Abstract
We demonstrated that a single copy of insertion sequence ISPme1 can mobilize adjacent segments of genomic DNA of Paracoccus methylutens DM12, which leads to the generation of diverse transposable elements of various size and DNA contents. All elements (named transposable modules [TMos]) contain ISPme1 (placed at the 5′ ends of the elements) and have variable 3′-end regions of between 0.5 and 5 kb. ISPme1 was shown to encode an outwardly oriented promoter, which may activate the transcription of genes transposed within TMos in evolutionarily distinct hosts. TMos may therefore be considered to be natural systems enabling gene capture, expression, and spread. However, unless these elements have been inserted into a highly conserved genetic context to enable a precise definition of their termini, it is extremely difficult or even impossible to identify them in bacterial genomes by in silico sequence analysis. We showed that TMos are present in the chromosome and plasmids of strain DM12. Sequence analysis of plasmid pMTH1 (32 kb) revealed that four TMos, previously identified with a trap vector, pMEC1, comprise 87% of its genome. Repeated TMos within pMTH1 may stimulate other structural rearrangements resulting from homologous recombination between long repeat sequences. This illustrates that TMos may play a significant role in shaping the structure of natural plasmids, which consequently may have a great impact on the evolution of plasmid genomes.
Sequencing projects have revealed that bacterial genomes are not static, monolithic structures. They can contain a number of different kinds of integrated mobile genetic elements (e.g., transposable elements, plasmids, bacteriophages, and integrative and conjugative elements) acquired by lateral gene transfer.
Insertion sequences (ISs), which are the simplest forms of transposable elements (TEs), are components of nearly all bacterial genomes. To date, more than 1,500 ISs have been identified in over 295 bacterial and archaeal species (21). The transposition of ISs promotes structural changes in DNA that lead to the formation of various mutations (insertions, deletions, inversions, translocations, and replicon fusion). These elements are therefore considered to be the major recombinogenic factors in bacterial genomes. Their activity results in the shuffling of genetic information among various replicons present in a bacterial cell (chromosomes, plasmids, and bacteriophages), which may ultimately enable its spread by lateral gene transfer. These elements thus play the role of a factor that significantly enhances variability and, consequently, the adaptive and evolutionary capacities of their hosts.
ISs have a very simple structure, since they carry only the genetic information necessary for their own transposition. Most ISs encode only a single gene for transposase (Tnp) bordered by inverted repeats (IRs), the sites for Tnp binding and action (7). However, ISs are also able to form composite transposons, which consist of random segments of genomic DNA, bordered by a pair of ISs. The transposition of these transposons is initiated by the interaction of the IS-encoded transposase with the extreme IRs flanking the complete element. Interestingly, it has recently been reported that during the transposition of just a single copy of an IS, resistance genes adjacent to the IS can also be translocated (19, 28). In addition, it has been shown that the IS231 transposase is able to mobilize segments of genomic DNA of Bacillus cereus that are bordered by naturally occurring sequences resembling the IRs of IS231 (8). The above-described examples provide evidence that ISs can efficiently enrich the pool of mobile DNA, which may have a great impact on lateral gene transfer and the evolution of bacterial genomes.
Due to the great diversity of IS-mediated TEs, it is not possible to distinguish them in bacterial genomes simply by classical in silico sequence analyses. For this reason, various entrapment vectors have been used for the identification of functional TEs. These are convenient tools, enabling the direct identification of even phenotypically silent elements (4, 16, 23). In this report, we present the characterization of “atypical” transposable elements “captured” by entrapment vector pMEC1 (4) in a methylotrophic bacterium utilizing dichloromethane, Paracoccus methylutens DM12 (Alphaproteobacteria) (10). We also show that these elements are generated frequently and that their activity is able to significantly shape the structures of genomes of plasmids naturally occurring in this strain.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. P. methylutens DM12 (10) was the host strain of the analyzed TEs. The related strain Paracoccus pantotrophus KL100 (2) was used for β-galactosidase assays and for analysis of transposition activities of transposable modules (TMos). Escherichia coli TG1 was used for plasmid construction, and strain DH5Δlac was used for β-galactosidase assays. All strains were grown in Luria-Bertani (LB) medium (20) at 30°C (Paracoccus sp. strains) or 37°C (E. coli). Where necessary, the medium was supplemented with antibiotics at the following concentrations: 50 μg ml−1 kanamycin, 50 μg ml−1 rifampin, and 20 μg ml−1 (E. coli) or 1 μg ml−1 (Paracoccus spp.) tetracycline. Paracoccus spp. formed colonies on solid medium after 48 h of incubation.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or feature(s) | Reference or source |
|---|---|---|
| Strains | ||
| P. methylutens DM12R | Rifr derivative of wild-type strain DM12; contains plasmids pMTH1 (32 kb), pMTH4 (22 kb), pMTH2 (approximately 200 kb), and pMTH3 (greater than 650 kb) | 2 |
| P. pantotrophus KL100 | Rifr derivative of wild-type strain DSM 11073; deprived of indigenous plasmid pKLW1 | 2 |
| E. coli DH5Δlac | deoR thi1 relA1 supE44 endA1 gyrA96 recA1 hsdR17Δ(argF lac)U169 Nalr | M. Yarmolinsky |
| E. coli TG1 | supE hsdΔ5 thi Δ(lac proAB), F′ (traD36 proAB+lacIqlacZΔM15) | Laboratory collection |
| Plasmids | ||
| pMEC1 | Kmr; mobilizable shuttle (E. coli-Paracoccus spp.) trap plasmid carrying cI-tetA selection cartridge | 4 |
| pDS132 | Cmr; mobilizable cloning vector; contains sacB gene; oriT RK2 | 18 |
| pBBR1MCS-3 | Tcr; ori pBBR1; cloning broad-host-range vector; oriT RK2 | 13 |
| pEBB10 | Tcr; pBBR1MCS-3 carrying sacB | This study |
| pABW1 | Kmr; ori pMB1; mobilizable cloning vector; oriT RK2 | 3 |
| pABW12 | pABW1 carrying a 0.7-kb HindIII fragment of pMTH4 | This study |
| pMTH4::pABW12 | Cointegrate of pMTH4 and pABW12 | This study |
| pCM132 | Kmr; ori RK2; lacZ reporter gene fusion vector | 14 |
| pCM132TC | pCM132 carrying promoterless tetA | This study |
| pRS551 | Kmr; ori pMB1; lacZ reporter gene fusion vector | 22 |
| pRK2013 | Kmr; helper plasmid carrying genes for conjugal transfer of RK2 | 9 |
Plasmid construction.
