Abstract
Integrating conjugative elements (ICEs) are self-transmissible mobile elements that transfer between bacteria via conjugation and integrate into the host chromosome. SXT and related ICEs became prevalent in Asian Vibrio cholerae populations in the 1990s and play an important role in the dissemination of antibiotic resistance genes in V. cholerae. Here, we carried out genomic and functional analyses of ICEPdaSpa1, an SXT-related ICE derived from a Spanish isolate of Photobacterium damselae subsp. piscicida, the causative agent of fish pasteurellosis. The ∼102-kb DNA sequence of ICEPdaSpa1 shows nearly 97% DNA sequence identity to SXT in genes that encode essential ICE functions, including integration and excision, conjugal transfer, and regulation. However, ∼25 kb of ICEPdaSpa1 DNA, including a tetracycline resistance locus, is not present in SXT. Most ICEPdaSpa1-specific DNA is inserted at loci where other SXT-related ICEs harbor element-specific DNA. ICEPdaSpa1 excises itself from the chromosome and is transmissible to other Photobacterium strains, as well as to Escherichia coli, in which it integrates into prfC. Interestingly, the P. damselae virulence plasmid pPHDP10 could be mobilized from E. coli in an ICEPdaSpa1-dependent fashion via the formation of a cointegrate between pPHDP10 and ICEPdaSpa1. pPHDP10-Cm integrated into ICEPdaSpa1 in a non-site-specific fashion independently of RecA. The ICEPdaSpa1::pPHDP10 cointegrates were stable, and markers from both elements became transmissible at frequencies similar to those observed for the transfer of ICEPdaSpa1 alone. Our findings reveal the plasticity of ICE genomes and demonstrate that ICEs can enable virulence gene transfer.
Integrating conjugative elements (ICEs) are mobile elements that can be excised from their host's chromosome, transfer via conjugation into a new host, and then reintegrate into the chromosome (8, 12). SXT is an ICE that carries multiple antibiotic resistance genes; it was originally discovered in MO10, one of the initial Vibrio cholerae O139 clinical isolates from India (35). Prior to the emergence of this novel V. cholerae serogroup, SXT was rarely if ever detected in V. cholerae O1 isolates (2). Since the emergence and spread of V. cholerae O139, ICEs closely related to the MO10 SXT (SXTMO10) have been identified in most V. cholerae O1 and O139 clinical isolates from Asia (2, 18, 20, 23). In addition, SXT-related ICEs in African V. cholerae O1 isolates (15), as well as in other vibrios (1) and in other Gammaproteobacteria (20, 33), have been described previously. SXT is genetically and functionally related to the IncJ element R391, which was derived from a South African Providencia rettgeri strain isolated in 1967. Although R391 was initially thought to be an R factor (14), subsequent studies have demonstrated that R391 is an ICE that is related to SXT (3, 6, 19). The SXT-R391 family of ICEs is now known to have at least 25 members derived from diverse gram-negative organisms and locations (7).
All members of the SXT-R391 family of ICEs encode nearly identical tyrosine recombinases (Int) that mediate the site-specific integration of the elements into prfC (7, 22). Another protein, Xis, which acts as a recombination directionality factor, is required in addition to Int for efficient SXT excision (10). The SXT conjugation genes are related to those found in plasmids pCAR1, derived from Pseudomonas resinovorans (27), and Rts1, derived from Proteus vulgaris (30). The conditions that promote SXT transfer are not fully understood, but it is known that transfer is stimulated by the host SOS response via a pathway that resembles the pathway governing the lytic development of phage lambda (5).
Comparative analyses of the genome sequences of SXTMO10 (99.5 kb) and R391 (89 kb) have revealed that these ICEs consist of a conserved set of “backbone” genes that mediate the essential functions of the elements, including their regulation, excision and integration, and conjugative transfer (3, 4, 6). There is more than 95% nucleotide sequence identity between these two elements in the ∼65 kb of DNA sequence they share. Both elements contain insertions into this backbone that confer element-specific properties, such as antibiotic resistance. SXTMO10 carries genes that mediate resistance to sulfamethoxazole, trimethoprim, chloramphenicol, and streptomycin, whereas R391 mediates resistance to kanamycin and mercury. In some cases, insertions of antibiotic resistance genes appear to have been mediated by transposons (20). Comparative sequence analyses of SXTMO10 and R391 also led to the identification of four sites that appear to correspond to hot spots for the insertion of accessory DNA into these elements (3, 7). These four sites contain totally unrelated sequences in these two ICEs; the recently described ICESpuPO1 from Shewanella putrefaciens also contains element-specific DNA in three of four of these hot spots (33). Recombination between tandem arrays of ICEs may also contribute to the generation of ICE diversity (7, 9, 11).
Juiz-Rio et al. recently reported that a Photobacterium damselae subsp. piscicida isolate derived from a diseased sole (Solea senegalensis) from a fish farm in Galicia, Spain, appears to contain an SXT-like ICE (24). They found that they could amplify five SXT backbone genes from this virulent P. damselae subsp. piscicida isolate (PC554.2) and showed that this strain's prfC locus contains SXT-related DNA. P. damselae subsp. piscicida is the causative agent of fish pasteurellosis in both wild and cultured warm-water marine fish (26, 29). The mechanisms of P. damselae subsp. piscicida pathogenicity are multifactorial and incompletely understood. One of the major virulence factors of P. damselae subsp. piscicida is the AIP56 toxin, which induces apoptosis in fish neutrophils and macrophages (17). This toxin is encoded in pPHDP10, a 9.6-kb plasmid that is present in most P. damselae subsp. piscicida strains isolated in Europe. A high-affinity siderophore-mediated iron acquisition system encoded by a genomic island similar to the Yersinia high-pathogenicity island is also involved in P. damselae subsp. piscicida virulence (32).
