Abstract
Inspection of the genome sequence of Lactobacillus casei ATCC 334 revealed two operons that might dissimilate the five isomers of sucrose. To test this hypothesis, cells of L. casei ATCC 334 were grown in a defined medium supplemented with various sugars, including each of the five isomeric disaccharides. Extracts prepared from cells grown on the sucrose isomers contained high levels of two polypeptides with Mrs of ∼50,000 and ∼17,500. Neither protein was present in cells grown on glucose, maltose or sucrose. Proteomic, enzymatic, and Western blot analyses identified the ∼50-kDa protein as an NAD+- and metal ion-dependent phospho-α-glucosidase. The oligomeric enzyme was purified, and a catalytic mechanism is proposed. The smaller polypeptide represented an EIIA component of the phosphoenolpyruvate-dependent sugar phosphotransferase system. Phospho-α-glucosidase and EIIA are encoded by genes at the LSEI_0369 (simA) and LSEI_0374 (simF) loci, respectively, in a block of seven genes comprising the sucrose isomer metabolism (sim) operon. Northern blot analyses provided evidence that three mRNA transcripts were up-regulated during logarithmic growth of L. casei ATCC 334 on sucrose isomers. Internal simA and simF gene probes hybridized to ∼1.5- and ∼1.3-kb transcripts, respectively. A 6.8-kb mRNA transcript was detected by both probes, which was indicative of cotranscription of the entire sim operon.
Comparative genomics and phylogenetic analyses of the lactic acid bacteria (LAB) have provided evidence that the order Lactobacillales comprises the families Lactobacillaceae, Streptococcaceae, Enterococcaceae, and Leuconostocaceae (17, 25; http://www.ncbi.nlm.nih.gov/Taxonomy/). These generally fastidious gram-positive organisms are used extensively for the fermentation of dairy, meat, and vegetable products. The industrial importance of LAB, and Lactobacillus casei strains in particular, results from the capacity of these microorganisms to metabolize carbohydrates rapidly (and primarily) to lactic acid. Prior to their homolactic fermentation via the glycolytic pathway, many carbohydrates are accumulated simultaneously with phosphorylation via sugar-specific phosphoenolpyruvate (PEP)-dependent phosphotransferase systems (PTSs) (7, 24, 29). The multicomponent PEP-dependent PTSs comprise membrane-localized, sugar-specific transporters (IICB enzymes) that may be fused or associated with a third protein (EIIA), as well as two general cytoplasmic proteins (EI and HPr). Collectively, these interactive proteins constitute a five-stage phosphorelay that, via transfer of the high-energy phosphoryl moiety from PEP, catalyzes the simultaneous phosphorylation and translocation of sugars through the cytoplasmic membrane. Biochemical and physiological studies performed during the past 25 years have established the presence of a variety of sugar PEP-dependent PTSs in L. casei, including PTSs for glucose (41, 45), galactose (2, 6), lactose (4, 5, 8, 9), sorbose (46), and pentitols (15, 16). Significantly, in the past decade, the development and application of techniques for manipulation of LAB genomes have provided extensive (and in some instances unexpected) insight into the molecular basis for regulation of PEP-dependent PTSs in L. casei (18, 42, 45).
Many bacterial species, including L. casei, have the capacity to transport sucrose via an inducible sucrose-specific PEP-dependent PTS (GenBank accession no. YP_807293). Intracellular sucrose-6-phosphate is subsequently hydrolyzed to glucose-6-phosphate (G6P) and fructose by sucrose-6-phosphate hydrolase (GenBank accession no. YP_807296). The latter cofactor-independent 6-phosphoglucosyl hydrolase has been assigned to family 32 of the 110-member glycosyl hydrolase (GH) superfamily (11, 12; Carbohydrate-Active Enzyme Database [http://www.cazy.org/fam/acc_GH.html]). In contrast to the commonly observed dissimilation of sucrose, it was assumed for a long time that microorganisms are unable to metabolize the five linkage-isomeric α-d-glucosyl-d-fructose isomers of sucrose, namely, trehalulose, turanose, maltulose, leucrose, and palatinose. However, studies in our laboratory have proved that this assumption is invalid by showing that several species belonging to both gram-positive and gram-negative genera readily utilize these isomeric disaccharides as energy sources for growth (23, 30, 34, 36, 37). Remarkably, besides the five sucrose isomers, these organisms also transport a variety of related O-α-linked glycosides, including maltose, isomaltose, maltitol, and α-methyl-d-glucoside, by the α-glucoside-specific PEP-dependent PTS. Furthermore, the accumulated α-glucoside-6-phosphates are hydrolyzed by a novel NAD+- and Mn2+-dependent phospho-α-glucosidase (Pagl) (EC 3.2.1.122) that is assigned to the unique GH family 4 (GH4) (3, 14, 27, 34, 36, 39).
Thus far, there have been no reports of the utilization of sucrose isomers by representatives of the families Streptococcaceae and Lactobacillaceae, including the species residing in plaque (at the tooth surface) or in biofilms in the oral cavity (20, 21). Consequently, these “noncariogenic” and comparatively sweet disaccharides are used increasingly as substitutes for dietary sucrose (10). Our interest in this area of carbohydrate dissimilation by LAB stemmed from in silico mining of data in the recently published genome sequence of L. casei strain ATCC 334 (GenBank accession no. NC_008526). Inspection of this chromosomal DNA sequence revealed two putative PTS operons (see Fig. 2A and B) that have genes at the LSEI_2684 and LSEI_0369 loci. The deduced amino acid sequences encoded by the two genes showed unusually high levels of identity with the NAD+- and Mn2+-dependent Pagls described previously for other bacteria. From the comparative alignments shown in Fig. 1, we hypothesized that expression of the products of one (or both) of these operons might also facilitate growth of L. casei ATCC 334 on sucrose isomers, maltose, and related α-linked glucosides. A collaborative study was initiated to test this hypothesis, and our findings are presented in this paper.
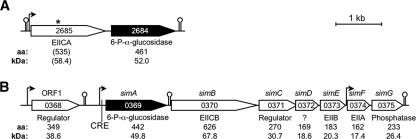
FIG. 2.
Gene organization of the two operons which encode the putative NAD+- and Mn2+-dependent Pagls in L. casei ATCC 334. The predicted function, amino acid (aa) chain length, and protein molecular mass are indicated under each gene. Potential promoters (bent arrows) and terminators (lollipop symbols) were predicted in silico. (A) Operon A. The LSEI_2684 gene encoding a putative Pagl is predicted to be transcribed from a promoter upstream of LSEI_2685. In the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov) LSEI_2685 is annotated as a pseudogene. The asterisk indicates a frameshift mutation that may prevent expression of the EII(CA) component of the PTS. (B) Operon B hypothesized to participate in sucrose isomer metabolism (sim). The LSEI_0369 (simA), LSEI_0370 (simB), and LSEI_0374 (simF) genes putatively encode a Pagl, a membrane-localized PTS transporter complex [EII(CB)], and an intracellular EIIA component of the PTS, respectively.
FIG. 1.
Comparative sequence alignment of NAD+- and metal ion-dependent Pagls belonging to the GH4 family. Genes in L. casei ATCC 334 are numbered in accordance with the annotation in the GenBank database (accession no. NC_008526). The sequences of the following proteins are shown: putative Pagls encoded by L. casei ATCC 334 LSEI_0369 (simA) (accession no. YP_805670) (L.c. SimA) and LSEI_2684 (YP_807845) (L.c. 2684) and enzymes purified from C. acetobutylicum (Swiss-Prot accession no. Q97LM4) (C.a. MalH), B. subtilis (accession no. P54716) (B.s. GlvA), K. pneumoniae (accession no. Q9AGA7) (K.p. AglB), and F. mortiferum (accession no. O06901) (F.m. MalH). The numbers on the right indicate the numbers of amino acids per protein, and conserved amino acids are highlighted. The filled triangles indicate (L. casei simA numbering) residues that comprise the NAD+-binding domain of the βαβ Rossmann fold (residues 6 to 72; importantly, G12, G14, S15, and D41), proton donor residue D172, proton acceptor residue Y265, residues that coordinate with the Mn2+ ion (including A151, C171, and H202), and amino acids that participate in substrate binding (including R95, N149, and R285). Residue E111 increases the basicity of the catalytically essential residue Y265 (28).
MATERIALS AND METHODS
Materials.
High-purity sugars, including glucose, sucrose [α-d-glucopyranosyl-β-d-fructofuranoside, α(1→2) linkage], maltose (4-O-α-d-glucopyranosyl-d-glucopyranose), and 1-O-methyl-α-d-glucopyranoside, were purchased from Pfanstiehl Laboratories, Inc. Maltitol (4-O-α-glucopyranosyl-d-sorbitol), chromogenic p-nitrophenyl-α-d-glucopyranosides, and isomaltose (6-O-α-d-glucopyranosyl-d-glucose) were obtained from Sigma and TCI America. The five linkage isomers of sucrose were obtained from the following sources: trehalulose (α, 1→1) was a generous gift from Südzucker, Germany; turanose (α, 1→3) was obtained from Pfanstiehl; maltulose (α, 1→4) was obtained from TCI America; leucrose (α, 1→5) was obtained from Fluka; and palatinose (α, 1→6) was purchased from Wako Chemicals. Phosphorylated derivatives, including trehalulose-6′-phosphate, sucrose-6-phosphate, turanose-6′-phosphate, maltulose-6′-phosphate, leucrose-6′-phosphate, palatinose-6′-phosphate, maltose-6′-phosphate, isomaltose-6′-phosphate, and maltitol-6-phosphate, were prepared enzymatically (38). Chromogenic phosphate derivatives, including p-nitrophenyl-α-glucopyranoside-6-phosphate (pNPαG6P), were prepared by selective phosphorylation with phosphorus oxychloride in trimethyl phosphate containing small proportions of water (33). Phosphorylated β-linked disaccharides, including 4-O-β-d-glucopyranosyl-d-glucopyranoside-6′-phosphate (cellobiose-6′-phosphate) and 6-O-β-d-glucopyranosyl-d-glucopyranose-6′-phosphate (gentiobiose-6′-phosphate), were prepared by phosphorylation of the parent compounds with purified ATP-dependent β-glucoside kinase (EC 2.7.1.85) from Klebsiella pneumoniae (35). NADP+, Ultrogel AcA-44, Tris-Acryl M-DEAE, and other materials were supplied by Sigma-Aldrich. Glucose-6-phosphate dehydrogenase (G6PDH) (EC 1.1.1.49) was obtained from Roche Molecular Biochemicals.