Plasmids used in this study are listed in Table 1. Entrapment vector pCM132TC was constructed by PCR amplification of the tetA gene of pMEC1 (primers LTETMET [5′-GTGGATCCGGATGGGCAGTGATAGAGAA-3′] and RTETMET [5′-TGGCATGCTTCCTGGATGCCGACGGATT-3′] [introduced restriction sites for BamHI and SphI are underlined]) and subsequently cloning the promoterless gene into BclI and SphI sites within the lacZ reporter gene of promoter probe vector pCM132. Entrapment vector pEBB10 was constructed by cloning an XbaI-EcoRV DNA fragment (containing the sacB gene) derived from plasmid pDS132 into compatible sites in pBBR1MCS-3. The pMTH4::pABW12 cointegrate was constructed by (i) cloning of a 0.7-kb HindIII restriction fragment of pMTH4 (contains part of a type A TMo) into the compatible site of vector pABW1 (E. coli specific; unable to replicate in Paracoccus spp.), (ii) introduction of the resulting suicide plasmid, pABW12, from E. coli TG1 into strain DM12 by conjugation, and (iii) selection of Kmr clones containing pMTH4::pABW12 cointegrates, which arose by means of homologous recombination.
Plasmid DNA isolation.
Plasmid DNA was isolated using a standard procedure (6) and, when required, purified by CsCl-ethidium bromide density gradient centrifugation. Megaplasmid visualization was achieved by in-gel lysis and DNA electrophoresis according to a method described previously by Wheatcroft et al. (31). Total DNA from P. methylutens DM12 was isolated by phenol extraction (32). Common DNA manipulation methods were performed as described previously by Sambrook and Russell (20).
DNA-DNA hybridization.
Molecular probes specific for all classes of individual types of transposable modules of P. methylutens DM12 were prepared by PCR amplification of DNA fragments of TMos directly adjacent to ISPme1. The primer pairs used were (i) LATYPE (5′-AAGGTTGGCTTTCTCGGGTT-3′) and RATYPE (5′-CCAGCTTGAGGTCCTTCAG-3′) (type A), (ii) LBTYPE (5′-GGCCGATCTCGACGACTGA-3′) and RBTYPE (5′-TCCATGTATCGCAGGTCGCA-3′) (type B), (iii) LCTYPE (5′-GCGGAAATTGAGCTGGCGT-3′) and RCTYPE (5′-TGACACACTCATCTGGCTAC-3′) (type C), and (iv) LDTYPE (5′-CCGGACCATTACCATGAACA-3) and RDTYPE (5′-GATACAGGATGAGCGCGGA-3′) (type D). The amplified DNA fragments were labeled with digoxigenin (DIG) (Roche). Hybridization and visualization of bound DIG-labeled probes were carried out as recommended by the supplier.
Introduction of DNA into bacterial cells.
DNA was introduced by triparental mating (into P. methylutens DM12 and P. pantotrophus KL100), transformation (into E. coli strains), or electroporation (into P. pantotrophus KL100) as previously described (11).
Assay for β-galactosidase activity.
β-Galactosidase activities in E. coli DH5Δlac and P. pantotrophus KL100 were measured by the conversion of o-nitrophenyl-β-d-galactopyranoside into nitrophenol as described previously by Miller (17). Assays for β-galactosidase activity were repeated three times.
Sequence analyses and annotation.
Nucleotide sequences of ISs, TMos, pMTH1, and pMTH4 were determined using a dye terminator sequencing kit and an automatic sequencer (ABI 377; Perkin-Elmer). A combination of vector-derived primers and primer walking was used to obtain the entire nucleotide sequences. Similarity searches were performed using the ISFinder (http://www.ncbi.nlm.nih.gov/) and the BLAST (1) programs provided by the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/).
Nucleotide sequence accession numbers.
The nucleotide sequences of ISPme1, ISPme2, TMo(ISPme1)C1, TMo(ISPme1)D1, and pMTH1 have been submitted to the GenBank database under accession numbers EF585233, EU016112, EU016113, EU016114, and EU043115, respectively.
RESULTS
Identification of TEs of P. methylutens DM12.
