Abstract
“His book was known as the Book of Sand, because neither the book nor the sand have any beginning or end.” — Jorge Luis Borges
The human genome is a three billion-letter recipe for the genesis of a human being, directing development from a single-celled embryo to the trillions of adult cells. Since the sequencing of the human genome was announced in 2001, researchers have an increased ability to discern the genetic basis for diseases. This reference genome has opened the door to genomic medicine, aimed at detecting and understanding all genetic variations of the human genome that contribute to the manifestation and progression of disease. The overarching vision of genomic (or “personalized”) medicine is to custom-tailor each treatment for maximum effectiveness in an individual patient. Detecting the variation in a patient’s deoxyribonucleic acid (DNA), ribonucleic acid (RNA), and protein structures is no longer an insurmountable hurdle. Today, the challenge for genomic medicine lies in contextualizing those myriad genetic variations in terms of their functional consequences for a person’s health and development throughout life and in terms of that patient’s susceptibility to disease and differential clinical responses to medication. Additionally, several recent developments have complicated our understanding of the nominal human genome and, thereby, altered the progression of genomic medicine. In this brief review, we shall focus on these developments and examine how they are changing our understanding of our genome.
The Five Modalities of Genetic Variation
Large-scale genomic variation
In the few years since the release of the draft human genome sequence, our understanding of the importance of this “reference” genome has changed. Indeed, molecular characterization of karyotypes in the late 1990s already revealed substantial chromosomal abnormalities in phenotypically normal people, a surprising degree of variation that seemingly conferred no negative consequences [1]. After a decade of subsequent work in fine-mapping the variation present in the human genome, these variable regions of DNA (called structural variants) now are thought to cover as much as 20 percent of the length of the human genome. Thus, depending on the variant present, some tens of millions of nucleotides can be missing or duplicated — even quadrupled — in any one person [2]. We now know everyone has extra copies or missing copies of large parts of the genome, called copy number variants (CNVs), and that such large-scale variation apparently leaves us no worse off.
Rampant small-scale variation
The first diploid sequence of a single human being has been published [3], further expanding the amount of observed variation in a single human genome. Perhaps fittingly, the genome in question belongs to Craig Venter, a genome sequencing pioneer and one of the leaders of the private human genome consortium that produced a draft sequence in 2001. Venter’s genome shows the initial estimate of similarity between the genomes of two randomly selected individuals was too high — each person is now thought to be a mere 99.5 percent similar rather than the often-quoted 99.9 percent [3]. In total, Venter’s genome has some 4.1 million variations with respect to the reference, totaling 12.3 million base pairs (MB) of sequence. This massive amount of divergent sequence includes hundreds of thousands of homozygous insertions and deletions (called “indels”), previously thought to be very rare.
Rogue agents: autonomous, repeated, and mitochondrial elements
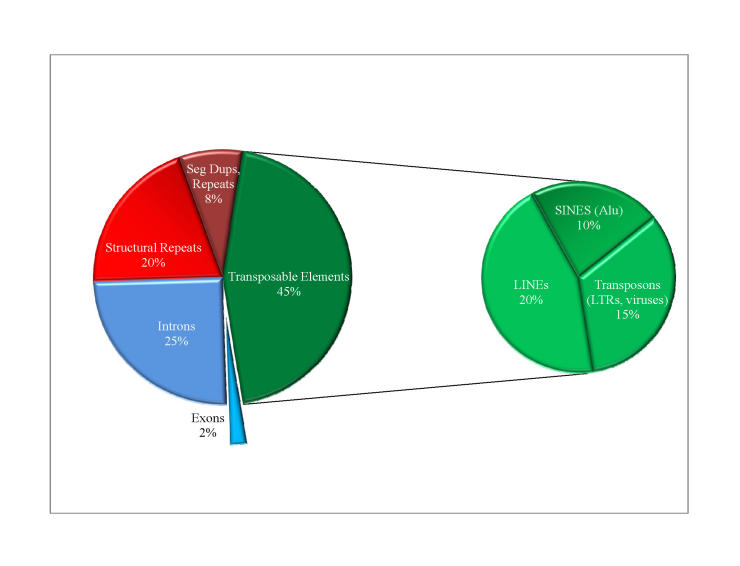
The organization of the human genome is more complex than initially believed (Figure 1) [4,5]. A quarter of the genome is dedicated to introns, the transcribed but noncoding parts of genes. Another third of the genome consists of repeats of varying kinds. Some repeated sections of the human genome serve structural purposes (for instance, those located at the ends and in the middle of chromosomes), but most repeats are of unknown function and take the form of simple sequence repeats or larger segmental duplications (SegDups). Counting toward the one-third of the genome that is repeated, SegDups (defined as segments at least 1,000bp long and appear at least twice throughout the genome) represent 5 percent of the human genome overall and 15 percent of all repeated sequence. Some of these segmental duplications hold extra copies of entire genes.