Organism, maintenance, and growth medium.
L. casei ATCC 334 was obtained from the American Type Culture Collection (Manassas, VA) and was maintained in MRS broth (Difco) supplemented with 0.2% (wt/vol) glucose. The medium used for growth of L. casei ATCC 334 contained (per liter) 10 g Trypticase (BBL), 0.5 g yeast extract (Difco), 3 g tryptone peptone (Difco), 3 g KH2PO4, 3 g K2HPO4, 1.7 g sodium acetate·3H2O, 1.2 g sodium citrate·2H2O, and 1 ml Tween 80. Prior to autoclaving, the pH of the medium was adjusted to 7 by addition of 5 N NaOH. After this, 100 ml of a filter-sterilized salts solution was prepared, which contained 11.6 g MgSO4·7H2O, 2.4 g MnSO4·2H2O, and 0.6 g FeSO4·7H2O. Prior to inoculation, 5 ml liter−1 of the salts solution was added to the medium, and filter-sterilized sugar solutions were added to obtain a final concentration of 0.4% (wt/vol). Organisms were grown to stationary phase in 1-liter capped bottles at 37°C (in the absence of a fermentable carbohydrate, there is not significant growth of L. casei ATCC 334 in this medium).
Analytical methods.
The molecular weight of denatured Pagl (subunit) was determined by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) using the Novex XCell mini-cell system (Invitrogen). Novex NuPage (4 to 12% acrylamide) bis-Tris gels and morpholineethanesulfonic acid (MES)-SDS running buffer (pH 7.3) were used together with Novex Mark12 protein standards. Polypeptides were visualized by staining with Coomassie blue R-250. For Western blots, proteins and SeeBlue-prestained standards were transferred to nitrocellulose membranes using NuPage transfer buffer. Immunodetection of Pagl was performed by sequential incubation of the membrane with (i) polyclonal antibody to Pagl (MalH) (EC 3.2.1.122) from Fusobacterium mortiferum and (ii) goat anti-rabbit horseradish peroxidase-conjugated antibody as described previously (33). The native molecular weight of Pagl was determined by gel filtration on the same AcA-44 column used for purification. The column was calibrated with the following standards: alcohol dehydrogenase (150 kDa), bovine serum albumin (66 kDa), ovalbumin (43 kDa), and chymotrypsinogen A (25 kDa). Protein concentrations of cell extracts were determined with a BCA assay kit (Pierce Chemical Co.). The N-terminal sequence of Pagl was determined with an ABI 477A protein sequencer (Applied Biosystems Inc.) with an online ABI 120A phenylthiohydantoin analyzer. Two-dimensional (2D) PAGE for proteomic analyses was carried out by Kendrick Laboratories, Inc., and protein spots (see Fig. 4A and B) were identified by matrix-assisted laser desorption ionization-time of flight mass spectrometry (Applied Biosystems Voyager DE Pro instrument) by M. A. Gawinowicz, Protein Core Facility, Columbia University, New York, NY.
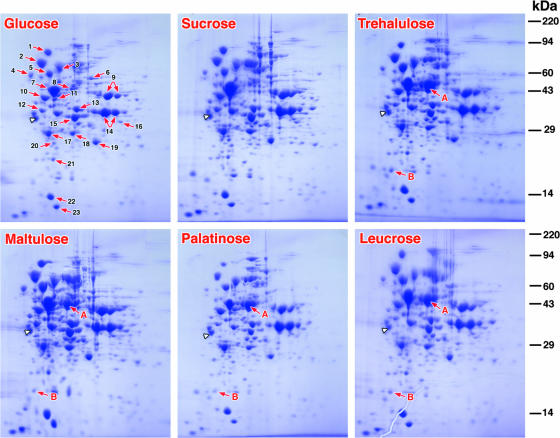
FIG. 4.
Proteomic analyses by 2D PAGE of extracts of L. casei ATCC 334 cells grown separately on glucose, sucrose, and four isomers of sucrose. 2D electrophoresis revealed a remarkable similarity in the polypeptide composition, and 23 highly conserved proteins were identified by MS-MS in all extracts (Table 2). However, extracts prepared from cells grown on the four sucrose isomers contained high levels of two proteins (spots A and B) that were not detected in extracts from either glucose- or sucrose-grown cells. The identities of the two “spots” were determined after excision, tryptic digestion, and MS-MS. The extensive peptide coverage established that proteins A (Mr, ∼50,000) and B (Mr, ∼17,000) were encoded by LSEI_0369 (simA) and LSEI_0374 (simF), respectively, of operon B (Fig. 2). The arrowhead indicates the position of the internal standard tropomyosin (32.7 kDa; pI 5.2) included in each of the gels. The molecular weight standards used were myosin (220,000), phosphorylase A (94,000), catalase (60,000), actin (43,000), carbonic anhydrase (29,000), and lysozyme (14,000).
For identification of major proteins (see Fig. 4, spots 1 to 23), the Coomassie blue-stained spots were excised and destained prior to reduction with dithiothreitol, alkylation with iodoacetamide, and digestion with trypsin. Tryptic peptides were reconstituted in formic acid (6.4 μl, 0.1% [vol/vol]) and concentrated on a C18 Optipak (Bodman Industries) trap column. After a 10-min delay, the sample was directed to a Vydac C18 column (100 mm by 150 μm [inside diameter]; 5 μm; Microtech Scientific) for separation. Solvent A was a mixture of 98.8% water, 1% acetonitrile, and 0.1% formic acid, and solvent B was a mixture of 98.8% acetonitrile, 1% water, and 0.1% formic acid. The gradient was increased from 5% solvent B to 95% solvent B in 40 min at a flow rate of 50 μl min−1 and was split at a 10:1 ratio on the column. The eluent from the liquid chromatography system was introduced into a linear ion trap Fourier transform ion cyclotron mass spectrometer (Thermo Electron, San Jose, CA) by electrospray ionization. The following instrument settings were used: source voltage, 3.05 kV; capillary voltage, 40.00 V; tube lens voltage, 80.00 V; and capillary temperature, 200°C. Tryptic fragments were generated by collision-induced dissociation at a normalized collision energy of 35% and an activation Q of 0.250. Tandem mass spectrometry (MS-MS) data were acquired with the Xcalibur 2.0 software and processed using Bioworks to create data (dta) files. These files were merged to obtain mfg files suitable for database searching using the MASCOT database search engine (www.matrixscience.com). The following parameters were used: database, SPTREMBLE; taxonomy, Firmicutes (gram-positive bacteria); one missed cut cleavage; iodoacetamidation of cysteines; and charge states of +2, +3, and +4. A window of 10 ppm for mass accuracy for precursor ions and 0.6-Da mass accuracy for MS-MS data was chosen. The results were parsed on Scaffold (Proteome Software, Portland, OR). Only proteins that were identified by at least two peptides and >95% peptide probability (based on MASCOT and Protein Prophet scores) were chosen.
Assay of Pagl activity.
The activity of the NAD+- and Mn2+-dependent Pagl in cell extracts and during purification was measured colorimetrically with pNPαG6P as the substrate. Enzyme activity was determined using a discontinuous assay and a mixture that contained (in 2 ml) Tris-HCl buffer (50 mM, pH 7.5), MnCl2 (1 mM), NAD+ (1 mM), and pNPαG6P (0.5 mM). After equilibration at 37°C, the reactions were initiated by addition of an enzyme preparation. Samples (0.25 ml) were removed at 20-s intervals and immediately injected into 0.75 ml of a 0.5 M Na2CO3 solution containing 0.1 M EDTA to stop the reaction. The absorbance at 400 nm was measured, and the rates of p-nitrophenol formation were calculated from progress plots, assuming that the molar extinction coefficient (ɛ) at pH 10.2 for the yellow p-nitrophenolate anion was 18,300 M−1 cm−1. Pagl activity was expressed in nanomoles (or micromoles) of p-nitrophenol formed per minute per milligram of protein. A continuous spectrophotometric method was used to measure the rates of hydrolysis of α-glucoside-6-phosphate substrates. This G6PDH/NADP+-coupled assay monitored the release of G6P in a 1-ml reaction mixture that contained Tris-HCl buffer (0.1 M, pH 7.5), MnCl2 (1 mM), MgCl2 (1 mM), NAD+ (1 mM), NADP+ (1 mM), substrate (1 mM), and ∼3 U of G6PDH. Reactions were started by adding the enzyme preparation, and the increase in A340 was monitored with a Beckman DU 640 recording spectrophotometer. The initial rates of G6P formation were calculated using the kinetics program of the instrument. A molar extinction coefficient (ɛ) of 6,220 M−1 cm−1 was assumed for calculation of the amount of NADPH formed (amount of G6P released). Pagl activity was expressed in nanomoles of phosphorylated substrate hydrolyzed per minute per milligram of protein.