The shuttle entrapment vector pMEC1 (Kmr) carries a selective cartridge composed of a silent tetA gene under the control of the pR promoter of bacteriophage λ and the gene coding for the λ CI repressor. Inactivation of the repressor gene (e.g., through the insertion of a TE) results in the constitutive expression of tetracycline resistance. Vector pMEC1 was introduced into P. methylutens DM12, and Tcr clones were selected on LB medium supplemented with tetracycline at a frequency of 4.4 × 10−5. The plasmid pattern of 100 Tcr clones was analyzed, and the sizes of the inserts were estimated by restriction analysis (data not shown). Three classes of plasmids were distinguished, carrying (i) potential ISs (inserts of <3 kb) (47% of tested plasmids), (ii) putative transposons (inserts of >3 kb) (39% of plasmids), and (iii) point mutations (plasmids of the size of pMEC1) (14% of plasmids). The locations of the TEs within the cI gene of pMEC1 were confirmed by PCR using a set of cartridge-specific primers, as previously described (4).
ISs of P. methylutens.
Detailed restriction and hybridization analyses led to the identification of three different ISs entrapped in pMEC1 (data not shown). One was the previously described ISPpa2 of P. pantotrophus DSM 11072 (4), while the remaining two were novel ISs. Analysis of their nucleotide sequences permitted the identification of (i) the Tnp gene(s), (ii) IRs, and (iii) the target sequence, which was duplicated (direct repeats [DRs]) upon insertion (summarized in Table 2). The novel elements were designated ISPme1 and ISPme2, and comparison with the ISFinder database revealed that they are members of different IS families: IS1380 (ISPme1) and IS5 (ISPme2).
TABLE 2.
Characteristics of ISs of P. methylutens DM12
| IS | Length (bp) | IR (bp) | Target sequence duplicated upon transposition | Transposase (aa)a | IS family/group |
|---|---|---|---|---|---|
| ISPme1 | 1,197 | 16 | ATGCA | ORF1 (451) | IS1380 |
| ISPme2 | 851 | 14 | TA | ORF1 (143) | IS5/IS427 |
| ORF2 (200) | |||||
| ISPpa2 | 832 | 14 | TA | ORF1 (115) | IS5/IS427 |
| ORF2 (189) |
aa, amino acid.
Transposable modules.
The sizes of the putative transposons were highly variable. To determine whether these putative transposons represented composite transposons generated by the above-mentioned ISs, hybridization analysis was performed. Isolated DNA of the pMEC1 derivatives carrying the putative transposons was probed with DIG-labeled internal fragments of ISPme1, ISPme2, or ISPpa2, respectively. All the plasmids gave a positive hybridization signal exclusively with the ISPme1-specific probe (data not shown). To reveal the genetic organization of the “transposons,” the nucleotide sequences of 16 randomly chosen elements were determined. All the elements had similar but atypical structures. They were composed of a single copy of ISPme1 (always placed in the same orientation at the 5′ end of the elements) with adjacent DNA fragments of lengths varying from 0.5 kb to 5 kb (called 3′-end DNA regions) encoding genes conserved in the chromosomes of many bacteria (Fig. 1). Colocalization of ISPme1 and the transposed 3′-end regions in the DM12 genome was confirmed by PCR analysis (data not shown).
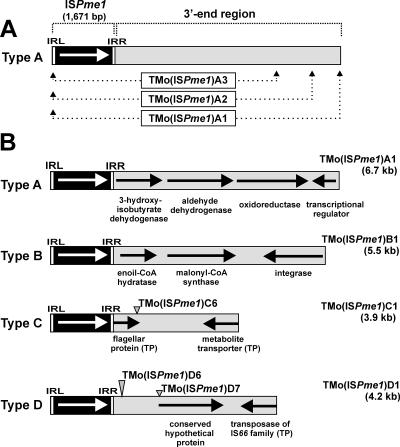
FIG. 1.
(A) Genetic structure of TMos composed of ISPme1 and variable 3′-end regions. The white arrow shows the direction of transcription of the transposase gene (tnp). Below, the proposed nomenclature of different size classes of TMos of one type is shown. (B) Genetic maps of four types of TMos identified by transposition into pMEC1 (only the longest class of a given type is presented). Endpoints of TMo(ISPme1)C6, TMo(ISPme1)D6, and TMo(ISPme1)D7, described further in this paper, are marked by gray arrowheads. Black arrows indicate the transcriptional orientations of the genes encoded by TMos. The putative function of the predicted ORFs is based on BLAST searches and a protein domain search. TP, terminal part; CoA, coenzyme A.
These observations strongly suggest that this group of P. methylutens transposable elements has arisen as the result of the ISPme1-mediated mobilization of large DNA segments adjacent to the IS primary target site. Insertion of these elements into the selective cartridge resulted in the generation of AT-rich 5-bp DRs (data not shown), which confirms the acquisition of these elements as a result of transposition events.
We analyzed a pool of approximately 200 elements identified with pMEC1 in several independent experiments. These diverse elements have been classified into different types and classes (Fig. 1). Within a given type, all the TEs carried homologous DNA segments adjacent to ISPme1, although the lengths of these segments varied in individual elements (size classes within a type). Interestingly, some of the identified elements were identical but had inserted at different sites in the selective cartridge (data not shown). As a result of our analyses, we distinguished four types of ISPme1-mediated TEs, designated types A, B, C, and D (Fig. 1B), and in each type, we identified different classes. We propose to name these diverse elements TMos and to designate them, e.g., TMo(ISPme1)A1, for transposable module class 1 of type A generated by ISPme1 (Fig. 1A).
Localization of TMos in the P. methylutens genome.