Figure 1.
Structural Divisions of the Human Genome. Most of the human genome is repetitive DNA sequences (red) and transposable elements (green, LINES, SINEs, LTRs, and viruses). Very little of the genome is coding sequence (blue, exons), but there is great room for gene flexibility and change with many of the gene’s long intron sequences (blue, introns).
Though the majority of the human genome is repetitive, not all of these repeats are functionless or inactive. The most ubiquitous repeated genetic elements in the genome are transposable elements (sometimes called TEs, or transposons), so called because they can transpose, or jump, from one place to another in the genome. When these elements jump, they sometimes create new forms of genes that may prove useful to humans; however, depending upon the precise site of insertion, such genomic shuffling instead may damage existing genes (for instance, the transposable element might land in the middle of a functional gene sequence, disrupting it). Transposable elements exist in various forms, including Long Interspersed Elements (LINEs), Short Interspersed Elements (SINEs), and the small 300bp Alu element (considered a SINE). The Alu element is present in 75 percent of introns and accounts for 10 percent of the entire genome, whereas the 6,000 bp (6 kb) LINEs account for 20 percent of the human genome and are present in most human genes. Indeed, when adding the sequences from repetitive elements into the size of the genes, introns account for 37 percent of the human genome, showing just how prevalent and successful these transposable elements have been at inserting themselves throughout the human genome over several million years.
Other transposable elements have invaded our genome more recently [6]. Since our divergence from the common ancestor we shared with the chimpanzee, approximately 98,000 viruses have invaded our genome and are now a part of our species’ genetic code, busily copying themselves and then re-inserting back into the human genome. These old viruses, called endogenous retroviruses, total 8 percent of the human genome (24 percent of all repeated sequence). These viruses include Long Terminal Repeats (LTRs), which reverse transcribe themselves (from RNA → DNA, as the AIDS virus does), DNA transposons, and some viruses that lie dormant and can no longer replicate themselves (totaling about 4 percent of the human genome, or 12 percent of repeated sequence).
The human genome has also exchanged genetic information with the mitochondrial genome [7]. To date, the human genome has been colonized by nearly 300 genes from mitochondria; these genes, called nuclear DNA of mitochondrial origin (NUMTs), are expressed by the human genome for the use of the mitochondria. Further, 27 of these NUMTs do not appear in the chimpanzee or other genomes, meaning they have been incorporated into the human genome within the last 4 to 6 million years (that is, since the divergence from our last common ancestor with chimpanzees). Most of these unique NUMTs (23 of 27) are present within known or predicted human genes, indicating that the symbiosis between these two genomes is very fast evolving and very gene-centered.
Most of the (nonrepetitive) genome is transcribed
It has been known since the 1960s that a significant amount of non-coding DNA (ncDNA) is transcribed and some of this leads to the production of functional ncRNAs (like ribosomal RNA). However, until recently, it was not clear what fraction of the genome was actively processed or how much of this transcription might contribute directly to biological function. To address this and other questions, the ENCODE project (the ENCyclopedia of DNA Elements) was launched in 2003 to identify all the functional elements in the human genome. The pilot phase of the ENCODE project was completed in 2007, and the published results show that an astonishing 93 percent of the non-repetitive sequence studied was either transcriptionally active or otherwise functionally relevant [8,9]. This surpassed earlier estimates, which had suggested that between 30 percent and 60 percent of the genome was transcriptionally active [10,11,12].
Epigenomics: The mutable backbone of the genome
The fifth and final modality of variation for the human genome is that the epigenome (“epi” is Latin for “above” or “upon”) has a large role to play in human genetic variation. Either with slight chemical modification to the DNA (methylation) or various changes to the histones supporting the DNA (methylation, acetylation, phosphorylation, and ubiquitination), the activity of a gene can be altered by inhibiting or allowing various transcription factors access to the targeted gene sequence. Some diseases now are being defined not only by catalogued sequence mutations, but also by the “epimutations” that appear to accompany the disease. These modifications to the epigenome can be transmitted to offspring in surprisingly simple ways, such as by a mother’s behavior during child-rearing [13] or general errors during egg and sperm production [14]. Because technologies and methodologies for studying epigenetics are still in the early stages, it remains unknown how much these type of modifications contribute to human phenotypic variation.