Cloning and expression of the L. casei ATCC 334 Pagl gene (simA) in Escherichia coli TOP10.
Based on the complete genome sequence of L. casei ATCC 334, the following pair of primers was designed to amplify the simA gene: forward primer 5′-GGGCCCCATGGATGATCGGAAGTTTTCAGTTTTAATTGC-3′ (the simA sequence is in boldface type, and the NcoI restriction site is underlined) and reverse primer 5′-CGAGCAGAATTCCTACTTCAGTTCTGGCCAGTAGTC-3′ (the sequence complementary to the downstream region of simA is in boldface type, and the EcoRI restriction site is underlined). PCR amplification was carried out with a thermal cycler (GeneAmp 9700PCR system; PE Applied Biosystems) using Pfu high-fidelity DNA polymerase from Stratagene. The components of the amplification mixture (100 μl) were as follows: 5 U of Pfu DNA polymerase, 1× reaction buffer provided by the manufacturer, 20 mM each of the four deoxynucleoside triphosphates, 250 ng of each primer, 100 ng of L. casei ATCC 334 chromosomal DNA, and 1% (vol/vol) dimethyl sulfoxide. After an initial 2-min incubation at 95°C, the mixture was subjected to 30 cycles of amplification under the following conditions: denaturation at 95°C for 1 min, annealing at 50°C for 1 min, and extension at 72°C for 2 min/kb of insert. This procedure was followed by a 10-min runoff at 72°C. The amplicon was digested with restriction endonucleases NcoI and EcoRI, electrophoresed in a 1% agarose gel, and purified (QIAquick gel extraction kit). The purified ∼1.3-kbp PCR product was ligated to similarly digested and purified expression vector pTrcHis2B (Invitrogen) to form the recombinant plasmid pTrcHis2BsimA (designated pAPsimA). In this construct, the simA gene is under control of the trc promoter. Since pTrcHis vectors also contain a copy of the lacIq gene, expression of the simA gene is fully induced in the presence of isopropyl-β-d-thiogalactopyranoside (IPTG). Recombinant plasmid pAPsimA was transformed into E. coli TOP10 (Invitrogen) competent cells, and colonies were selected on Luria-Bertani (32) agar plates containing 150 μg ml−1 ampicillin. It should be noted that in the pTrcHis2B vector the fusion peptide is located at the C terminus rather than the N terminus, as is the case for pTrcHis vectors. However, the stop codon included in our reverse primer prevented the expression of the fusion peptide in the pTrcHis2B vector.
Purification of Pagl from E. coli TOP10(pAPsimA).
The plasmid-containing cells were grown in 800 ml Luria-Bertani broth containing ampicillin (150 μg ml−1) in 2-liter baffled flasks at 37°C on a rotary shaker at 200 rpm. At an A600 of 0.4 to 0.6 U, IPTG (0.5 mM) was added to each culture, and the culture was grown for 3 h. The culture was harvested by centrifugation (13,000 × g for 10 min at 5°C), and the cells (2.4 g liter−1) were washed by resuspension and centrifugation from Tris-HCl buffer (25 mM, pH 7.5) containing MnCl2 (1 mM), NAD+ (0.1 mM), and dithiothreitol (1 mM) (TMND buffer).
(i) Preparation of cell extract.
Washed cells (18 g, wet weight) were resuspended in 45 ml of TMND buffer, and the organisms were disrupted (at 0°C) by 1.5 min of sonic oscillation twice using a Branson instrument (model 350) operating at ∼75% of the maximum power. The extract was clarified by two centrifugations; the first centrifugation was a low-speed centrifugation (25,000 × g for 30 min at 5°C) and was followed by ultracentrifugation at 180, 000 × g for 2 h at 5°C. The high-speed supernatant was transferred to sacs and dialyzed in a cold room for 16 h against 4 liters of TMND buffer. Pagl was purified to homogeneity by a simple two-stage chromatographic procedure.
(ii) Step 1: Tris-Acryl M-DEAE anion exchange.
Dialyzed high-speed supernatant (56 ml) was transferred at a flow rate of 0.5 ml min−1 to a column of Tris-Acryl M-DEAE (2.6 by 15 cm) previously equilibrated with TMND buffer containing dithiothreitol (5 mM). Nonadsorbed material was removed by passage of this buffer through the column. After this, Pagl was eluted with 500 ml of a linear increasing-concentration gradient of NaCl (0 to 0.4 M) in TMND buffer. Fractions (5 ml) were collected, and Pagl activity was detected by the intense yellow color formed upon addition of fraction samples (10 μl) to microtiter wells containing 100 μl of pNPαG6P assay solution. The fractions with the greatest activity (fractions 52 to 60) were pooled and concentrated to 6.5 ml by pressure filtration (50 lb/in2) through an Amicon PM-10 membrane.
(iii) Step 2: Ultrogel AcA gel filtration.
Approximately 2 ml of DEAE concentrate was applied at a flow rate of 0.15 ml min−1 to a column of Ultrogel AcA-44 (1.6 by 94 cm) previously equilibrated with TMND buffer containing 0.1 M NaCl. Fractions (2 ml) were collected, and the fractions containing the maximum Pagl activity (fractions 49 to 55) were pooled. This procedure was twice repeated with the remaining two 2-ml portions of DEAE concentrate. The appropriate fractions (fractions 49 to 55) from each of the three runs were pooled and concentrated with an Amicon pressure cell to a final volume of 10 ml. Assuming that the Mr of the Pagl monomer was 49,711 and that the extinction coefficient (ɛ) was 73,145 M−1 cm−1, the concentration of highly purified Pagl in the sample was calculated to be 13.9 mg ml−1 and the specific activity was calculated to be 3.78 μmol of pNPαG6P hydrolyzed min−1 mg protein−1.
Extraction of RNA from L. casei ATCC 334.
Total RNA was extracted from exponential-phase cells (optical density at 600 nm, ∼0.8) cultured in medium containing glucose, sucrose, or sucrose isomers as the sole energy source. Two volumes of RNAprotect (Qiagen) was added to 1 volume of cells, and samples were mixed and then incubated at 25°C for 5 min. Cells were harvested by centrifugation at 3,500 × g at 25°C for 15 min, and following removal of the supernatant by aspiration, cells were stored at −70°C for 24 h. Cell pellets were thawed, resuspended in 1 ml Trizol reagent (Invitrogen), and transferred to FastPrep blue tubes containing lysing matrix B (Qbiogene). Cells were mechanically disrupted with a FastPrep bead beater (Qbiogene) for 40 s at speed setting 6 and allowed to stand at 25°C for 10 min. Chloroform (0.2 ml) was added, and samples were mixed and incubated for 3 min at 25°C. Following centrifugation at 12,000 × g for 15 min, the aqueous phase was collected and mixed with 0.5 ml isopropanol to precipitate RNA. Samples were incubated at 25°C for 10 min, and RNA precipitates were recovered by centrifugation at 12,000 × g for 10 min. Pellets were washed twice in 75% (vol/vol) ethanol and air dried. Purified RNA was dissolved in 50 μl H2O and stored at −70°C.
Northern blot procedures.
Expression of specific transcripts was assessed by Northern blotting using a NorthernMax kit (Ambion) according to the manufacturer's instructions. Briefly, RNA was thawed on ice, and 5 μg of each RNA sample was subjected to electrophoresis through a 1% (wt/vol) agarose gel. RNA was transferred to a BrightStar Plus membrane (Ambion) under a vacuum pressure of 5 mm Hg for 1.5 h in a model 785 vacuum blotter (Bio-Rad). Membranes were fixed using a Stratalink UV cross-linker (Stratagene) and prehybridized for 30 min at 42°C in ULTRAhyb buffer (Ambion). Oligonucleotide DNA probes internal to the simA (5′-GCAAAGACTCATCGCTTCCCAAAGTTTGGTGTACGATTGC-3′) and simF (5′-CGAGTGGCATAAGGATGCCCGTGACAGGGGCAATAATCGGCTGTCC-3′) genes were synthesized and 5′ end labeled with 32P (Lofstrand Labs). Following prehybridization, probes were added at a final concentration of 106 cpm ml−1 and incubated for 16 h at 42°C. Membranes were washed twice at 25°C for 5 min in low-stringency wash solution no. 1 (Ambion) and once at 42°C in high-stringency wash solution no. 1. Washed membranes were exposed to Kodak BioMax MR film for 10 h.
Sequence alignment and operon prediction.
Protein sequences were aligned by using the Clustal W algorithm in the MegAlign software (DNASTAR Inc.). The structure of operons was predicted by automated genome annotation using the fgenesB module of the MolQuest software (Softberry Inc.). Putative promoter and terminator elements were identified as part of the annotation pipeline. A potential catabolite response element (CRE box) was found in the promoter region of simA by using the Virtual Footprint software program (http://prodoric.tu-bs.de/vfp/) to search the simABCDEFG operon and upstream sequence for close matches to a position weight matrix based on the CRE box elements upstream of 13 Bacillus subtilis genes.
RESULTS
Identification of putative α-glucoside PTS operons in L. casei ATCC 334.
