Abstract
The opportunistic human pathogen Acinetobacter baumannii strain M2 was found to produce distinct acyl-homoserine lactone (AHL) signals based on the use of an Agrobacterium tumefaciens traG-lacZ biosensor. An A. baumannii gene, designated abaI, was cloned and directed AHL production in recombinant Escherichia coli. The AbaI protein was similar to members of the LuxI family of autoinducer synthases and was predicted to be the only autoinducer synthase encoded by A. baumannii. The primary AHL signal directed by AbaI was identified by mass spectrometry as being N-(3-hydroxydodecanoyl)-l-HSL (3-hydroxy-C12-HSL). Minor amounts of at least five additional AHLs were also identified. The expression of abaI at the transcriptional level was activated by ethyl acetate extracts of culture supernatants or by synthetic 3-hydroxy-C12-HSL. An abaI::Km mutant failed to produce any detectable AHL signals and was impaired in biofilm development.
Acinetobacter baumannii is a nonmotile, gram-negative member of the class Pseudomonadales of the Gammaproteobacteria. A. baumannii is an important nosocomial pathogen and can cause a variety of infections including wound infections, bloodstream infections, ventilator-acquired pneumonia, and urinary tract infections (2, 5, 6, 11, 13, 14, 21, 23, 31, 41, 43). In addition, A. baumannii has been extremely problematic for U.S. military personnel in Iraq and Afghanistan, where this pathogen has been responsible for a large number of infections (1, 3, 8, 19). Of particular concern with A. baumannii is the increasing frequency of multidrug-resistant strains that leave few, if any, therapeutic options for treatment (16, 19, 24, 32).
In gram-negative bacteria, cell-to-cell signaling is often mediated by the production of N-acyl-homoserine lactone (AHL) signaling molecules (12). To detect these signals, a variety of bacterial biosensor strains have been constructed (27, 35, 44). These strains detect AHL signals by the activation of a reporter gene such as lacZ or lux or by the production or inhibition of a purple pigment in Chromobacterium violaceum. Using these biosensors, previous studies showed AHL production by Acinetobacter spp. (15). In a variety of gram-negative bacteria, it has also been demonstrated that biofilm development can be dependent on AHL signaling (7, 18, 26). In A. baumannii, very little is known regarding factors required for biofilm formation (25, 39, 40). A putative chaperone that is required for this process has been identified (39). In addition, a homolog of a staphylococcal biofilm-associated protein (Bap) has been characterized in A. baumannii, where it appears to act as an extracellular adhesin (25).
In this study, we report the identification and characterization of an autoinducer synthase from A. baumannii that was designated AbaI. Mass spectrometry was used to identify AHL signals that were directed by AbaI. The abaI gene was activated in a positive-feedback loop by an AbaI-dependent AHL signal(s). An abaI mutant was impaired in the later stages of biofilm development, and this phenotype was rescued by ethyl acetate extracts of cell supernatants from a wild-type strain.
A. baumannii produces AHL signals.
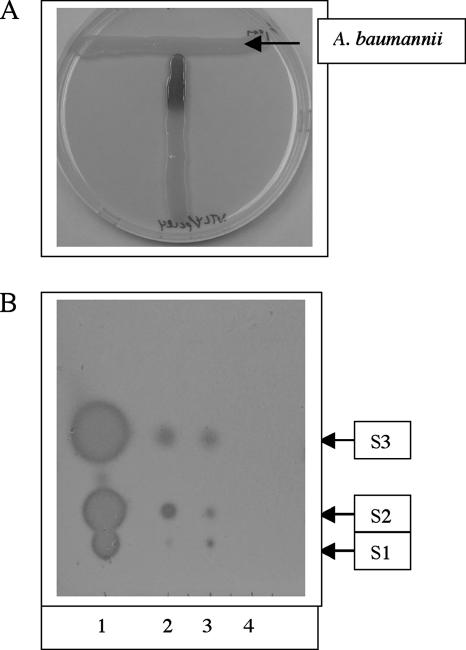
An A. tumefaciens traG::lacZ fusion, responsive to a variety of 3-oxo, 3-hydroxy, and 3-unsubstituted AHL signals of various acyl chain lengths (35), was strongly activated by a diffusible factor from an A. baumannii clinical isolate designated M2 that was obtained from the MetroHealth Medical Center, Cleveland, OH (Fig. 1A). To further characterize the extracellular signal(s) produced by A. baumannii M2, cell-free supernatants were prepared from cells grown to an optical density at 600 nm (OD600) of at least 1.1 in 500 ml of M9 minimal salts medium containing 1% glucose and 0.3% Casamino Acids (28). Cell-free supernatants were prepared by pelleting cells and filter sterilizing the resulting supernatant with a 0.22-μm filter. Supernatants were then extracted twice with an equal volume of acidified ethyl acetate (0.1 ml acetic acid per liter), and the extracts were dried to a volume of 50 μl.
FIG. 1.