The larger picture: What it means for the genome
Taken together, these five discoveries complicate traditional notions of the gene and our understanding of the genome. While a clear translational distinction remains between coding and non-coding genes, we now know that a large amount of the genome is transcriptionally active and produces RNAs that do not fit into current functional categories. As we look closer at the structure of the human genome, we find it resembles a patchwork manuscript: a palimpsest of genetic elements (new and old, foreign and endogenous) constantly adapting to fast evolutionary selection pressures, as opposed to an eternal, unchanging “Book of Life” for our species. Indeed, when viewed this way, it seems almost miraculous that this “Book of Sand” [15] functions at all. For the structurally dynamic human genome to remain operational, given our complex neurophysiology and precise developmental plan, the system must contain either extensive functional redundancy or a scarcity of critical genetic architectures. Given the work of the ENCODE Consortium in humans and corroborating evidence from other species, it now seems likely that massively parallel functional redundancy is the most likely explanation. Could critical genetic architectures — those absolutely intolerant of disruption — really be that scarce, or are there other reasons why disruptions of these areas are not seen?
Tests of these two genomic hypotheses (rarity vs. redundancy as an explanation for our high tolerance for variation) will occur within our lifetimes, and these newfound reservoirs of variation will be needed to generate an accurate phenotype-to-genotype map. Indeed, the next step for Craig Venter’s genome is to probe its cellular function: Levy and colleagues plan to sample tissues from each part of Venter’s body and test the expression of all exons (i.e. the “exome”) [16]. (Suddenly, volunteering your genome doesn’t sound like such a good idea.) Compiling all this information on genotype (genetic sequence variants such as SNPs, CNVs, and mutations) and relating it to phenotype (enzyme levels, gene expression, and severity of a disease) is a mammoth undertaking. This information will need to be contextualized and understood before it can be used to implement genomic medicine, and the current paucity of such genotype-to-phenotype databases — as well as the large amount of work needed to create them — likely will delay the application of such personalized care for some time.
However, one thing that can be done very well with currently available sequence variants is to trace an individual’s family history and ancestral roots. As mentioned previously, it is now estimated that any person is 99.5 percent identical, on average, to anyone else at the genomic level, which translates to a difference of about one nucleotide per 500 bp. This means an average of six million single nucleotide polymorphisms (SNPs) and other sequence variants should exist between any two randomly selected human individuals. The hunt to characterize these SNPs and their distribution in various populations spurred the creation of the haplotype mapping project (HapMap). HapMap has thus far shown that SNP variation is not random, but instead can be traced to the likely migration patterns of Homo sapiens (modern humans) during the last 100,000 years. Certain SNPs and CNVs have been found only within certain populations — this is particularly noticeable among isolated peoples who have not interbred with other groups, like the African Yorubi. More commonly, genetic variants within a group of people are not binary (i.e., simply present or absent), but rather show an increased or decreased frequency. Using only 10 of these types of markers, called Ancestry-Informing Markers (AIMs), virtually anyone can be genotyped into a stereotype.
Pharmacogenomics and Personalized Medicine
The five new data and genetic categories detailed above have created pharmacogenomics, a new discipline that draws upon both pharmacology and genomics. Researchers in this nascent area aim to understand a person’s risk for disease based upon his or her ancestral genetic roots, plus the accrued genetic variation within that person’s lifetime (i.e., mutations since birth). They then hope to tailor medications to the individual in consideration of these genetic variations. Because many commonly used drugs are not equally effective in all patients, it is hoped that a more thorough understanding of the genetic underpinnings to a given disease will enable more accurate diagnosis and treatment. For example, two commonly used cancer drugs, Gleevec [17] and Herceptin [18], have been shown to be substantially more effective if certain genes are being expressed or protein conformations are present. The promise of personalized genomic medicine has prompted the creation of the Personal Genome Project at Harvard University (as distinguished from the Human Genome Project, which is concerned with the genetic component of our species as a whole), and volunteers are encouraged to submit genetic samples for whole-genome sequencing, which is accompanied by a rigorous examination and physical characterization (MRIs, blood tests, medical records). It also prompted action in the United States Senate by Senator Barack Obama, who introduced a bill to procure more federal funding for such research [19].