A BLAST search (1) of the L. casei ATCC 334 genome with the gene encoding the NAD+- and Mn2+-dependent Pagl (GlvA) of B. subtilis as the probe revealed two sequences encoding proteins with strikingly high homology to this enzyme. The LSEI_2684 gene encodes a 461-amino-acid protein that exhibits 60% identity with GlvA, whereas LSEI_0369 (simA) encodes a smaller protein (442 residues) with 62% sequence identity with GlvA. The translation products of the two L. casei genes, which exhibit 59% sequence identity (Fig. 1), were also remarkably similar to the sequences of Pagls recently purified from Fusobacterium mortiferum (MalH) (3), K. pneumoniae (AglB) (37), and Clostridium acetobutylicum (MalH) (34). These NAD+- and Mn2+-requiring enzymes are assigned to the unique GH4 family of the GH superfamily, and the comparative sequence alignment of the six proteins revealed strict conservation of the 14 active site residues known to participate in binding of the nucleotide, metal ion, and substrate during catalysis (28, 34, 36).
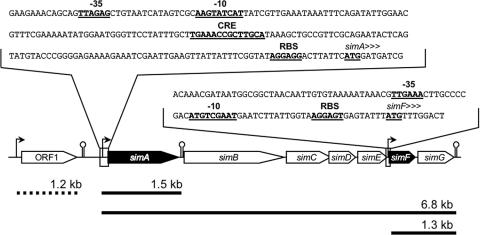
To identify genes that might be cotranscribed with LSEI_2684 or LSEI_0369 and might be subject to similar transcriptional regulation, the genome of L. casei ATCC 334 was scanned for predicted promoters and terminators using automated annotation software (Fig. 2). No promoter was identified immediately upstream of LSEI_2684, and the gene was predicted to be cotranscribed with LSEI_2685, which encodes EIICA components of the PTS (Fig. 2A). In the L. casei genome annotation, LSEI_2685 is designated a pseudogene because it contains a frameshift mutation that likely causes premature truncation during translation. A σ70 consensus promoter was identified in the upstream noncoding region of LSEI_0369, and a Rho-independent transcription terminator was predicted downstream of the LSEI_0369 stop codon (Fig. 2B). No other consensus promoter was located in the sequence containing LSEI_0369 to LSEI_0373. Therefore, expression of LSEI_0370 to LSEI_0373 requires either an atypical promoter element or transcription readthrough from LSEI_0369. A σ70-type promoter was predicted upstream of LSEI_0374, and a characteristic Rho-independent transcription terminator was identified downstream of LSEI_0375. The putative proteins encoded by LSEI_0370 to LSEI_0375 inclusive are shown in Fig. 2B. A CRE box sequence was identified between the −10 promoter element and the ATG start codon of LSEI_0369, suggesting that this promoter may be under carbon catabolite (CcpA) control. Upstream of LSEI_0369 there was an open reading frame (LSEI_0368) that was predicted to encode a transcriptional regulator. From these in silico analyses, we hypothesized that the operon shown in Fig. 2B might facilitate growth of L. casei ATCC 334 on α-glucoside substrates, including the isomers of sucrose. For convenience, genes in this multicistronic operon were designated sim (sucrose isomer metabolism) genes.
Pagl activity in cell extracts of L. casei ATCC 334.
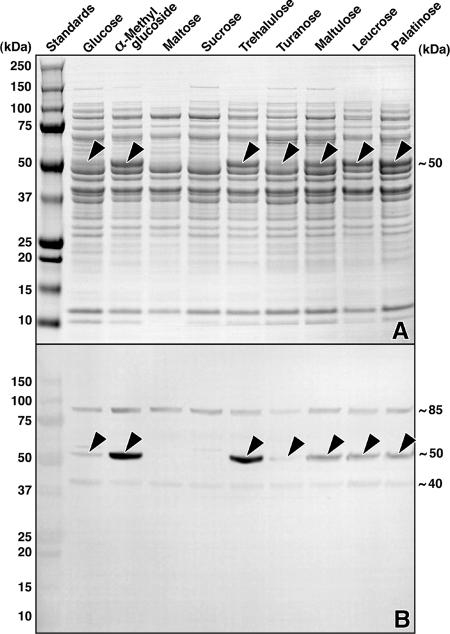
L. casei ATCC 334 grew readily on the 12 sugars tested, including all five isomers of sucrose (Table 1). Cell extracts were prepared, and the Pagl activity of these extracts was assayed using chromogenic pNPαG6P as the substrate. Comparatively high levels of enzyme activity were found in organisms grown previously on the α-glucosides described above, whereas little or no Pagl activity was detectable in extracts of cells grown previously on glucose, sucrose, melezitose, or maltose (the significance of the latter result is described in the Discussion). SDS-PAGE analysis revealed high levels of expression of a ∼50-kDa protein in the extracts that contained Pagl activity (Fig. 3A) and that cross-reacted strongly with polyclonal antibody against purified Pagl (MalH) from F. mortiferum (Fig. 3B).
TABLE 1.
Pagl activity in extracts prepared from cells of L. casei 334 grown on various sugars
| Sugar in growth mediuma | Pagl activity (nmol pNPαG6P hydrolyzed min−1 mg protein−1)b |
|---|---|
| Glucose | ∼0.4 |
| Methyl-α-d-glucopyranoside | 22.6 |
| Melezitose | NDAc |
| Trehalose | NDA |
| Maltose | NDA |
| Isomaltose | 15.8 |
| Sucrose | NDA |
| Trehalulosed | 15.8 |
| Turanosed | 2.1 |
| Maltulosed | 10.0 |
| Leucrosed | 15.0 |
| Palatinosed | 15.2 |
Organisms were grown in a medium containing individual sugars at a final concentration of 0.4% (wt/vol).
The values are averages of at least two determinations, and the variation was 5 to 10%.
NDA, no detectable activity.
The compound is an isomer of sucrose.
FIG. 3.
SDS-PAGE and Western blot analyses of extracts from cells of L. casei ATCC 334 grown on various sugars. (A) SDS-PAGE and visualization of proteins by Coomassie blue R-250 staining. Approximately 30 μg protein was applied per lane. Note the expression (arrowheads) of a ∼50-kDa protein induced by growth on the five sucrose isomers. This polypeptide is either absent or barely detectable in similarly prepared extracts from organisms grown previously on glucose, sucrose, or maltose. (B) Western blot of a duplicate gel, showing the cross-reactivity of the induced protein (arrowheads) with polyclonal antibody raised against the NAD+- and Mn2+-dependent Pagl (MalH) from F. mortiferum. The identities of the ∼40 and ∼85-kDa immunoreactive polypeptides are unknown.
Proteomic analyses.
Because of the similarities in the theoretical masses of the proteins encoded by LSEI_0369 (Mr, 48,842) and LSEI_2684 (Mr, 52,030), neither the results of the enzymatic analyses nor the results of Western blotting permitted differentiation between the putative Pagl enzymes. To differentiate these enzymes, proteomic analyses of cell extracts were performed by using 2D PAGE (Fig. 4). Because the Pagl expression was minimal in glucose- or sucrose-grown organisms, the proteomic profiles of these extracts served as references for comparison with protein expression in cells grown on four sucrose isomers. The results of 2D PAGE revealed generally similar proteomic profiles, and 23 common proteins were readily identified in all cell extracts (Table 2). However, two polypeptides, polypeptides A (Mr, ∼50,000) and B (Mr, ∼17,000), were present in the proteome of cells grown on the sucrose isomers. Neither protein was detectable in extracts from glucose- or sucrose-grown organisms. For identification, spots A and B were excised from the gels and treated with trypsin, and peptide fragments in the digests were characterized by liquid chromatography-MS-MS. Peptides identified in the digest of spot A accounted for 77% of the predicted sequence of the Pagl encoded by LSEI_0369 (simA) of the operon (Fig. 2B). Peptides recovered from the digest of spot B comprised 92% of the amino acid sequence deduced by translation of LSEI_0374 (simF), confirming that polypeptide B was the EIIA component of an α-glucoside PEP-dependent PTS. In the NCBI database, the accession number of the phosphoglycosyl hydrolase (442 residues; Mr, 49,842) is YP_805670, and the accession number of the EIIA protein (162 amino acids; Mr, 17,351) is YP_805674.
TABLE 2.