AHL signal production by A. baumannii. (A) A line of A. baumannii cells (horizontal top line) was placed perpendicular to a line of A. tumefaciens cells containing an AHL-responsive traG::lacZ fusion. Plates were incubated at 28°C for 12 h. (B) For each condition, ethyl acetate extracts were prepared from 500-ml cultures, dried by rotary evaporation, and resuspended to a final concentration of 8,000×. For all samples, 1 μl of concentrated extract was applied to a C18 TLC plate and developed with methanol-water (60:40). Plates were dried at room temperature for 2 h and overlaid with a soft agar suspension containing the A. tumefaciens traG-lacZ biosensor as described previously (35). Plates were then incubated overnight at 28°C until blue spots appeared (typically 24 h). Samples are as follows: E. coli MG1655/pSK.abaI (lane 1), a 1/1,000 dilution of E. coli MG1655/pSK.abaI extract (lane 2), A. baumannii M2 wild-type extract (lane 3), and A. baumannii M2 abaI::Km (lane 4).
Concentrated extracts were analyzed on a C18 reverse-phase thin-layer chromatography (TLC) plate (JT Baker) using a method described previously by Shaw et al. (35), with a few modifications. A concentrated extract (1 μl) was applied onto a C18 TLC plate and developed in a glass chamber with methanol-water (60:40). The TLC plate was developed overnight and then overlaid with a soft agar lawn containing the Agrobacterium tumefaciens traG::lacZ indicator strain. The plate was sandwiched between two Pyrex glass containers, one containing water at the bottom to maintain humidity, and sealed with plastic wrap. Plates were incubated at 28°C until blue spots appeared, which indicated the presence of putative AHL signals.
As shown Fig. 1B, lane 3, there are three putative AHL signals (S1, S2, and S3) produced by A. baumannii that activate the traG-lacZ biosensor. All three A. baumannii signals lacked the trailing, comet tail pattern characteristic of 3-oxo derivatives (35), suggesting that they were 3-hydroxy or 3-unsubstituted AHLs.
Cloning of an A. baumannii gene that directs AHL signal production.
To identify a gene that directed the production of the AHL signal(s), we employed a screening procedure that allowed us to directly identify Escherichia coli transformants containing an autoinducer synthase gene. For this screen, LB agar plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) were overlaid with approximately 105 cells of the A. tumefaciens traG-lacZ biosensor. Next, a pooled recombinant library of A. baumannii Sau3AI DNA fragments in pACYC184 (4) transformed into E. coli MC4100 cells was directly plated onto the A. tumefaciens lawn. After overnight growth at 28°C, E. coli transformants were visible within the A. tumefaciens lawn. Colonies containing recombinant plasmids with a putative autoinducer synthase were identified by a blue halo that resulted from the activation of the A. tumefaciens traG-lacZ fusion in the indicator lawn.
Using the above-described screen, we isolated a pACYC184 derivative that encoded a putative autoinducer synthase. Retransformation of the plasmid from positive colonies verified that the recombinant plasmid directed the production of the activating signal. A recombinant plasmid with an approximately 5-kb insert was subjected to transposon mutagenesis using the GPS Tn7 in vitro mutagenesis system (New England BioLabs) to identify the putative autoinducer synthase encoded within the inserted DNA fragment. Insertions that inactivated the gene responsible for signal production were identified by cross-streaking colonies from an insertional library against the A. tumefaciens indicator strain. Tn7Km insertions that abolished signal production were then sequenced using the N and S primers that read outward from the ends of Tn7Km. All insertions that prevented signal production mapped to the same open reading frame encoding a 129-amino-acid protein with extensive homology to autoinducer synthases of the LuxI family, which was therefore designated abaI (Acinetobacter baumannii LuxI).
The abaI gene was recloned from the chromosome of A. baumannii M2 as a 2.0-kb chromosomal EcoRI fragment. This was necessary because the original 5-kb Sau3AI fragment was found to contain a scrambled insert. However, this did not alter the interpretation of the DNA sequence. A pBluescript SK derivative containing this 2-kb insert (pSK.abaI) was used for all further experiments. E. coli MG1655 containing pSK.abaI produced AHL signal profiles in the TLC overlay assay that were essentially identical to those of native A. baumannii M2, with three areas of signal activity (Fig. 1B, lanes 1 and 2). However, the relative amounts of each signal varied between E. coli/pSK.abaI and A. baumannii M2. The amount of S2 was greater in E. coli/pSK.abaI (Fig. 1B, lane 2) than in A. baumannii M2 (Fig. 1B, lane 3), and the amount of S1 was greater in A. baumannii M2. In addition, quantitative analysis of signal production revealed that E. coli/pSK.abaI produced approximately 1,000-fold more signal than did A. baumannii M2 (Fig. 1B, compare lanes 1 and 3).
Organization of the abaI region in A. baumannii.