Bio-patenting: An obstacle to personalized medicine?
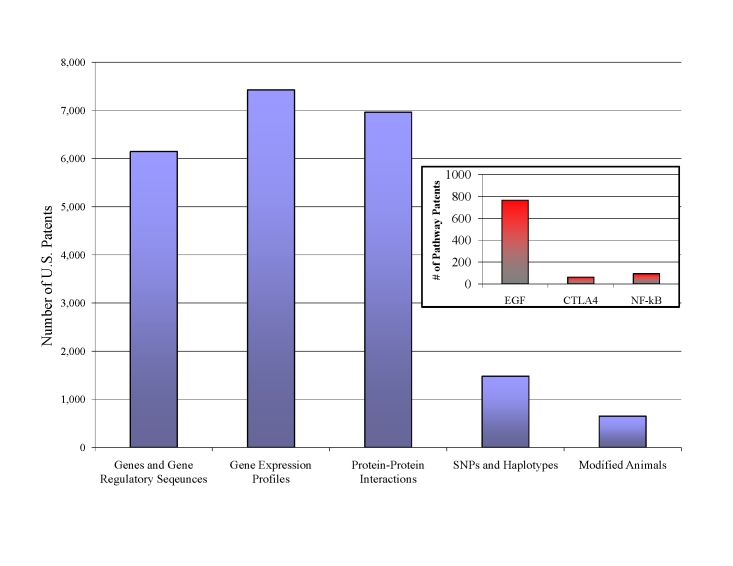
Pharmacogenomics research does present problems, however. Whenever any significant disease marker is found, it is generally patented immediately, potentially frustrating future research. Notable examples of this patent rush include the BRCA gene (Myriad Genetics), Canavan Disease gene (MCH), and Hepatitis C marker genotypes (Roche). If researchers from another laboratory or organization wish to look for additional markers or mutations within these genes, they may be infringing these patents and could face legal action. Unless a specific policy of open licensing for a patent is created, downstream research on these genes may effectively be blocked. The number of human genes patented today exceeds 20 percent of all known genes in the genome [20], and already many patents have been issued on entire biochemical pathways, such as NF-κB [21] (Figure 2). Some preliminary evidence suggests that such “patent thickets” may negatively impact further research and knowledge dissemination, which are, of course, critical to scientific progress [22].
Figure 2.
Number of Life-Based Patents. Genes, their regulatory sequences, and gene expression profiles account for bulk of life-based patents, followed by protein-protein interactions. Haplotypes and SNPs, which will become critical for pharmacogenomics, are just beginning to be patented. Nearly 1,000 patents have already issued on three major biochemical pathways (inset) [23].
Summary
The technologies and discoveries discussed here open the door to new questions in genomics. Along with incremental knowledge of genotype-to-phenotype matches comes a new set of issues: scientific, legal, and ethical. At a crime scene, for instance, should sweeps of genomic DNA samples be permitted, as we now demand of fingerprints? Will the ability to “read” the genetic probabilities in an individual’s genome provide fodder for discrimination? Should it? Will such knowledge transform parenting into a series of checkboxes to craft the ideal child? Vague fear of such eventualities has been lingering for years, highlighted by the Genetic Information and Nondiscrimination Act (GINA), which has been introduced into Congress every year since 1995. It has yet to pass both the House and the Senate.
Personalized genomic medicine holds great promise, but is not without its risks. Any partitioning of human genetic variation into separate states (e.g., disease vs. normal) could be of great benefit to both medicine and science. With such information, the population-specific effects of any ailment could be modeled and appropriately treated, and those whose DNA does not predispose them to such diseases can be given the happy news. Still, it remains unclear how well these ongoing studies will translate into actual, available treatments for patients. The difficulties for pharmacogenomics should not be understated, given the emerging complexities of the genome (massive structural variation, large-scale sequence variation, pervasive expression of ncDNA, autonomous transposable elements, and epigenetic changes). To fully understand the genotypic risks for a disease, particularly one with links to multiple interacting loci, scientists will need to account and correct for all of these sources of variation. How many of these solutions will be found and how many obstacles will appear remains to be seen — but as we consider these questions, the dawn of personalized medicine is showing its first light.