Identification by MS-MS of prominent proteins present in all cell extracts of L. casei 334 revealed by 2D PAGE (Fig. 4)
| Spot | Protein | Accession no. | Mol wt (103) | pI |
|---|---|---|---|---|
| 1 | Translation elongation factor (GTPase) | YP_807686 | 76.7 | 4.77 |
| 2 | Chaperone protein (HSP70) | YP_806781 | 67.4 | 4.77 |
| 3 | PTS enzyme I (EC 2.7.3.9) | YP_806973 | 63.3 | 5.00 |
| 4 | Pyruvate kinase (EC 2.7.1.40) | YP_806585 | 62.7 | 5.20 |
| 5 | Chaperonin GroEL (HSP 60) | YP_807425 | 57.4 | 4.89 |
| 6 | AICAR transformylase (EC 2.1.2.3) | YP_806961 | 54.7 | 5.40 |
| 7 | GTPase elongation factor(s) | YP_806555 | 43.4 | 4.87 |
| 8 | Oxaloacetate decarboxylase (EC 4.1.1.3) | YP_807056 | 51.8 | 5.10 |
| 9 | 3-Phosphoglycerate kinase (EC 2.7.2.3) | YP_806208 | 42.1 | 5.64 |
| 10 | Enolase (EC 4.2.1.11) | YP_806210 | 47.0 | 4.73 |
| 11 | DNA polymerase/clamp unit (EC 2.7.7.7) | YP_805310 | 41.3 | 4.64 |
| 12 | Elongation factor fragment | YP_805520 | 40.1 | 4.84 |
| 13 | Tagatose-1,6-bisphosphate aldolase (EC 4.1.2.40) | YP_807760 | 36.2 | 5.25 |
| 14 | Glyceraldehyde-3-phosphate dehydrogenase (EC 1.2.1.12) | YP_806207 | 36.6 | 5.68 |
| 15 | l-Lactate dehydrogenase (EC 1.1.1.27) | YP_807714 | 35.4 | 5.24 |
| Catabolite control protein (CcpA) | YP_806087 | 36.3 | 5.26 | |
| 16 | UDP-glucose-4-epimerase (EC 5.1.3.2) | YP_805947 | 36.2 | 5.69 |
| 17 | Triose phosphate isomerase (EC 5.3.1.1) | YP_806209 | 26.8 | 4.87 |
| 18 | dTDP-glucose pyrophosphorylase (EC 2.7.7.24) | YP_807212 | 32.2 | 5.48 |
| 19 | Phosphoglycerate mutase (EC 5.4.2.1) | YP_807326 | 25.8 | 5.43 |
| 20 | DNA-binding protein (Ssb) | YP_805318 | 21.0 | 5.14 |
| 21 | ATP-dependent Clp protease fragment/subunit | YP_806204 | 21.4 | 5.09 |
| 22 | PTS phosphocarrier protein (HPr) | YP_806974 | 9.1 | 4.91 |
| 23 | Chaperonin GroES (Cpn 10) | YP_807426 | 9.9 | 4.93 |
Expression, purification, and properties of Pagl (simA).
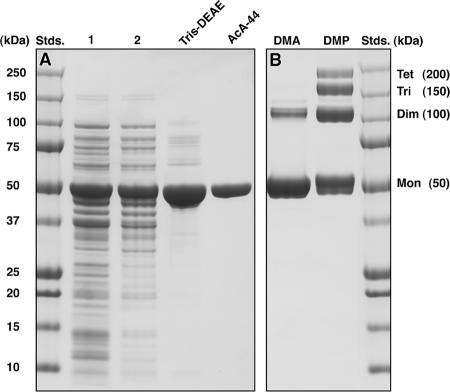
The procedures used for expression and purification of Pagl from the transformant E. coli TOP10(pAPsimA) are described in Materials and Methods. The four-step purification procedure yielded 140 mg of Pagl (specific activity, 3.78 μmol of pNPαG6P hydrolyzed min−1 mg protein−1), and SDS-PAGE of the denatured enzyme revealed a single polypeptide with an Mr of ∼50,000 (Fig. 5A). The homogeneity of the preparation was confirmed by unambiguous determination of the first 30 residues from the N terminus by microsequence analysis: MDDRKFSVLIAGGGSTYTPGIVLTLLDHIQ. This sequence is the same as the sequence deduced by translation of the Pagl gene (simA). The molecular weights of Pagl determined from two electrospray ionization-mass spectrometry measurements (48,837 and 49,851) compared favorably with the theoretical average molecular weight (49,842) deduced from the amino acid sequence encoded by simA. However, passage of the native protein through a calibrated gel filtration column (AcA-44; exclusion limit, ∼200 kDa) yielded an Mr of 95,000 to 120,000 for Pagl. Furthermore, the results obtained from cross-linking experiments with homobifunctional imidoesters provided evidence of formation of a dimeric protein that was a similar size (∼100 kDa), as well as trimeric and tetrameric oligomers (Fig. 5B). It is likely that in solution Pagl exists primarily in the homodimeric form, but our data do not preclude the possibility that there is a mixture of catalytically active dimer and tetramer species.
FIG. 5.
(A) SDS-PAGE of samples from each of the four stages of the Pagl purification procedure. TMND buffer was used throughout the purification. The chromogenic analog pNPαG6P was used as the substrate for the assay of Pagl activity. Analysis of purified Pagl by SDS-PAGE revealed a single polypeptide at an Mr of ∼50,000 (lane AcA-44). (B) Cross-linking of the ∼50-kDa subunits of Pagl in the presence of the homobifunctional imidoesters dimethyl adipimidate (lane DMA) and dimethyl pimelimidate (lane DMP) revealed formation of dimeric, trimeric, and tetrameric forms of the enzyme. Whether these oligomeric species exhibit catalytic activity is unknown. Stds., standards.
Cofactor requirements and substrate specificity of Pagl.
The GH4 family comprises a variety of hydrolases, including 6-phospho-α-glucosidases, 6-phospho-β-glucosidases, α-glucosidases, and α-galactosidases (http://www.cazy.org). All members of this unique family require a nucleotide (NAD+), a divalent metal ion, and usually reducing conditions for optimum activity. Table 3 shows that Pagl from L. casei ATCC 334 is also dependent on the same cofactors for hydrolysis of pNPαG6P. The importance of a metal ion (e.g., Mn2+) in catalysis is evident from the complete loss of Pagl activity when a chelating agent (EDTA) is added to the reaction mixture. α-Glucosides transported via the PEP-dependent PTS are accumulated as phosphorylated derivatives, and to verify the function of Pagl, it was necessary to demonstrate the enzyme-catalyzed cleavage of these compounds. Investigation of the substrate specificity of Pagl was possible because of our previous syntheses of a variety of phosphorylated disaccharides with both α and β conformations (35, 38). Consistent with its purported physiological role, Pagl hydrolyzed all phosphorylated α-glucosides tested, including the five isomeric derivatives of sucrose (Table 4). Importantly, neither sucrose-6-phosphate nor O-β-linked phosphorylated conformers were hydrolyzed by the enzyme. Experiments conducted with the three chromogenic compounds revealed the requirement for equatorial orientation of the OH groups at C-2 and C-4 of the G6P moiety for cleavage of the glycosidic linkage. Whereas pNPαG6P was readily hydrolyzed by Pagl, neither the C-2 nor the C-4 axial OH isomer (p-nitrophenyl-α-mannopyranoside-6-phosphate and p-nitrophenyl-α-galactopyranoside-6-phosphate, respectively) was a substrate for the enzyme.
TABLE 3.
Cofactor requirements for activity of purified Pagl from L. casei 334
| Addition(s) to basal assay mixturea | Pagl activity (μmol pNPαG6P hydrolyzed min−1 mg protein−1)b |
|---|---|
| None | 0.22 ± 0.04 |
| NAD+ | 0.72 ± 0.12 |
| Mn2+ | 1.74 ± 0.15 |
| NAD+ + Mn2+ | 2.68 ± 0.25 |
| NAD+ + Mn2+ + EDTA (5 mM) | NDAc |
| NAD+ + Co2+ | 2.32 ± 0.18 |
| NAD+ + Ni2+ | 2.23 ± 0.23 |
| NAD+ + Fe2+ | 2.16 ± 0.11 |
| NAD+ + Mg2+ | 1.33 ± 0.09 |
| NAD+ + Zn2+ | NDA |
The basal assay mixture (2 ml) contained 50 mM Tris-HCl buffer (pH 7.5) and 0.5 mM pNPαG6P. Cofactors were included at a final concentration of 1 mM. After 3 min of incubation at 37°C, the reaction was initiated by addition of 5 μl (69.5 μg) of purified enzyme that previously had been dialyzed for 16 h at 4°C against 25 mM Tris-HCl buffer (pH 7.5). Substrate hydrolysis was monitored as described in Materials and Methods.
The values are averages ± standard deviations of at least three independent measurements.
NDA, no detectable activity.
TABLE 4.
Substrate specificity of purified Pagl from L. casei 334a
| Substrateb | Rate of substrate hydrolysisc |
|---|---|
| Trehalulose-6′-phosphate (α, 1-1f)d | 267 |
| Sucrose-6-phosphate (α, 1-2f) | NDAe |
| Turanose-6′-phosphate (α, 1-3f)d | 135 |
| Maltulose-6′-phosphate (α, 1-4f)d | 411 |
| Leucrose-6′-phosphate (α, 1-5f)d | 62 |
| Palatinose-6′-phosphate (α, 1-6f)d | 256 |
| Maltose-6′-phosphate (α, 1-4g) | 54 |
| Trehalose-6-phosphate (α, 1-1g) | 6 |
| Methyl-α-G6P | 298 |
| Methyl-β-G6P | NDA |
| Cellobiose-6′-phosphate (β, 1-4g) | NDA |
| Gentiobiose-6′-phosphate (β, 1-6g) | NDA |
| Cellobiitol-6-phosphate (β) | NDA |
| pNPαG6P | 3.78 |
| p-Nitrophenyl-α-galactopyranoside-6-phosphate | NDA |
| p-Nitrophenyl-α-mannopyranoside-6-phosphate | NDA |
The NADP+/G6PDH-coupled spectrophotometric assay used with sugar 6-phosphate derivatives and the chromogenic assay employed with phosphorylated p-nitrophenyl substrates are described in Materials and Methods.
The substrates were added to all reaction mixtures at a final concentration of 1 mM.
The rates of hydrolysis of phosphorylated sugars are expressed in nanomoles of G6P formed per minute per milligram of protein. The rates of hydrolysis of chromogenic (p-nitrophenyl) substrates are expressed in micromoles of p-nitrophenol formed per minute per milligram protein. The rates are the means of three separate determinations.
The compound is an isomer of sucrose.
NDA, no detectable activity.
Northern blot analysis of the sim operon.