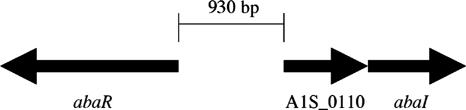
Compared to the recently published A. baumannii ATCC 17978 genome (36), the AbaI protein was 96% identical to the corresponding protein (GenBank accession number NC_009085.1). However, the AbaI protein identified from strain M2 was predicted to be 56 amino acids shorter at the amino terminus. Using the BLAST analysis program, the AbaI protein exhibited 54% identity and 69% similarity to the AfeI protein of Acidithiobacillus ferrooxidans and to LuxI proteins from Burkholderia cepacia (46% identity and 64% similarity) and Ralstonia solanacearum (41% identity and 65% similarity). Sequence analysis of the M2 genome revealed that immediately upstream from the abaI gene, an open reading frame (A1S_0110) encoding a protein of unknown function was present and was also seen in the 17978 genome (Fig. 2). Interestingly, an open reading frame similar to A1S_0110 was present immediately upstream of putative autoinducer synthases from A. ferrooxidans (37% identity and 48% similarity) and Burkholderia pseudomallei (38% identity and 62% similarity) (9, 33). Based on the 17989 genome, an open reading frame divergently transcribed from the A1S_0110 and abaI open reading frames was predicted to encode a LuxR protein designated AbaR (Fig. 2). The AbaR protein was 198 amino acids long and exhibited the best match to AfeR from A. ferrooxidans (46% identity) and to a LuxR-type protein from Burkholderia pseudomallei (38% identity). The similarity of gene organizations in A. baumannii, A. ferrooxidans, and B. pseudomallei suggests a common origin for these genes.
FIG. 2.
Organization of the abaI region of A. baumannii. The genomic region surrounding abaI in both A. baumannii M2 and ATCC 17978 is shown. The arrows above each gene indicate the orientation of each gene. The coding regions of abaR and the open reading frame of unknown function (A1S_0110) are separated by 930 bp. The abaR location is based on the published sequence of the ATCC 17978 genome (36).
Structural identification of the AHL signals produced by A. baumannii.
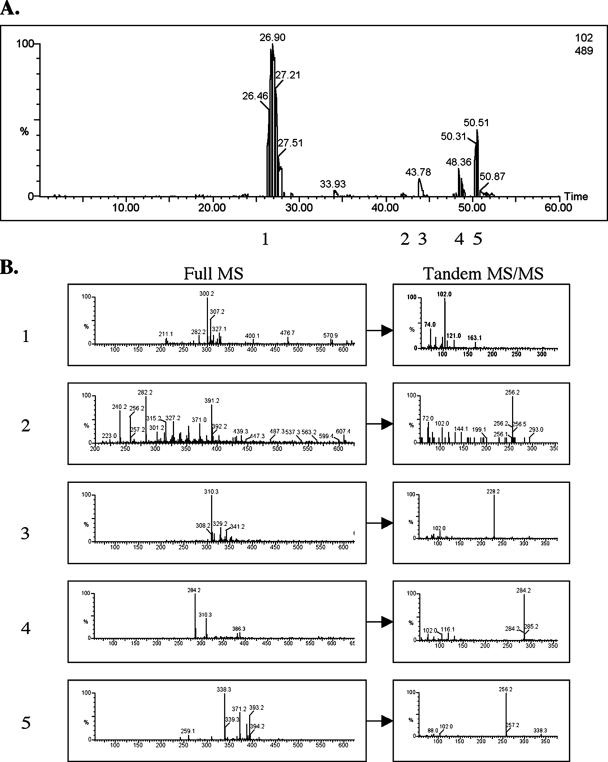
To characterize the AHL signals produced by A. baumannii, concentrated ethyl acetate extracts of spent culture supernatants were analyzed by liquid chromatography (LC)/mass spectrometry (MS) and LC/tandem MS (MS/MS) (Fig. 3). Initially, each parent ion was screened by MS/MS fragmentation for an ion with an m/z of 102, characteristic of the homoserine moiety (17, 29, 35). Putative AHLs with a breakdown ion with an m/z of 102 were observed and centered at 27, 42, 44, 48, and 50.5 min of elution from the reversed-phase C18 column (Fig. 3A). These areas were designated areas 1 to 5, respectively. The peaks at 33.93 and after 50.87 did not contain any ions that were identifiable as being HSL derivatives and were not examined further. For each peak, a full MS profiling of the parent ion was performed (Fig. 3B), and the profile was compared to those of known 3-oxo-, hydroxy-, and unsubstituted AHL signals (17, 29, 35). Based on this analysis, the primary AHL signal produced was 3-hydroxy-C12-HSL (m/z of 300.2) (Fig. 3B1) and was present in the material that eluted from 26 to 28 min (Fig. 3A). Synthetic 3-hydroxy-C12-HSL migrated in the same position in the reversed-phase column and had the same m/z of 300.2. In addition, at this time, a peak with an m/z of 282.2 was present but in significantly smaller amounts (Fig. 3B1). This species could correspond to the unsaturated form of C12-HSL or unsaturated 3-oxo-C11-HSL. At position 2 in Fig. 1A, two ions were identified, with m/z values of 256.2 and 282.2, that matched the values for unsubstituted C10-HSL (m/z of 256.2) and unsaturated C12-HSL or unsaturated 3-oxo-C11-HSL (m/z of 282.2) (Fig. 3B2). The ion at m/z 256.2 fragmented in MS/MS to m/z 102 (Fig. 3B2). However, MS/MS analysis of the m/z 282.2 ion did not give an obvious fragment with an m/z of 102. Additional AHL signals that were identified at other elution times in significantly smaller amounts than 3-hydroxy-C12-HSL included unsaturated C14-HSL or unsaturated 3-oxo-C13-HSL (m/z 310.3; 44 min) (Fig. 3B3), unsubstituted C12-HSL (m/z 284.3; 48 min) (Fig. 3B4), and unsaturated C16-HSL or unsaturated 3-oxo-C15-HSL (m/z 338.3; 50.1 min) (Fig. 3B5). Subsequent MS/MS analyses of all the above-described ions indicated the presence of the m/z 102 ion, verifying the presence of the HSL moiety (Fig. 3B).