Here, we carried out genomic and functional analyses of the PC554.2-derived ICE. We found that this ICE, dubbed ICEPdaSpa1, is indeed closely related to SXT and R391. The ICEPdaSpa1 genome contained all of the genes known to be required for SXT transfer, excision, and integration. ICEPdaSpa1 proved to be self-transmissible and to be excised from and integrate into prfC. Remarkably, we also found that ICEPdaSpa1 could mobilize the pPHDP10 virulence plasmid via the formation of an ICEPdaSpa1::pPHDP10 cointegrate. Thus, ICEPdaSpa1 may contribute to the dissemination of virulence genes.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The strains used in this study are listed in Table 1. P. damselae subsp. piscicida strains were routinely grown at 25°C in brain heart infusion (BHI; Difco) supplemented with 1% NaCl. Escherichia coli strains were routinely grown in Luria-Bertani (LB) broth at 37°C. Bacterial strains were stored frozen at −70°C in LB broth containing 20% (vol/vol) glycerol. Antibiotics were used at the following concentrations: ampicillin, 100 mg liter−1; kanamycin, 50 mg liter−1; rifampin, 50 mg liter−1; tetracycline, 4 mg liter−1 (Photobacterium strains) or 12 mg liter−1 (E. coli strains); chloramphenicol, 20 mg liter−1; spectinomycin, 100 mg liter−1; streptomycin, 200 mg liter−1; and nalidixic acid, 40 mg liter−1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| P. damselae subsp. piscicida | ||
| PC554.2 | Virulent strain; prfC::ICEPdaSpa1 Tcr | 24 |
| DI21 | Virulent strain | 28 |
| CS31 | DI21 derivative; Rfr Knr | 32 |
| E. coli | ||
| DH5α | Cloning strain | Laboratory stock |
| XL1-Blue MR | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac | Stratagene |
| MC1061 | F−araD139 Δ(ara-leu)7696 galE15 galK16 Δ(lac)X74 rpsL (Smr) hsdR2(rK− mK+) mcrA mcrB1 | 36 |
| CAG18420 | MG1655 lacZU118 lacI42::Tn10kan | 34 |
| BI533 | MG1655 Nxr | 21 |
| BW25113 | lacIqrrnBT14 ΔlacZWJ16hsdR514 ΔaraBADAH33 ΔrhaBAADLD78 | 16 |
| VI499 | BW25113 derivative; recA | |
| CCW026 | ICEPdaSpa1 exconjugant of MC1061; Smr Tcr | This study |
| CCW01 | BW25113 prfC::ICEPdaSpa1 | This study |
| CCW02 | BW25113 prfC::ICEPdaSpa1 Δint | This study |
| CCW03 | VI499 prfC::ICEPdaSpa1 | This study |
| CCW069 | MC1061 transformed with pPHDP10-Cm; Smr Cmr | This study |
| CCW077 | CCW026 transformed with pPHDP10-Cm; Smr Tcr Cmr | This study |
| CCW088 | Nxr Tcr Cmr exconjugant from CCW077 × BI533 | This study |
| CCW129 | Tcr Cmr Apr exconjugant from PC554.2 × VI499 transformed with pPHDP10-Cm and with pGG2B; recA | This study |
| Plasmids | ||
| pSuperCos1 | Cosmid cloning vector; Apr Knr | Stratagene |
| pPHDP10 | Native virulence plasmid of P. damselae subsp. piscicida DI21 strain | 17 |
| pPHDP10-Cm | pPHDP10 with a Cmr cassette from pKD3 inserted in BamHI site; Cmr | This study |
| pKD3 | PCR template for one-step chromosomal gene inactivation; Cmr | 16 |
| pGG2B | setCD genes from SXTMO10 cloned into pBAD30; Apr | V. Burrus |
DNA preparation and manipulation.
Total genomic DNA was prepared with G Nome DNA kits (Q-Biogene). Plasmid and cosmid DNA purification and DNA extraction from agarose gels were accomplished using kits from Qiagen. Recombinant DNA manipulations were carried out by standard procedures. The TA cloning kit (Invitrogen) was used for the cloning of PCR products. The deletion of the ICEPdaSpa1 int gene was accomplished using the one-step chromosomal gene inactivation technique (16). The ECL direct nucleic acid labeling and detection system (Amersham Biosciences) was used for the Southern blot analyses.
Determination of the ICEPdaSpa1 DNA sequence.
To obtain template DNA to determine and assemble the entire ICEPdaSpa1 DNA sequence, we initially constructed a cosmid library from PC554.2 and then isolated a set of overlapping cosmids that hybridized with SXT genes. The cosmid vector SuperCos1 (Stratagene) was used to construct a library of partially Sau3AI-digested DNA isolated from P. damselae subsp. piscicida PC554.2. The cosmid library was propagated in E. coli XLI-Blue MR and screened by colony PCR using primers for the previously described ICEPdaSpa1 int, traI, traC, and traN genes (24).
Five cosmids, pCW010, pCW013, pCW017, pCW021, and pCW050, were found to span the complete element. A combination of primers previously used to sequence SXT (4), along with several new primers, enabled us to obtain the complete ICEPdaSpa1 DNA sequence. Automated DNA sequencing was carried out at the Tufts Medical School DNA Sequencing Core Facility. Vector NTI (Invitrogen) was used to assemble DNA sequences. Open reading frames (ORFs) were determined using BioEdit version 7.0.4.1. The FASTA3 and BLAST algorithms were used to assess similarities between the sequences of products of ICEPdaSpa1 DNA and putative protein sequences listed in the European Bioinformatics Institute and NCBI databases.
Bacterial conjugation.
Conjugation assays were performed by mixing equal volumes of log-phase cultures of donor and recipient strains. Cell mixtures were concentrated by centrifugation, resuspended in a 0.1 volume of LB or BHI broth, and then applied to 0.45-μm-pore-size membrane filters (Millipore) on LB or BHI agar plates. Matings were performed for 4 h at 25°C when P. damselae subsp. piscicida was used either as a donor or as a recipient; the remainder of the matings were performed at 37°C. Cells were collected in 2 ml of LB or BHI broth, and serial dilutions were plated onto the appropriate selective media to determine the numbers of donors, recipients, and exconjugants. The transfer frequency was calculated as the number of exconjugants observed per donor cell.
Real-time quantitative PCR assay for relative quantification of ICEPdaSpa1 attB.
A real-time quantitative PCR assay was used to measure the percentage of PC554.2 cells that contained unoccupied ICEPdaSpa1 attB sites. The amount of attB DNA in each sample was normalized to the amount of chromosomal DNA in the sample (determined using a real-time quantitative PCR assay for the P. damselae subsp. piscicida tonB gene). Primer design, reactions, and analysis were performed as described previously (10). Primers to amplify the attB region of P. damselae subsp. piscicida were QattBF (5′ TCACGCTAATGTCGAACAGTTATCA 3′) and QattBR (5′ GCGTTTCCGAATAATAGAACTTTTTC 3′), and primers to amplify the tonB locus were QtonBF (5′ CCAAGCCAAAACGCAATAGC 3′) and QtonBR (5′ TGACTTCTGCTTTTGCAACATCTT 3′).
Analysis of the ICEPdaSpa1 insertion site.
PCR assays were used to assess whether P. damselae subsp. piscicida CS31 or E. coli CAG18420 harbored ICEPdaSpa1 in prfC. A primer, VISLR (5′ GCTGCCATCTTTTATTCTTC 3′), that targets the ICEPdaSpa1 int gene was used in combination with a specific primer for the host prfC gene. We used primer QattBF (see above) for P. damselae subsp. piscicida prfC and primer EattBF (11) for E. coli prfC.
Construction of a marked pPHDP10.
