Abstract
Type IV fimbriae are essential virulence factors of Dichelobacter nodosus, the principal causative agent of ovine foot rot. The fimA fimbrial subunit gene is required for virulence, but fimA mutants exhibit several phenotypic changes and it is not certain if the effects on virulence result from the loss of type IV fimbria-mediated twitching motility, cell adherence, or reduced protease secretion. We showed that mutation of either the pilT or pilU gene eliminated the ability to carry out twitching motility. However, the pilT mutants displayed decreased adhesion to epithelial cells and reduced protease secretion, whereas the pilU mutants had wild-type levels of extracellular protease secretion and adherence. These data provided evidence that PilT is required for the type IV fimbria-dependent protease secretion pathway in D. nodosus. It was postulated that sufficient fimbrial retraction must occur in the pilU mutants to allow protease secretion, but not twitching motility, to take place. Although no cell movement was detected in a pilU mutant of D. nodosus, aberrant motion was detected in an equivalent mutant of Pseudomonas aeruginosa. These observations explain how in D. nodosus protease secretion can occur in a pilU mutant but not in a pilT mutant. In addition, virulence studies with sheep showed that both the pilT and pilU mutants were avirulent, providing evidence that mutation of the type IV fimbrial system affects virulence by eliminating twitching motility, not by altering cell adherence or protease secretion.
Foot rot is a highly contagious and economically significant bacterial disease affecting sheep in most countries. Type IV fimbriae, extracellular proteases, and several genomic islands are potential virulence factors of Dichelobacter nodosus, the principal causative agent of ovine foot rot (3, 23, 43). Type IV fimbriae are major surface antigens of D. nodosus, and more than 30 potential orthologs of type IV fimbrial machinery genes have been identified in the D. nodosus genome (17, 35). By analyzing specific mutants constructed by allelic exchange, we have shown that the expression of the fimbrial subunit gene fimA is mediated by an RpoN-dependent PilS/PilR two-component signal transduction system (39) and that FimN, FimO, FimP, and PilE are required for fimbrial biogenesis and function (17). Previous studies have shown that the fimA gene is required for virulence in sheep (23). However, fimA mutants of D. nodosus exhibited several phenotypic changes: they were nonpiliated and defective in twitching motility, and they secreted reduced amounts of extracellular proteases. Therefore, it was not certain that the effects on virulence resulted from reduced fimbria-mediated cell adherence, loss of twitching motility, or reduced protease secretion.
Type IV fimbriae mediate twitching motility, a process that enables bacteria to move along solid surfaces (27, 52). Twitching motility involves fimbrial extension and retraction, which reflects fimbrial subunit polymerization and depolymerization events (30, 47) that involve conserved cytosolic hexameric ATPases (21, 25, 31, 45). One of these proteins, PilT, drives fimbrial retraction (33), generating forces of more than 100 pN to power twitching motility (26, 32). By contrast, the FimN protein of D. nodosus (17) or the orthologous PilB protein of Pseudomonas aeruginosa (36) acts as an antagonist of PilT by promoting fimbrial polymerization. A third ATPase, PilU, which has a high level amino acid sequence similarity to PilT, is also required for fimbrial retraction in P. aeruginosa (54). Both pilT and pilU null strains of P. aeruginosa are piliated but are not motile (53, 54).
Orthologs of PilT and PilU have been identified in other organisms, such as Neisseria gonorrhoeae and Neisseria meningitidis (38, 42, 56), as well as Pseudomonas stutzeri (15). The PilT proteins of N. gonorrhoeae and P. stutzeri are also required for twitching motility. However, in contrast to P. aeruginosa pilU mutants, pilU mutants of N. gonorrhoeae and P. stutzeri still exhibit twitching motility. Mutation of pilT leads to resistance to the fimbria-specific bacteriophage PO4 in P. aeruginosa or to a defect in DNA uptake in N. gonorrhoeae and P. stutzeri, whereas the equivalent pilU mutants still exhibit PO4 phage susceptibility and DNA uptake phenotypes (15, 38, 53, 54). Other studies have shown that P. aeruginosa PilT and PilU are required for corneal infections (60) and for virulence in a mouse model of acute pneumonia (7). Twitching motility mediated by PilT also appears to be essential for invasion by the plant pathogen Azoarcus sp. (4).
In this paper, we describe characterization of two D. nodosus genes, pilT and pilU, which encode orthologs of the P. aeruginosa PilT and PilU proteins. We used natural transformation and homologous recombination to isolate several chromosomal pilT and pilU mutants, and we describe the effects of the mutations in these mutants on twitching motility, natural competence, in vitro cell adherence, protease secretion, and virulence in sheep. Our results revealed significant differences between D. nodosus and other type IV fimbriate organisms, including Pseudomonas and Neisseria spp. In particular, we showed that PilT, but not PilU, is required for extracellular protease secretion and that twitching motility is essential for virulence.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli DH5α was used for plasmid propagation and cloning experiments and was grown at 37°C in 2×YT medium (44). D. nodosus strain VCS1703A and its derivatives were grown in a Coy anaerobic chamber (Coy Laboratory Products Inc.) as previously described (17, 23). D. nodosus strains used in the sheep virulence trial were grown on hoof agar (49). P. aeruginosa strains were grown at 37°C in Luria-Bertani (LB) medium (44).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Characteristics | Source or reference |
|---|---|---|
| E. coli DH5α | F−endA1 hsdR17(rK− mK−) thi-1 λ−recA1 gyrA96 relA1 rhoA supE44 deoR φ80dlacZΔM15 Δ(lacZYA argF)U169 | Invitrogen |
| D. nodosus strains | ||
| VCS1703A | Serogroup G, transformable virulent isolate | 23 |
| JIR3727 | VCS1703A fimA Ωtet(M) | 23 |
| JIR3770 | VCS1703A pilU Ωerm(B) (pilU1) | Natural transformation with pJIR2581 |
| JIR3771 | VCS1703A pilU Ωerm(B) (pilU2) | Natural transformation with pJIR2581 |
| JIR3880 | VCS1703A pilT Ωerm(B) (pilT1) | Natural transformation with pJIR2662 |
| JIR3881 | VCS1703A pilT Ωerm(B) (pilT2) | Natural transformation with pJIR2662 |
| JIR3898 | JIR3771 rrnA ΩpilTU+ (pilU2/pilU+) | Natural transformation with pJIR2864 |
| JIR3943 | JIR3771 rrnA ΩpilT+ | Natural transformation with pJIR3528 |
| P. aeruginosa strains | ||
| PAK | Wild type | D. Bradley, Memorial University of Newfoundland, St. John's, Canada |
| PAKpilA | PAK pilA::TcR; previously referred to as AWK | 51 |
| R364 | PAK pilT::Tn5-B21 | 53 |
| S237 | PAK pilU::Tn5-B21 | 54 |
| Plasmids | ||
| pBluescript II SK+ | Apr, lacZ+ cloning vector | Stratagene |
| pWSK29 | Apr, lacZ+ cloning vector | 50 |
| pUC18K | pUC18 SmaI ΩaphA-3, nonpolar base vector | 28 |
| pJIR1532 | pBluescript II SK+ containing tet(M) located between D. nodosus rrnA promoter and terminator | 22 |
| pJIR2549 | pBluescript II SK+ XbaI/BamHI Ω(1.4-kb PCR product containing pilU+) | Recombinant |
| pJIR2581 | pJIR2549 T4-filled NarI Ωerm(B) | Recombinant |
| pJIR2582 | pUC18K XbaI/BamHI Ω(650-bp PCR product containing 3′ fragment of pilT) | Recombinant |
| pJIR2611 | pJIR2582 T4-filled EcoRI Ω(680-bp PCR product containing 5′ fragment of pilT) | Recombinant |
| pJIR2660 | pBluescript II SK+ XhoI/BamHI Ω(1.5-kb PCR product containing pilT+) | Recombinant |
| pJIR2662 | pJIR2660 T4-filled EcoRI Ωerm(B) | Recombinant |
| pJIR2691 | pWSK29 containing tet(M) located between D. nodosus rrnA promoter and terminator | 17 |
| pJIR2864 | pJIR2691 EcoRV Ω(2.6-kb PCR product containing pilTU operon) | Recombinant |
| pJIR3528 | pJIR2691 EcoRV Ω(1.6-kb PCR product containing pilT) | Recombinant |
DNA manipulations and molecular techniques.
Unless otherwise stated, standard procedures were used for DNA manipulation and molecular techniques, as described elsewhere (17, 44). Nucleotide and amino acid sequence comparisons were performed by using the National Center for Biotechnology Information BLAST server (http://www.ncbi.nlm.nih.gov/BLAST). Amino acid sequences were aligned by using the Network Protein Sequence Analysis server provided by Pole Bioinformatique lyonnais (http://npsa-pbil.ibcp.fr). Reverse transcriptase PCR (RT-PCR) and quantitative RT-PCR were performed as described previously (17, 39). The sequences of synthetic oligonucleotides are shown in Table S1 in the supplemental material.
Mutant construction and confirmation.