FIG. 3.
MS profiles of AHL signals. A 10,000-fold-concentrated ethyl acetate extract of A. baumannii M2 supernatant was diluted 1:3 in 30 μl of methanol for LC/MS and LC/MS/MS analysis. Ten microliters was injected onto a 3-μm, 100- by 2.1-mm, C18 reverse-phase column (Restek) installed on a Waters Alliance 2695 high-performance liquid chromatograph (Waters Corporate, Milford, MA) operated at a flow rate of 200 μl/min with the effluent flowing directly into the mass spectrometer. Solvent A consisted of water with 0.1% formic acid, and solvent B consisted of acetonitrile with 0.1% formic acid. A gradient elution method was utilized, which started at 5% solvent B for 5 min, went to 95% solvent B over 30 min, and remained isocratic at 95% solvent B for 15 min. Electrospray ionization-MS was performed by using a Waters Q-TOF micro-MSD instrument (Waters Corporate, Milford, MA). The instrument parameters were as follows: capillary voltage of 2,800 V, cone voltage of 25 V, and collision energy of 25 V. Nitrogen was used as the collision gas. All signal peaks containing fragments with an m/z of 102 were analyzed using MassLynx 4.0 software. The elution times (min) off the C18 column of species with an MS/MS breakdown ion at m/z 102 are shown (A). Each of the five peaks was then analyzed by MS to identify the parent ion, and the full MS profile for each ion is shown. The MS/MS profile of each parent ion is shown on the right.
To further verify the role of AbaI in AHL production, concentrated ethyl acetate extracts were prepared from supernatants of E. coli MG1655 containing the cloned abaI gene on a high-copy plasmid (pSK.abaI). The prominent AHL in E. coli/pSK.abaI had an m/z of 300.2, consistent with 3-hydroxy-C12-HSL. Ions of m/z 256.2 and m/z 284.2 were also present in minor amounts, corresponding to unsubstituted C10-HSL and C12-HSL, respectively (data not shown). E. coli/pSK.abaI also produced a AHL with an m/z of 272.2 that was not observed in A. baumannii. This corresponded to 3-hydroxy-C10-HSL.
We attempted to assign each signal to the corresponding spots (S1 to S3) observed in the TLC plates (Fig. 1B). Each spot was individually isolated from the C18 TLC matrix. The spot corresponding to S1 contained three AHL signals, and the primary product exhibited an m/z of 300.2, consistent with 3-hydroxy-C12-HSL. Moreover, synthetic 3-hydroxy-C12-HSL exhibited an Rf value identical to that of the material in S1 (data not shown). Minor peaks at m/z 284 and m/z 312 that were consistent with unsubstituted C12-HSL and unsubstituted C14-HSL, respectively, were also present in S1. The S2 spot contained a signal with an m/z of 284, corresponding to unsubstituted C12-HSL. In addition, the material eluted from the S2 spot exhibited an Rf value on TLC plates identical to that of a synthetic C12 standard (data not shown). Lastly, the material in S3 exhibited an m/z of 256, consistent with unsubstituted C10-HSL. However, the material purified from this spot did not comigrate with a C10-HSL standard. One explanation for this result is that a second AHL that accounts for the activity in the S3 spot is present but that the concentration of this AHL was too low to detect by mass spectrometry.
Role of the abaI gene in AHL signal production.
To inactivate the chromosomal copy of abaI, plasmid pSK.abaI was mutagenized with Tn7Km, and insertions that prevented the production of the AHL signal were identified using the A. tumefaciens biosensor containing traG-lacZ. An insertion that disrupted abaI in the middle of the coding region was cloned along with flanking chromosomal DNA as an SalI-XbaI fragment into suicide vector pKNG101 (22). This plasmid, pKNG101.abaI::Km, was introduced into the A. baumannii chromosome by filter mating with E. coli SM10/pKNG101.abaI::Km, followed by selection for kanamycin- and streptomycin-resistant exconjugants. These exconjugants contained pKNG101.abaI::Km integrated at the wild-type abaI gene, an event that results in a wild-type copy of abaI and a second copy containing the abaI::Kmr disruption. To excise the integrated plasmid, cells were grown for 5 to 10 generations without antibiotic selection, and dilutions were plated onto LB plates without sodium chloride and containing 10% sucrose, followed by incubation at room temperature. All sucrose-resistant colonies were streptomycin sensitive, indicating that they had lost the integrated pKNG101 plasmid. These colonies were then tested for kanamycin resistance, and all colonies with this phenotype contained the abaI::Km disruption as determined by Southern blot analysis (37) using a probe internal to abaI generated by PCR using primers 5′-GTACAGTCGACGTATTTGTTGAATATTTGGG-3′ and 5′-CGTACGTCTAGAGTAATGAGTTGTTTTGCGCC-3′.