To construct a marked version of pPHDP10, we first purified the plasmid from P. damselae subsp. piscicida strain DI21 (17). Plasmid DNA was digested with BamHI, which linearizes pPHDP10, and the 9.6-kb band was gel purified and ligated to a chloramphenicol resistance cassette obtained by PCR amplification of the cat gene from the pKD3 plasmid (16). The insertion of the cat gene into the BamHI site of pPHDP10 was confirmed by DNA sequencing. The cat-marked pPHDP10 was designated pPHDP10-Cm.
Nucleotide sequence accession number.
The DNA sequence described in this article has been deposited in the EMBL database under accession number AJ870986.
RESULTS AND DISCUSSION
General organization of the ICEPdaSpa1 genome.
Juiz-Rio et al. found that PC554.2, a highly virulent P. damselae subsp. piscicida isolate, contains homologues of SXT genes that are apparently inserted into prfC, the V. cholerae SXT integration site, and proposed that these genes are likely to be part of a P. damselae subsp. piscicida SXT-like ICE (24). We determined the complete DNA sequence of this SXT-like element (here named ICEPdaSpa1) to further comparative genomic analyses of the SXT-R391 family of ICEs. The ICEPdaSpa1 sequence is 102,985 bp in length and contains 86 putative ORFs (Fig. 1). Overall, our assembly and analysis of the complete ICEPdaSpa1 genome sequence confirmed the proposal of Juiz-Rio et al. (24) that this fish pathogen harbors an SXT-R391 family ICE.
FIG. 1.
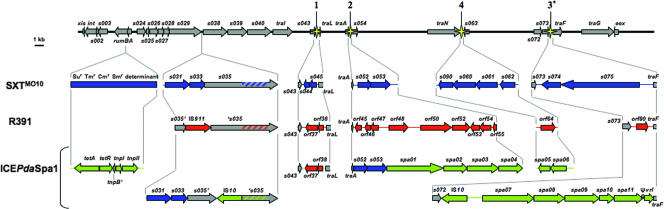
Schematic representation of portions of the ICEPdaSpa1, SXTMO10, and R391 genomes. Conserved genes are shown as gray arrows, and DNA initially identified in ICEPdaSpa1, SXTMO10, or R391 is shown in green, blue, or orange, respectively. The numbered yellow stars represent the sites of hot spots 1 to 4. Note that the scales used to represent the conserved and the nonconserved genes are different and that some conserved genes have been left out of the figure (as have regions without genes). The ICEPdaSpa1 and SXTMO10 insertions in rumB are shown in more detail in Fig. 2. The left border of hot spot 3 in ICEPdaSpa1 begins in the 3′ end of s072, as s073 is absent from this ICE. In R391 and ICEPdaSpa1, s035 is split by insertion sequence elements into two ORFs, noted as s035′ and ′s035. The striped sections in s035 and ′s035 correspond to an ICE-specific variable region.
The organization of the ICEPdaSpa1 genome proved to be very similar to those of the three other fully sequenced SXT-related ICEs, SXTMO10 (4), R391 (6), and ICESpuPO1 (33). All four ICEs consist of a highly conserved core set of genes, encompassing approximately 60 kb (Fig. 1), that mediate critical ICE functions, including regulation, integration and excision, and conjugative DNA processing and transfer, as well as variable numbers of additional genes (Fig. 1) that mediate ICE-specific properties, such as antibiotic resistance. The ICEPdaSpa1 core genes are arranged in the same order as those in the other elements and have 96 to 97% identity to the corresponding SXT and R391 sequences at the nucleotide level. Of the 86 predicted ICEPdaSpa1 ORFs, 69 are also present in SXTMO10, in R391, or in both. ICEPdaSpa1 contains all the genes that are thought to be required for element transfer and regulation (4). Interestingly, s073, a gene previously thought of as a gene common to SXT-related ICEs, is missing in ICEPdaSpa1 (Fig. 1). This gene is apparently not part of the core set of genes required for ICE transfer, since we found that ICEPdaSpa1 is capable of transfer (see below).
Seventeen ICEPdaSpa1 ORFs are reported here for the first time to be present in an SXT-related ICE (Table 2). Nearly all of the ∼25 kb of ICEPdaSpa1-specific DNA is found in the same sites that harbor element-specific DNA in other SXT-related ICEs (Fig. 1). ICEPdaSpa1 contains the same four hot spots (Fig. 1) that exhibit variable DNA sequences in SXTMO10 and R391. The two other ICEPdaSpa1 sites that harbor additional ICEPdaSpa1-specific DNA correspond to SXTMO10 sites that contain element-specific DNA.
TABLE 2.
Predicted products of ICEPdaSpa1-specific genes
| Genome region and predicted ORF or product (no. of amino acids) | Positions of ORF in ICEPdaSpa1a | Gene or protein homologue | GenBank accession no. for homologue gene | % Identity/% similarity to homologueb |
|---|---|---|---|---|
| rumB region | ||||
| TetA (400) | 11676-12878 (compl) | P. damselae subsp. piscicida tetracycline resistance protein, class D | BAA03719 | 66/78 |
| TetR (200) | 12959-13561 | E. coli Tet repressor, class D | 1A6I | 74/85 |
| TnpI (96) | 13951-14241 | Psychromonas ingrahamii IS3/IS911 transposase | ZP_01350213 | 62/80 |
| TnpII (243) | 14373-15104 | Psychromonas ingrahamii integrase | ZP_01350212 | 64/80 |
| Region between s035′ and ′s035 | ||||
| IS10 transposase (402) | 28188-29396 (compl) | Salmonella enterica serovar Typhi IS10 transposase | NP_058298 | 100/100 |
| Hot spot 2 | ||||
| Spa01 (909) | 52041-54770 | Psychromonas sp. strain CNPT3 putative helicase | ZP_01215992 | 40/60 |
| Spa02 (369) | 54767-55876 | No database matches with significant similarity | ||
| Spa03 (510) | 55889-57421 | Acidovorax sp. strain JS42 phospholipase D/transphosphatidylase | ZP_01383961 | 28/47 |
| Spa04 (377) | 57736-58869 | Desulfovibrio desulfuricans two-component transcriptional regulator, Fis family | ABB37621 | 38/61 |
| Hot spot 4 | ||||
| Spa05 (175) | 68950-69474 (compl) | No database matches with significant similarity | ||
| Spa06 (257) | 69487-70257 (compl) | Salmonella enterica hypothetical protein | AAG03008 | 38/52 |
| Hot spot 3 | ||||
| IS10 transposase (402) identical to the IS10 transposase listed above | 81904-83112 (compl) | Salmonella enterica serovar Typhi IS10 transposase | NP_058298 | 100/100 |
| Spa07 (831) | 83835-86330 | Deltaproteobacterium MLMS-1 heat shock protein Hsp70 | EAT04212 | 80/88 |
| Spa08 (489) | 86334-87803 | Deltaproteobacterium MLMS-1 AAA ATPase, central region | EAT04211 | 82/91 |
| Spa09 (553) | 87816-89477 | Deltaproteobacterium MLMS-1 hypothetical protein | EAT04215 | 68/82 |
| Spa10 (235) | 89470-90177 | No database matches with significant similarity | ||
| Spa11 (464) | 90286-91680 | Deltaproteobacterium MLMS-1 peptidoglycan-binding domain 1 | EAT04214 | 39/51 |
| ψvrlR | 91687-92181 | Desulfococcus multivorans vrlR gene | CT009609 | 88 |
Numbers are nucleotide positions. Compl, complementary strand.