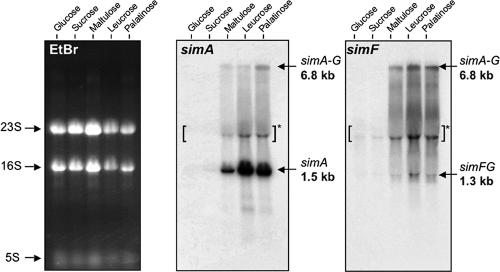
To study regulation of the sim operon, RNA was extracted from exponential-phase cells of L. casei ATCC 334 cultured with glucose, sucrose, or a sucrose isomer as the sole energy source, and the RNA was analyzed by Northern hybridization with simA- or simF-specific probes (Fig. 6). No simA mRNA was detected in RNA preparations from glucose- or sucrose-grown cells. However, a strong band at ∼1.5 kb, the predicted size of a monocistronic simA transcript, was observed in extracts from cells cultured on maltulose, leucrose, and palatinose (Fig. 6). A band at ∼6.8 kb was also detected in these samples and likely corresponded to a seven-gene simABCDEFG transcript. The 6.8-kb band was also observed in preparations from sucrose isomer-grown cells by hybridization with the simF probe. Additionally, a ∼1.3-kb band, corresponding to a bicistronic simFG transcript, was detected with the simF probe. This transcript was not present in RNA from cells cultured with glucose or sucrose and thus appeared to be under regulation similar to that of simA transcripts. The Northern blot data, together with a transcriptional map of the sim operon, are shown in Fig. 7.
FIG. 6.
Northern blot analyses of the sim operon. Total RNA was extracted from cells of L. casei ATCC 334 growing exponentially on the five sugars indicated. Hybridizations were performed using 32P-labeled probes corresponding to internal DNA regions of simA and simF. A 6.8-kb mRNA transcript was detected by both probes, which was indicative of cotranscription of all seven genes of the sim operon during growth on three sucrose isomers. The simA and simF probes also hybridized with 1.5- and 1.3-kb transcripts, respectively. The apparent band at ∼3 kb (brackets and asterisk) is a commonly observed compression artifact caused by the 23S rRNA. Ethidium bromide (EtBr)-stained rRNAs (23S, 16S, and 5S) served as loading controls (left blot).
FIG. 7.
Map of the sim operon, showing the promoter regions of simA and simF. The −35 and −10 boxes of potential promoters, the translational start codons, the putative CRE site, and the ribosomal binding sites (RBS) are indicated by boldface type and underlining. The heavy solid lines indicate RNA transcripts found only in L. casei ATCC 334 cells grown on the isomers of sucrose. The heavy dashed line indicates the ORF1 transcript.
DISCUSSION
Recent publications have described the growth of several species of bacteria (K. pneumoniae, F. mortiferum, B. subtilis, and C. acetobutylicum) on O-α-linked glucosides. However, to our knowledge, this is the first report of the metabolism of the five isomers of sucrose by an LAB. Physiological, biochemical, and genetic evidence supports our hypothesis that expression of proteins encoded by a seven-gene (sim) operon (Fig. 2B) facilitates the transport, phosphorylation, and hydrolysis of sucrose isomers (but not sucrose per se) by L. casei ATCC 334. By contrast, the functions of polypeptides encoded by the two genes of a smaller operon (Fig. 2A) are unknown, and we obtained no evidence of their expression in our studies. Because of a midgene frameshift, the putative EII(CA) transporter is unlikely to be functional, and while the LSEI_2684 gene almost certainly encodes an NAD+- and metal-dependent Pagl, the properties and substrate specificity of this GH4 enzyme cannot be determined until it is expressed and purified.
Catalytic mechanism of Pagl from L. casei ATCC 334.
Many disaccharide phosphate hydrolases have been characterized (43, 44), including 6-phospho-β-galactosidase (EC 3.2.1.85), 6-phospho-β-glucosidase (EC 3.2.1.86), and sucrose-6-phosphate hydrolase (EC 3.2.1.26). These hydrolases do not require cofactors for activity, and on the basis of primary sequence alignment, the first two enzymes are assigned to family 1 and the third enzyme is assigned to family 32 of the GH superfamily. In a landmark article in 1953, Koshland formulated two classic mechanisms for glycoside hydrolysis (13). These mechanisms have withstood the test of time and have received copious support from structural and mechanistic analyses of enzymes throughout the 109 members of the GH superfamily (31, 50, 51). In Koshland's scheme, two acidic residues (Glu and Asp) acting as a general acid or base catalyze hydrolysis of the glycosidic linkage such that (i) the reaction product has an anomeric configuration opposite that of the substrate (direct displacement or inversion) or (ii) the reaction product maintains the same C-1 configuration as the substrate (double displacement or retention).
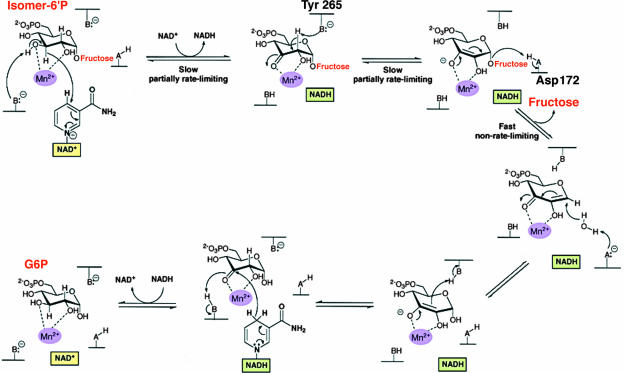
The requirements of family GH4 enzymes (including Pagl from L. casei ATCC 334) for NAD+ and a divalent metal ion are unprecedented for glycosidases, but for some time it was not clear whether these two cofactors play structural and/or catalytic roles in enzyme activity (14, 26, 27, 36, 39). The requirement for NAD+ suggested that there is a redox step in the hydrolytic process, a possibility inconsistent with either of Koshland's general mechanisms. Equally perplexing was the finding that the dinucleotide cofactor was not consumed during hydrolysis, and no NADH was formed at the end of the catalytic cycle. It was only in 2004, after the solution of the structures of the first GH4 Pagl (GlvA from B. subtilis; Protein Data Bank code 1u8x) and phospho-β-glucosidase (BglT from Thermotoga maritima; Protein Data Bank code 1up6) and extensive kinetic analyses, that a catalytic mechanism could be proposed for these unique enzymes (28, 40, 47-49). In this mechanism, hydrolysis proceeds via a sequence of oxidation-elimination-addition and reduction reactions, which result in overall retention of the configuration at the anomeric center. Figure 1 shows that the 14 catalytically functional residues of GlvA are strictly conserved in Pagl, and it is reasonable to assume that there is a similar six-step mechanism for cleavage of the phosphorylated isomers of sucrose by the L. casei enzyme (Fig. 8). We believe that in the initial oxidative step, Pagl catalyzes the extraction of the hydride from C-3 by NAD+, yielding enzyme-bound NADH. The attendant base-catalyzed deprotonation of the C-3 OH group (possibly effected by a metal-bound hydroxide) forms a ketone at the C-3 position of the G6P moiety. Ketone formation causes acidification of the C-2 proton and its subsequent removal by a suitably situated base, Tyr 265. Proton extraction at C-2 is also facilitated by the Mn2+ ion, which polarizes the carbonyl at C-3 and stabilizes the resultant enolate species. Cleavage of the anomeric C-1—O-fructose bond by acid-catalyzed assistance from Asp 172 causes the elimination of the fructose moiety of the disaccharide and formation of an enzyme-bound α,β-unsaturated ketone intermediate. A water molecule is then added to the anomeric center of this unsaturated Michael acceptor, and reprotonation at C-2 is catalyzed by Tyr 265 via reversal of the abstraction process. In the final step, the C-3 keto intermediate is reduced by the “on-board” NADH to form G6P. The reaction cycle, resulting in the production of G6P and fructose from the sucrose-6-phosphate isomer, is now complete, and Pagl assumes its original NAD+-activated state.
FIG. 8.
Proposed mechanism for the hydrolysis of phosphorylated isomers of sucrose by the NAD+- and Mn2+-dependent Pagl from L. casei ATCC 334. (Adapted from reference 47 with permission of the publisher.)
Genetic composition and regulation of the sim operon in L. casei ATCC 334.
The α-glucoside PEP-dependent PTS operons of F. nucleatum (3, 23), K. pneumoniae (37, 38), B. subtilis (36), and C. acetobutylicum (34) consist of only three genes that encode a transcriptional regulatory protein (RpiR/GntR), an NAD+- and Mn2+-dependent Pagl (designated MalH, AglB, or GlvA), and a membrane-localized EII(CB) transporter of the PTS. It is noteworthy that all of these operons lack a gene encoding EIIA, the small intracellular phosphotransfer protein that is a prerequisite for complementation and functional activity of the PTS. Previously, we hypothesized that an EIIA component from a separate PTS must substitute for the “missing” protein in the species mentioned above (23, 34). This hypothesis was tested and verified by the finding that plasmid-mediated transfer and expression of the aglA [EII(CB)] and aglB (Pagl) genes from K. pneumoniae to E. coli K-12 conferred upon the latter strain the capacity to metabolize a wide variety of α-glucosides provided that the E. coli strain contained a functional EIIAglc of the glucose PTS (22).
