed] [Google Scholar]
- Keating P, Cambrosio A. Signs, markers, profiles, and signatures: Clinical hematology meets the new genetics (1980-2000) New Genetics and Society. 2004;2:15–45. [Google Scholar]
- Mogoutov A, Cambrosio A, Keating P, Mustar P. Biomedical innovation at the laboratory, clinical and commercial interface: mapping research projects, publications and patents in the field of microarrays. 6th Biennial International Triple Helix Conference on University-Industry Government-Links; 2007 May 16-18; Singapore. 2007. [Google Scholar]
- Frost and Sullivan. U.S. DNA Microarray Markets. 2005. Dec 29,
- Frost and Sullivan. Strategic Analysis of World DNA Microarray Markets. 2004. Mar 1,
- Datamonitor Company Profiles. Document Code DCC06282-9FEA-4BD4-A49B-DCB7E4C2E4F4 (Affymetrix/April 2007) and A8DD7B13-69B1-4126-A04C-71F7920B1876 (Illumina/May 2007)
- Fodor SP, Read JL, et al. Light-directed, spatially addressable parallel chemical synthesis. Science. 1991;251(4995):767. doi: 10.1126/science.1990438. [DOI] [PubMed] [Google Scholar]
- Microarray analysis resource: Microarray basics. Microarrays vs. macroarrays. Ambion; [cited 2007 Nov 1]. Available from: http://www.ambion.com/techlib/resources/microarray/basics2.html. [Google Scholar]
- Lopez MF, Pluskal MG. Protein micro-and macroarrays: digitizing the proteome. J Chromatography A. 2003;787(1):19–27. doi: 10.1016/s1570-0232(02)00336-7. [DOI] [PubMed] [Google Scholar]
- Lipshutz RJ, Morris D, et al. Using oligonucleotide probe arrays to access genetic diversity. Biotechniques. 1995;19(3):442–447. [PubMed] [Google Scholar]
- Gerhold D, Rushmore T, Caskey CT. DNA chips: promising toys have become powerful tools. Trends Biochem Sci. 1999;24:168–173. doi: 10.1016/s0968-0004(99)01382-1. [DOI] [PubMed] [Google Scholar]
- Schena M, Shalon D, et al. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270(5235):467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Michael M. Futures of the present: From performativity to prehension. In: Brown N, Rappert B, Webster A, editors. Contested futures: A sociology of prospective techno-science. Aldershot: Ashgate; 2000. pp. 21–42. [Google Scholar]
- Lesko LJ, Woodcock J. Pharmacogenomic-guided drug development: regulatory perspective. Pharmacogenomics J. 2002;2:20–24. doi: 10.1038/sj.tpj.6500046. [DOI] [PubMed] [Google Scholar]
- Nowak R. Entering the postgenome era. Science. 1995;270:368–371. doi: 10.1126/science.270.5235.368. [DOI] [PubMed] [Google Scholar]
- DeRisi J, et al. Use of cDNA Microarray to analyse gene expression patterns in human cancer. Nat Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- Star R, Rasooly RS. Searching for array standards in Rockville. Nat Biotechnol. 2001;19:418–419. doi: 10.1038/88070. [DOI] [PubMed] [Google Scholar]
- Bammler T, Beyer RP, et al. Standardizing global gene expression analysis between laboratories and across platforms. Nature Methods. 2005;2:477. doi: 10.1038/nmeth754. [DOI] [PubMed] [Google Scholar]
- Brazma A. On the importance of standardization in the life sciences [editorial] Bioinformatics. 2001;17(2):113–114. doi: 10.1093/bioinformatics/17.2.113. [DOI] [PubMed] [Google Scholar]
- Botstein D. Editorial; Mol Biol Cell. 1998;9:7. [PMC free article] [PubMed] [Google Scholar]
- Afshari CA. Perspective: microarray technology, seeing more than spots. Endocrinology. 2002;143(6):1983–1989. doi: 10.1210/endo.143.6.8865. [DOI] [PubMed] [Google Scholar]
- Knight J. Minimum standards set out for gene-expression data. Nature. 2002;415:946. doi: 10.1038/415946b. [DOI] [PubMed] [Google Scholar]
- Cambrosio A, Keating P. Of lymphocytes and pixels: The techno-visual production of cell populations. Studies in History and Philosophy of Biological and Biomedical Sciences. 2000;31:233–270. [Google Scholar]
- National Institutes of Health. Simon R. Dr. Richard Simon outlines Microarray Myths and Truths. Division of Cancer Treatment and Diagnosis, Program Accomplishments [Internet] 2006:20. Available from: http://dctd.cancer.gov/includefiles/DCTD.pdf. [Google Scholar]
