Abstract
Streptococcus pyogenes (group A streptococcus [GAS]), a catalase-negative gram-positive bacterium, is aerotolerant and survives H2O2 exposures that kill many catalase-positive bacteria. The molecular basis of the H2O2 resistance is poorly known. Here, we demonstrate that serotype M49 GAS lacking the Rgg regulator is more resistant to H2O2 and also decomposes more H2O2 than the parental strain. Subgenomic transcriptional profiling and genome-integrated green fluorescent protein reporters showed that a bicistronic operon, a homolog of the Streptococcus mutans ahpCF operon, is transcriptionally up-regulated in the absence of Rgg. Phenotypic assays with ahpCF operon knockouts demonstrated that the gene products decompose H2O2 and protect GAS against peroxide stress. In a murine intraperitoneal-infection model, Rgg deficiency increased the virulence of GAS, although in an ahpCF-independent manner. Rgg-mediated repression of H2O2 resistance is divergent from the previously characterized peroxide resistance repressor PerR. Moreover, Rgg-mediated repression of H2O2 resistance is inducible by cellular stresses of diverse natures—ethanol, organic hydroperoxide, and H2O2. Rgg is thus identified as a novel sensoregulator of streptococcal H2O2 resistance with potential implications for the virulence of the catalase-negative GAS.
Lactic acid bacteria, like Streptococcus pyogenes (group A streptococcus [GAS]), lack cytochromes and other heme-containing compounds. Their energy metabolism is based on glycolysis, since the crucial heme-cofactored multiprotein complexes for oxidative phosphorylation are not assembled. In addition, the lack of heme biosynthesis leads to a lack of the heme-cofactored catalase, one of the most conserved antioxidant enzymes. This is in striking contrast with the aerotolerance of lactic acid bacteria, and especially GAS, as it is believed that the main reservoir of GAS in nature is the human upper respiratory tract. In addition, GAS face H2O2 during the invasive state within the host leukocytes, in the form of the oxidative burst and possibly also from antibodies (39). Furthermore, in vitro studies have shown that GAS itself produces significant quantities of H2O2, mainly by an enzymatic lactate oxidase activity (37). Even mM concentrations of this highly membrane-diffusible oxidant have been measured from the culture supernatants (14, 22, 30, 37). GAS may therefore face H2O2 at concentrations that are lethal to many catalase-positive bacteria.
The H2O2 resistance of GAS can be increased by exposing the bacteria to sublethal doses of H2O2 or by growing the bacteria under vigorous aeration prior to challenge with a lethal amount of H2O2 (26, 36). This is a clear indication that GAS has a sensoregulatory system for H2O2 resistance. Serotype M1 and M6 GAS strains that lack a homolog of the Bacillus subtilis peroxide resistance repressor PerR tolerate H2O2 better than the parental strains (26, 36). In contrast to that of the parental strain, the H2O2 hyperresistance of serotype M1 PerR-deficient GAS cannot be increased by growth of the bacteria under vigorous aeration (36). In serotype M6 PerR-deficient GAS, exposure to sublethal H2O2 prior to challenge with a lethal concentration of H2O2 does not increase H2O2 resistance, again in contrast to the parental strain (26). These findings have created a model in which PerR is the oxidative-stress-sensing repressor of H2O2 resistance in GAS. More support for this model came when PerR was shown to repress the expression of an important streptococcal H2O2 resistance factor, Dpr/MrgA (4). Dpr/MrgA acts as a scavenger of excess cytosolic Fe(II) and therefore inhibits the Fe(II)-catalyzed formation of highly toxic hydroxyl radicals from H2O2 [H2O2 + Fe(II) → OH + −OH + Fe(III); the Fenton reaction] (34, 41). However, recent microarray analysis of the PerR regulon failed to identify additional PerR-regulated H2O2 resistance factors, although it reinforced the importance of iron homeostasis control (6). It therefore seems evident that GAS has an additional, as-yet-unidentified H2O2 resistance regulon(s).