Data refer to identity and similarity at the amino acid level, except the value for vrlR, which refers to the similarity at the nucleotide sequence level.
ICEPdaSpa1 antibiotic resistance genes.
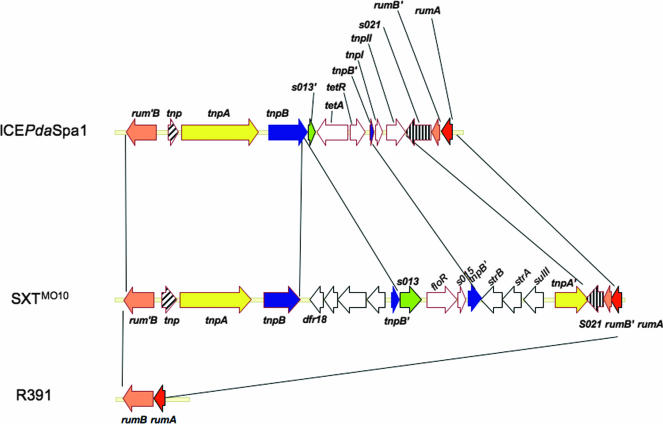
As SXTMO10, ICEPdaSpa1 contains genes that mediate resistance to antibiotics inserted into the rumB locus (Fig. 1 and 2). The rumB locus of ICEPdaSpa1 is disrupted at exactly the same base pair as that of SXTMO10, and in contrast, the R391 rumB is not disrupted (20). The sequences inserted at rumB in ICEPdaSpa1 and SXT are related. The genes at the boundaries of the insertions (tnp and s021) in the two ICEs are identical (Fig. 2). The ICEPdaSpa1-specific DNA includes a tetracycline resistance locus that comprises an active (see below) class D tetA gene, the tetR repressor gene, and two ORFs that appear to encode intact transposases, tnpI and tnpII (Fig. 2 and Table 2). ICEPdaSpa1 shares additional genetic features with SXTMO10 in this region. Interestingly, in both of these ICEs, tnpB (or a tnpB fragment) brackets antibiotic resistance genes, perhaps suggesting that this gene promotes the acquisition of antibiotic resistance genes (Fig. 2). This possibility is reinforced by the finding that tnpB or truncated tnpB fragments are closely linked to antibiotic resistance genes in other bacterial species and in large P. damselae subsp. piscicida plasmids (13, 25). Altogether, the structure of the DNA insertion in the rumB locus suggests that in this region SXT and ICEPdaSpa1 share a common evolutionary history.
FIG. 2.
Comparison of the rumB regions in ICEPdaSpa1, SXTMO10, and R391.
Gene contents of the ICEPdaSpa1 hot spots.
As those in other SXT- and R391-related ICEs (3, 9, 33), most of the none-core genes in ICEPdaSpa1 are found in four hot spots (Fig. 1) where SXT-related ICEs appear to acquire additional DNA. Hot spot 1, between s043 and traL, contains two genes, orf37 and orf38, that are also present in this location in R391 but are not found in SXTMO10 (Fig. 1). The identity of the contents of hot spots 1 in R391 and ICEPdaSpa1 suggests that these regions of these two ICEs are likely to be derived from a common ancestor. The partial identity of ICEPdaSpa1 to SXTMO10 in the rumB region and to R391 in hot spot 1 indicates that the ICEPdaSpa1 genome has a mosaic structure and provides further evidence supporting the idea that recombination events occur between ICEs belonging to the SXT-R391 family (7, 11). The content of hot spot 2 in ICEPdaSpa1 also suggests that recombination between SXT-related ICEs shaped ICEPdaSpa1's genome, as this region is composed of sequences found in SXTMO10 (s052 and s053) and sequences unique to ICEPdaSpa1 (spa01, spa02, spa03, and spa04) (Fig. 1 and Table 2).
An analysis of the boundaries of hot spot 2 revealed that ICEPdaSpa1, SXTMO10, and R391 share the same nucleotide sequence up to the traA stop codon (Fig. 3). At the right end of hot spot 2, downstream of spa04, ICEPdaSpa1 lacks a 19-bp sequence present in SXTMO10 and R391 (Fig. 3) that was previously described as the 3′ boundary of hot spot 2 (3). This 19-bp sequence was presumably deleted during the acquisition of the novel sequences found in ICEPdaSpa1's hot spot 2.
FIG. 3.
DNA sequences found at the boundaries of the four hot spots in ICEPdaSpa1, SXTMO10, and R391. Conserved DNA is shown in gray, ICEPdaSpa1-specific DNA is shown in green, SXT-specific DNA is shown in blue, and R391-specific DNA is shown in orange. Nucleotide differences in the conserved DNA are indicated in black. The box represents the stop codon in traA.
The boundaries and content of hot spot 3 in ICEPdaSpa1 differ from those in SXTMO10 and R391 (Fig. 1 and 3; Table 2). The 5′ ends of the corresponding hot spots in the latter two ICEs are located downstream of s073; however, this gene is not present in ICEPdaSpa1. The 5′ boundary of ICEPdaSpa1's hot spot 3 is found just downstream from s072 (Fig. 1 and 3), and the 3′ boundary is 27 bp downstream of the corresponding boundaries found in SXTMO10 and R391. Since s073 is not present in ICEPdaSpa1 but this ICE is functional (see below), s073 should not be considered to be a part of the functional core set of genes in SXT-related ICEs. The new locations of the hot spot 3 boundaries in ICEPdaSpa1 may have arisen from the insertion of the IS10-like transposase gene found in this region (Table 2). Aside from the IS10-like sequence, the predicted proteins encoded in ICEPdaSpa1's hot spot 3 have similarity to Hsp70 and an AAA ATPase, as well as to hypothetical proteins of unknown functions (Table 2). Interestingly, the 3′ end of the ICEPdaSpa1 hot spot 3 DNA, between spa11 and traF, contains 495 bp of an apparently noncoding sequence that is 88% identical to a portion of the so-called vrl island of Desulfococcus multivorans at the nucleotide level (Fig. 1; Table 2).
ICEPdaSpa1's hot spot 4, between traN and s063, contains two genes whose predicted protein products have unknown functions (Fig. 1 and Table 2). Although the contents of hot spots 4 in ICEPdaSpa1 and R391 differ, the 5′ ends of these hot spots are nearly identical (Fig. 3).