To construct a pilT suicide vector, a 1.5-kb fragment containing the pilT gene was amplified from VCS1703A chromosomal DNA with primers JRP1795 and JRP1628 and cloned into the XhoI/BamHI sites of pBluescript II SK+ to form pJIR2660. The suicide plasmid pJIR2662 was constructed by inserting a 1.1-kb SmaI/EcoRI fragment that contained an erythromycin resistance gene, erm(B), into the T4 polymerase-filled EcoRI site of pJIR2660. In this construct, the erm(B) gene was flanked on each side by 750-bp fragments of the contiguous D. nodosus sequence. To construct a nonpolar pilT suicide vector, a 650-bp PCR product that was amplified with primers JRP1644 and JRP1645 and contained the 3′ end of pilT was cloned into XbaI/BamHI sites of pUC18K, which carries a nonpolar cassette that contains the kanamycin resistance gene aphA-3 (28). The 680-bp 5′ pilT fragment then was amplified using primers JRP1642 and JRP1643 and inserted into the T4 polymerase-filled EcoRI site of the resultant plasmid, pJIR2582, to obtain the nonpolar pilT suicide vector pJIR2611.
To construct a pilU suicide vector, a 1.4-kb PCR fragment containing the pilU gene was amplified with primers JRP1627 and JRP1629 and cloned into the XbaI/BamHI sites of pBluescript II SK+ to construct plasmid pJIR2549. The SmaI/EcoRI erm(B) cassette was cloned into the T4 polymerase-filled NarI site of pJIR2549 to form the pilU suicide vector pJIR2581. There was a 750-bp fragment upstream and a 650-bp fragment downstream of erm(B) in pJIR2581.
Mutants (Table 1) were constructed by homologous recombination using natural transformation (23). Transformants were initially screened by determining resistance to the appropriate antibiotics and then performing a capillary PCR analysis (17). Potential mutants were further analyzed by PCR using 16S rrnA gene-, relevant antibiotic resistance gene-, and pilT- and pilU-specific primers (see Table S1 in the supplemental material). Southern hybridization analysis and omp1 PCR-restriction fragment length polymorphism analysis were performed as described previously (14, 17, 22).
Natural transformation and complementation of pilU mutants.
Plasmids pJIR1532 and pJIR2691 (Table 1), which are suicide vectors containing the tet(M) tetracycline resistance gene flanked by the D. nodosus rrnA promoter and terminator regions, were used to test the abilities of the mutants to undergo natural transformation (17). To complement the pilU mutation, a 2.6-kb PCR product produced with primers JRP1795 and JRP1627 and containing the wild-type pilTU region was cloned into the EcoRV site of pJIR2691 to construct the pilU complementation plasmid pJIR2864, which was subsequently introduced into the pilU mutants JIR3770 and JIR3771 by natural transformation. Complementation derivatives were selected on medium containing both kanamycin and tetracycline and confirmed by PCR analysis using the pilU primers JRP1627 and JRP1629 and the tet(M) primers JRP1138 and JRP1139. Plasmid pJIR3528, which contained the wild-type pilT gene, was constructed by cloning a 1.6-kb PCR product generated with primers JRP1795 and JRP1645 into the EcoRV site of pJIR2691 and was introduced into the pilU mutant JIR3771 to construct a pilT-complemented pilU strain. The resultant derivatives were selected and confirmed using the methodology described above.
Biological analysis of mutants.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western immunoblotting analysis were performed with whole-cell extracts of D. nodosus strains as described previously (17, 23). Twitching motility assays on 1% TAS agar with stab-inoculated cultures were carried out as described previously (23). Caseinase activity was detected qualitatively on EYE agar plates containing 2% skim milk powder, and elastase activity was detected on 0.3% elastin agar (48). Total protease activity was subsequently determined quantitatively using azocasein (Sigma) as the substrate, as previously described (23). Protease zymogram analyses were carried out as previously described (24). Transmission electron microscopy was performed using 3-day TAS agar cultures as previously described (23). CHO-K1 epithelial cell adherence assays were carried out in triplicate with at least three independent D. nodosus cultures, using semiconfluent CHO-K1 cells, as previously described (39). CHO-K1 cells, grown in 24-well tissue culture plates, were washed three times with 500 μl of 37°C prewarmed phosphate-buffered saline (PBS) before addition of 500 μl of prewarmed antibiotic-free α minimal essential medium plus 0.5% mannose. D. nodosus cells were incubated for 3 days and harvested from one EYE or TAS agar plate with 2 ml of prewarmed PBS. To each well, 50 μl of a D. nodosus culture (turbidity at 600 nm, 0.4) was added, and the mixture was incubated for 6 h at 37°C anaerobically. After incubation, the cells were washed four times with prewarmed PBS and then resuspended in 100 μl of 0.1% (wt/vol) digitonin in PBS and incubated for 5 min to lyse the CHO-K1 cells. After serial dilution, the mixture was plated onto EYE agar and incubated for 5 days at 37°C in the anaerobic chamber. The number of D. nodosus colonies was then compared to the viable counts obtained for the original cultures. Adhesion ratios were calculated by comparison with wild-type levels. To obtain single colonies for still photography, cells in 3-day D. nodosus cultures were removed from agar plates with 2 ml of PBS, serially diluted, plated onto TAS agar, and incubated for 5 days at 37°C anaerobically.
Video microscopy.
To examine D. nodosus subsurface colony expansion microscopically, 2.5 ml of TAS agar (containing 0.9% agar [grade A; BDH]) was poured into 35-mm tissue culture petri dishes (Falcon) so that it formed a thin layer. D. nodosus strains were stab inoculated into the bottom of the petri dishes and incubated at 37°C anaerobically for 1 day to ensure that the environment in the petri dishes was anaerobic. The petri dishes then were sealed with Parafilm to maintain the humidity and incubated for 1 day for wild-type strain VCS1703A to obtain a reasonable-size twitching motility zone (diameter, ∼1.0 to 1.2 cm) and for 2 days for the pilT and pilU mutants to produce visible growth beneath the stab inoculum. To maintain anaerobic conditions, the petri dishes, without lids, were place into anaerobic incubation bags (AnaerocultP; Merck) and sealed before they were removed from the anaerobic chamber.
To examine P. aeruginosa subsurface colony expansion microscopically, 10 ml of LB medium with 1% agar (Oxoid) was poured into standard 90-mm petri dishes (BioLab) so that it formed a thin layer. P. aeruginosa strains were stab inoculated into the bottom of the petri dishes and incubated at 37°C overnight.
Bacterial movement at the agar-plastic interface was observed at 37°C with an Olympic IX71 inverted microscope equipped with phase objectives and a heated stage. Time-lapse images were recorded with either an FViewII or CC12 digital camera using AnalySIS Research software (Olympus Soft Imaging Systems). At least 10 rate measurements were obtained for each of two time-lapse experiments for each strain examined. Time-lapse images were compiled into movie files using Quicktime Pro software.
Proteomics.
The proteins present in culture supernatants derived from the wild-type and mutant strains were precipitated with trichloroacetic acid, separated by two-dimensional gel electrophoresis, and analyzed by mass spectrometry as previously described (17).
Sheep pen virulence trials.
Standard diagnostic elastase (48) and gelatin gel (37) tests were performed to characterize the strains prior to in vivo virulence testing. Virulence tests were carried out as previously described (9, 23), except that six groups of eight sheep were challenged blind with the wild-type strain VCS1703A, the pilT mutants JIR3880 and JIR3881, the pilU2 mutant JIR3771, the complemented pilU2/pilU+ strain JIR3898, and a negative control that did not contain any bacteria. All animals were examined and foot rot lesion scores were determined at the start of the trial and then at weekly intervals using the standard method (8, 55). The total weighted foot score, which is the sum of the scores for the four feet after raising scores of 3 and 4 by the power of 2, was used as an overall disease score for each animal. Blood samples were collected from the jugular vein of sheep at the start of the trial and at each time that the feet were scored. These experiments were carried out in a PC2 containment facility in accordance with the guidelines of the Australian Government Office of the Gene Technology Regulator and the Elizabeth Macarthur Agricultural Institute Animal Ethics Committee.
Histopathological examination.
The two most severely affected feet from each sheep that had gross foot rot lesions, two feet from two sheep in the negative control group, and one foot from the sheep with no gross lesions were subjected to histopathological examination. After the sheep were euthanized, the feet were gently washed in running water to remove surface debris. Each foot was then removed at the metacarpal/metatarsal-phalangeal joint with a scalpel and fixed overnight in 10% neutral buffered formalin. The two digits from each fixed foot were separated in the midline of the limb with a scalpel, and a single block of tissue was selected from each digit; this tissue extended from the interdigital skin across the hard horn of the heel just posterior to the sole in order to encompass the full extent of the grossly underrun horn in an attempt to include the leading edge of the lesion. The tissues were processed routinely for histopathological examination and embedded in paraffin. Sections (5 μm) were cut and stained with hematoxylin and eosin and Gram's stain. Sections in which bacteria with morphology suggestive of D. nodosus were present were mounted on Superfrost plus adhesive glass slides and labeled with antibody against D. nodosus.
RESULTS
pilT gene is essential for twitching motility, natural transformation, adherence to CHO-K1 cells, and protease secretion, whereas pilU is essential only for twitching motility.