Rgg, also known as RopB, was identified as a DNA-binding positive transcriptional regulator of secreted cysteine proteinase (SpeB) in GAS (10, 29). The current view of Rgg as a global transcriptional regulator started to emerge from proteomic studies. Rgg was shown to influence the expression of several extracellular proteins, especially in starvation, at the level of transcription (13). Phenotypic studies of the Rgg regulon have been scarce. However, it has been reported that Rgg is involved in resistance to paraquat (a compound that produces free radicals) and puromycin (a compound eliciting a heat shock-like response) (9). The aim of the current study was to evaluate the role of the Rgg regulator under physiologically significant stress conditions for catalase-negative GAS—survival under H2O2 stress. The results indicate that Rgg is a novel sensoregulator of H2O2 resistance in GAS.
MATERIALS AND METHODS
Materials.
H2O2 was purchased from Merck and beef liver catalase from Roche. t-Butyl hydroperoxide (t-BHP), horseradish peroxidase type II, 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt (ABTS), and all antibiotics were from Sigma. DNA-modifying enzymes were from either Promega or Fermentas. PCRs were conducted with either Vent (New England Biolabs) or Phusion (Finnzymes) DNA polymerase. Plasmid DNA and DNA fragments from enzymatic reactions or agarose gels were purified with kits purchased from Qiagen, Sigma, and Bio-Rad. All of the primers (sequences are shown in Table 1) were constructed based on the genomic sequence of the serotype M1 GAS strain (20) and were purchased from Thermo Electron Corporation.
TABLE 1.
Oligonucleotide primers
| Primer | Sequence 5′→3′a |
|---|---|
| pSF151-MCS | TTAGCTCACTCATTAGGCAC |
| Dpr-GFP-5′ | TTTTTTCTG CAGAGAAATCAAATACTTGCA (PstI) |
| Dpr-GFP-3′ | TTTTTTGGTACCAATAACATCTCCTTACTT (KpnI) |
| 5′-PYO-Dpr | GATCAGTATAGTAGAAGTC |
| AhpC-GFP-5′ | TTTTTTCTGCAGTAACAGCCATTCCCTGAT (PstI) |
| AhpC-GFP-3′ | TTTTTTGGTACCATAGTTGTCCTCCTTTTT (KpnI) |
| AhpC-GFPtest | TCCGTCAAGCACTATTTGC |
| AhpC-KOIN-5′ | TTTTTTCTGCAGGAAATTGCTGAATTTTCAGCTCAA (PstI) |
| AhpC-KOIN-3′ | TTTTTTGAATTCCTAGCGTCACGTCCAATACCATCA (EcoRI) |
| AhpC-KOtest | TAACAGCCATTCCCTGATCT |
| PerR-5′-KOIN | TTTTTTCTGCAGGACATTCATTCACATCAGCAA (PstI) |
| PerR-3′-KOIN | TTTTTTGAATTCATCCATAACATCAACATCCATAAA (EcoRI) |
| PerR-KOA-5′ | CTTACTTGTTATTTCTCATTTTGACAA |
| Spy1840-5′ | CCTCATTCACATCTATTTGA |
| Spy1840-3′ | ACATTGACAACTGACAAAG |
| Dpr-5′ | TTAGATGAAATGAGCGAGCGT |
| Dpr-3′ | CTCTGCTTGGAGCATCCAAAT |
| Spy1374-5′ | GCATGCAGTGCAAAATGAC |
| Spy1374-3′ | CAATTCTTTTAACTTTGCAGG |
| GpoA-5′ | CTATGATTTTACGGTTAAGGC |
| GpoA-3′ | AGTGAAATTCCATTCGATGCG |
| SodA-5′ | ACGCGTATGATGCTCTTGAAC |
| SodA-3′ | GTTAGCGGTTGATGTGATTTC |
| Spy1835-5′ | GCTTTCAATTGGTCTTTAGT |
| Spy1835-3′ | TAGAAGTAACAGATGCTACA |
| PerR-5′ | CCTAGAGCATCTACGGGA |
| PerR-3′ | CCGGACAAATCCCGTAAG |
| Rgg-5′ | TGGTGAAACCGTTGAATTC |
| Rgg-3′ | AGCACAGTCTCATAGTGAC |
| GroEL-5′ | GAGCATCCTCAATGCGAA |
| GroEL-3′ | TGCCACTACTTGAGGAAG |
| CovR-5′ | GTATGAAGTCATTGTTGAGG |
| CovR-3′ | AGTGAGAGAAATCTCATCG |
| DnaK-5′ | GTGGGATTCCACGAGGT |
| DnaK-3′ | TTCTGCCGGTCCTCTTCA |
| AhpC-5′ | CTGAACTCGGTGACCTTC |
| AhpC-3′ | CAAACTTCACCTGGATGTT |
| AhpF-5′ | TCTGGTCGCGCTACCATT |
| AhpF-3′ | TGTTACGCCATTTGGCCC |
| 16S-PYO-5′ | GATACATAGCCGACCTGA |
| 16S-PYO-3′ | GTTACAGACCAGAGAGCC |
Restriction sites are underlined, with the corresponding enzyme in parentheses.