ICEPdaSpa1 is excised from the chromosome and can be conjugally transferred to other bacteria.
The known SXT-related ICEs can be excised from the chromosomes of their respective hosts to become extrachromosomal circular forms. We tested whether ICEPdaSpa1 is excised from the PC554.2 chromosomal prfC locus by using a PCR assay that detects the unoccupied prfC locus. This assay, which relies on primers that flank the ICEPdaSpa1 integration site, revealed an ∼120-bp product when PC554.2 DNA was used as the template, suggesting that ICEPdaSpa1 was excised from the chromosomes in a fraction of the cells in the culture, yielding unoccupied attB sites. Using a real-time quantitative PCR assay (10), we determined that ICEPdaSpa1 was excised from approximately 0.4% of cells in an overnight culture, a percentage that is similar to that observed for SXTMO10 excision from the V. cholerae chromosome (10).
Plate mating assays were used to test whether ICEPdaSpa1 could be transferred to other P. damselae subsp. piscicida strains, as well as to E. coli. These assays were facilitated by our discovery that the ICEPdaSpa1 genome contains a tet locus (see above) and, thus, we could use tetracycline to select for exconjugants containing ICEPdaSpa1. When we carried out mating experiments with PC554.2 and CS31, a rifampin-resistant (Rfr) and kanamycin-resistant (Knr) P. damselae subsp. piscicida strain lacking ICEPdaSpa1, tetracycline-resistant (Tcr) Rfr Knr exconjugants were obtained at a frequency of 1.3 × 10−4 exconjugants/donor (Table 3). Using a PCR-based assay (see Materials and Methods), we found that all 50 exconjugants tested contained ICEPdaSpa1 integrated into the prfC gene of the recipient. ICEPdaSpa1 could also be transferred from PC554.2 to E. coli K-12 strains CAG18420 and MC1061, though at somewhat lower frequencies than that of the transfer to P. damselae subsp. piscicida CS31 (Table 3 and data not shown). All E. coli exconjugants tested contained ICEPdaSpa1 integrated into the E. coli prfC gene. Finally, E. coli ICEPdaSpa1 exconjugants could serve as donors to transfer ICEPdaSpa1 to other E. coli strains at high frequencies (Table 3); in the latter E. coli exconjugants, ICEPdaSpa1 was integrated at prfC. Together, these observations demonstrate that ICEPdaSpa1 is a functional, self-transmissible ICE capable of conjugal transfer and integration into and excision from the chromosome.
TABLE 3.
ICEPdaSpa1 is transmissible
| Donor strain namea | Donor strain description | Recipient strain name | Recipient strain description | Transfer frequencyb |
|---|---|---|---|---|
| PC554.2 | P. damselae | CS31 | P. damselae | 1.3 × 10−4 |
| PC554.2 | P. damselae | CAG18420 | E. coli; Knr | 1.5 × 10−5 |
| CCW01 | E. coli BW25113 prfC::ICEPdaSpa1 | CAG18420 | E. coli; Knr | 2.2 × 10−3 |
| CCW02 | E. coli BW25113 prfC::ICEPdaSpa1Δint | CAG18420 | E. coli; Knr | 4.1 × 10−8 |
| CCW03 | E. coli BW25113 recA prfC::ICEPdaSpa1 | CAG18420 | E. coli; Knr | 3.7 × 10−5 |
All donor strains harbored ICEPdaSpa1.
Transfer frequencies were calculated as the numbers of exconjugants divided by the numbers of donors. The values shown are the means of results from three independent assays.
int and recA are critical for ICEPdaSpa1 transfer.
The SXTMO10 integrase is crucial for SXTMO10 transmission since this recombinase is essential for the element's excision and integration (22). As expected, the ICEPdaSpa1 int, which is nearly identical to the SXTMO10 int, proved to be critical for ICEPdaSpa1 transfer. The deletion of int from ICEPdaSpa1 reduced the frequency of transfer of this element by nearly 5 orders of magnitude (Table 3, compare lines 3 and 4). The very low frequency of transfer of the ICEPdaSpa1 int may be attributable to the Hfr-like transfer of the ICEPdaSpa1 markers as previously observed with SXTMO10 (21). Similarily, given the identity of the predicted amino acid sequences of the ICEPdaSpa1 and SXTMO10 SetR repressors, we expected that recA would be important for ICEPdaSpa1 transfer. RecA is thought to promote the autocleavage of SetR, thereby alleviating its repression of setDC, the activators of SXTMO10 transfer (5). The frequency of the transfer of ICEPdaSpa1 from a recA E. coli donor was nearly 2 orders of magnitude lower than the frequency of transfer from an isogenic recA+ E. coli donor (Table 3). This observation suggests that the regulation of ICEPdaSpa1 transfer is similar to that of SXTMO10 transfer.
ICEPdaSpa1 can mobilize a virulence plasmid and acquire new DNA.
Some P. damselae subsp. piscicida strains, including PC554.2, harbor pPHDP10, an ∼10-kb plasmid that encodes an important P. damselae subsp. piscicida virulence factor, AIP56. This toxin induces apoptosis in fish macrophages and neutrophils (17). We constructed pPHDP10-Cm, a pPHDP10 derivative containing a chloramphenicol resistance cassette, to test whether E. coli harboring the ICEPdaSpa1 element could mobilize this virulence plasmid. Isogenic E. coli strains CCW069 (MC1061 pPHDP10-Cm) and CCW077 (MC1061 prfC::ICEPdaSpa1 pPHDP10-Cm) were used as donors in conjugation experiments, and BI533, a nalidixic acid-resistant (Nxr) derivative of E. coli K-12 strain MG1655, was used as a recipient. While no exconjugants were detected when CCW069 was used as a donor, the marked virulence plasmid was transferred at a low but reproducible frequency (∼1.3 × 10−8 exconjugants/donor) from CCW077 (Table 4). Given this low frequency of pPHDP10-Cm transfer, we were concerned that the apparent Nxr chloramphenicol-resistant (Cmr) exconjugants were donors that had become spontaneously resistant to nalidixic acid; however, we excluded this possibility by identifying additional markers characteristic of the recipient cells in all exconjugants tested (data not shown). Thus, ICEPdaSpa1 can enable the horizontal transmission of pPHDP10-Cm. All of the Cmr exconjugants containing pPHDP10-Cm also contained ICEPdaSpa1 and were resistant to tetracycline (Table 4), suggesting that pPHDP10-Cm transfer depends on the cotransfer of ICEPdaSpa1 from the donor and not merely the presence of ICEPdaSpa1 in the donor. The transfer of ICEPdaSpa1 did not appear to be influenced by the presence of pPHDP10-Cm in donor cells. The frequency of ICEPdaSpa1 transfer from CCW077 was ∼1.7 × 10−3 exconjugants/donor, similar to the frequency of ICEPdaSpa1 transfer from an E. coli donor lacking pPHDP10-Cm (Table 3) and ∼5 orders of magnitude greater than the frequency of the transfer of pPHDP10-Cm (Table 4).