To examine the localization of the TMos in the P. methylutens DM12 genome, hybridization analysis was performed. Four probes specific for each type of TMo were hybridized with (i) DNA of two plasmids naturally occurring in the host strain, pMTH1 (32 kb) and pMTH4 (22 kb), and (ii) DNA of two megasized replicons, pMTH2 (approximately 200 kb) and pMTH3 (>650 kb), visualized by in-gel cell lysis and electrophoresis. We detected TMos of type A in pMTH1, pMTH4, and pMTH2 and TMos of type B in all tested replicons. TMos of types C and D were absent in the plasmids, which suggests their chromosomal localization (data not shown). Plasmids pMTH1 and pMTH4 appeared to contain several copies of integrated TMos, as judged by restriction analysis (data not shown), which suggested that these elements might significantly influence the structure of plasmid genomes.
Insight into pMTH1 structure.
In order to define the TMos of pMTH1, the complete nucleotide sequence of this plasmid was determined. It consisted of 31,999 bp with an average GC content of 64.1%, which is close to that of the P. methylutens chromosome (67%) (10). pMTH1 was predicted to encode 27 putative open reading frames (ORFs) (Fig. 2A). A summary of the predicted ORFs, including their positions, the sizes of the encoded proteins, and their closest homologs, is presented in the supplemental material.
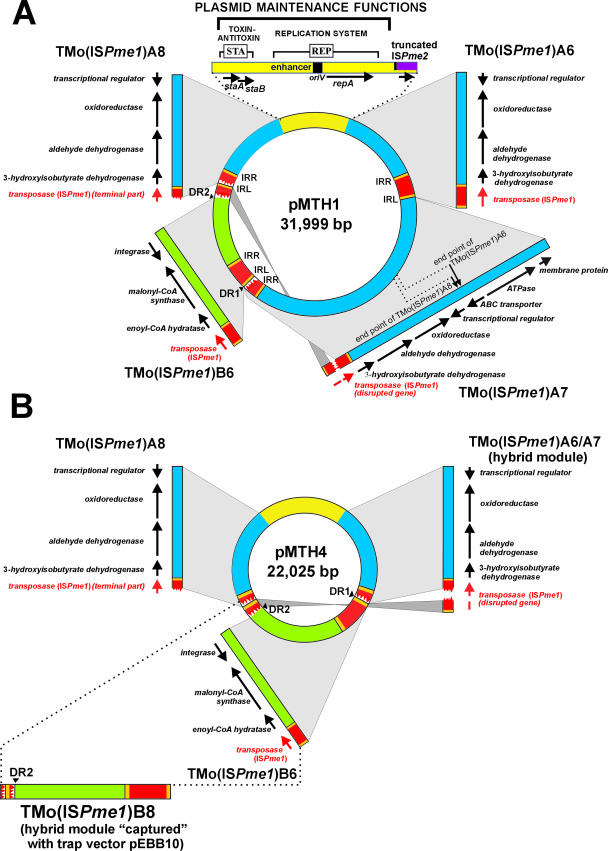
FIG. 2.
Genetic organization of plasmids pMTH1 and pMTH4 of P. methylutens DM12. (A) Plasmid pMTH1. The region responsible for the stable maintenance of the plasmid, coding for replication (REP) and stabilization (STA) systems, is shown in yellow. The proximal part of the truncated ISPme2-like element is marked. ISPme1 is marked in red with thick yellow lines at both ends, corresponding to the IRR and IRL. The predicted TMos are indicated. The 3′-end regions of the A-type TMos are shown in blue, and those of the type B module are shown in green. The sites corresponding to the 3′ endpoints of TMo(ISPme1)A6 and TMo(ISPme1)A8 are marked by arrows within TMo(ISPme1)A7 (carries the longest 3′-end region of TMos of type A). The putative function of the predicted ORFs is based on a protein domain search (see the supplemental material for details). (B) Plasmid pMTH4, a deletion derivative of pMTH1. The hybrid TMos of pMTH4 are indicated. TMo(ISPme1)A6/7 resulted from homologous recombination between TMo(ISPme1)A6 and TMo(ISPme1)A7, and TMo(ISPme1)B8 was captured by entrapment vector pEBB10. CoA, coenzyme A.
Interestingly, sequence analysis revealed that the DNA region of pMTH1 coding for putative replication and stabilization systems was identical to the previously analyzed minireplicon of coresiding plasmid pMTH4 (24). The replication system of pMTH1/pMTH4 comprises a rep gene coding for replication initiation protein and an iteron-like origin of replication and enhancer, which is necessary to maintain the correct plasmid copy number (25). The only stabilization system of pMTH1/pMTH4 consists of two genes and is based on the toxin-and-antitoxin principle (24).
Comparative analysis of the nucleotide sequences of pMTH1 and TMos “captured” by pMEC1 revealed that four TMos [three classes of the A type, TMo(ISPme1)A6, TMo(ISPme1)A7, TMo(ISPme1)A8, and one of the B type, TMo(ISPme1)B6] comprise 87% of the plasmid genome (Fig. 2A). Among the TMos of type A, only one [TMo(ISPme1)A7] contains a complete copy of ISPme1. TMo(ISPme1)A8 carries only the terminal part of the IS; no putative remnants of the absent proximal part of the IS were identified in pMTH1. On the other hand, ISPme1 of TMo(ISPme1)A7 was disrupted by the transposition of TMo(ISPme1)B6 (Fig. 2A), which led to the generation of 5-bp repeat sequences (DRs) flanking the inserted element. TMo(ISPme1)A7 contains the longest 3′-end DNA region of 8,303 bp.