By contrast, the sim operon of L. casei ATCC 334 comprises seven genes, one of which (simF) encodes an EIIA protein of the PTS. The fact that this EIIA protein is expressed during growth on the isomers of sucrose suggests that the ∼17-kDa polypeptide plays an important and perhaps essential role in α-glucoside PEP-dependent PTS activity in L. casei ATCC 334. Details of the mechanism(s) for the regulation of expression of the sim operon have not been elucidated yet, but two findings are of interest. First, typical −35 and −10 promoter regions, a ribosome binding site, and a sequence consisting of 14 nucleotides that exhibit the consensus of a CRE occur immediately upstream of the first gene (simA) of the operon (Fig. 7). Second, results from Northern blot experiments revealed an mRNA fragment whose size (6.8 kb) is consistent with transcription of the entire seven-gene operon during growth of L. casei ATCC 334 on sucrose isomers (Fig. 6 and 7). These results, together with the lack of transcription during growth on rapidly metabolized substrates (glucose and sucrose), suggest that regulation of the sim operon is mediated via carbon catabolite repression. This mode of regulation of catabolic operons, particularly those involved in the dissimilation of sugars, is found in many species of gram-positive bacteria, including strains of L. casei (7, 18, 41, 45). It is likely that the CRE region is the target for a multicomponent complex (phosphorylated disaccharide, catabolite control protein A [CcpA], and phospho-seryl-HPr) that modulates expression of the sim operon in L. casei ATCC 334.
Metabolism of maltose by L. casei ATCC 334.
The α-glucoside PEP-dependent PTS operon that facilitates the metabolism of sucrose isomers in F. mortiferum, K. pneumoniae, B. subtilis, and C. acetobutylicum is also induced by, and allows the dissimilation of, maltose in these bacteria. Surprisingly, this is not the case in L. casei ATCC 334, and cells grown on the α(1→4)-linked diglucoside contain neither the NAD+- and Mn2+-dependent Pagl nor the EIIA protein encoded in the sim operon (Table 1 and Fig. 3).
There must be an alternate route for transport and catabolism of maltose in L. casei, and in recent reports Monedero et al. (18, 19) showed that the dissimilation of maltose in L. casei strain BL23 is facilitated by the products encoded by two non-PTS mal operons. The larger operon (mal1) comprises 10 genes, which are directionally opposed to a smaller five-gene operon (mal2). The first gene in the mal1 operon (malR) encodes a transcriptional regulator, and this gene is followed sequentially by malL, nplT, mapA, pgmA, malK1, dexB, malE1, malF1, and malG1. Comparative genomic analyses also revealed the presence of mal2 in the L. casei ATCC 334 strain used by Monedero et al. However, in the mal1 operon of this strain there is a 52-bp deletion in nplT (pseudogene LSEI_0981), and a 6.3-kb excision caused the loss of dexB, malE1, malF1, malG1, and a portion of the malK1 gene. Because of these extensive deletions, the strain of L. casei ATCC 334 used by Monedero et al. (in contrast to our strain) is unable to grow on maltose. Clarification of these conflicting observations and of other questions pertaining to the regulation of maltose metabolism in L. casei strains must await future investigations.
Summary: sucrose metabolism and etiology of dental caries.
For numerous reasons, sucrose is a “problem child” in oral biology. For example, this disaccharide provides the molecular precursors for the synthesis of a glycan(s) that facilitates adherence of streptococci to the tooth surface. The subsequent fermentation of sucrose to lactic acid causes the localized demineralization of tooth enamel and initiation of dental caries. Prior to our studies, it was generally assumed that oral microorganisms (including mutans streptococci and lactobacilli) are unable to metabolize the isomers of sucrose. The fact that these isomeric compounds are comparatively sweet and are produced on an industrial scale has encouraged the use of these purportedly “noncariogenic” disaccharides as substitutes for sucrose in various food products. The genes required for the utilization of sucrose isomers are not present in the genome of Streptococcus mutans, but whether oral strains of lactobacilli can metabolize these compounds has yet to be determined. L. casei ATCC 334 is used extensively in the dairy industry as a starter for cheese manufacture and is not a common resident of the oral microflora. However, in view of the potential for interspecies transfer of genetic material, we wonder if the widespread use of sucrose isomers will “encourage” dissimilation of this metabolic trait in oral bacteria. Selection of extant lactobacilli or of new species capable of utilizing the isomers of sucrose could conceivably change the composition and metabolic activities of oral biofilms.
Acknowledgments
We thank Rick Dreyfuss for assistance with photography and computer graphics, Nga Nguyen for microsequence analyses, and Mary Ann Gawinowicz for mass spectrometry fingerprinting. Experiments by Sonja Hess and Bindu Abraham were conducted at the Proteomics and Mass Spectrometry Facility of NIDDK.
This work was supported by the Intramural Research Programs of the NIDCR and NIDDK, National Institutes of Health, Bethesda, MD.
Footnotes
Published ahead of print on 29 February 2008.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bettenbrock, K., U. Siebers, P. Ehrenreich, and C.-A. Alpert. 1999. Lactobacillus casei 64H contains a phosphoenolpyruvate-dependent phosphotransferase system for uptake of galactose, as confirmed by analysis of ptsH and different gal mutants. J. Bacteriol. 181225-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouma, C. L., J. Reizer, A. Reizer, S. A. Robrish, and J. Thompson. 1997. 6-Phospho-α-d-glucosidase from Fusobacterium mortiferum: cloning, expression, and assignment to family 4 of the glycosylhydrolases. J. Bacteriol. 1794129-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chassy, B. M., and C.-A. Alpert. 1989. Molecular characterization of the plasmid-encoded lactose-PTS of Lactobacillus casei. FEMS Microbiol. Rev. 63157-165. [DOI] [PubMed] [Google Scholar]
- 5.Chassy, B. M., and J. Thompson. 1983. Regulation of lactose-phosphoenolpyruvate dependent phosphotransferase system and β-d-phosphogalactoside galactohydrolase activities in Lactobacillus casei. J. Bacteriol. 1541195-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chassy, B. M., and J. Thompson. 1983. Regulation and characterization of the galactose-phosphoenolpyruvate-dependent phosphotransferase system in Lactobacillus casei. J. Bacteriol. 1541204-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deutscher, J., C. Francke, and P. W. Postma. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70939-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gosalbes, M. J., V. Monedero, C.-A Alpert, and G. Pérez-Martínez. 1997. Establishing a model to study the regulation of the lactose operon in Lactobacillus casei. FEMS Microbiol. Lett. 14883-89. [DOI] [PubMed] [Google Scholar]
- 9.Gosalbes, M. J., V. Monedero, and G. Pérez-Martínez. 1999. Elements involved in catabolite repression and substrate induction of the lactose operon in Lactobacillus casei. J. Bacteriol. 1813928-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamada, S. 2002. Role of sweeteners in the etiology and prevention of dental caries. Pure Appl. Chem. 741293-1300. [Google Scholar]
- 11.Henrissat, B., and A. Bairoch. 1996. Updating the sequence-based classification of glycosyl hydrolases. Biochem. J. 316695-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henrissat, B., and G. Davies. 1997. Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 7637-644. [DOI] [PubMed] [Google Scholar]
- 13.Koshland, D. E., Jr. 1953. Stereochemistry and the mechanism of enzymatic reactions. Biol. Rev. 28416-436. [Google Scholar]
- 14.Lodge, J. A., T. Maier, W. Liebl, V. Hoffmann, and N. Sträter. 2003. Crystal structure of Thermotoga maritima α-glucosidase AglA defines a new clan of NAD+-dependent glycosidases. J. Biol. Chem. 27819151-19158. [DOI] [PubMed] [Google Scholar]
- 15.London, J., and N. M. Chace. 1979. Pentitol metabolism in Lactobacillus casei. J. Bacteriol. 140949-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.London, J., and S. Z. Hausman. 1983. Purification and characterization of the IIIXtl phospho-carrier protein of the phosphoenolpyruvate-dependent xylitol:phosphotransferase found in Lactobacillus casei Cl83. J. Bacteriol. 156611-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makarova, K., A. Slesarev, Y. Wolf, A. Sorokin, B. Mirkin, E. Koonin, A. Pavlov, N. Pavlova, V. Karamychev, N. Polouchine, V. Shakhova, I. Grigoriev, Y. Lou, D. Rohksar, S. Lucas, K. Huang, D. M. Goodstein, T. Hawkins, V. Plengvidhya, D. Welker, J. Hughes, Y. Goh, A. Benson, K. Baldwin, J.-H. Lee, I. Diaz-Muniz, B. Dosti, V. Smeianov, W. Wechter, R. Barabote, G. Lorca, E. Altermann, R. Barrangou, B. Ganesan, Y. Xie, H. Rawsthorne, D. Tamir, C. Parker, F. Breidt, J. Broadbent, R. Hutkins, D. O'Sullivan, J. Steele, G. Unlu, M. Saier, T. Klaenhammer, P. Richardson, S. Kozyavkin, B. Weimer, and D. Mills. 2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. USA 10315611-15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monedero, V., A. Mazé, G. Boël, M. Zuniga, S. Beaufils, A. Hartke, and J. Deutscher. 2007. The phosphotransferase system of Lactobacillus casei: regulation of carbon metabolism and connection to cold shock response. J. Mol. Microbiol. Biotechnol. 1220-32. [DOI] [PubMed] [Google Scholar]