TABLE 4.
ICEPdaSpa1 enables the transfer of pPHDP10
| Mating | Donor strain | Relevant property(ies) of donor strain | Frequency of transfer of following selected marker into exconjugantsa,b:
|
% Of exconjugants with cotransfer of following unselected marker:
|
||
|---|---|---|---|---|---|---|
| Cmr | Tcr | Tcr | Cmr | |||
| 1 | E. coli CCW069 | pPHDP10-Cm | <1.0 × 10−9c | NAd | NA | NA |
| 2 | E. coli CCW077 | pPHDP10-Cm ICEPdaSpa1 | 1.3 × 10−8 | 1.7 × 10−3 | 100 | <0.01c |
| 3 | E. coli CCW088 | Exconjugant from mating 2 (Cmr Tcr) | 2.6 × 10−3 | 2.9 × 10−3 | 100 | 100 |
The plasmid pPHDP10-Cm encodes resistance to chloramphenicol (Cmr) and ICEPdaSpa1 encodes resistance to tetracycline (Tcr).
Transfer frequencies were calculated as the numbers of exconjugants with the indicated marker divided by the numbers of donors. The values shown are the means of results from at least three independent assays; standard errors were ≤20%.
Assay limit.
NA, not applicable.
Since there was strict linkage of pPHDP10-Cm transfer to the transfer of ICEPdaSpa1, we speculated that pPHDP10-Cm and ICEPdaSpa1 formed a cointegrate at some point prior to or during transfer from the donor to the recipient. In support of this idea, we found that pPHDP10-Cm was not recoverable as a plasmid from exconjugants derived from the mating of CCW077 with BI533, even though this plasmid was detectable in the donor (data not shown). To further explore this possibility, we tested whether exconjugants that contained both pPHDP10-Cm and ICEPdaSpa1 transferred both elements at similar frequencies. One of the BI533-derived Nxr Cmr Tcr exconjugants (designated CCW088) isolated as described above was used as a donor of ICEPdaSpa1 and pPHDP10-Cm in plate mating experiments with E. coli CAG18420 (Knr) as the recipient. In these experiments, we independently selected for the transfer of ICEPdaSpa1 (yielding Tcr Knr exconjugants) and the transfer of pPHDP10-Cm (yielding Cmr Knr exconjugants). Remarkably, the frequency of the transfer of pPHDP10-Cm (2.6 × 10−3 exconjugants/donor) was approximately the same as the frequency of the transfer of ICEPdaSpa1 (2.9 × 10−3 exconjugants/donor) (Table 4). The similar transfer frequencies of ICEPdaSpa1 and pPHDP10-Cm from CCW088 were in marked contrast to what we observed when the original host of these two elements (CCW077) was tested as the donor (Table 4). Using replica plating, we found that 100% of Tcr exconjugants (containing ICEPdaSpa1) were also Cmr (i.e., contained pPHDP10-Cm) and that 100% of Cmr exconjugants (containing pPHDP10-Cm) were also Tcr (i.e., contained ICEPdaSpa1) (Table 4), establishing that pPHDP10-Cm and ICEPdaSpa1 were cotransferred from CCW088. PCR analyses verified that for all exconjugants tested, Cmr colonies contained additional pPHDP10-Cm sequences (including the gene encoding the AIP56 toxin) and all Tcr colonies contained additional ICEPdaSpa1 sequences. Taken together, these observations provide strong evidence that in CCW088, pPHDP10-Cm is integrated into ICEPdaSpa1. Thus, independent selection for the transfer of either element invariably results in the cotransfer of the two elements. Independently isolated Cmr Nxr exconjugants from the CCW077 × BI533 mating also harbored ICEPdaSpa1::pPHDP10-Cm cointegrates (data not shown).
The integration of pPHDP10-Cm into ICEPdaSpa1 might have been expected to compromise this ICE's transmissibility, but this was not the case. The frequencies of the transfer of hybrid ICEs from CCW088 were similar to the frequencies of the transfer of ICEPdaSpa1 alone from CCW077 and from other donors (data not shown). Finally, the ICEPdaSpa1::pPHDP10-Cm hybrid was found to integrate into prfC. Thus, the integration of pPHDP10-Cm into ICEPdaSpa1 yielded a functional hybrid ICE capable of disseminating a potent toxin.
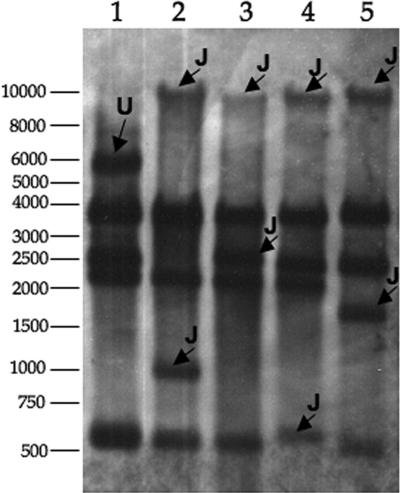
We analyzed the sites of pPHDP10-Cm integration in four independently isolated Cmr Tcr Knr exconjugants to begin to address the mechanism of ICEPdaSpa1::pPHDP10-Cm cointegrate formation. For these experiments, SacI-digested pPHDP10-Cm was used as a probe to investigate whether this plasmid was integrated into the same or different locations in the chromosomes of the four exconjugants. The probe hybridized to the four SacI pPHDP10-Cm restriction fragments of the correct predicted sizes (Fig. 4, lane 1). The bands detected in the SacI-digested DNA from the four exconjugants revealed that pPHDP10-Cm was integrated into the chromosome. In the DNA from each of the exconjugants, one of the bands observed in the SacI-digested pPHDP10-Cm DNA was absent and two new bands (junction fragments) were apparent (Fig. 4, lanes 2 to 5). The junction fragments in each exconjugant differed from those in the other exconjugants, indicating that the site of pPHDP10-Cm integration differed in each exconjugant. Thus, pPHDP10-Cm does not integrate into ICEPdaSpa1 in a site-specific fashion. In one of the exconjugants, we cloned the pPHDP10-Cm chloramphenicol resistance gene along with adjacent DNA. The DNA sequence of this cloned fragment revealed that pPHDP10-Cm was integrated into s040, an ICEPdaSpa1 gene of unknown function. Previous studies have revealed that s040 is not required for SXTMO10 excision, integration, or transfer (4).
FIG. 4.
Southern blot analysis of chromosomal DNA isolated from four independently derived Cmr Tcr Knr exconjugants from CCW088 × CAG18420 matings by using pPHDP10-Cm DNA as a probe. Lane 1, SacI-digested pPHDP10-Cm (the U refers to undigested plasmid DNA); lanes 2 to 5, SacI-digested DNA from four Cmr Tcr Knr exconjugants. The band patterns observed for the four exconjugants reveal that the pPHDP10-Cm integration site in each exconjugant differs from those in the other exconjugants; the J's mark junction fragments. Numbers at the left are molecular weight markers.