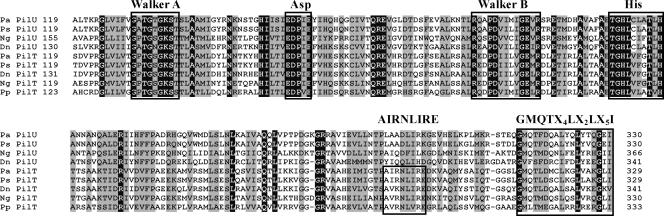
The D. nodosus type G strain VCS1703A used in this study was isolated from an outbreak of virulent ovine foot rot, is naturally transformable, and produces virulent foot rot in sheep (23). Analysis of the genome sequence of VCS1703A (35) revealed the presence of two putative genes encoding proteins with sequence identity to PilT (69%) and PilU (38%) from P. aeruginosa. The products of these D. nodosus genes had 31% identity and contained conserved Walker box A and B nucleotide-binding motifs and the aspartate and histidine boxes that are conserved in GspE-like ATPases (2). Their C-terminal domains contained a GMQTX4LX2LX5I motif, which is conserved in PilT and PilU proteins (38), and the PilT protein also had a PilT-specific AIRNLIRE motif (Fig. 1), which is required for PilT function (2). These data suggested that the two genes from the D. nodosus genome sequence that were identified encoded typical PilT- and PilU-like proteins that were likely to be part of the type IV fimbrial system.
FIG. 1.
Amino acid sequence alignment of the conserved regions of PilT and PilU proteins. Identical residues are indicated by a black background, similar residues are indicated by a gray background, and conserved domains are enclosed in boxes. The alignment starts with the Walker A box, spans the Asp box, the Walker B box, the His box, and the AIRNLIRE motif in PilT, and is truncated at the end of the GMQTX4LX2LX5I motif 3 (Pseudomonas putida PilT) to 65 (D. nodosus PilU) amino acid residues from the carboxy termini of the proteins. The sequences of P. aeruginosa PilU (Pa PilU) (accession no. AAG03785), P. stutzeri PilU (Ps PilU) (accession no. CAB56296), N. gonorrhoeae PilU (Ng PilU) (accession no. YP_208935), D. nodosus PilU (Dn PilU) (accession no. CP000513), P. aeruginosa PilT (Pa PilT) (accession no. AAG03784), P. stutzeri PilT (Ps PilT) (accession no. CAB56295), D. nodosus PilT (Dn PilT) (accession no. CP000513), N. gonorrhoeae PilT (Ng PilT) (accession no. AAB30824), and P. putida PilT (Pp PilT) (accession no. AAN70658) were aligned by using the CLUSTAL W program at the NPS@ server.
The D. nodosus pilT and pilU genes were tandemly arranged, with 12 nucleotides between the pilT termination codon and the pilU initiation codon. RT-PCR carried out using multiple primer pairs spanning the pilT and pilU region (see Table S1 in the supplemental material) confirmed that the pilT and pilU genes comprised an operon (data not shown).
To determine the function of PilT and PilU in D. nodosus, insertionally inactivated double-crossover mutants for each gene were constructed and confirmed by PCR and Southern hybridization (data not shown). Since disruption of pilT may have polarity effects on the downstream pilU gene, both nonpolar and polar pilT mutants were constructed.
To see if mutation of pilT had polar effects on pilU expression, RT-PCR was carried out using two different primer pairs (JRP1629/JRP1918 and JRP1917/JRP1918 [see Table S1 in the supplemental material]). Both products were observed in the wild-type strain and in both the nonpolar and polar pilT mutants but not in a pilU mutant (data not shown). Therefore, it appears that none of the pilT mutations disrupted the transcription of pilU; in the polar mutants pilU must be expressed either as a result of read-through from the inserted erm(B) gene or from a secondary promoter. In further studies we focused on two independently derived erythromycin-resistant pilT mutants, JIR3880 (pilT1) and JIR3881 (pilT2).
In a similar manner, transformation of strain VCS1703A with the pilU suicide vector pJIR2581 led to the production of erythromycin-resistant pilU mutants. PCR analysis and Southern hybridization confirmed that these mutants resulted from double-crossover events, and RT-PCR analysis confirmed that the two independently derived pilU mutants chosen for further analysis, JIR3770 (pilU1) and JIR3771 (pilU2), did not produce pilU-specific transcripts (data not shown).
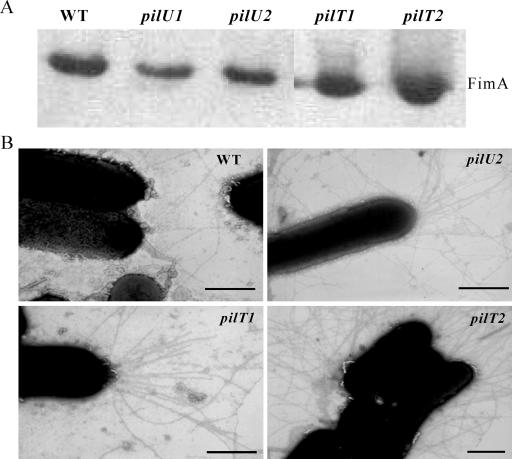
To determine if the pilT and pilU mutants still produced the fimbrial subunit protein FimA (18, 22), Western immunoblotting using antisera raised against serogroup G fimbriae was performed using whole-cell lysates of the wild-type strain and the mutants. The results showed that like the wild-type strain, the pilT and pilU mutants produced the 17-kDa FimA protein (Fig. 2A). Transmission electron microscopy confirmed that type IV fimbriae were produced by both types of mutants (Fig. 2B), indicating that in D. nodosus PilT and PilU are not required for fimbrial biogenesis. These cells did not have increased amounts of either surface fimbriae or surface-exposed FimA subunits (data not shown).
FIG. 2.
Fimbrial analysis of pilT and pilU mutants. (A) Samples (5 μl) of whole-cell lysates of wild-type strain VCS1703A (WT), the pilU mutants JIR3770 (pilU1) and JIR3771(pilU2), and the pilT mutants JIR3880 (pilT1) and JIR3881 (pilT2) were separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the production of FimA was determined by Western immunoblotting with fimbrial antisera at a 1:1,000 dilution. (B) Transmission electron microscopy of D. nodosus cells. Three-day-old cultures of wild-type strain VCS1703A and the pilU2, pilT1, and pilT2 mutants were removed from TAS agar with PBS, negatively stained, and analyzed by transmission electron microscopy. The profile of the pilU1 mutant appeared to be identical to that of the pilU2 mutant. Bars = 1 μm.
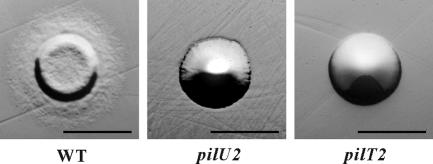
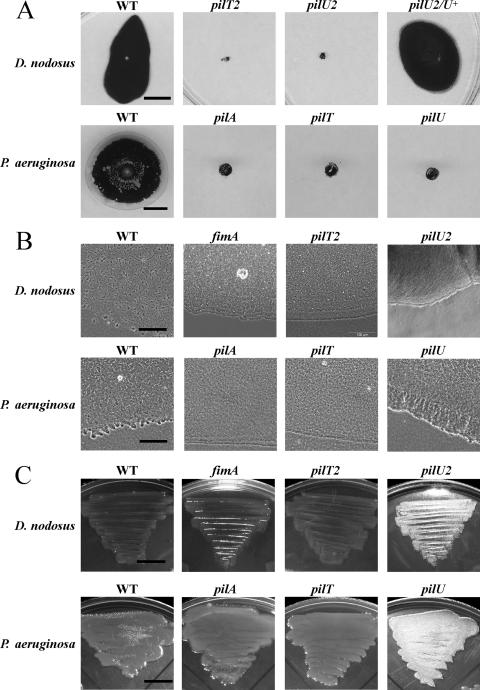
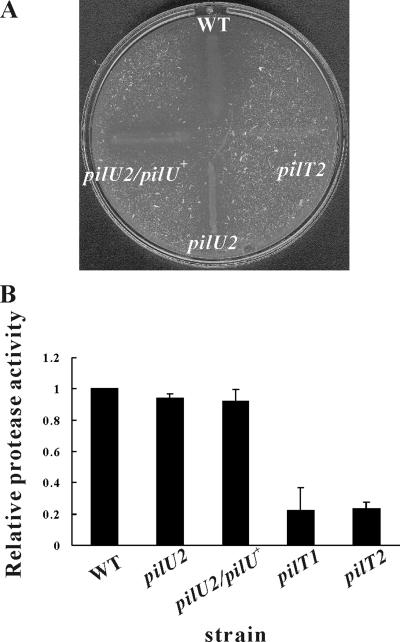
Although there was a discernible difference in colony morphology between the pilT and pilU mutants, unlike the diffuse colony morphology observed for the wild-type strain, these mutants had nonspreading colonies with smooth edges (Fig. 3), indicating that they may be defective in twitching motility. Analysis of the mutants using an agar stab twitching motility assay (23) showed that unlike the wild-type strain, neither the pilT nor pilU mutants exhibited the spreading zones typical of twitching motility (Fig. 4A). Based on these observations, we concluded that both the PilT and PilU proteins are essential for twitching motility in D. nodosus.