Bacterial strains, plasmids, media, and growth conditions.
Wild-type (wt) strain NZ131 of serotype M49 and its Rgg-deficient derivative (NZ131Δrgg) have been described previously (10). No genetic-complementation experiments could be conducted in the current study due to apparent dose-dependent toxicity effects of the proteins and/or instability problems of the Escherichia coli-streptococcus shuttle plasmid used, pLZ12-Km (34, 35). Instability of plasmid DNA has been witnessed in the NZ131 strain background (10), which might explain the lack of genetic-complementation experiments in a number of other studies using this strain (2, 9, 11-13, 18). GAS strains were grown in Todd-Hewitt broth medium (Difco) supplemented with 0.5% (wt/vol) yeast extract (Biokar Diagnostics) (THY) as standing cultures at 37°C in either polypropylene tubes or glass flasks. The E. coli general cloning host DH5α was cultured in Luria-Bertani medium with vigorous shaking at 37°C. When needed, the media were solidified with 1.5% agar. Bacteria were stored at −70°C in their respective liquid growth media containing 15% (vol/vol) glycerol. Antibiotics were used at the following concentrations: (i) E. coli, 50 μg/ml of kanamycin and 50 μg/ml of spectinomycin; (ii) GAS, 7.5 μg/ml of erythromycin and 500 μg/ml of kanamycin.
Western analysis of Dpr expression.
The method to prepare total cellular protein extracts of streptococci by sonication, the Dpr-specific polyclonal antibodies, and the Dpr-specific Western analysis have been described previously (34).
Green fluorescent protein (GFP) reporter strains for dpr and the ahpCF operon.
To create a GAS-compatible suicide plasmid with a promoterless gfp gene, a gfpmut2 gene (15) was cleaved from pNE1orfRgfp (1) with EcoRI. The resulting 750-bp fragment was cloned into the EcoRI site of pSF151 (38) to create pSF151-GFP. The upstream regions of dpr and ahpC were amplified from NZ131 genomic DNA by PCR using primer pairs DPR-GFP-5′/DPR-GFP-3′ and AhpC-GFP-5′/AhpC-GFP-3′, respectively. Genomic DNA was isolated from GAS as previously described for Streptococcus suis (34). The resulting 210-bp dpr-specific and 310-bp ahpC-specific fragments were digested with PstI and KpnI and cloned into PstI-KpnI-digested pSF151-GFP. The resulting plasmids (2.5 μg), pDpr-GFP and pAhpC-GFP, were transformed into GAS as previously described for S. suis (34), and Kanr colonies (for NZ131Δrgg, Kanr and Eryr) were selected. Insertion of the reporter plasmids into the target genomic sites was verified by PCR using the primers 5′-PYO-Dpr or AhpC-GFPtest (annealing upstream of dpr or aphC, respectively) and pSF151-MCS (annealing to the vector DNA).
Inactivation of the ahpCF operon by insertional inactivation.
An internal 400-bp fragment of ahpC was amplified from NZ131 genomic DNA by PCR using the primers AhpC-KOIN-5′ and AhpC-KOIN-3′. The resulting PCR product was digested with EcoRI and PstI and cloned into the EcoRI-PstI-digested pSF151 to generate pAhpC-KO. pAhpC-KO was transformed into GAS as described above, and Kanr colonies (for NZ131Δrgg, Kanr and Eryr) were selected. Insertion of pAhpC-KO into the genomic ahpC was verified by PCR using primers AhpC-KOtest (annealing upstream of ahpC) and pSF151-MCS (annealing to the vector DNA). Inactivation of ahpC and the downstream ahpC by a polar effect was further verified by Northern hybridization (data not shown).