- Bijker W, Law J, editors. Shaping technology /building society. Studies in sociotechnical change. Cambridge: MIT Press; 1994. [Google Scholar]
- Cambrosio A, Keating P, Mogoutov A. Mapping Collaborative Work and Innovation in Biomedicine: A Computer-Assisted Analysis of Antibody Reagent Workshops. Social Studies of Science. 2004;34:325–364. [Google Scholar]
- McMeekin A, Harvey M. The Formation of Bioinformatic Knowledge Markets: An ‘Economies of Knowledge’ Approach. Revue d’Économie Industrielle. 2002;101:47–64. [Google Scholar]
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, et al. Minimum Information About a Microarray Experiment (MIAME). Toward Standards for Microarray Data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- Davenport RJ. Data standards on the horizon. Science. 2001;292(5516):414–415. doi: 10.1126/science.292.5516.414b. [DOI] [PubMed] [Google Scholar]
- Brazma A, Robinson A, et al. One-stop shop for microarray: Is a universal, public DNA-microarray database a realistic goal? Nature. 2000;403:600–700. doi: 10.1038/35001676. [DOI] [PubMed] [Google Scholar]
- Ball CA, Sherlock G, et al. A guide to microarray experiments-an open letter to the scientific journals. Lancet. 2002;360(9338):1019. [PubMed] [Google Scholar]
- Microarray standards at last [Editorial] Nature. 2002;419:323. doi: 10.1038/419323a. [DOI] [PubMed] [Google Scholar]
- Shields R. MIAME, we have a problem [Editorial] Trends Genet. 2006;22:65–66. doi: 10.1016/j.tig.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Taylor CF, Hermjakob H, Julian RK Jr, Garavelli JS, Aebersold R, Apweiler R. The work of the Human Proteome Organisation's Proteomics Standards Initiative (HUPO PSI) OMICS. 2006;10(2):145–151. doi: 10.1089/omi.2006.10.145. [DOI] [PubMed] [Google Scholar]
- Taylor CF, Paton NW, et al. The minimum information about a proteomics experiment. Nat Biotechnol. 2007;25(8):887–893. doi: 10.1038/nbt1329. [DOI] [PubMed] [Google Scholar]
- Orchard S, Salwinski L, Kerrien S, Montecchi-Palazzi L, Oesterheld M, Stümpflen V, et al. The minimum information required for reporting a molecular interaction experiment (MIMIx) Nat Biotechnol. 2007;25:894–898. doi: 10.1038/nbt1324. [DOI] [PubMed] [Google Scholar]
- Jones AR, Miller M, Aebersold R, Apweiler R, Ball CA, Brazma A, et al. The Functional Genomics Experiment model (FuGE): an extensible framework for standards in functional genomics. Nat Biotechnol. 2007;25:1127–1133. doi: 10.1038/nbt1347. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Boststein D, Butler G, Cherry JM, et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, et al. The MicroArray Quality Control (MAQC) project shows inter-and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry A. Political machines. Governing a technological society. London: Athlone Press; 2001. [Google Scholar]
- Jasanoff S, editor. States of knowledge. The co-production of science and social order. London: Routledge; 2004. [Google Scholar]
- Funkhouser J. Reinventing pharma: The theranostic revolution. Current Drug Discovery. 2002;2:17–19. [Google Scholar]
- Warner S. Diagnostics+therapy=theranostics. The Scientist. 2004;18(16):38–39. [Google Scholar]
- Petrone J. SNP chip prices appear to have plateaued, but some predict decrease in coming years. BioArray News [Internet] 2007;7(38) Available from: http://www.bioarraynews.com/issues/7_38/features/142501-1.html. [Google Scholar]
- Curtin C. Get ready for microarray diagnostics. Genome Technology [Internet] 2007 Sep; Available from: http://www.genome-technology.com/issues/2_7/coverstory/141883-1.html. [Google Scholar]
- Jarvis N, Centola M. Gene-espression profiling: Time for clinical application? Lancet. 2005;365:199–200. doi: 10.1016/S0140-6736(05)17754-X. [DOI] [PubMed] [Google Scholar]
- Febbo PG, Kantoff PW. Noise and bias in microarray analysis of tumor specimens. Am J Clin Oncol. 2006;24:3719–3721. doi: 10.1200/JCO.2006.06.7942. [DOI] [PubMed] [Google Scholar]
- Krislov S. How nations choose product standards and standards change nations. Pittsburgh: University of Pittsburgh Press; 1997. [Google Scholar]