FIG. 3.
Colony morphology of pilT and pilU mutants. Colonies of the wild-type strain VCS1703A (WT), the pilU2 mutant JIR3771 (pilU2), and the pilT2 mutant JIR3881 (pilT2) grown on EYE blood agar plates are shown. Bars = 1 mm.
FIG. 4.
Loss of twitching motility in pilT and pilU mutants. (A) Twitching stab assays were carried out on 1% TAS agar with stab-inoculated D. nodosus cultures that were incubated for 5 to 7 days under anaerobic conditions or on 1% LB agar with stab-inoculated P. aeruginosa cultures that were incubated aerobically overnight. Twitching motility was indicated by a large dark zone after staining with Coomassie brilliant blue. Bars = 1 cm. (B) Light microscopy of twitching stab zones of D. nodosus and P. aeruginosa strains. D. nodosus cultures were stab inoculated into 1% TAS agar and incubated anaerobically at 37°C for 1 to 2 days. P. aeruginosa cultures were stab inoculated into 1% LB agar and incubated aerobically overnight. Bars = 100 μm. (C) Agar plate morphology of D. nodosus and P. aeruginosa cultures. D. nodosus strains were patched onto TAS agar and incubated anaerobically at 37°C for 3 days. P. aeruginosa strains were patched on LB agar and incubated aerobically at 37°C overnight. Bars = 1 cm. The D. nodosus strains used were wild-type strain VCS1703A (WT), the pilU2 mutant JIR3771, the pilU2/pilU+ strain JIR3898, the pilT2 mutant JIR3881, and a fimA mutant (JIR3727). The results obtained for the D. nodosus pilT1 and pilU1 mutants were not different than the results shown here for the pilT2 and pilU2 mutants. The P. aeruginosa strains used were wild-type strain PAK (WT), the pilA mutant PAKpilA, the pilT mutant strain R364, and the pilU mutant S237.
To determine the effect of pilT and pilU mutations on natural transformability, transformation experiments were carried out with the wild-type strain and the pilT and pilU mutants, using the rrnA-specific D. nodosus suicide vectors pJIR1532 and pJIR2691 (Table 1), which carry a tet(M) tetracycline resistance cassette. Natural transformation of wild-type strain VCS1703A with these plasmids consistently produced low numbers of tetracycline-resistant colonies (average, ca. 8 × 101 transformants/μg DNA) that resulted from homologous recombination between the suicide vectors and one of the three chromosomal rrnA operons. However, despite eight attempts, no transformants were obtained with either of the pilT mutants. Therefore, we concluded that the PilT protein was essential for natural transformation in D. nodosus. By contrast, transformation of the pilU mutants did produce some transformants (average, ca. 8 × 10−1 transformant/μg DNA).
Natural transformation is the only gene transfer system available for genetic manipulation in D. nodosus; it is not yet possible to introduce recombinant plasmids by either electroporation or conjugation. Therefore, since a functional natural transformation system is a prerequisite for the complementation of D. nodosus chromosomal mutants (17, 23), it was not possible to complement the pilT mutants by introducing a wild-type copy of the pilT gene into the chromosome. Consequently, to rule out the possibility that the phenotypic changes observed with the pilT mutants were the result of mutations at other sites in the genome, in subsequent studies both of the independently derived pilT mutants, JIR3880 (pilT1) and JIR3881 (pilT2), were analyzed in all subsequent studies, including virulence studies with sheep.
An attempt was made to complement a pilU mutation by natural transformation, even though the transformation frequency was at the limit of detection. A 2.6-kb PCR product encompassing the entire pilTU operon, including the potential promoter region, was cloned into the EcoRV site of pJIR2691 to construct the pilU complementation plasmid pJIR2864. Allelic exchange between this plasmid and an rrnA operon would introduce the intact pilTU region into the chromosome and thereby complement the pilU mutation. Transformation of JIR3770 and JIR3771 with pJIR2864 and selection for erythromycin resistance (original pilU mutation) and tetracycline resistance (complementation cassette) led to the isolation of several independently derived complemented pilU/pilU+ derivatives after approximately 10 separate experiments. PCR analysis confirmed the predicted genotype of four of these derivatives. Colony morphology differences between the pilU mutants and the pilU/pilU+ strains were readily discernible, and the complemented strains had diffuse, spreading wild-type colonies (data not shown). Further studies demonstrated that twitching motility was restored in the complemented mutants (Fig. 4A). One of the pilU/pilU+ strains, JIR3898, which was a derivative of JIR3771, was chosen for further analysis. To exclude the possibility that the duplicated pilT gene in JIR3898 may complement the pilU mutation, a pilT-complemented pilU strain JIR3943, which contained two copies of pilT, was constructed and characterized. The results showed that JIR3943 had the same characteristics as JIR3771 (data not shown). Specifically, the twitching motility defect which was observed in the pilU mutant JIR3771 and which was complemented by the wild-type pilTU operon was not complemented by just the wild-type pilT gene.
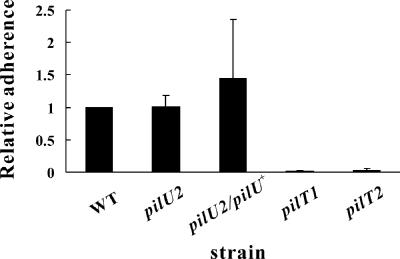
D. nodosus type IV fimbriae are required for adherence to epithelial CHO-K1 cells (39). In N. meningitidis (42) and N. gonorrhoeae (29), pilT mutants adhere to epithelial cells, whereas N. gonorrhoeae pilU mutants show increased adherence (38). By contrast, P. aeruginosa pilT or pilU mutants show reduced adherence to some epithelial cell types (7) but not to other types (60). The results of D. nodosus adherence assays showed that both pilT mutants, JIR3880 (pilT1) and JIR3881 (pilT2), had a reduced ability to adhere to the CHO-K1 cells in vitro (P < 0.05, Student's t test) compared to the wild-type strain (Fig. 5). The adhesion assays revealed phenotypic differences between the pilT and pilU mutants; the pilU mutant and the pilU/pilU+ strain adhered to CHO-K1 cells at the wild-type level (Fig. 5). Based on these results, we concluded that notwithstanding the fact that mutation of the pilT gene and mutation of the pilU gene had similar effects on fimbria-mediated twitching motility, unlike PilT, PilU did not play a significant role in fimbria-mediated epithelial cell adhesion.
FIG. 5.
Adherence of the pilT and pilU mutants to CHO-K1 epithelial cells. The wild-type strain VCS1703A (WT), the pilU2 mutant JIR3771, the pilU2/pilU+ strain JIR3898, and the pilT mutants JIR3880 (pilT1) and JIR3881 (pilT2) were incubated with CHO-K1 monolayer cells for 6 h anaerobically. The monolayers were then washed, and cell-associated bacteria were quantified. Relative adherence is expressed as the ratio of the mutant value to the wild-type value. Means and standard deviations from at least three independent experiments are shown.
Previous studies showed that mutation of the fimA gene (23), the pilR and rpoN genes (39), and the fimN, fimO, fimP, and pilE genes (17) resulted in a reduced ability to secrete extracellular serine proteases. To examine the effect of mutation of pilT and pilU on protease secretion, an agar plate elastase test, which has been routinely used as a diagnostic foot rot protease test, and caseinase assays were performed initially. The results showed that the wild-type strain and the pilU mutants were elastase positive after 7 days of incubation, whereas both pilT mutants were elastase negative even after 28 days of incubation (Fig. 6A). Furthermore, the pilT mutants, but not the pilU mutants, exhibited greatly reduced caseinase activity on 1% skim milk agar plates compared to the wild-type strain (data not shown). These results suggested that the pilT mutants did not produce wild-type levels of extracellular protease. Quantitative protease assays were subsequently carried out with culture supernatants, using azocasein as the substrate (23). The results were consistent with the initial observations, with the pilT mutants having significantly lower levels of extracellular protease activity (P < 0.05, Student's t test) than the wild-type strain (Fig. 6B) and the pilU mutants having wild-type activity levels. Once more a significant phenotypic difference between the pilT and pilU mutants was demonstrated, and we concluded that unlike PilT, PilU does not play a significant role in fimbria-mediated protease secretion.
FIG. 6.
Extracellular protease activity of pilT and pilU mutants. The strains used were the wild-type strain VCS1703A (WT), the pilU2 mutant JIR3771, the pilU2/pilU+ strain JIR3898, and the pilT mutants JIR3880 (pilT1) and JIR3881 (pilT2). (A) Qualitative elastase assay on TAS agar containing 0.3% (wt/vol) elastin incubated for 28 days. The particulate matter in the agar was the insoluble elastin substrate. Elastase activity was indicated by a zone of clearing around the colony streak. The zone for the pilT1 mutant JIR3880 was identical to that for JIR3881. (B) Quantitative protease activity assay. The protease activities of culture supernatants of bacteria were determined in triplicate using azocasein as the substrate. The relative protease activity is expressed as the ratio of the mutant value to the wild-type strain value. Means and standard deviations from at least three independent experiments are shown.