Inactivation of perR by insertional inactivation.
An internal 360-bp fragment of perR was amplified from NZ131 genomic DNA by PCR using the primers PerR-5′-KOIN and PerR-3′-KOIN. The resulting PCR product was digested with EcoRI and PstI and cloned into the EcoRI-PstI-digested pSF151 to generate pPerR-KO. pPerR-KO was transformed into GAS as described above, and Kanr colonies (for NZ131Δrgg, Kanr and Eryr) were selected. Insertion of pPerR-KO into the genomic perR was verified by PCR using the primers PerR-KOA-5′ (annealing upstream of perR) and pSF151-MCS (annealing to the vector DNA). Inactivation of the perR gene was further verified by Northern hybridization (data not shown).
H2O2 and t-BHP sensitivity assays. (i) Growth in the presence of H2O2 or t-BHP.
GAS strains were grown overnight (O/N) at 37°C as standing cultures in 30 ml of THY. The strains were subcultured in quadruplicate by diluting the O/N cultures 20-fold in fresh THY. The bacteria were grown at 37°C as standing cultures to early stationary phase (e-stat) (an optical density at 600 nm [OD600] of 0.8 to 0.9; 4 h after subculturing). At this stage, the bacteria were pooled and diluted 12.5-fold in quadruplicate in fresh THY either with or without H2O2 and t-BHP. The bacteria were grown at 37°C as standing cultures, and the ODs of the cultures were recorded at 600 nm either at 1-h intervals up to 8 h or after an O/N incubation (20 h). The results for any given time point were calculated as percentages ± standard deviations (SD) of the OD600 in the presence of H2O2 compared to the averaged OD600 from the control cultures without H2O2 or t-BHP (growth yield percentage).
(ii) Survival in an H2O2-containing buffer.
GAS strains were grown O/N as described above. The strains were subcultured by diluting them 25-fold in fresh THY. The bacteria were grown at 37°C as standing cultures. Ten-milliliter aliquots were taken from the cultures at exponential growth phase (exp) (OD600 = 0.3 to 0.4; 2 h after subculturing), e-stat, and stationary phase (stat) (7 h after subculturing). When the modulation of H2O2 resistance by sublethal cellular stress was studied, the toxic compounds were included in sub-growth-inhibitory amounts (250 μM H2O2, 500 μM t-BHP, and 4% ethanol) in the exp cultures, and the bacteria were allowed to grow for 2 h at 37°C as standing cultures to reach e-stat. The bacteria were harvested and washed once with 10 ml of ice-cold phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4). The bacteria were resuspended in PBS to an OD600 of ∼0.8. The suspensions were divided into 1-ml aliquots. When appropriate, chloramphenicol was added at this stage at a concentration of 50 μg/ml to inhibit de novo protein synthesis during the H2O2 stress. After 30 min of incubation at 37°C without agitation, the cells were exposed to 4.0 mM H2O2 for 2 h at 37°C without agitation. To terminate the H2O2 exposure, beef liver catalase was added at 10,000 U/ml, and the suspensions were incubated for 10 min at room temperature. Subsequently, the bacteria were diluted in PBS and plated for viability counts on THY agar. The colonies were counted after O/N growth under 6% CO2 at 37°C.
Secretion and decomposition of H2O2 by GAS.