We also detected the proximal part of a putative IS within pMTH1 (77% identity to ISPme2, identified in this study), which was most probably disrupted upon the transposition of TMo(ISPme1)A6 (Fig. 2A). However, the terminal part of this IS is not present in the pMTH1 genome.
Structure of plasmid pMTH4.
Analysis of the complete nucleotide sequence of pMTH4 (22,025 bp) revealed that the whole plasmid is identical to part of pMTH1 (Fig. 2). This plasmid therefore represents a deletion derivative of pMTH1, most probably the result of homologous recombination between two TMos of type A, present in the same orientation within pMTH1 [TMo(ISPme1)A6 and TMo(ISPme1)A7]. Such a recombination event would result in the loss of a region of pMTH1 containing a terminal part of the 3′ end of TMo(ISPme1)A7 as well as the complete ISPme1 of TMo(ISPme1)A6, which are both lacking in pMTH4 (Fig. 2B). Therefore, TMo(ISPme1)A6/A7 of pMTH4 has a hybrid structure containing a 3′-end DNA sequence identical to that of TMo(ISPme1)A6 and the truncated IS of TMo(ISPme1)A7 (Fig. 2). Plasmids pMTH1 and pMTH4 are incompatible and are randomly segregated upon cell division, since it was possible to obtain individual clones containing either pMTH1 or pMTH4 (data not shown).
Generation of hybrid TMos.
pMTH4 contains only one potentially functional module, TMo(ISPme1)B6, which is also present in pMTH1 (Fig. 2). To exclude the possibility that the generation of TMos in P. methylutens DM12 is a host-specific phenomenon, we tested the transposition of these elements in P. pantotrophus KL100, a strain that does not contain ISs homologous to ISPme1 (as judged by DNA-DNA hybridization analysis) (data not shown).
A plasmid cointegrate composed of cryptic pMTH4 joined to Kmr plasmid pABW12 (unable to replicate in Paracoccus spp.) was constructed in strain DM12 (see Materials and Methods for details). The pMTH4::pABW12 fusion was introduced by electroporation into strain KL100, carrying an entrapment vector, pEBB10 (Tcr broad-host-range vector pBBR1MCS-3 containing the sacB gene of Bacillus subtilis). The expression of sacB is lethal for the bacterial host in the presence of sucrose (5, 12). This allows the direct selection of sacB mutants (e.g., carrying inserted TEs), whose growth under these conditions is not affected. The sacB gene serves, therefore, as a cassette enabling the positive selection of transposition mutants. A culture of KL100 cells carrying both plasmids was plated onto LB medium containing sucrose and grown overnight, and approximately 200 colonies were selected. A detailed analysis of plasmids isolated from several randomly chosen clones revealed that the analyzed TMo was able to transpose from pMTH4::pABW12 into the sacB gene of pEBB10. As expected, different classes of TMos could be distinguished. All these elements were bordered by 5-bp-long DRs. Although most of the TMos were shorter than TMo(ISPme1)B6, we also identified a longer element [designated TMo(ISPme1)B8 (6.5 kb)] with a mosaic structure. Its 3′-end region carried an additional DNA segment from pMTH4 (933 bp) directly adjacent to the 3′ terminus of TMo(ISPme1)B6 (Fig. 2B). In subsequent experiments, we also showed that the captured TMos of type B were able to transpose from one entrapment vector (pEBB10 insertion derivatives) into another (pMEC1) in the KL100 strain lacking pMTH4::pABW12 (data not shown).
The above-described experiments showed that the transposition of TMos is not restricted exclusively to strain DM12, and it is not dependent on the presence of plasmids pMTH1 and pMTH4. The results also provided evidence for the generation of hybrid TMos, whose 3′-end DNA regions were composed of DNA segments acquired during successive transposition events.
Transcriptional activation by ISPme1.
The highly conserved orientation of ISPme1 within TMos suggests the possibility of the activation of transcription of downstream genes by IS-encoded promoter(s). To identify the promoter(s), DNA sequences upstream and downstream of the ISPme1 transposase gene were separately amplified by PCR and inserted into a promoter probe vector to generate transcriptional fusions with a promoterless lacZ reporter gene. Two test vectors were used for the analysis: pRS551 (specific for E. coli) and pCM132 (functional in Paracoccus spp.). The resulting plasmid constructs were introduced into the appropriate hosts (E. coli DH5Δlac or P. pantotrophus KL100), and β-galactosidase activity assays were used to examine promoter strength. The results suggested that ISPme1 encodes two promoters (functional in both tested hosts), termed P1, a weak promoter for the transposase gene (β-galactosidase activity of 97 ± 2.1 Miller units), and P2, an outwardly oriented strong promoter (β-galactosidase activity of 14,500 ± 1,500 Miller units) located close to the right IR (IRR) of ISPme1. The localization of the promoters is shown in Fig. 3.
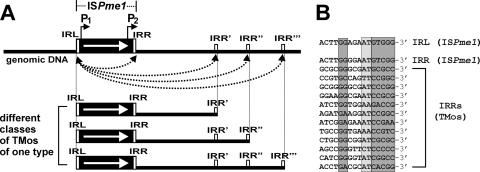
FIG. 3.
Probable mechanism of the formation of various classes of a single type of TMo. (A) The white arrows show the direction of transcription of the transposase gene (tnp). The potential secondary IRR sequences (IRR′, IRR", and IRR‴) occurring in the adjacent DNA fragments are marked. Black arrowheads and broken lines indicate the potential sites of transposase action. The localization of the P1 and P2 promoters of ISPme1 is indicated. (B) Comparison of IRL and IRR sequences of ISPme1 and IRRs of TMos captured by pMEC1. The short conserved GC- or AT-rich sequences are shown on dark gray and light gray backgrounds, respectively.