The abaI::Km mutant failed to produce any detectable signals in the TLC overlay using the A. tumefaciens biosensor (Fig. 1B, lane 4). In addition, there were no detectable ions present at an m/z of 256, 284, or 300 in ethyl acetate extracts from the abaI::Km mutant (data not shown).
abaI is transcriptionally activated by AHL signals in a positive-feedback loop.
Transcription of an autoinducer synthase is often positively regulated by the accumulation of the cognate AHL signal (12), although exceptions to this can exist (38). To determine if abaI expression was altered by AHL signals, a promoterless lacZ cassette obtained from pQF50 (10) as an SmaI-ScaI fragment was inserted into the abaI coding region by ligation with pKNG101.abaI::Km digested with PmeI, which excises out the kanamycin resistance cassette within abaI. The introduction of the pKNG101.abaI-lacZ fusion into the M2 chromosome was done by a conjugal mating with E. coli SM10 containing this plasmid. A. baumannii exconjugants were then selected for sucrose resistance to excise the integrated plasmid by a second crossover. The presence of the correct single-copy abaI-lacZ fusion in A. baumannii M2 was verified by PCR and DNA sequence analysis of the PCR product. It is important that this lacZ cassette creates a null allele in abaI, a phenotype that was verified by the lack of AHL production in cross-streak assays with the A. tumefaciens traG-lacZ biosensor (data not shown). When cells were harvested at an OD600 of 0.5, the expression levels of abaI-lacZ in the presence of 1 μM and 10 μM 3-hydroxy-C12-HSL were increased 2.1- and 4.9-fold, respectively, above the levels in cells grown in LB only (Table 1). The addition of an ethyl acetate extract of a culture supernatant from wild-type M2 activated the abaI-lacZ fusion fourfold. However, an ethyl acetate extract from the abaI::Km mutant failed to activate the abaI-lacZ fusion. When cells were harvested at a higher density (OD600 of 1.2), the abaI-lacZ fusion was activated to a greater extent by AHL signals, 5.8- and 13.7-fold in the presence of 1 μM and 10 μM, respectively, of 3-hydroxy-C12-HSL. In addition, ethyl acetate extracts of wild-type M2 activated the abaI-lacZ fusion 8.9-fold. These results indicate that the transcription of the abaI gene is positively activated by the AHL signals directly resulting from AbaI enzymatic activity.
TABLE 1.
Activation of abaI-lacZ by AHL signals
| Growth conditiona | Relative level of expression of abaI-lacZ (Miller units) ± SDb |
|---|---|
| OD600 of 0.5 | |
| LB + EtAc control | 1.0 ± 0 |
| LB + 1 μM 3OH-C12-HSL | 2.1 ± 0.2 |
| LB + 10 μM 3OH-C12-HSL | 4.9 ± 0.7 |
| LB + wild-type EtAc extract | 4.0 ± 1.1 |
| LB + abaI::Km EtAc extract | 1.1 ± 0.005 |
| OD600 of 1.2 | |
| LB + EtAc control | 1.0 ± 0.02 |
| LB + 1 μM 3OH-C12-HSL | 5.8 ± 0.55 |
| LB + 10 μM 3OH-C12-HSL | 13.7 ± 1.45 |
| LB + wild-type EtAc extract | 8.9 ± 0.1 |
| LB + abaI::Km EtAc extract | 1.1 ± 0.005 |
Cells were grown in LB broth, and ethyl acetate (EtAc) extracts of culture supernatants were added at a 10× concentration.
Relative expression values are reported as Miller units of β-galactosidase activity in cells grown under the indicated growth conditions divided by the values in cells grown in LB broth only.
AHL-dependent quorum sensing is required for biofilm development in A. baumannii.
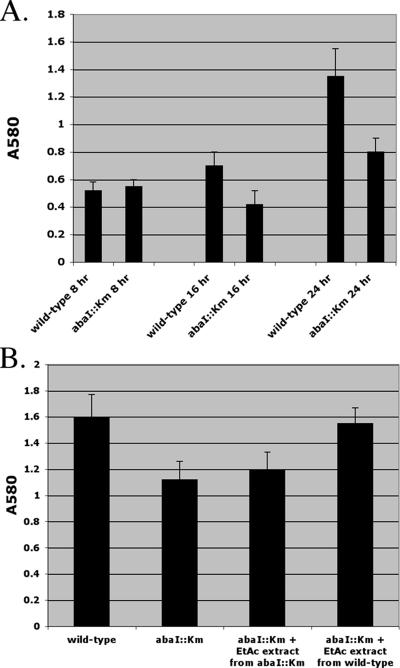
The ability of the wild type and the isogenic abaI::Km mutant to form biofilms was examined in cells grown for 8, 16, and 24 h in wells of a microtiter plate. The growth rates of the wild type and the abaI::Km mutant were identical (31-min generation time) and were at similar optical densities after each of the indicated times. For cells grown for 8 h, the biofilm-forming abilities were similar for the wild type and the abaI::Km mutant as measured by crystal violet staining of adherent cells (A580 values of 0.52 and 0.55, respectively) (Fig. 4A). However, the biofilm-forming ability of the abaI::Km mutant was markedly lower at 16 and 24 h, at 40% and 41%, respectively, than that of the wild type (Fig. 4A). To directly demonstrate that this biofilm defect was due to a loss of AHL signal production directed by AbaI, we assayed the biofilm formation of the abaI::Km mutant in the presence of exogenously added AHL signal provided as an ethyl acetate extract from wild-type cells or the abaI::Km mutant. Ethyl acetate alone was added in negative control samples. The addition of ethyl acetate extracts from wild-type cells restored the ability of the abaI::Km mutant to form normal biofilms (Fig. 4B). However, the addition of ethyl acetate extracts from the abaI::Km mutant did not rescue the defect in biofilm formation exhibited by the abaI::Km mutant (Fig. 4B).