GAS strains were grown O/N as described above. The strains were subcultured by diluting them 25-fold in fresh THY and grown at 37°C as standing cultures. Aliquots were taken from the cultures at e-stat and stat. The bacteria were harvested and washed once by PBS with vigorous vortexing. The bacteria were resuspended in PBS to an OD600 of 0.8 and divided into 1-ml aliquots. H2O2 was added at 40 μM, and the cells were incubated at 37°C without agitation. Control experiments included the following: (i) PBS incubated with 40 μM H2O2, (ii) PBS with 10,000 U/ml of beef liver catalase incubated with 40 μM H2O2, (iii) bacterial suspensions without added H2O2 (secretion controls), and (iv) suspensions of heat-killed bacteria (90°C; 60 min) incubated with 40 μM H2O2. At different time points (0, 30, 60, or 120 min), 250-μl aliquots were taken from the incubations and the bacteria were removed by centrifugation (15,000 × g; 2 min; 22°C). Two hundred microliters of the cleared incubations was mixed with 50 μl of freshly prepared H2O2 indicator solution (30 mg/ml of ABTS and 2 mg/ml of horseradish peroxidase in 100 mM Na phosphate buffer [pH 6.0]). After 5 min of incubation at 22°C, the amount of oxidized ABTS formed was measured at 405 nm (ɛmax = 36.8 mM−1 cm−1).
Slot blot Northern analysis.
NZ131 and NZ131Δrgg were grown O/N as described above. The strains were subcultured in triplicate by diluting the O/N cultures 20-fold in fresh THY. At e-stat, bacteria from 3 ml of the cultures were harvested by centrifugation, immediately snap-frozen in liquid N2, and stored at −70°C. The cells were later thawed, and total RNA was extracted by using the RNeasy Mini kit (Qiagen) according to the instructions of the manufacturer. Isolated RNAs from three independent e-stat cultures were pooled and slot blotted in triplicate onto Hybond-N+ membranes (Amershamn Biosciences) at 5 μg/slot by using a Minifold II apparatus (Schleicher & Schuell) according to the instructions of the manufacturer. DNA probes were labeled radioactively with [α-32P]CTP (Amersham) using the Prime-a-Gene Labeling System (Promega) according to the instructions of the manufacturer. DNA templates for labeling were generated by PCR using gene-specific primers as described in Table 1. Hybridization and washing conditions were as previously reported (34). The Fujifilm BA-2500 Phosphor Imaging Plate System (Fuji Photo Film Co.) was used to acquire the autoradiographs, and the hybridization signal intensities were quantified with TINA 2.0 software. The data were processed for each gene as follows: (i) signal intensities from each three slots for wt and Δrgg were normalized to the signal intensities of the same slots subsequently probed with 16S rRNA-specific probe, (ii) the normalized signal intensities for the wt slots were averaged, and (iii) the normalized signal intensities of the Δrgg slots were individually divided by the normalized and averaged signal intensities from the corresponding wt slots.
GFP reporter assays.
GAS strains were grown O/N as described above. The strains were subcultured in quadruplicate by diluting the O/N cultures 20-fold in fresh THY. The bacteria were grown to e-stat at 37°C as standing cultures. The bacteria were harvested and washed three times with 20 ml of PBS, including vigorous vortexing. The bacteria were resuspended in PBS to an OD600 of ∼1.0. The bacterial suspensions were applied as 200-μl aliquots in triplicate onto black Nunc F96 MicroWell plates. The bacterium-associated fluorescence was measured using a Victor2 Multilabel Counter (Wallac). GAS autofluorescence was measured from GFP-negative strains grown and processed as described above. The acquired mean autofluorescence counts were subtracted from the individual fluorescence counts of the corresponding GFP-positive strains to get the final GFP reporter-specific fluorescence counts.
Animal experiments.