To prove that the P2 promoter can activate the transcription of downstream genes in vivo, a novel entrapment vector, pCM132TC, containing a promoterless tetracycline resistance gene, tetA, inserted within the lacZ gene of promoter probe vector pCM132 (Kmr) was constructed (see Materials and Methods for details). pCM132TC was transferred into P. methylutens DM12, assuming that the transposition of ISPme1 upstream of the tetA gene might initiate its expression, resulting in tetracycline resistance in cells carrying such mutated plasmids. As anticipated, Tcr clones were obtained (at a frequency of 5 × 10−7), four of which (randomly selected) were analyzed in detail. Only one of the tested pCM132TC derivatives appeared to carry ISPme1 alone, inserted 713 bp upstream of the ATG start codon of the tetA gene. In the other clones, we found TMos inserted in an orientation enabling the activation of transcription of the resistance gene from P2 of ISPme1. However, the possibility that the 3′-end regions of the TMos contained additional promoter sequences cannot be excluded. Two of the transposed TMos were of type D [TMo(ISPme1)D7 (742-bp-long 3′-end region) and TMo(ISPme1)D6 (177-bp-long 3′-end region)], and one was of type C [TMo(ISPme1)C6 (725-bp-long 3′-end region)] (Fig. 1). The TMos were inserted 318 bp, 73 bp, and 234 bp, respectively, upstream of the ATG start codon of the tetA gene. Interestingly, TMo(ISPme1)C6 was identical to the element previously “captured” by pMEC1.
DISCUSSION
In this study, we have demonstrated mobilization by a single-copy IS (ISPme1) of segments of genomic DNA adjacent to the IS primary target site. These ISPme1-mediated TMos were identified in P. methylutens DM12 by transposition into entrapment vector pMEC1. We have shown that the transposition of these elements is not a rare phenomenon but occurs in strain DM12 even more frequently than the transposition of the ISPme1 itself.
The DNA content of TMos depends on the location of ISPme1 in the host genome, and their sizes are highly variable. For these reasons, the genetic information carried by TMos is much more diverse than is the case for regular transposons. Additionally, we showed the possibility of the in vivo generation of elements, which contain hybrid 3′-end regions, composed of DNA segments captured from the primary and then secondary target sites. Such hybrid TMos can potentially contain combinations of genes captured from different locations in the genome, which (when transposed into other mobile elements, e.g., plasmids) may be propagated by lateral gene transfer.
We found that the outwardly directed P2 promoter of ISPme1, which is functional in Paracoccus spp. (Alphaproteobacteria) and in E. coli (Gammaproteobacteria), is able to activate the transcription of downstream genes. Therefore, this broad-host-range promoter can ensure efficient transcription of TMo-encoded genes in evolutionarily distinct hosts. It is noteworthy that within TMos identified in P. methylutens, several of the “passenger” genes are placed in an orientation to enable their transcription from the P2 promoter (Fig. 1). This promoter is located close to the IRR of ISPme1; therefore, it should promote gene expression even in truncated or disrupted elements, which have lost their transposition activities. This is the case for two TMos located in plasmid pMTH1 [TMo(ISPme1)A7 and TMo(ISPme1)A8], which contain only the terminal parts of ISPme1 together with the intact P2 promoter.
Based on a comparative analysis, ISPme1 has been classified within the IS1380 family, which is comprised of only 17 elements (ISFinder database). Although little is known about transposition of these elements, it has been shown that one of them (ISEcp1) is able to transfer an adjacent β-lactamase gene (blaCTX-M) by transposition in a manner similar to that of ISPme1 (19, 28). In addition, detailed inspection of the nucleotide sequence surrounding another member of the IS1380 family (IS1247) identified a putative TMo, which has been inserted into the conserved ereA2 gene cassette of an integron harbored by plasmid pMPDHA of Klebsiella oxytoca (30). This predicted TMo (3.9 kb) is bordered by 4-bp DRs and contains IS1247 (placed analogously to ISPme1 in TMos of P. methylutens) followed by two ORFs coding for a putative aminoglycoside acetyltransferase and rifampin ADP-ribosyl-transferase, respectively (30). Another example comes from studies described previously by van der Ploeg et al. (29), who fortuitously identified large inserts of chromosomal origin transposed into residing plasmid pPJ20 in Xanthobacter autotrophicus GJ10. Sequencing of the termini of these elements (6 to 10.5 kb) revealed the presence of only one terminal copy of IS1247 (IS1380 family). These putative TMos were surrounded by 4-bp DRs, representing a trace of their transposition activities.
These examples strongly suggest that the ability to generate TMos is not limited to ISPme1 but is shared by other members of the IS1380 family. When we consider that these ISs might be present in a large number of copies in the host genome (e.g., Acetobacter pasteurianus carries approximately 100 copies of IS1380) (26), it seems highly probable that the transposition of TMos and possible TMo-mediated DNA rearrangements might be a significant factor influencing the structure of bacterial genomes. Unfortunately, none of the completely sequenced bacterial genomes is rich in IS1380-like elements. On the other hand, it should be kept in mind that unless TMos have been inserted into a highly conserved genetic context so that their termini can be defined, it is extremely difficult and often impossible to distinguish such diverse elements in bacterial genomes by classical in silico sequence analysis. For this reason, the identification of functional TEs using entrapment vectors may result in many unexpected and interesting findings even when carried out in bacteria whose genomes have been fully sequenced.