To determine if the reduction in extracellular protease activity in the pilT mutants was due to a defect in secretion or to down-regulation of protease gene expression, quantitative real-time RT-PCR experiments were performed with mRNA molecules derived from each of three genes encoding the closely related extracellular proteases AprV2, AprV5, and BprV (17). The results showed that for each gene there was no significant difference between the wild-type strain and the pilT mutants (data not shown), suggesting that the pilT mutations had no effect on protease gene expression and that PilT was required for extracellular protease secretion. Protease zymogram analysis was also carried out with culture supernatants from these isogenic strains. The results showed that each of the pilT mutants had greatly reduced AprV2 and AprV5 activities (data not shown). The basic protease BrpV cannot be detected by this method.
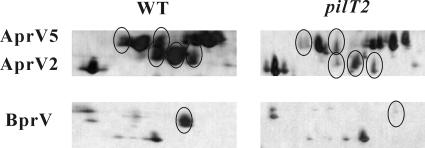
Comparative proteomic analysis coupled with mass spectrometry was used to identify proteins affected by the pilT and pilU mutations. Analysis of the extracellular proteome revealed that the intensities of only six protein spots (Fig. 7 and data not shown), all of which were variants of AprV2, AprV5, and BprV, were lower in the pilT mutants than in the wild-type strain, resulting in a profile similar to that of the previously described fimP mutants (17).
FIG. 7.
Reduced secretion of the D. nodosus proteases. Trichloroacetic acid-precipitated culture supernatants (35 μg protein per sample) were analyzed by two-dimensional gel electrophoresis. Sections of representative gels for the wild-type strain VCS1703A (WT) and the pilT2 mutant JIR3881 are shown. Reduced protein spots for AprV5, AprV2, and BprV are circled.
Finally, we observed that after extended passage the cell surface fimbriae of the pilT mutants were lost and that these derivatives did not produce FimA. We analyzed two nonfimbriated derivatives of pilT mutants and showed that they had mutations in either pilR or pilS, which are regulatory genes required for FimA expression (39). Note that in all of the phenotypic studies of the pilT mutants reported here, the pilT cultures were confirmed to produce fimbriae and therefore not to have undergone secondary mutations that affect FimA production.
Microscopic analysis of twitching motility in pilT and pilU mutants of D. nodosus and P. aeruginosa.
Microscopic analysis showed that in stab-inoculated cultures of the wild-type strain many individual cells were present at the edge of the twitching motility zone, whereas the pilT, pilU, and fimA mutants had no obvious twitching motility zone. With the pilT and fimA mutants there were very few or no individual cells past the edge of the colony, whereas the pilU mutants produced thick, dense subsurface colonies in which individual cells extruded past the colony edge more frequently (Fig. 4B). To rule out the possibility that these individual motile cells were evidence of secondary mutations, they were subcultured, and the resultant cultures were shown to be identical to the original pilU cultures (data not shown).
Based on these results, the results of the protease secretion experiments, and previous evidence showing that the type IV fimbrial system of D. nodosus represents the only type II secretion system in this bacterium (17, 35), it was postulated that in the pilU mutants there must be sufficient PilT-mediated fimbrial retraction to allow subsequent protease secretion with the next round of fimbrial polymerization but not enough retraction to observe twitching motility-mediated colony expansion. Time-lapse video microscopy showed that within 2 h the leading edge of the wild-type VCS1703A colony, which was composed of individual cells, moved away from the original inoculum at an average rate of 2.6 ± 0.2 μm per min (see Video S1 in the supplemental material). However, no obvious cellular movement was observed for the pilT mutants (see Video S2 in the supplemental material) or pilU mutants (see Video S3 in the supplemental material) over 4 h. However, on closer examination, there did appear to be some slight movement of individual cells at the periphery of a pilU mutant stab colony (see Video S3 in the supplemental material). Due to the constraints involved in maintaining anaerobic conditions under the microscope, it was not possible to analyze the movement for a longer time period.
The surface colony morphologies of wild-type D. nodosus and P. aeruginosa strains and mutants with mutations in the pilin subunit genes (fimA or pilA), pilT, and pilU were strikingly similar, including the previously reported (6) matte appearance of pilU mutants when they were grown on the surface of nutrient agar compared with the glossy appearance of wild-type, pilin subunit, or pilT mutants (Fig. 4C). Twitching motility stab assays revealed similar gross colony expansion phenotypes at the agar-plastic interface for the D. nodosus and P. aeruginosa wild-type strains and pilT, pilU, or pilin subunit mutants (Fig. 4A). Microscopic analysis of subsurface colony stabs showed that both wild-type strains produced groups of cells at the leading edge of the expanding colony, followed by a loose network of cells, whereas the nontwitching pilin subunit (fimA or pilA) and pilT mutants produced colonies with smooth edges (Fig. 4B). In addition, the pilU mutants of both D. nodosus and P. aeruginosa produced thick subsurface colonies with a distinctive fringe and evidence of some migration of cells beyond the leading edge (Fig. 4B).
Since P. aeruginosa is significantly more motile, we reasoned that we would be more likely to detect type IV fimbria-mediated cellular movements with a P. aeruginosa pilU mutant than with a D. nodosus pilU mutant. We found that in P. aeruginosa wild-type strain PAK twitching motility-mediated colony expansion was extremely rapid and occurred at a rate of 10.9 ± 1.6 μm per min (see Video S4 in the supplemental material), which was approximately four times faster than wild-type D. nodosus colony expansion. A P. aeruginosa pilT mutant (R464) exhibited colony expansion through cell division identical to that observed with a pilA mutant (PAK pilA::TcR) (see Video S5 in the supplemental material; data not shown) at a rate of only 0.11 ± 0.03 μm per min, which was approximately 100-fold slower than the wild-type strain colony expansion. P. aeruginosa pilU mutant S237 had an overall colony expansion rate that was slightly higher than that of the the pilT or pilA mutants (0.17 ± 0.08 μm per min). Furthermore, unlike the latter mutants, this pilU mutant showed numerous cellular translocation events, particularly in the region immediately behind the outermost edge of the expanding colony (see Video S6 in the supplemental material). It appears that the P. aeruginosa pilU mutant had cellular translocation ability but that this activity was uncoordinated and did not result in significant overall expansion of the colony but rather resulted in piling up of the cells, producing a thick colony edge. These observations provide preliminary evidence that limited cellular translocation occurs in pilU mutants of P. aeruginosa, presumably through retraction of type IV fimbriae. It has been reported previously that pilU mutants of P. aeruginosa are hyperpiliated but are sensitive to type IV fimbria-specific bacteriophages, which is also consistent with the presence of retractile type IV fimbriae in these mutants (54).
Both PilT and PilU are essential for virulence in sheep.
Virulence testing with sheep was carried out with the wild-type strain VCS1703A, the pilT mutants JIR3880 and JIR3881, the pilU2 mutant JIR3771, and the complemented derivative JIR3898, using a standardized method as previously described (23). Under high-security biocontainment conditions groups of eight sheep were challenged by applying agar cultures of D. nodosus to the interdigital skin of all four feet. Over a period of 30 days, each foot was quantitatively scored for foot rot lesions at weekly intervals, and the values were converted to a total weighted foot score, which represents an objective and quantitative analysis of the severity of the foot rot lesions in a sheep (8, 55).
The results (Table 2) showed that only sheep challenged with the wild-type strain and the complemented pilU2/pilU+ strain JIR3898 contracted virulent foot rot. There was no sign of foot rot in the groups challenged with the pilT or pilU mutants at day 30 postinfection or in the negative control group, which was not challenged with D. nodosus. For ethical reasons, after 30 days the animals were treated with antibiotics to eliminate the infection and the trial was terminated. Prior to challenge, foot swabs from the sheep were cultured, and the results showed that they were all free of D. nodosus. Culturing of foot swabs taken 4 weeks after challenge resulted in D. nodosus isolates only for the sheep challenged with the wild-type and the pilU complemented strains. All 71 isolates obtained from infected sheep were identical to the wild-type strain in diagnostic serogrouping, elastase, gelatin gel, and omp1 PCR-restriction fragment length polymorphism analysis tests.
TABLE 2.