FIG. 4.
Biofilm formation of the wild type and an abaI mutant. Biofilm assays were done according to method a described previously by O'Toole and Kolter (30). Exponential-phase cultures of M2 (wild type) and the abaI::Km mutant were diluted in LB broth to an OD600 of 0.02, and 100 μl of each culture was applied to wells of a microtiter plate (Fisher). After incubation at 30°C for various times, growth at an OD600 was measured. Ten microliters of 0.4% crystal violet was then added to each well and incubated for 15 min, and wells were rinsed thoroughly with sterile H2O to remove nonadherent cells. After rinsing, 100 μl of 33% acetic acid was added into each well, and plates were vortexed for 30 s to solubilize the crystal violet. Biofilm formation was measured by reading the absorbance at an OD580. The values reported in both panels represent the averages of 12 individual wells. (B) Ethyl acetate (EtAc) extracts of wild-type and the abaI::Km mutant wells at a 5× concentration relative to the original volume of spent culture supernatant that was extracted. For the growth of the wild type and the abaI::Km mutant, the same volume of ethyl acetate alone was added.
Concluding remarks.
The gene for an autoinducer synthase, designated abaI, has been cloned and characterized in this study. The deduced AbaI protein was highly similar to members of the LuxI family. Analysis of the recently completed A. baumannii 17978 genome sequence (36) did not reveal additional genes that encode members of the LuxI or LuxM/AinS family of autoinducer synthases. Therefore, based on the AHL profiles determined in Fig. 3, and the fact that AbaI is probably the only autoinducer synthase, the primary AHL signal in A. baumannii M2 is 3-hydroxy-C12-HSL. However, since AHL signals can be produced by acyltransferases that do not have similarity to LuxI or LuxM/AinS (34), it cannot be ruled out that additional AHL signals are present in A. baumannii M2. To determine if AHL quorum sensing was utilized by other clinical isolates of A. baumannii, we tested 19 additional isolates (10 from military infections in Iraq and 9 from U.S. hospitals) for AHL production. All isolates produced AHL signals that activated the A. tumefaciens traG-lacZ biosensor (data not shown). However, in two cases, TLC analysis indicated that a unique AHL signal not produced by M2 was present. The nature of this signal is under investigation.
The abaI gene was activated by ethyl acetate extracts of culture supernatants and by synthetic 3-hydroxy-C12-HSL (Table 1). Therefore, abaI is regulated by a positive-feedback loop involving self-produced AHL signal and a cognate R protein. The activation of abaI-lacZ by the addition of AHL signal in the abaI null background was greater in cells at high density (Table 1). This indicates that an inhibitor of quorum sensing is present in LB broth and depleted during cell growth. A similar situation was previously reported for Pseudomonas aeruginosa (45). The role of the LuxR homolog AbaR in the activation of abaI in the presence of 3-hydroxy-C12-HSL is unknown at present, as repeated attempts to create an abaR null allele have failed for unknown reasons. However, given that the only predicted LuxR protein encoded in A. baumannii is AbaR, it is likely that this is required for the activation of abaI. Although the abaI promoter has not yet been identified, a putative Lux box (CTGTAAATTCTTACAG) is centered 67 bp upstream of the putative ATG start for AbaI and may represent a binding site for AbaR.
The AbaI autoinducer synthase was required for normal biofilm development (Fig. 4). The differences between the wild type and the abaI::Km mutant were evident after 16 and 24 h of development, but there was no difference at 8 h. This suggests that the abaI-directed quorum-sensing pathway is required for the later stages of biofilm maturation. At present, the only known determinants required for biofilm formation in A. baumannii are the csu-ecoded chaperone-usher pilus assembly system (39) and the Bap protein (25). It is possible that quorum sensing influences the expression of these genes, but this remains to be determined. The identification of abaI and the corresponding AHL signals will now allow the identification of signal antagonists that inhibit biofilm development. These antagonists may also reduce the ability of A. baumannii to survive on environmental surfaces for extended periods, a key component of its ability to persist in intensive care wards (20, 42).
Nucleotide sequence accession number.
The abaI sequence from A. baumannii M2 has been deposited in the GenBank database under accession number EU334497.
Acknowledgments
We are grateful to Stephen Farrand at the University of Illinois for providing the A. tumefaciens traG-lacZ fusion and for helpful advice and to Paul Williams at the University of Nottingham for generously providing us with synthetic 3-hydroxy-C12-HSL. In addition, we are grateful to Fred Stroebel at Emory University and Binghe Wang and Siming Wang at Georgia State University for the high-performance liquid chromatography and mass spectrometry analyses.