So far, the proposed molecular mechanism of generation of TMos by ISPme1 has been based on assumptions alone. We observed that after the transposition of TMos into pMEC1 (followed by a spontaneous loss of the TMo-containing entrapment vector), the hybridization patterns of the TMos were identical to those observed in wild-type strain DM12 (data not shown). This indicates that the copies of the transposed elements remain intact at their original positions within the host genome, which suggests the replicative transposition of TMos. This supposition needs to be experimentally confirmed by further detailed studies.
It is probable that TMos are generated by a process called one-ended transposition, which may produce different random endpoints at one end of the transposed element. Such a mechanism has been described for members of the IS91 family (5), which were also shown to be able to generate TMo-like elements (28). As shown by Mendiola et al. (15), the IRs of IS91 play a different role in the transposition of this element. The left IR (IRL) seems to be dispensable for transposition; therefore, derivatives of IS91 lacking the IRL were capable of efficient one-ended transposition at frequencies similar to that of wild-type IS91 (15). Similar results were obtained for IS1294 (IS91 family member), where an intact copy of the element was able to transpose genes adjacent to its IRL frequently (IS1294 transposed at a frequency of approximately 10−4, while TMo-like elements were detected at a frequency of 10−6 to 10−5). Thus, in this case, the degree of nonrecognition of the IRL by transposase was calculated to be between 1 and 10% (27). In the case of ISPme1, the calculated value was 80%, since we have shown that the generation of TMos occurs in strain DM12 more frequently than does transposition of ISPme1 itself (39% of analyzed pMEC1 mutants carried TMos, while only 10% contained ISPme1 alone).
The above-described data show that in general, the frequency of generation of TMos by ISPme1 is in agreement with the frequency of one-ended transposition events detected for the elements of the IS91 family. However, since the IS1380 and IS91 families vary in many of their properties, the mechanism for the mobilization of genomic DNA might not be common. It is possible that the ISPme1-encoded transposase might mistakenly recognize genomic sequences, which are functional analogs of the IRR of ISPme1 (Fig. 3A). In support of this hypothesis, comparative analysis of the nucleotide sequences of the 3′ termini of ISPme1 and of TMos did show some similarities, mainly in the location of short GC-rich and AT-rich regions (Fig. 3B). Moreover, in our experiments, we have “captured” several identical TMos inserted into different entrapment vectors, which strongly suggests that the IRRs of these elements might not be randomly selected by the transposase. Further experiments are needed to elucidate the mechanism of transposition of ISPme1, which is essential for understanding the generation of TMos. This is an immediate goal of our future studies.
TMos were shown to reside within the chromosome and all plasmids (including megaplasmids) of P. methylutens. It is clear that the repeated transposition of TMos of the same type into a single replicon may stimulate other structural rearrangements resulting from homologous recombination between long repeat sequences. Depending on the orientation of the inserted elements, such recombination might potentially result in the deletion or inversion of DNA segments placed between the inserted TMos. We speculate that the former process led to a loss of an approximately 10-kb region of plasmid pMTH1, which resulted in the generation of the deletion derivative pMTH4.
Plasmid pMTH1 has a very unusual structure, with 87% of its genome composed of functional and truncated TMos. As shown in Fig. 2, the genome of pMTH1 has been subjected to various insertions and deletions. As a result, the plasmid backbone is limited exclusively to replication and stabilization systems, which are absolutely necessary for its stable maintenance (25; unpublished results). These two systems are apparently the only remnants of an ancestor replicon whose structure remains a mystery. Plasmids pMTH1 and pMTH4 are spectacular examples of the significant role of TMos in shaping the structure of natural replicons of P. methylutens DM12, which may have great evolutionary implications.
In conclusion, the TMos described here may be considered to be natural IS-mediated systems for gene capture, expression, and spread, which may significantly enhance variability and consequently the adaptive and evolutionary capacities of their hosts. Their identification is a spectacular demonstration of how transposition and lateral gene transfer could have contributed to genome evolution in bacteria.
Supplementary Material
Acknowledgments
This work was supported by the State Committee for Scientific Research, Poland (grant 2 P04A 019 26).
Footnotes
Published ahead of print on 22 February 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartosik, D., J. Baj, A. A. Bartosik, and M. Wlodarczyk. 2002. Characterization of the replicator region of megaplasmid pTAV3 of Paracoccus versutus and search for plasmid-encoded traits. Microbiology 148871-881. [DOI] [PubMed] [Google Scholar]
- 3.Bartosik, D., A. Bialkowska, J. Baj, and M. Wlodarczyk. 1997. Construction of mobilizable cloning vectors derived from pBGS18 and their application for analysis of replicator region of a pTAV202 mini-derivative of Paracoccus versutus pTAV1 plasmid. Acta Microbiol. Pol. 46387-392. [PubMed] [Google Scholar]
- 4.Bartosik, D., M. Sochacka, and J. Baj. 2003. Identification and characterization of transposable elements of Paracoccus pantotrophus. J. Bacteriol. 1853753-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernales, I., M. V. Mendiola, and F. de la Cruz. 1999. Intramolecular transposition of insertion sequence IS91 results in second-site simple insertions. Mol. Microbiol. 33223-234. [DOI] [PubMed] [Google Scholar]
- 6.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 71513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandler, M., and J. Mahillon. 2002. Insertion sequences revised, p. 305-366. In N. L. Craig, M. Craigie, M. Gellert, and A. M. Lambovitz (ed.), Mobile DNA II. ASM Press, Washington, DC.