CD-1 male mice (Harlan) were housed according to standard laboratory animal conditions and handled by protocols approved by the local Laboratory Animal Care and Use Committee, University of Turku, Turku, Finland. The mice were monitored daily and, if they showed signs of severe illness (significantly reduced movement or reduced capacity to respond to physical stimulus), were sacrificed by CO2 overdose. GAS strains were grown O/N as described above. The strains were subcultured in triplicate by diluting the O/N cultures 25-fold in 100 ml of fresh THY. The bacteria were grown to exp at 37°C as standing cultures. The bacteria were pooled, harvested, and washed twice with 20 ml of PBS with vigorous vortexing. Bacteria from pooled 300-ml culture volumes were resuspended in 20 ml of PBS (OD600, ∼0.35 as measured from 10-fold dilutions). In the first reported experiment, 10 mice in two separate cages (average infection weight, 28.3 ± 2.4 g) were intraperitoneally infected with either 100 μl of PBS or wt or Δrgg suspensions. The mice received different dilutions of GAS as follows: wt 1:1 infection, 7.3 × 108 CFU/mouse; Δrgg 1:1 infection, 6.7 × 108 CFU/ml; Δrgg 1:2 infection, a twofold dilution from the 1:1 suspension; Δrgg 1:5 infection, a fivefold dilution from the 1:1 suspension. The mice were weighed daily for up to 1 week. Weight indexes were calculated for each mouse by dividing the daily weight by the weight of the same animal at the time of infection. The reported values for each time point represent the medians of the weight indexes. The surviving animals were killed by CO2 overdose at day 7. In the second reported experiment, 10 mice in two separate cages (average infection weight, 25.3 ± 1.6 g) were intraperitoneally infected with 100 μl of bacterial suspensions in PBS as follows: Δrgg, 1.5 × 108 CFU/mouse; Δrgg ΔahpCF, 1.6 × 108 CFU/mouse. The mice were monitored and weighed daily for up to 2 weeks to acquire the weight indexes as described above. The surviving animals were killed by CO2 overdose at day 14. SPSS for Windows 11.0.1 software (SPSS Inc.) was used to create Kaplan-Meier survival curves and to calculate the log rank test statistics for comparison of different infections.
RESULTS
Rgg is involved in repression of H2O2 resistance.
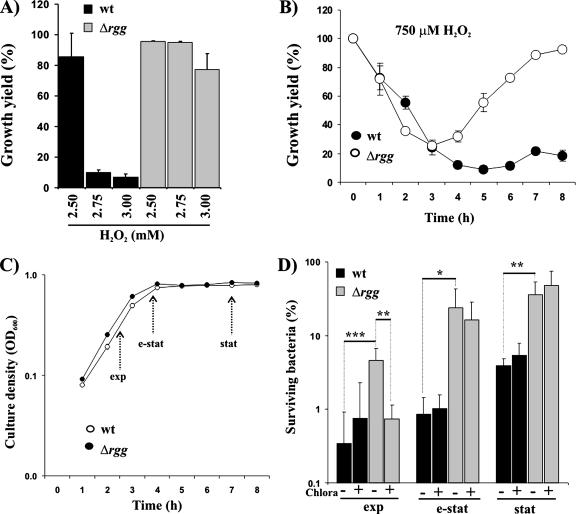
The sensitivity of GAS to H2O2 was assayed by measuring the ODs of O/N cultures that were incubated in the absence or presence of H2O2. As shown in Fig. 1A, cultures of Rgg-deficient GAS grew to higher ODs than the parental strain in the presence of 2.75 and 3 mM H2O2. Rgg-deficient GAS could also establish a replicative state faster than the parental strain in the presence of 750 μM H2O2 (Fig. 1B) in a second type of H2O2 sensitivity assay used in the current study. Importantly, the inactivation of rgg did not affect the replication of GAS in plain THY growth medium (Fig. 1C). In an independent H2O2 sensitivity assay, the wt and Δrgg were grown to different growth phases (as defined in Fig. 1C), and after being washed with PBS, the bacteria were stressed with 4 mM H2O2 in PBS. In the absence of exogenous energy sources and bacterial replication during the H2O2 exposure, the assay measures the intrinsic capacity of GAS grown to a defined growth phase to respond and to survive under H2O2 exposure. As shown in Fig. 1D, the growth phase modulated the strength of H2O2 resistance. H2O2 resistance was most efficient in bacteria grown to stat. Most importantly, the Δrgg strain was more resistant than the wt in each of the growth phases studied (13, 28, and 9 times more resistant in exp, e-stat, and stat, respectively). Coincubation with a translational protein synthesis inhibitor, chloramphenicol, decreased the H2O2 resistance of exp Δrgg to the wt level, but it had no effect on e-stat or stat Δrgg bacteria. The chloramphenicol data indicate that the increased H2O2 resistance of exp Δrgg bacteria was dependent on de novo protein synthesis, taking place even under mM level H2O2 stress, but the factors causing the increased H2O2 resistance of e-stat and stat Δrgg bacteria were already present in the bacteria at the point of bacterial harvest. Therefore, the lack of Rgg appears to influence the capacity of GAS to tolerate H2O2 starting from the early states of bacterial growth in THY. Most importantly, the data indicate that Rgg is involved in repression of H2O2 resistance in GAS.