- 19.Monedero, V., M. J. Yebra, S. Poncet, and J. Deutscher. 4 November 2007, posting date. Maltose transport in Lactobacillus casei and its regulation by inducer exclusion. Res. Microbiol. doi: 10.1016/j.resmic.2007.10.002. [DOI] [PubMed]
- 20.Ooshima, T., A. Izumitani, S. Sobue, N. Okahashi, and S. Hamada. 1983. Non-cariogenicity of the disaccharide palatinose in experimental dental caries of rats. Infect. Immun. 3943-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ooshima, T., A. Izumitani, T. Minami, T. Fujiwara, Y. Nakajima, and S. Hamada. 1991. Trehalulose does not induce dental caries in rats infected with mutans streptococci. Caries Res. 25277-282. [DOI] [PubMed] [Google Scholar]
- 22.Pikis, A., S. Hess, I. Arnold, B. Erni, and J. Thompson. 2006. Genetic requirements for growth of Escherichia coli K12 on methyl-α-d-glucopyranoside and the five α-d-glucosyl-d-fructose isomers of sucrose. J. Biol. Chem. 28117900-17908. [DOI] [PubMed] [Google Scholar]
- 23.Pikis, A., S. Immel, S. A. Robrish, and J. Thompson. 2002. Metabolism of sucrose and its five isomers by Fusobacterium mortiferum. Microbiology 148843-852. [DOI] [PubMed] [Google Scholar]
- 24.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57543-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pridmore, R. D., B. Berger, F. Desiere, D. Vilanova, C. Barretto, A.-C. Pittet, M.-C. Zwahlen, M. Rouvet, E. Altermann, R. Barrangou, B. Mollet, A. Mercenier, T. Klaenhammer, F. Arigoni, and M. A. Schell. 2004. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc. Natl. Acad. Sci. USA 1012512-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raasch, C., M. Armbrecht, W. Streit, B. Höcker, N. Sträter, and W. Liebl. 2002. Identification of residues important for NAD+ binding by the Thermotoga maritima α-glucosidase AglA, a member of the glycoside hydrolase family 4. FEBS Lett. 517267-271. [DOI] [PubMed] [Google Scholar]
- 27.Raasch, C., W. Streit, J. Schanzer, M. Bibel, U. Gosslar, and W. Liebl. 2000. Thermotoga maritima AglA, an extremely thermostable NAD+-, Mn2+-, and thiol-dependent α-glucosidase. Extremophiles 4189-200. [DOI] [PubMed] [Google Scholar]
- 28.Rajan, S. S., X. Yang, F. Collart, V. L. Y. Yip, S. G. Withers, A. Varrot, J. Thompson, G. J. Davies, and W. F. Anderson. 2004. Novel catalytic mechanism of glycoside hydrolysis based on the structure of an NAD+/Mn2+-dependent phospho-α-glucosidase from Bacillus subtilis. Structure 121619-1629. [DOI] [PubMed] [Google Scholar]
- 29.Reizer, J., M. H. Saier, Jr., J. Deutscher, F. Grenier, J. Thompson, and W. Hengstenberg. 1988. The phosphoenolpyruvate:sugar phosphotransferase system in gram-positive bacteria: properties, mechanism, and regulation. Crit. Rev. Microbiol. 15297-338. [DOI] [PubMed] [Google Scholar]
- 30.Robrish, S. A., H. M. Fales, C. Gentry-Weeks, and J. Thompson. 1994. Phosphoenolpyruvate-dependent maltose: phosphotransferase activity in Fusobacterium mortiferum ATCC 25557: specificity, inducibility, and product analysis. J. Bacteriol. 1763250-3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rye, C. S., and S. G. Withers. 2000. Glycosidase mechanisms. Curr. Opin. Chem. Biol. 4573-580. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 33.Thompson, J., C. R. Gentry-Weeks, N. Y. Nguyen, J. E. Folk, and S. A. Robrish. 1995. Purification from Fusobacterium mortiferum ATCC 25557 of a 6-phosphoryl-O-α-d-glucopyranosyl:6-phosphoglucohydrolase that hydrolyzes maltose 6-phosphate and related phospho-α-d-glucosides. J. Bacteriol. 1772505-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson, J., S. Hess, and A. Pikis. 2004. Genes malH and pagl of Clostridium acetobutylicum ATCC 824 encode NAD+- and Mn2+-dependent phospho-α-glucosidase(s). J. Biol. Chem. 2791553-1561. [DOI] [PubMed] [Google Scholar]
- 35.Thompson, J., F. W. Lichtenthaler, S. Peters, and A. Pikis. 2002. β-Glucoside kinase (BglK) from Klebsiella pneumoniae: purification, properties, and synthesis of 6-phospho-β-d-glucosides. J. Biol. Chem. 27734310-34321. [DOI] [PubMed] [Google Scholar]
- 36.Thompson, J., A. Pikis, S. B. Ruvinov, B. Henrissat, H. Yamamoto, and J. Sekiguchi. 1998. The gene glvA of Bacillus subtilis 168 encodes a metal-requiring, NAD(H)-dependent 6-phospho-α-glucosidase. J. Biol. Chem. 27327347-27356. [DOI] [PubMed] [Google Scholar]
- 37.Thompson, J., S. A. Robrish, S. Immel, F. W. Lichtenthaler, B. G. Hall, and A. Pikis. 2001. Metabolism of sucrose and its five linkage-isomeric α-d-glucosyl-d-fructoses by Klebsiella pneumoniae: participation and properties of sucrose-6-phosphate hydrolase and phospho-α-glucosidase. J. Biol. Chem. 27637415-37425. [DOI] [PubMed] [Google Scholar]
- 38.Thompson, J., S. A. Robrish, A. Pikis, A. Brust, and F. W. Lichtenthaler. 2001. Phosphorylation and metabolism of sucrose and its five linkage-isomeric α-d-glucosyl-d-fructoses by Klebsiella pneumoniae. Carbohydr. Res. 331149-161. [DOI] [PubMed] [Google Scholar]
- 39.Thompson, J., S. B. Ruvinov, D. I. Freedberg, and B. G. Hall. 1999. Cellobiose-6-phosphate hydrolase (CelF) of Escherichia coli: characterization and assignment to the unusual family 4 of glycosylhydrolases. J. Bacteriol. 1817339-7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varrot, A., V. L. Y. Yip, Y. Li, S. S. Rajan, X. Yang, W. F. Anderson, J. Thompson, S. G. Withers, and G. J. Davies. 2005. NAD+ and metal-ion dependent hydrolysis by family 4 glycosidases: structural insight into specificity for phospho-β-glucosides. J. Mol. Biol. 346423-435. [DOI] [PubMed] [Google Scholar]
- 41.Veyrat, A., V. Monedero, and G. Pérez-Martinez. 1994. Glucose transport by the phosphoenolpyruvate:mannose phosphotransferase system in Lactobacillus casei ATCC 393 and its role in carbon catabolite repression. Microbiology 1401141-1149. [DOI] [PubMed] [Google Scholar]
- 42.Viana, R., V. Monedero, V. Dossonnet, C. Vadeboncoeur, G. Pérez-Martínez, and J. Deutscher. 2000. Enzyme I and HPr from Lactobacillus casei: their role in sugar transport, carbon catabolite repression and inducer exclusion. Mol. Microbiol. 36570-584. [DOI] [PubMed] [Google Scholar]
- 43.Wilson, G., and C. F. Fox. 1974. The β-glucoside system of Escherichia coli. IV. Purification and properties of phospho-β-glucosidases A and B. J. Biol. Chem. 2495586-5598. [PubMed] [Google Scholar]
- 44.Witt, E., R. Frank, and W. Hengstenberg. 1993. 6-Phospho-β-galactosidases of Gram-positive and 6-phospho-β-glucosidase B of Gram-negative bacteria: comparison of structure and function by kinetic and immunological methods and mutagenesis of the lacG gene of Staphylococcus aureus. Protein Eng. 6913-929. [DOI] [PubMed] [Google Scholar]
- 45.Yebra, M. J., V. Monedero, M. Zuniga, J. Deutscher, and G. Pérez-Martínez. 2006. Molecular analysis of the glucose-specific phosphoenolpyruvate:sugar phosphotransferase system from Lactobacillus casei and its links with the control of sugar metabolism. Microbiology 15295-104. [DOI] [PubMed] [Google Scholar]
- 46.Yebra, M. J., A. Veyrat, M. A. Santos, and G. Pérez-Martínez. 2000. Genetics of l-sorbose transport and metabolism in Lactobacillus casei. J. Bacteriol. 182155-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yip, V. L. Y., J. Thompson, and S. G. Withers. 2007. Mechanism of GlvA from Bacillus subtilis: a detailed kinetic analysis of a 6-phospho-α-glucosidase from glycoside hydrolase family 4. Biochemistry 469840-9852. [DOI] [PubMed] [Google Scholar]
- 48.Yip, V. L. Y., A. Varrot, G. J. Davies, S. S. Rajan, X. Yang, J. Thompson, W. F. Anderson, and S. G. Withers. 2004. An unusual mechanism of glycoside hydrolysis involving redox and elimination steps by a family 4 β-glycosidase from Thermotoga maritima. J. Am. Chem. Soc. 1268354-8355. [DOI] [PubMed] [Google Scholar]
- 49.Yip, V. L. Y., and S. G. Withers. 2006. Family 4 glycoside hydrolases are special: the first β-elimination mechanism amongst glycoside hydrolases. Biocatal. Biotransform. 24167-176. [Google Scholar]
- 50.Zechel, D. L., and S. G. Withers. 2000. Glycosidase mechanisms: anatomy of a finely tuned catalyst. Acc. Chem. Res. 3311-18. [DOI] [PubMed] [Google Scholar]
- 51.Zechel, D. L., and S. G. Withers. 2001. Dissection of nucleophilic and acid-base catalysis in glycosidases. Curr. Opin. Chem. Biol. 5643-649. [DOI] [PubMed] [Google Scholar]