- 8.De Palmenaer, D., C. Vermeiren, and J. Mahillon. 2004. IS231-MIC231 elements from Bacillus cereus sensu lato are modular. Mol. Microbiol. 53457-467. [DOI] [PubMed] [Google Scholar]
- 9.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 777347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doronina, N. V., Y. A. Trotsenko, V. I. Krausova, and N. E. Suzina. 1998. Paracoccus methylutens sp. nov.—a new aerobic facultatively methylotrophic bacterium utilizing dichloromethane. Syst. Appl. Microbiol. 21220-236. [DOI] [PubMed] [Google Scholar]
- 11.Dziewit, L., M. Jazurek, L. Drewniak, J. Baj, and D. Bartosik. 2007. The SXT conjugative element and linear prophage N15 encode toxin-antitoxin-stabilizing systems homologous to the tad-ata module of the Paracoccus aminophilus plasmid pAMI2. J. Bacteriol. 1891983-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gay, P., D. Le Coq, M. Steinmetz, T. Berkelman, and C. I. Kado. 1985. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J. Bacteriol. 164918-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166175-176. [DOI] [PubMed] [Google Scholar]
- 14.Marx, C. J., and M. E. Lidstrom. 2001. Development of improved versatile broad-host-range vectors for use in methylotrophs and other gram-negative bacteria. Microbiology 1472065-2075. [DOI] [PubMed] [Google Scholar]
- 15.Mendiola, M. V., I. Bernales, and F. de la Cruz. 1994. Differential roles of the transposon termini in IS91 transposition. Proc. Natl. Acad. Sci. USA 911922-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mikosa, M., M. Sochacka-Pietal, J. Baj, and D. Bartosik. 2006. Identification of a transposable genomic island of Paracoccus pantotrophus DSM 11072 by its transposition to a novel entrapment vector pMMB2. Microbiology 1521063-1073. [DOI] [PubMed] [Google Scholar]
- 17.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor laboratory Press, Cold Spring Harbor, NY.
- 18.Philippe, N., J. P. Alcaraz, E. Coursange, J. Geiselmann, and D. Schneider. 2004. Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 51246-255. [DOI] [PubMed] [Google Scholar]
- 19.Poirel, L., M. F. Lartigue, J. W. Decousser, and P. Nordmann. 2005. ISEcp1B-mediated transposition of blaCTX-M in Escherichia coli. Antimicrob. Agents Chemother. 49447-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Siguier, P., J. Filee, and M. Chandler. 2006. Insertion sequences in prokaryotic genomes. Curr. Opin. Microbiol. 9526-531. [DOI] [PubMed] [Google Scholar]
- 22.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 5385-96. [DOI] [PubMed] [Google Scholar]
- 23.Solyga, A., and D. Bartosik. 2004. Entrapment vectors—how to capture a functional transposable element. Pol. J. Microbiol. 53139-144. [PubMed] [Google Scholar]
- 24.Szymanik, M., D. Bartosik, and M. Wlodarczyk. 2004. Genetic organization of the basic replicon of plasmid pMTH4 of a facultatively methylotrophic bacterium Paracoccus methylutens DM12. Curr. Microbiol. 48291-294. [DOI] [PubMed] [Google Scholar]
- 25.Szymanik, M., R. Welc-Faleciak, D. Bartosik, and M. Wlodarczyk. 2006. Replication system of plasmid pMTH4 of Paracoccus methylutens DM12 contains an enhancer. Pol. J. Microbiol. 55261-270. [PubMed] [Google Scholar]
- 26.Takemura, H., S. Horinouchi, and T. Beppu. 1991. Novel insertion sequence IS1380 from Acetobacter pasteurianus is involved in loss of ethanol-oxidizing ability. J. Bacteriol. 1737070-7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tavakoli, N., A. Comanducci, H. M. Dodd, M. C. Lett, B. Albiger, and P. Bennett. 2000. IS1294, a DNA element that transposes by RC transposition. Plasmid 4466-84. [DOI] [PubMed] [Google Scholar]
- 28.Toleman, M. A., P. M. Bennett, and T. R. Walsh. 2006. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol Rev. 70296-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Ploeg, J., M. Willemsen, G. van Hall, and D. B. Janssen. 1995. Adaptation of Xanthobacter autotrophicus GJ10 to bromoacetate due to activation and mobilization of the haloacetate dehalogenase gene by insertion element IS1247. J. Bacteriol. 1771348-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verdet, C., Y. Benzerara, V. Gautier, O. Adam, Z. Ould-Hocine, and G. Arlet. 2006. Emergence of DHA-1-producing Klebsiella spp. in the Parisian region: genetic organization of the ampC and ampR genes originating from Morganella morganii. Antimicrob. Agents Chemother. 50607-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wheatcroft, R., G. D. McRae, and R. W. Miller. 1990. Changes in the Rhizobium meliloti genome and the ability to detect supercoiled plasmids during bacteroid development. Mol. Plant-Microbe Interact. 39-17. [Google Scholar]
- 32.Williams, D. R., D. P. Macartney, and C. M. Thomas. 1998. The partitioning activity of the RK2 central control region requires only incC, korB and KorB-binding site O(B)3 but other KorB-binding sites form destabilizing complexes in the absence of O(B)3. Microbiology 1443369-3378. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.