Virulence of D. nodosus mutants in sheep
| Strain | Genotype | No. of infected sheep/total no. on day 30 | No. of infected feet/total no. on day 30 | Total weighted foot score on day 21 | Total weighted foot score on day 30 |
|---|---|---|---|---|---|
| None | 0/8 | 0/32 | 0 | 0 | |
| VCS1703A | Wild type | 6/8 | 12/32 | 0.38 | 23.25 |
| JIR3880 | pilT1 | 0/8 | 0/32 | 0 | 0 |
| JIR3881 | pilT2 | 0/8 | 0/32 | 0 | 0 |
| JIR3771 | pilU2 | 0/8 | 0/32 | 0 | 0 |
| JIR3898 | pilU2/pilU+ | 5/8 | 8/32 | 0.13 | 8.63 |
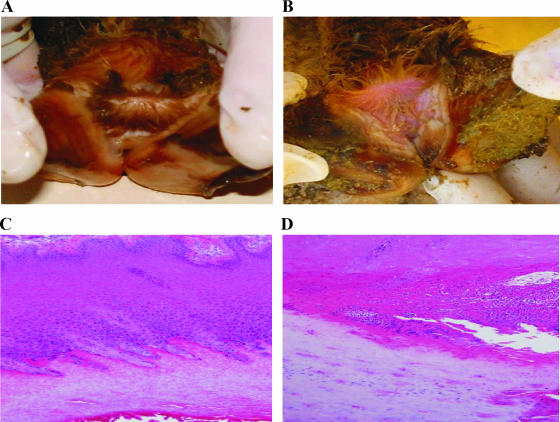
For each sheep that had gross foot rot lesions (Fig. 8B), the two most severely affected feet were subjected to histopathological examination. Two feet from two sheep from the negative control group (Fig. 8A) and one foot from each of the other groups (all with no gross lesions) were also examined. Microscopic analysis of hematoxylin- and eosin-stained sections revealed that in the feet with clinical lesions underrunning commenced at the junction between the interdigital skin and the hard horn of the hoof, usually in the outer part of the stratum granulosum (Fig. 8D). Sometimes multiple fronts of underrunning were present, and the most recent front always commenced at the interdigital area. In affected feet, the keratohyalin granules of the stratum granulosum of underrun areas were smaller and less obvious. At the end of the lesion there were few bacteria, but sometimes D. nodosus-like organisms (large gram-negative rods, often slightly curved with prominent poles) were present on the surface or within the adjacent palely stained epithelial cells, singly or in small clumps.
FIG. 8.
Pathological examination of infected sheep feet. Sections of control feet (A) and feet with typical foot rot lesions (B) were stained with hematoxylin and eosin and subjected to microscopic analysis (C and D). Underrunning occurred at the junction between the interdigital skin and the hard horn of the hoof, usually in the outer part of the stratum granulosum (D).
Although surface maceration was a consistent finding for all of the feet that were examined, no histopathological evidence of disease was apparent in sections from the control animals (Fig. 8C) or the animals infected with the pilT or pilU mutants. Based on both the gross pathological and the detailed histopathological analyses and on the fact that the effect of the pilU mutation on virulence was reversed by complementation with the wild-type pilU gene, we concluded that mutation of either the pilT or pilU genes eliminated the ability of D. nodosus to cause ovine foot rot.
DISCUSSION
D. nodosus is the essential causative pathogen of ovine foot rot, and it was previously shown that the type IV fimbrial subunit protein is essential for virulence, although it was not clear if the effects on virulence resulted from reduced fimbria-mediated cell adherence, loss of twitching motility, or reduced protease secretion (23). The results presented in this paper provide evidence that type IV fimbria-mediated twitching motility is essential for D. nodosus cells to cause ovine foot rot. Virulence studies carried out with the native ovine host showed that two independently derived pilT mutants and a pilU mutant all failed to cause disease, unlike the wild-type strain. Furthermore, complementation of the pilU mutant restored the ability to cause virulent disease. The pilT mutants had reduced abilities to secrete extracellular proteases and to adhere to epithelial cells, but these parameters could not have been responsible for the loss of virulence in these mutants since the pilU mutants had unaltered secretion and adhesion phenotypes. The common feature of the pilU and pilT mutants was their inability to undergo twitching motility even though they still produced type IV fimbriae on the surface. We postulate that the role of twitching motility in the disease process is to enable D. nodosus cells to move to a deeper location within the developing lesion, thereby increasing the probability that the cells will find a more suitable anaerobic environment for growth and extracellular protease production.
The genetic organization of the pilT and pilU genes in D. nodosus (35) is similar to that in P. aeruginosa (54), P. stutzeri (15), and N. gonorrhoeae (38), with pilU located downstream of pilT. Our results showed that the pilT gene was required for virulence, twitching motility, natural transformation, epithelial cell adherence in vitro, and protease secretion. By contrast, the pilU gene was required only for virulence and twitching motility; it was not essential for protease secretion, and it was not required for epithelial cell adherence. Orthologs of pilT have been found in several other gram-negative bacteria, and mutations in these genes are always associated with a piliated but nonmotile phenotype due to the loss of twitching motility or social gliding motility (15, 53, 56, 58). However, PilU is not always required for twitching motility; for example, pilU mutants of P. stutzeri (15) and N. gonorrhoeae (38) still undergo twitching motility or have other characteristics correlated with type IV fimbriae, such as phage sensitivity (54).
The type IV fimbrial biogenesis system consists of three closely related ATPases, designated PilB, PilT, and PilU in P. aeruginosa (52). These enzymes appear to be hexameric proteins that act as rotary molecular machines or switches, in a manner similar to F1-ATPase, to power the aggregation and disaggregation of the fimbrial subunits (19, 27, 45). It has been shown that the PilB protein (36) and its orthologs, such as FimN in D. nodosus (17) and PilF in Neisseria (12), are required for assembly of the fimbrial subunits into fimbriae and their extrusion through the outer membrane secretin PilQ. By contrast, PilT has been shown to be responsible for the opposite event, fimbrial retraction, which requires the disaggregation of the polymerized subunits (11, 57). PilT from N. gonorrhoeae has been shown to generate substantive forces (>100 pN) that drive twitching motility as part of this retraction process (26, 32). In addition, it has been shown that inactivation of pilT or pilE leads to upregulation of the antimicrobial gene mtrF and two ABC transporter operons in N. gonorrhoeae (13).
The precise biological role of the third ATPase, PilU, is not known. pilU mutants of P. aeruginosa do not exhibit normal twitching motility, and it has been postulated that PilU may cooperatively enhance the action of PilT in this organism (27). Localization of PilU to the twitching pole of the cell suggests that PilU may be required for twitching motility because it pushes the balance between PilB-mediated fimbrial extension and PilT-mediated retraction toward retraction (5, 6). By contrast, pilU mutants of N. gonorrhoeae exhibit twitching motility, and it appears that PilU is an antagonist of PilT in this species (38), acting downstream of PilT at the mechanistic level and perhaps competing for a common binding site or complex. The results obtained in this study are more consistent with the hypothesis that in D. nodosus PilU acts in concert with PilT, since we showed that although PilU is also essential for twitching motility, mutation of pilU had fewer phenotypic effects than the corresponding pilT mutation.
Comparisons of type IV fimbrial biogenesis and type II secretion systems have revealed significant similarities (10, 18, 40, 41, 46). The current model for type II secretion involves the formation of a pseudopilus, a pilus-like secreton that is formed by pseudopilins with similarity to fimbrial subunits, and the expulsion of folded proteins through an outer membrane secretin channel by cycles of extension and retraction (5, 10, 20, 40). Interestingly, no PilT or PilU orthologs have been identified in type II secretion systems, although orthologs of PilB (GspE family), which is postulated to mediate fimbrial elongation and therefore to be a functional antagonist of PilT, are present (5, 10, 20, 40). It has been postulated (17) that in D. nodosus extracellular proteases primarily are exported from the cell by the type IV fimbrial system, which also acts as the sole type II secretion system in this bacterium. This hypothesis is based on the observation that protease secretion requires the type IV fimbrial subunit protein FimA (23) and the type IV fimbrial biogenesis components FimN, FimO, FimP, and PilE (17). In addition, analysis of the D. nodosus genome sequence did not reveal any other type II secretion system (17, 35). In this paper we provide evidence that PilT is involved in both type IV fimbria-mediated twitching motility and protease secretion and that pilT mutants have greatly decreased extracellular protease activity compared to the wild-type strain, a reduction that was not due to altered transcription of the three extracellular protease genes, aprV2, aprV5, and bprV.
To explain the role of PilT in protease secretion in D. nodosus, we propose that the extracellular protease precursors bind to the inner surface of the putative PilQ outer membrane channel complex and are secreted when they are pushed through PilQ and out of the cell by the growing pseudopilus or pilus (17). It appears that in D. nodosus both PilQ and the fimbrial biogenesis apparatus are required for both fimbrial extrusion and protein secretion. Therefore, pilus or pseudopilus retraction mediated by PilT would be important to provide newly synthesized protease precursors continued access to PilQ. We suggest that since retraction does not occur in a pilT mutant, the PilQ complex is blocked by the protruded fimbriae, preventing further protease secretion. This hypothesis would explain the essential role in protease secretion of PilT, which is not needed for classical type II secretion (5, 10, 20, 40). This process would also explain our inability to transform the pilT mutants since DNA uptake presumably also requires the PilQ outer membrane channel and pilus retraction, as previously suggested for P. stutzeri (15). Note that since recent studies have suggested that PilT-mediated fimbrial retraction may be involved in the regulation of glutamine transport genes in N. gonorrhoeae (13), we cannot completely rule out the possibility that there is an alternative explanation for the dependence of protease secretion on the presence of functional type IV fimbriae that undergo PilT-dependent fimbrial retraction.