This work was supported in part by the Atlanta Research and Education Foundation to P.N.R. and by R01AI072219-01A1 from the National Institutes of Health.
Footnotes
Published ahead of print on 15 February 2008.
REFERENCES
- 1.Aronson, N. E., J. W. Sanders, and K. A. Moran. 2006. In harm's way: infections in deployed American military forces. Clin. Infect. Dis. 431045-1051. [DOI] [PubMed] [Google Scholar]
- 2.Bergogne-Berezin, E., and K. J. Towner. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 9148-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2004. Acinetobacter baumannii infections among patients at military medical facilities treating injured U.S. service members, 2002-2004. MMWR Morb. Mortal. Wkly. Rep. 531063-1066. [PubMed] [Google Scholar]
- 4.Chang, A. C. Y., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 1341141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chastre, J. 2003. Infections due to Acinetobacter baumannii in the ICU. Semin. Respir. Crit. Care Med. 2469-78. [DOI] [PubMed] [Google Scholar]
- 6.Chen, M. Z., P. R. Hsueh, L. N. Lee, C. J. Yu, P. C. Yang, and K. T. Luh. Severe community-acquired pneumonia due to Acinetobacter baumannii. Chest 1201072-1077. [DOI] [PubMed]
- 7.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280295-298. [DOI] [PubMed] [Google Scholar]
- 8.Davis, K. A., K. A. Moran, C. K. McAllister, and P. J. Gray. 2005. Multidrug-resistant Acinetobacter extremity infections in soldiers. Emerg. Infect. Dis. 111218-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farah, C., M. Vera, D. Morin, D. Haras, C. A. Jerez, and N. Guiliani. 2005. Evidence for a functional quorum sensing type AI-1 system in the extremophilic bacterium Acidothiobacillus ferrooxidans. Appl. Environ. Microbiol. 717033-7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farinha, M. A., and A. M. Kropinski. 1990. Construction of broad-host-range plasmid vectors for easy visible selection and analysis of promoters. J. Bacteriol. 1723496-3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fournier, P. E., and H. Richet. 2006. The epidemiology and control of Acinetobacter baumannii in health care facilities. Clin. Infect. Dis. 42692-699. [DOI] [PubMed] [Google Scholar]
- 12.Fuqua, C., S. C. Winans, and E. P. Greenberg. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum sensing transcriptional regulators. Annu. Rev. Microbiol. 50727-751. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Garmendia, J. L., C. Ortiz-Leyba, J. Garnacho-Montero, F. J. Jimenez-Jimenez, C. Perez-Paredes, A. E. Barrero-Almodovar, and M. Gili-Miner. 2001. Risk factors for Acinetobacter baumannii nosocomial bacteremia in critically ill patients: a cohort study. Clin. Infect. Dis. 33939-946. [DOI] [PubMed] [Google Scholar]
- 14.Gaynes, R., and J. R. Edwards. 2005. Overview of nosocomial infections caused by gram-negative bacilli. Clin. Infect. Dis. 41848-854. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez, R. H., A. Nusblat, and B. C. Nudel. 2001. Detection and characterization of quorum sensing signal molecules in Acinetobacter strains. Microbiol. Res. 155271-277. [DOI] [PubMed] [Google Scholar]
- 16.Goossens, H. 2005. European status of resistance in nosocomial infections. Chemotherapy 51177-181. [DOI] [PubMed] [Google Scholar]
- 17.Gould, T. A., J. Herman, J. Krank, R. C. Murphy, and M. E. Churchill. 2006. Specificity of acyl-homoserine lactone synthases examined by mass spectrometry. J. Bacteriol. 188773-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huber, B., K. Riedel M. Hentzer, A. Heydorn, A. Gotschlich, M. Givskov, S. Molin, and L. Eberl. 2001. The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology 1472517-2528. [DOI] [PubMed] [Google Scholar]
- 19.Hujer, K. M., A. M. Hujer, E. A. Hulten, S. Bajaksouzian, J. M. Adams, C. J. Donskey, D. J. Ecker, C. Massire, M. W. Eshoo, R. Sampath, J. M. Thomson, P. N. Rather, D. W. Craft, J. T. Fishbain, A. J. Ewell, M. R. Jacobs, D. L. Paterson, and R. A. Bonomo. 2006. Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob. Agents Chemother. 504114-4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jawad, A., H. Seifert, A. M. Snelling, J. Heritage, and P. M. Hawkey. 1998. Survival of Acinetobacter baumannii on dry surfaces: comparison of outbreak and sporadic isolates. J. Clin. Microbiol. 361938-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joly-Guillou, M. L. 2005. Clinical impact and pathogenicity of Acinetobacter. Clin. Microbiol. Infect. 11868-873. [DOI] [PubMed] [Google Scholar]
- 22.Kaniga, K., I. Delor, and G. R. Cornelis. 1991. A wide host range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109137-141. [DOI] [PubMed] [Google Scholar]