We introduced pPHDP10-Cm into a recA derivative of MC1061 (yielding strain CCW129) to explore if homologous recombination was required for the formation of ICEPdaSpa1::pPHDP10-Cm cointegrates. Since recA plays a critical role in promoting ICEPdaSpa1 transfer, for these experiments a setCD expression vector, pGG2B, was introduced into CCW129 to bypass the role of recA in ICEPdaSpa1 transfer (5). Remarkably, the marked virulence plasmid was transferred at approximately the same frequencies from CCW129 (5 × 10−7 exconjugants/recipient) and from CCW077 (5 × 10−7 exconjugants/recipient). (Since the overexpression of setCD is toxic for the donors, for these experiments, transfer frequencies are expressed as numbers of exconjugants per recipient rather than per donor.) As described above for exconjugants derived from CCW077 donors, all exconjugants derived from CCW129 donors were resistant to tetracycline as well as to chloramphenicol, suggesting that ICEPdaSpa1::pPHDP10-Cm cointegrates had formed. Additional pPHDP10-Cm sequences besides the Cmr gene were detected in the exconjugants, providing additional evidence that cointegrates had formed. These observations indicate that recA is not required in the formation of ICEPdaSpa1::pPHDP10-Cm cointegrates. As ICEPdaSpa1 and pPHDP10-Cm do not show long regions of nucleotide identity, it is possible that the insertion sequences in pPHDP10-Cm and/or ICEPdaSpa1 promote cointegrate formation. Alternatively, the ICEPdaSpa1 homologues of the phage lambda bet and exo genes may facilitate the formation of cointegrates.
Summary and conclusions.
Our findings show that there is considerable conservation in the genomes of SXT-related ICEs. The ICEPdaSpa1 genome is the fourth sequenced SXT-related ICE genome. The sequenced ICEs are derived from four different Gammaproteobacteria: P. damselae subsp. piscicida (ICEPdaSpa1), V. cholerae (SXTMO10), Providencia rettgeri (R391), and Shewanella putrefaciens (ICESpuPO1). They all contain a set of highly conserved genes that are required for essential element functions, including genes for regulation (such as setR and setDC), excision and integration (xis and int), and conjugative functions (14 tra genes). These conserved core genes are arranged in the same order in the different ICEs.
Our findings also highlight the considerable plasticity of the genomes of SXT-related ICEs. Although we found that the ICEPdaSpa1 genome contains nearly all of the conserved genes present in SXTMO10 and R391, this fish-pathogen-derived ICE also harbors 25 kb of DNA that is not found in other SXT-related ICEs. Nearly all of the ICEPdaSpa1-specific DNA is found in the same locations as the ICE-specific DNA found in SXTMO10 and R391. The conservation of the insertion sites may simply reflect that these sites can accommodate additional DNA without altering ICE function. However, it is also possible that there are specific mechanisms that promote the integration of foreign DNA at these sites. Recombination between SXT-related ICEs appears to be another mechanism for generating diversity in this group of mobile elements.
Remarkably, the ICEPdaSpa1 genome remains plastic. We found that ICEPdaSpa1 could mobilize a P. damselae subsp. piscicida virulence plasmid via the formation of a cointegrate. The transfer of this new ICEPdaSpa1::pPHDP10-Cm hybrid ICE did not appear to be compromised by the addition of pPHDP10-Cm, likely reflecting the fact that we identified hybrids based on their capabilities for exconjugant formation. Plasmid pPHDP10-Cm integrated into ICEPdaSpa1 in a non-site-specific fashion independently of RecA. Future studies to elucidate the mechanism of pPHDP10-Cm::ICEPdaSpa1 cointegrate formation should yield important insights into processes that govern ICE evolution. Previous work has revealed that SXT-related ICEs can mobilize chromosomal DNA in an Hfr-like manner (21, 31) and that SXT can mobilize a plasmid in trans (21). Our present findings suggest that SXT-related ICEs can still acquire DNA, an additional mechanism whereby these mobile elements can promote horizontal gene flux.
Acknowledgments
This work was supported by grants from Howard Hughes Medical Institute and the NIH (AI-42347) to M.K.W. and by grants AGL2006-00697 (cofunded by the FEDER Program from the EU) and CSD2007-00002 (Aquagenomics, Consolider-Ingenio 2010) from the Ministry of Education and Science of Spain to M.L.L. C.R.O. also thanks Xunta de Galicia, Spain, for a grant enabling him to come to the Waldor lab.
We are grateful to Brigid Davis for her helpful comments on the manuscript.
Footnotes
Published ahead of print on 7 March 2008.