In contrast to the active role of PilT in protease secretion, PilU was not essential for this process, except for a possible effect on BprV secretion. This result is in agreement with the hypothesis that the role of PilU in D. nodosus is simply to facilitate PilT-mediated retraction. Our model for protease secretion inherently implies that although twitching motility was not observed in the pilU mutants, some limited amount of retraction must take place, sufficient for the pili or pseudopili to clear the PilQ complex and to enable a subsequent round of protease secretion. The differences in colonial morphology between the pilU and pilT mutants, the observation that more individual cells emerged from occasional flares at the growth edge of the pilU mutants, and the observation that these isolated cells did not have wild-type-like movement supported this hypothesis. In addition, the evidence obtained from our transformation studies, in which a very low level of transformation was observed in the pilU mutants but not in the pilT mutants, is also in agreement with this hypothesis.
Attempts to use video microscopy to demonstrate cellular translocation in a pilU mutant of D. nodosus were unsuccessful, presumably because of technical difficulties inherent in maintaining anaerobic conditions under the microscope for more than 4 h. However, for the first time, limited motion was detected in a pilU mutant of P. aeruginosa, and the movement was clearly distinct from the movement due to cellular division observed in pilA or pilT mutants of this aerobic bacterium. This result is in agreement with the hypothesis that there is sufficient PilT-mediated retraction in a D. nodosus pilU mutant to allow protease secretion and limited DNA uptake through the type IV fimbrial apparatus.
In N. gonorrhoeae (56) and P. stutzeri (15), disruption of pilT eliminates the ability of these organisms to undergo natural transformation, providing evidence that there is a relationship between this process and fimbrial retraction. pilU mutants of P. stutzeri are still transformable, but at a greatly reduced level, as shown here for D. nodosus. Likewise, pilU mutants of P. aeruginosa are bacteriophage susceptible, implying that phage DNA can enter the cell in these mutants (54). Finally, recent studies with N. meningitidis have shown that DNA binds to the PilQ secretin (1). These observations all support the hypothesis that in D. nodosus natural transformation requires DNA uptake that is mediated by PilT-mediated retraction of the type IV fimbriae, a process that is facilitated by PilU and involves transport through the PilQ secretin. Again, it is necessary to postulate that in a pilU mutant sufficient fimbrial retraction must occur to allow at least some DNA to enter the cell. This suggestion is supported by the protease secretion data and is consistent with previous suggestions regarding the role of PilU in other organisms (5, 15, 54).
In other bacteria type IV fimbriae mediate the initial adherence to host cells (16, 34), which has been reported to be independent of PilT (29, 42, 60). Moreover, one recent study demonstrated that PilT negatively regulates the adhesion of N. meningitidis (59). However, in contrast to these results, our study showed that the D. nodosus pilT mutants had reduced adherence to CHO-K1 cells. Similar results were reported for P. aeruginosa, where pilT and pilU mutants showed decreased adherence to three different epithelial cell lines (7). By contrast, mutation of the pilU gene of D. nodosus had no significant effect on adherence. These results may be explained by once more postulating that in such mutants there is enough fimbrial retraction to allow the extrusion of putative adhesins either with the fimbriae or onto the cell surface.
In conclusion, we demonstrated that in D. nodosus, type IV fimbria-mediated twitching motility, not adhesion or protease secretion, is the key reason why the type IV fimbriae are essential for virulence in sheep, the major host of this bacterium. We differentiated the biological effects of the PilT and PilU proteins, two well-conserved ATPases that are both required for twitching motility, and obtained evidence that PilT is also involved in protease secretion and natural transformation in D. nodosus, probably by mediating fimbrial depolymerization. These results support the hypothesis that in D. nodosus, a relatively slowly growing anaerobic bacterium with a small genome (ca. 1.4 Mb), reductive evolution has led to a situation where the type IV fimbrial biogenesis system is also responsible for extracellular protein secretion and natural transformation.
Supplementary Material
Acknowledgments
We sincerely thank Craig Kristo of the Faculty of Veterinary Science, University of Sydney, and the technical staff of the Elizabeth Macarthur Agricultural Institute for invaluable assistance with the sheep virulence trials, Khim Hoe for assistance with electron microscopy, Ian Smith, David Steer, and Shane Reeve of the Department of Biochemistry and Molecular Biology, Monash University, for help with proteomics and mass spectrometry analysis, and Steven Morton of School of Physics, Faculty of Science, Monash University, for photographic assistance.
This research was supported by the Australian Research Council through funding of the Australian Research Council Centre of Excellence in Structural and Functional Microbial Genomics. Xiaoyan Han was supported by a postgraduate scholarship provided by the Centre of Excellence. Lynne Turnbull and Cynthia Whitchurch were supported by the National Health and Medical Research Council of Australia. We thank the Rebecca L. Cooper Foundation for a financial contribution toward the purchase of the microscope.
Footnotes
Published ahead of print on 29 February 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Assalkhou, R., S. Balasingham, R. F. Collins, S. A. Frye, T. Davidsen, A. V. Benam, M. Bjoras, J. P. Derrick, and T. Tonjum. 2007. The outer membrane secretin PilQ from Neisseria meningitidis binds DNA. Microbiology 1531593-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aukema, K., E. M. Kron, T. J. Herdendorf, and K. T. Forest. 2005. Functional dissection of a conserved motif within the pilus retraction protein PilT. J. Bacteriol. 187611-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Billington, S. J., J. L. Johnston, and J. I. Rood. 1996. Virulence regions and virulence factors of the ovine footrot pathogen, Dichelobacter nodosus. FEMS Microbiol. Lett. 145147-156. [DOI] [PubMed] [Google Scholar]
- 4.Bohm, M., T. Hurek, and B. Reinhold-Hurek. 2007. Twitching motility is essential for endophytic rice colonization by the N2-fixing endophyte Azoarcus sp. strain BH72. Mol. Plant-Microbe Interact. 20526-533. [DOI] [PubMed] [Google Scholar]
- 5.Burrows, L. I. 2005. Weapons of mass retraction. Mol. Microbiol. 57878-888. [DOI] [PubMed] [Google Scholar]
- 6.Chiang, P., M. Habash, and L. I. Burrows. 2005. Disparate subcellular localization patterns of Pseudomonas aeruginosa type IV pilus ATPases involved in twitching motility. J. Bacteriol. 187829-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Comolli, J. C., A. R. Hauser, L. Waite, C. B. Whitchurch, J. S. Mattick, and J. N. Engel. 1999. Pseudomonas aeruginosa gene products PilT and PilU are required for cytotoxicity in vitro and virulence in a mouse model of acute pneumonia. Infect. Immun. 673625-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egerton, J. R., and D. S. Roberts. 1971. Vaccination against ovine foot-rot. J. Comp. Pathol. 81179-185. [DOI] [PubMed] [Google Scholar]
- 9.Egerton, J. R., D. S. Roberts, and I. M. Parsonson. 1969. The aetiology and pathogenesis of ovine footrot. I. A histological study of the bacterial invasion. J. Comp. Pathol. 79207-216. [DOI] [PubMed] [Google Scholar]
- 10.Filloux, A. 2004. The underlying mechanisms of type II protein secretion. Biochim. Biophys. Acta 1694163-179. [DOI] [PubMed] [Google Scholar]
- 11.Forest, K. T., K. A. Satyshur, G. A. Worzalla, J. K. Hansen, and T. J. Herdendorf. 2004. The pilus-retraction protein PilT: ultrastructure of the biological assembly. Acta Crystallogr. Sect. D 60978-982. [DOI] [PubMed] [Google Scholar]
- 12.Freitag, N. E., H. S. Seifert, and M. Koomey. 1995. Characterization of the pilF-pilD pilus-assembly locus of Neisseria gonorrhoeae. Mol. Microbiol. 16575-586. [DOI] [PubMed] [Google Scholar]
- 13.Friedrich, A., C. G. Arvidson, W. M. Shafer, E. H. Lee, and M. So. 2007. Two ABC transporter operons and the antimicrobial resistance gene mtrF are pilT responsive in Neisseria gonorrhoeae. J. Bacteriol. 1895399-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghimire, S. C., and J. R. Egerton. 1999. PCR-RFLP of outer membrane proteins gene of Dichelobacter nodosus: a new tool in the epidemiology of footrot. Epidemiol. Infect. 122521-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graupner, S., N. Weger, M. Sohni, and W. Wackernagel. 2001. Requirement of novel competence genes pilT and pilU of Pseudomonas stutzeri for natural transformation and suppression of pilT deficiency by a hexahistidine tag on the type IV pilus protein PilAI. J. Bacteriol. 1834694-4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hahn, H. P. 1997. The type-4 pilus is the major virulence-associated adhesin of Pseudomonas aeruginosa—a review. Gene 19299-108. [DOI] [PubMed] [Google Scholar]