Abbreviations
- DNA
deoxyribonucleic acid
- RNA
ribonucleic acid
- CNVs
copy number variants
- ncDNA
non-coding DNA
- ENCODE
Encyclopedia of DNA Elements
- SegDups
segmental duplications
- TEs
transposable elements
- LINEs
Long Interspersed Elements
- SINEs
Short Interspersed Elements
- LTRs
Long Terminal Repeats
- NUMTs
nuclear DNA of mitochondrial origin
- SNPS
single nucleotide polymorphisms
- AIMs
Ancestry-Informing Markers
- GINA
Genetic Information and Nondiscrimination Act
References
- Bryndorf T, Kirchhoff M, Maahr J, Gerdes T, Karhu R, Kallioniemi A, et al. Comparative genomic hybridization in clinical cytogenetics. Am J Hum Genet. 1995;57:1211–1220. [PMC free article] [PubMed] [Google Scholar]
- Scherer S, Lee C, Birney E, Altshuler DM, Eichler EE, Carter NP, et al. Challenges and standards in integrating surveys of structural variation. Nat Genet. 2007;39:S7–S15. doi: 10.1038/ng2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, et al. The Diploid Genome Sequence of an Individual Human. PLoS Biology. 2007;5(10):e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Bannert N, Kurth R. Retroelements and the human genome: new perspectives on an old relation. PNAS. 2004;101:14572–14579. doi: 10.1073/pnas.0404838101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricchetti M, Tekaia F, Dujon B. Continued Colonization of the Human Genome by Mitochondrial DNA. PLoS Biology. 2007;2(9):e273. doi: 10.1371/journal.pbio.0020273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8(6):413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306(5705):2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Euskirchen G, Bertone P, Martone R, Luscombe NM, et al. The transcriptional activity of human Chromosome 22. Genes Dev. 2003;17(4):529–540. doi: 10.1101/gad.1055203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason C, Gauhar Z, Stolc V, Halasz G, van Batenburg MF, et al. A gene expression map for the euchromatic genome of Drosophila melanogaster. Science. 2004;306(5696):655–660. doi: 10.1126/science.1101312. [DOI] [PubMed] [Google Scholar]
- Weaver IC, Champagne FA, Brown SE, Dymov S, Sharma S, Meaney MJ, Szyf M. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: altering epigenetic marking later in life. J Neurosci. 2005;25(47):11045–11054. doi: 10.1523/JNEUROSCI.3652-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchins MP, Wong JJ, Suthers G, Suter CM, Martin DI, Hawkins NJ, Ward RL. Inheritance of a cancer-associated MLH1 germ-line epimutation. N Engl J Med. 2007;356(7):697–705. doi: 10.1056/NEJMoa064522. [DOI] [PubMed] [Google Scholar]
- Soares LM, Valcárcel J. The expanding transcriptome: the genome as the “Book of Sand.”. EMBO J. 2006;25:923–931. doi: 10.1038/sj.emboj.7601023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy Samuel. Conversation with: Christopher Mason. 2007. Oct 16,
- Hughes TP, Kaeda J, Branford S, Rudzki Z, Hochhaus A, et al. Frequency of major molecular responses to imatinib or interferon alfa plus cytarabine in newly diagnosed chronic myeloid leukemia. N Engl J Med. 2003;349(15):1423–1432. doi: 10.1056/NEJMoa030513. [DOI] [PubMed] [Google Scholar]
- Dowsett M, Bartlett J, Ellis IO, et al. Correlation between immunohistochemistry (HercepTest) and fluorescence in situ hybridization (FISH) for HER-2 in 426 breast carcinomas from 37 centres. J Pathol. 2003;199:418–423. doi: 10.1002/path.1313. [DOI] [PubMed] [Google Scholar]
- Obama B. (United States Senate). The Genomics and Personalized Medicine Act. S. 3822. 109th Cong. 2006
- Jensen K, Murray F. Intellectual property. Enhanced: intellectual property landscape of the human genome. Science. 2005;310(5746):239–240. doi: 10.1126/science.1120014. [DOI] [PubMed] [Google Scholar]
- Garber K. Decision on NFkappaB patent could have broad implications for biotech. Science. 2006;312(5775):827. doi: 10.1126/science.312.5775.827a. [DOI] [PubMed] [Google Scholar]
- Huang KG. Impact of intellectual property rights on scientific knowledge diffusion, accumulation and utilization [dissertation] Boston: Massachusetts Institute of Technology; 2006. [Google Scholar]
- Merrill SA, Mazza A, editors. Reaping the Benefits of Genomic and Proteomic Research: Intellectual Property Rights, Innovation, and Public Health. Washington DC: National Academies Press; 2006. [PubMed] [Google Scholar]