REFERENCES
- 1.Ahmed, A. M., S. Shinoda, and T. Shimamoto. 2005. A variant type of Vibrio cholerae SXT element in a multidrug-resistant strain of Vibrio fluvialis. FEMS Microbiol. Lett. 242241-247. [DOI] [PubMed] [Google Scholar]
- 2.Amita, S., R. Chowdhury, M. Thungapathra, T. Ramamurthy, G. B. Nair, and A. Ghosh. 2003. Class I integrons and SXT elements in El Tor strains isolated before and after 1992 Vibrio cholerae O139 outbreak, Calcutta, India. Emerg. Infect. Dis. 9500-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaber, J. W., V. Burrus, B. Hochhut, and M. K. Waldor. 2002. Comparison of SXT and R391, two conjugative integrating elements: definition of a genetic backbone for the mobilization of resistance determinants. Cell. Mol. Life Sci. 592065-2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beaber, J. W., B. Hochhut, and M. K. Waldor. 2002. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J. Bacteriol. 1844259-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beaber, J. W., B. Hochhut, and M. K. Waldor. 2004. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 42772-74. [DOI] [PubMed] [Google Scholar]
- 6.Boltner, D., C. MacMahon, J. T. Pembroke, P. Strike, and A. M. Osborn. 2002. R391: a conjugative integrating mosaic comprised of phage, plasmid, and transposon elements. J. Bacteriol. 1845158-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burrus, V., J. Marrero, and M. K. Waldor. 2006. The current ICE age: biology and evolution of SXT-related integrating conjugative elements. Plasmid 55173-183. [DOI] [PubMed] [Google Scholar]
- 8.Burrus, V., G. Pavlovic, B. Decaris, and G. Guedon. 2002. Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 46601-610. [DOI] [PubMed] [Google Scholar]
- 9.Burrus, V., R. Quezada-Calvillo, J. Marrero, and M. K. Waldor. 2006. SXT-related integrating conjugative element in New World Vibrio cholerae. Appl. Environ. Microbiol. 723054-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burrus, V., and M. K. Waldor. 2003. Control of SXT integration and excision. J. Bacteriol. 1855045-5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burrus, V., and M. K. Waldor. 2004. Formation of SXT tandem arrays and SXT-R391 hybrids. J. Bacteriol. 1862636-2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burrus, V., and M. K. Waldor. 2004. Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 155376-386. [DOI] [PubMed] [Google Scholar]
- 13.Cloeckaert, A., S. Baucheron, G. Flaujac, S. Schwarz, C. Kehrenberg, J. L. Martel, and E. Chaslus-Dancla. 2000. Plasmid-mediated florfenicol resistance encoded by the floR gene in Escherichia coli isolated from cattle. Antimicrob. Agents Chemother. 442858-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coetzee, J. N., N. Datta, and R. W. Hedges. 1972. R factors from Proteus rettgeri. J. Gen. Microbiol. 72543-552. [DOI] [PubMed] [Google Scholar]
- 15.Dalsgaard, A., A. Forslund, D. Sandvang, L. Arntzen, and K. Keddy. 2001. Vibrio cholerae O1 outbreak isolates in Mozambique and South Africa in 1998 are multiple-drug resistant, contain the SXT element and the aadA2 gene located on class 1 integrons. J. Antimicrob. Chemother. 48827-838. [DOI] [PubMed] [Google Scholar]
- 16.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.do Vale, A., M. T. Silva, N. M. dos Santos, D. S. Nascimento, P. Reis-Rodrigues, C. Costa-Ramos, A. E. Ellis, and J. E. Azevedo. 2005. AIP56, a novel plasmid-encoded virulence factor of Photobacterium damselae subsp. piscicida with apoptogenic activity against sea bass macrophages and neutrophils. Mol. Microbiol. 581025-1038. [DOI] [PubMed] [Google Scholar]
- 18.Ehara, M., B. M. Nguyen, D. T. Nguyen, C. Toma, N. Higa, and M. Iwanaga. 2004. Drug susceptibility and its genetic basis in epidemic Vibrio cholerae O1 in Vietnam. Epidemiol. Infect. 132595-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hochhut, B., J. W. Beaber, R. Woodgate, and M. K. Waldor. 2001. Formation of chromosomal tandem arrays of the SXT element and R391, two conjugative chromosomally integrating elements that share an attachment site. J. Bacteriol. 1831124-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hochhut, B., Y. Lotfi, D. Mazel, S. M. Faruque, R. Woodgate, and M. K. Waldor. 2001. Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob. Agents Chemother. 452991-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hochhut, B., J. Marrero, and M. K. Waldor. 2000. Mobilization of plasmids and chromosomal DNA mediated by the SXT element, a constin found in Vibrio cholerae O139. J. Bacteriol. 1822043-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hochhut, B., and M. K. Waldor. 1999. Site-specific integration of the conjugal Vibrio cholerae SXT element into prfC. Mol. Microbiol. 3299-110. [DOI] [PubMed] [Google Scholar]
- 23.Iwanaga, M., C. Toma, T. Miyazato, S. Insisiengmay, N. Nakasone, and M. Ehara. 2004. Antibiotic resistance conferred by a class I integron and SXT constin in Vibrio cholerae O1 strains isolated in Laos. Antimicrob. Agents Chemother. 482364-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juiz-Rio, S., C. R. Osorio, V. de Lorenzo, and M. L. Lemos. 2005. Subtractive hybridization reveals a high genetic diversity in the fish pathogen Photobacterium damselae subsp. piscicida: evidence of a SXT-like element. Microbiology 1512659-2669. [DOI] [PubMed] [Google Scholar]
- 25.Kim, E. H., and T. Aoki. 1994. The transposon-like structure of IS26-tetracycline, and kanamycin resistance determinant derived from transferable R plasmid of fish pathogen, Pasteurella piscicida. Microbiol. Immunol. 3831-38. [DOI] [PubMed] [Google Scholar]
- 26.Kitao, T. 1993. Pasteurellosis, p. 159-165. In V. Inglis, R. J. Roberts, and N. R. Bromage (ed.), Bacterial diseases of fish. Blackwell Science, Oxford, United Kingdom.
- 27.Maeda, K., H. Nojiri, M. Shintani, T. Yoshida, H. Habe, and T. Omori. 2003. Complete nucleotide sequence of carbazole/dioxin-degrading plasmid pCAR1 in Pseudomonas resinovorans strain CA10 indicates its mosaicity and the presence of large catabolic transposon Tn4676. J. Mol. Biol. 32621-33. [DOI] [PubMed] [Google Scholar]
- 28.Magarinos, B., J. L. Romalde, I. Bandin, B. Fouz, and A. E. Toranzo. 1992. Phenotypic, antigenic, and molecular characterization of Pasteurella piscicida strains isolated from fish. Appl. Environ. Microbiol. 583316-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magarinos, B., A. E. Toranzo, and J. L. Romalde. 1996. Phenotypic and pathobiological characteristics of Pasteurella piscicida. Annu. Rev. Fish Dis. 641-46. [Google Scholar]
- 30.Murata, T., M. Ohnishi, T. Ara, J. Kaneko, C. G. Han, Y. F. Li, K. Takashima, H. Nojima, K. Nakayama, A. Kaji, Y. Kamio, T. Miki, H. Mori, E. Ohtsubo, Y. Terawaki, and T. Hayashi. 2002. Complete nucleotide sequence of plasmid Rts1: implications for evolution of large plasmid genomes. J. Bacteriol. 1843194-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nugent, M. E. 1981. A conjugative ‘plasmid’ lacking autonomous replication. J. Gen. Microbiol. 126305-310. [DOI] [PubMed] [Google Scholar]
- 32.Osorio, C. R., S. Juiz-Rio, and M. L. Lemos. 2006. A siderophore biosynthesis gene cluster from the fish pathogen Photobacterium damselae subsp. piscicida is structurally and functionally related to the Yersinia high-pathogenicity island. Microbiology 1523327-3341. [DOI] [PubMed] [Google Scholar]
- 33.Pembroke, J. T., and A. V. Piterina. 2006. A novel ICE in the genome of Shewanella putrefaciens W3-18-1: comparison with the SXT/R391 ICE-like elements. FEMS Microbiol. Lett. 26480-88. [DOI] [PubMed] [Google Scholar]
- 34.Singer, M., T. A. Baker, G. Schnitzler, S. M. Deischel, M. Goel, W. Dove, K. J. Jaacks, A. D. Grossman, J. W. Erickson, and C. A. Gross. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol. Rev. 531-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waldor, M. K., H. Tschape, and J. J. Mekalanos. 1996. A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J. Bacteriol. 1784157-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wertman, K. F., A. R. Wyman, and D. Botstein. 1986. Host/vector interactions which affect the viability of recombinant phage lambda clones. Gene 49253-262. [DOI] [PubMed] [Google Scholar]