- 17.Han, X., R. M. Kennan, D. Parker, J. K. Davies, and J. I. Rood. 2007. Type IV fimbrial biogenesis is required for protease secretion and natural transformation in Dichelobacter nodosus. J. Bacteriol. 1895022-5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hobbs, M., and J. S. Mattick. 1993. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol. Microbiol. 10233-243. [DOI] [PubMed] [Google Scholar]
- 19.Hopper, S., B. Vasquez, A. Merz, S. Clary, J. S. Wilbur, and M. So. 2000. Effects of the immunoglobulin A1 protease on Neisseria gonorrhoeae trafficking across polarized T84 epithelial monolayers. Infect. Immun. 68906-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson, T. L., J. Abendroth, W. G. Hol, and M. Sandkvist. 2006. Type II secretion: from structure to function. FEMS Microbiol. Lett. 255175-186. [DOI] [PubMed] [Google Scholar]
- 21.Kaiser, D. 2000. Bacterial motility: How do pili pull? Curr. Biol. 10R777-780. [DOI] [PubMed] [Google Scholar]
- 22.Kennan, R. M., S. J. Billington, and J. I. Rood. 1998. Electroporation-mediated transformation of the ovine footrot pathogen Dichelobacter nodosus. FEMS Microbiol. Lett. 169383-389. [DOI] [PubMed] [Google Scholar]
- 23.Kennan, R. M., O. P. Dhungyel, R. J. Whittington, J. R. Egerton, and J. I. Rood. 2001. The type IV fimbrial subunit gene (fimA) of Dichelobacter nodosus is essential for virulence, protease secretion, and natural competence. J. Bacteriol. 1834451-4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, D., and W. K. Yong. 1993. Use of elastase test, gelatin gel test and electrophoretic zymogram to determine virulence of Dichelobacter nodosus isolated from ovine foot rot. Res. Vet. Sci. 55124-129. [DOI] [PubMed] [Google Scholar]
- 25.Maier, B. 2005. Using laser tweezers to measure twitching motility in Neisseria. Curr. Opin. Microbiol. 8344-349. [DOI] [PubMed] [Google Scholar]
- 26.Maier, B., L. Potter, M. So, H. S. Seifert, and M. P. Sheetz. 2002. Single pilus motor forces exceed 100 pN. Proc. Natl. Acad. Sci. USA 9916012-16017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56289-314. [DOI] [PubMed] [Google Scholar]
- 28.Menard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 1755899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merz, A. J., C. A. Enns, and M. So. 1999. Type IV pili of pathogenic Neisseriae elicit cortical plaque formation in epithelial cells. Mol. Microbiol. 321316-1332. [DOI] [PubMed] [Google Scholar]
- 30.Merz, A. J., and K. T. Forest. 2002. Bacterial surface motility: slime trails grappling hooks and nozzles. Curr. Biol. 12R297-303. [DOI] [PubMed] [Google Scholar]
- 31.Merz, A. J., and M. So. 2000. Interactions of pathogenic Neisseria with epithelial cell membranes. Annu. Rev. Cell Dev. Biol. 16423-457. [DOI] [PubMed] [Google Scholar]
- 32.Merz, A. J., M. So, and M. P. Sheetz. 2000. Pilus retraction powers bacterial twitching motility. Nature 40798-102. [DOI] [PubMed] [Google Scholar]
- 33.Morand, P. C., E. Bille, S. Morelle, E. Eugene, J. L. Beretti, M. Wolfgang, T. F. Meyer, M. Koomey, and X. Nassif. 2004. Type IV pilus retraction in pathogenic Neisseria is regulated by the PilC proteins. EMBO J. 232009-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morand, P. C., P. Tattevin, E. Eugene, J. L. Beretti, and X. Nassif. 2001. The adhesive property of the type IV pilus-associated component PilC1 of pathogenic Neisseria is supported by the conformational structure of the N-terminal part of the molecule. Mol. Microbiol. 40846-856. [DOI] [PubMed] [Google Scholar]
- 35.Myers, G. S., D. Parker, K. Al-Hasani, R. M. Kennan, T. Seemann, Q. Ren, J. H. Badger, J. D. Selengut, R. T. Deboy, H. Tettelin, J. D. Boyce, V. P. McCarl, X. Han, W. C. Nelson, R. Madupu, Y. Mohamoud, T. Holley, N. Fedorova, H. Khouri, S. P. Bottomley, R. J. Whittington, B. Adler, J. G. Songer, J. I. Rood, and I. T. Paulsen. 2007. Genome sequence and identification of candidate vaccine antigens from the animal pathogen Dichelobacter nodosus. Nat. Biotechnol. 25569-575. [DOI] [PubMed] [Google Scholar]
- 36.Nunn, D., S. Bergman, and S. Lory. 1990. Products of three accessory genes, pilB, pilC, and pilD, are required for biogenesis of Pseudomonas aeruginosa pili. J. Bacteriol. 1722911-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palmer, M. A. 1993. A gelatin test to detect activity and stability of proteases produced by Dichelobacter nodosus. Vet. Microbiol. 36113-122. [DOI] [PubMed] [Google Scholar]
- 38.Park, H. S. M., M. Wolfgang, and M. Koomey. 2002. Modification of type IV pilus-associated epithelial cell adherence and multicellular behavior by the PilU protein of Neisseria gonorrhoeae. Infect. Immun. 703891-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parker, D., R. M. Kennan, G. S. Myers, I. T. Paulsen, J. G. Songer, and J. I. Rood. 2006. Regulation of type IV fimbrial biogenesis in Dichelobacter nodosus. J. Bacteriol. 1884801-4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peabody, C. R., Y. J. Chung, M. Yen, D. Vidal-Ingigliardi, A. P. Pugsley, and M. H. Saier, Jr. 2003. Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology 1493051-3072. [DOI] [PubMed] [Google Scholar]
- 41.Pugsley, A. P. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 5750-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pujol, C., E. Eugene, M. Marceau, and X. Nassif. 1999. The meningococcal PilT protein is required for induction of intimate attachment to epithelial cells following pilus-mediated adhesion. Proc. Natl. Acad. Sci. USA 964017-4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rood, J. I. 2002. Genomic islands of Dichelobacter nodosus. Curr. Top. Microbiol. Immunol. 26447-60. [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 45.Satyshur, K. A., G. A. Worzalla, L. S. Meyer, E. K. Heiniger, K. G. Aukema, A. M. Misic, and K. T. Forest. 2007. Crystal structures of the pilus retraction motor PilT suggest large domain movements and subunit cooperation drive motility. Structure 15363-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sauvonnet, N., G. Vignon, A. P. Pugsley, and P. Gounon. 2000. Pilus formation and protein secretion by the same machinery in Escherichia coli. EMBO J. 192221-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skerker, J. M., and H. Berg, C. 2001. Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. USA 986901-6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stewart, D. J. 1979. The role of elastase in the differentiation of Bacteroides nodosus infections in sheep and cattle. Res. Vet. Sci. 2799-105. [PubMed] [Google Scholar]
- 49.Thomas, J. H. 1958. A simple medium for the isolation and cultivation of Fusiformis nodosus. Aust. Vet. J. 34411. [Google Scholar]
- 50.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100195-199. [PubMed] [Google Scholar]
- 51.Watson, A. A., J. S. Mattick, and R. A. Alm. 1996. Functional expression of heterologous type 4 fimbriae in Pseudomonas aeruginosa. Gene 175143-150. [DOI] [PubMed] [Google Scholar]
- 52.Whitchurch, C. B. 2006. Type IV pili in Pseudomonas species, p. 139-188. In J. L. Ramos and R. J. Levesque (ed.), Pseudomonas, vol. IV. Kluwer Academic/Plenum Publishers, New York, NY. [Google Scholar]
- 53.Whitchurch, C. B., M. Hobbs, S. P. Livingston, V. Krishnapillai, and J. S. Mattick. 1991. Characterisation of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialised protein export system widespread in eubacteria. Gene 10133-44. [DOI] [PubMed] [Google Scholar]
- 54.Whitchurch, C. B., and J. S. Mattick. 1994. Characterization of a gene, pilU, required for twitching motility but not phage sensitivity in Pseudomonas aeruginosa. Mol. Microbiol. 131079-1091. [DOI] [PubMed] [Google Scholar]
- 55.Whittington, R. J., and P. J. Nicholls. 1995. Grading the lesions of ovine footrot. Res. Vet. Sci. 5826-34. [DOI] [PubMed] [Google Scholar]
- 56.Wolfgang, M., P. Lauer, H. S. Park, L. Brossay, J. Hebert, and M. Koomey. 1998. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol. Microbiol. 29321-330. [DOI] [PubMed] [Google Scholar]
- 57.Wolfgang, M., J. P. van Putten, S. F. Hayes, D. Dorward, and M. Koomey. 2000. Components and dynamics of fiber formation define a ubiquitous biogenesis pathway for bacterial pili. EMBO J. 196408-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu, S., J. Wu, and D. Kaiser. 1997. The Myxococcus xanthus pilT locus is required for social gliding motility although pili are still produced. Mol. Microbiol. 23109-121. [DOI] [PubMed] [Google Scholar]
- 59.Yasukawa, K., P. Martin, C. R. Tinsley, and X. Nassif. 2006. Pilus-mediated adhesion of Neisseria meningitidis is negatively controlled by the pilus-retraction machinery. Mol. Microbiol. 59579-589. [DOI] [PubMed] [Google Scholar]
- 60.Zolfaghar, I., D. J. Evans, and S. M. Fleiszig. 2003. Twitching motility contributes to the role of pili in corneal infection caused by Pseudomonas aeruginosa. Infect. Immun. 715389-5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.