FIG. 1.
Rgg is involved in repression of H2O2 resistance. (A) Effects of H2O2 on the ODs of O/N cultures. Bacteria from e-stat cultures were inoculated into fresh THY at different concentrations of H2O2. The OD600s of standing cultures were measured after an O/N (20-h) incubation at 37°C. The results from five independent cultures are expressed as mean growth yield percentages ± SD (OD600s in the presence of H2O2 divided by the averaged OD600s without H2O2). (B) Ability of GAS to establish a replicative state in the presence of H2O2. Bacteria from e-stat cultures were inoculated into fresh THY containing 750 μM H2O2. The bacteria were incubated at 37°C as standing cultures, and the OD600s were measured at 1-hour intervals. The results from five independent cultures are expressed as defined for panel A. (C) Effect of Rgg deficiency on the microaerophilic growth of GAS. Bacteria from e-stat cultures were inoculated into fresh THY. The bacteria were incubated at 37°C as standing cultures, and the OD600s were measured at 1-hour intervals. The results from five independent cultures are expressed as mean OD600 ± SD. (D) H2O2 sensitivity of GAS in PBS. Bacteria were harvested from different growth phases (as defined panel C), and after being washed with PBS, they were exposed in PBS to 4 mM H2O2 for 2 h at 37°C. To study whether GAS expressed proteins during the H2O2 stress, the bacteria were coincubated with 50 μg/ml of chloramphenicol (Chlora). The numbers of viable bacteria were determined by plating dilution series onto THY agar. The values are mean survival percentages ± SD (shown here in log scale) of six independent determinations from two independent cultures (CFU in the presence of H2O2 divided by the averaged CFU in the absence of H2O2). Student's t test: *, P < 0.05; **, P < 0.01; ***, P < 0.005.
Rgg is involved in repression of H2O2 decomposition.
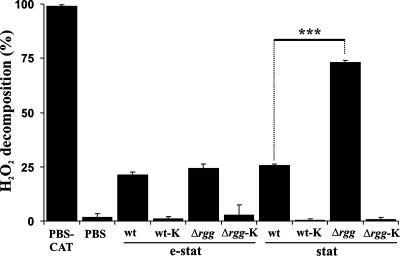
Since GAS does not express catalase, it is expected to rely on some other means to decompose H2O2. The expression of these proteins could be under the repression of Rgg and therefore could contribute to the increased H2O2 resistance of Rgg-deficient GAS. To initiate studies of this possibility, washed e-stat and stat bacteria were incubated in the presence of a sublethal concentration of H2O2 (40 μM) in PBS, and the nondecomposed H2O2 was measured at a defined time point (60 min). The H2O2 concentration was kept well below mM levels in order to retain full bacterial viability during the assay. Exp bacteria were excluded from the analysis due to the lack of up-regulated H2O2 resistance factors in Rgg-deficient GAS at the point of bacterial harvest (Fig. 1D). As shown in Fig. 2, e-stat and stat GAS decomposed H2O2, demonstrating that catalase-negative GAS may indeed decompose H2O2. Control experiments with heat-killed bacteria indicated that the detected H2O2 decomposition was dependent on bacterial viability and on a factor(s) that was heat labile. There were no significant differences in H2O2 decomposition between e-stat wt and Δrgg bacteria. However, H2O2 decomposition was markedly faster by stat Δrgg bacteria (see Fig. 4B for the kinetics). It is noteworthy that neither wt nor Δrgg bacteria secreted H2O2 during the H2O2 decomposition assay (see Fig. 4A for the kinetics). Taken together, it appears that in addition to the repression of H2O2 resistance, Rgg is also involved in the repression of H2O2 decomposition in GAS.
FIG. 2.