- 23.Leung, W. S., C. M. Chu, K. Y. Tsang, F. H. Lo, K. F. Lo, and P. L. Ho. 2006. Fulminant community-acquired Acinetobacter baumannii pneumonia as a distinct clinical syndrome. Chest 129102-109. [DOI] [PubMed] [Google Scholar]
- 24.Livermore, D. M. 2003. The threat from the pink corner. Ann. Med. 35226-234. [DOI] [PubMed] [Google Scholar]
- 25.Loehfelm, T. W., N. R. Luke, and A. A. Campagnari. 2008. Identification and characterization of an Acinetobacter baumannii biofilm-associated protein. J. Bacteriol. 1901036-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lynch, M. J., S. Swift, D. F. Kirke, C. W. Keevil, C. E. Dodd, and P. Williams. 2002. The regulation of biofilm development by quorum sensing in Aeromonas hydrophila. Environ. Microbiol. 418-28. [DOI] [PubMed] [Google Scholar]
- 27.McClean, K. H., M. K. Winson, L. Fish, A. Taylor, S. R. Chhabra, M. Camara, M. Daykin, J. H. Lamb, S. Swift, B. W. Bycroft, G. S. Stewart, and P. Williams. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 1433703-3711. [DOI] [PubMed] [Google Scholar]
- 28.Miller, J. H. 1972. Experiments in molecular genetics, p. 431. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Ortori, C. A., S. Atkinson, S. R. Chhabra, M. Camara, P. Williams, and D. A. Barrett. 2007. Comprehensive profiling of N-acyl homoserine lactones produced by Yersinia pseudotuberculosis using liquid chromatography coupled to hybrid quadrupole-linear ion trap mass spectrometry. Anal. Bioanal. Chem. 387497-511. [DOI] [PubMed] [Google Scholar]
- 30.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28449-461. [DOI] [PubMed] [Google Scholar]
- 31.Perez, F., A. M. Hujer, K. M. Hujer, B. K. Decker, P. N. Rather, and R. A. Bonomo. 2007. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 513471-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rice, L. B. 2006. Challenges in identifying new antimicrobial agents effective for treating infections with Acinetobacter baumannii and Pseudomonas aeruginosa. Clin. Infect. Dis. 43(Suppl. 2)S100-S105. [DOI] [PubMed] [Google Scholar]
- 33.Rivas, M., M. Seeger, D. S. Holmes, and E. Jedlicki. 2005. A lux-like quorum sensing system in the extreme acidophile Acidithiobacillus ferrooxidans. Biol. Res. 38283-297. [DOI] [PubMed] [Google Scholar]
- 34.Rivas, M., M. Seeger E. Jedlicki, and D. S. Holmes. 2007. Second acyl homoserine lactone production system in the extreme acidophile Acidithiobacillus ferrooxidans. Appl. Environ. Microbiol. 733225-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaw, P. D., G. Ping, S. L. Daly, C. Cha, J. E. Cronan, Jr., K. L. Rinehart, and S. K. Farrand. 1997. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl. Acad. Sci. USA 946036-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith, M. G., T. A. Gianoulis, S. Pukatzki, J. J. Mekalanos, L. N. Ornston, M. Gerstein, and M. Snyder. 2007. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 21601-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98503-517. [DOI] [PubMed] [Google Scholar]
- 38.Throup, J. P., M. Camara, G. S. Briggs, M. K. Winson, S. R. Chhabra, B. W. Bycroft, P. Williams, and G. S. Stewart. 1995. Characterisation of the yenI/yenR locus from Yersinia enterocolitica mediating the synthesis of two N-acylhomoserine lactone signal molecules. Mol. Microbiol. 17345-356. [DOI] [PubMed] [Google Scholar]
- 39.Tomaras, A. P., C. W. Dorsey, R. E. Edelmann, and L. A. Actis. 2003. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology 1493473-3484. [DOI] [PubMed] [Google Scholar]
- 40.Vidal, R., M. Dominguez, H. Urrutia, H. Bello, G. Gonzalez, A. Garcia, and R. Zemelman. 1996. Biofilm formation by Acinetobacter baumannii. Microbios 8649-58. [PubMed] [Google Scholar]
- 41.Villegas, M. V., and A. I. Hartstein. 2003. Acinetobacter outbreaks, 1977-2000. Infect. Control Hosp. Epidemiol. 24284-295. [DOI] [PubMed] [Google Scholar]
- 42.Wendt, C., B. Dietze, E. Dietz, and H. Ruden. 1997. Survival of Acinetobacter baumannii on dry surfaces. J. Clin. Microbiol. 351394-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson, S. J., C. J. Knipe, M. J. Zieger, K. M. Gabehart, J. E. Goodman, H. M. Volk, and R. Sood. 2004. Direct costs of multidrug-resistant Acinetobacter baumannii in the burn unit of a public teaching hospital. Am. J. Infect. Control 32342-344. [DOI] [PubMed] [Google Scholar]
- 44.Winson, M. K., S. Swift, L. Fish, J. P. Throup, F. Jorgensen, S. R. Chhabra, B. W. Bycroft, P. Williams, and G. S. Stewart. 1998. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol. Lett. 163185-192. [DOI] [PubMed] [Google Scholar]
- 45.Yarwood, J. M., E. M. Volper, and E. P. Greenberg. 2005. Delays in Pseudomonas aeruginosa quorum-controlled gene expression are conditional. Proc. Natl. Acad. Sci. USA 1029008-9013. [DOI] [PMC free article] [PubMed] [Google Scholar]