Rgg is involved in repression of H2O2 decomposition. Bacteria were harvested at e-stat or stat and, after being washed, were incubated in PBS in the presence of 40 μM of H2O2 for 1 h at 37°C. The incubations were cleared of bacteria by centrifugation, and the concentration of H2O2 in the supernatant was measured by a colorimetric method. The values represent mean percentages ± SD of decomposed H2O2 in five independent incubations. Control experiments included H2O2 decomposition in PBS containing 10,000 U/ml of beef liver catalase (PBS-CAT), in plain PBS, and in the presence of heat-killed (60 min; 90°C) bacteria (wt-K/Δrgg-K). Student's t test: ***, P < 0.005.
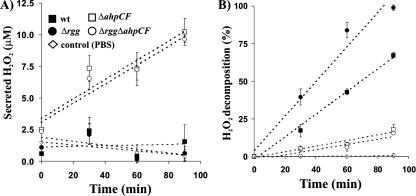
FIG. 4.
AhpCF mediates H2O2 decomposition. (A) Inactivation of the ahpCF operon converts the non-H2O2-secreting GAS into an H2O2 secretor. Bacteria were harvested at stat and, after being washed, were incubated in PBS at 37°C. At the indicated time points, the incubations were cleared of bacteria by centrifugation, and the concentrations of secreted H2O2 in the supernatants were measured by a colorimetric method. The values are mean percentages ± SD of secreted H2O2 in three independent incubations. Control experiments covered H2O2 accumulation in PBS. (B) Inactivation of the ahpCF operon converts the H2O2-decomposing GAS into an H2O2 nondecomposer. Bacteria were harvested at stat and, after being washed, were incubated in PBS in the presence of 40 μM of H2O2 at 37°C. At the indicated time points, the incubations were cleared of bacteria by centrifugation, and the concentrations of H2O2 in the supernatants were measured by a colorimetric method. The values are mean percentages ± SD of decomposed H2O2 in three independent incubations. Control experiments covered H2O2 decomposition in PBS. The amounts of secreted H2O2 (panel A) were subtracted from the measured amounts of H2O2 to get the final reported values for panel B.
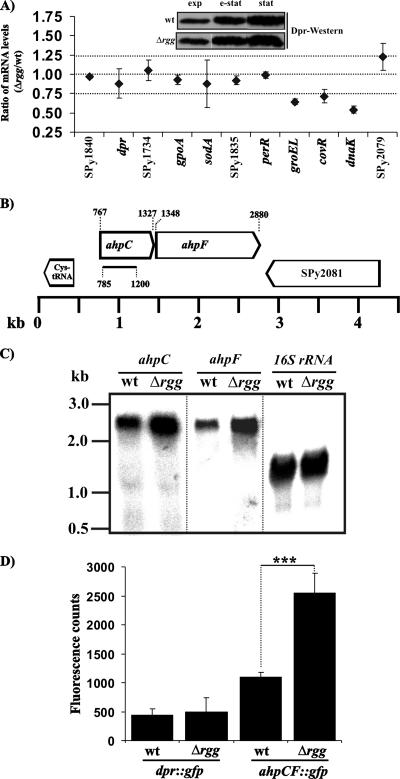
Transcriptional up-regulation of a bicistronic ahpCF operon in Δrgg bacteria.
Potentially, the increased H2O2 decomposition or H2O2 resistance of Δrgg bacteria could be traced to transcriptional up-regulation of a certain gene or a gene cluster. In addition, we reasoned that analysis of e-stat bacteria might best reflect the phenotypic differences of stat wt and Rgg-deficient GAS. Therefore, the mRNA levels of a number of different open reading frames (ORFs) with a known or a putative role in streptococcal (oxidative) stress responses were compared between the wt and Δrgg bacteria grown to e-stat. As shown in Fig. 3A, there were no significant differences in the mRNA levels of ORFs such as SPy1840 (a hypothetical gene), dpr (encoding a Dps-like peroxide resistance protein) (4), SPy1374 (encoding a putative glutathione reductase), gpoA (encoding glutathione peroxidase) (5), sodA (encoding superoxide dismutase) (22), SPy1835 (encoding a putative thioredoxin), or perR (encoding a peroxide resistance repressor) (26, 36). The mRNA levels for the general stress response components GroEL and DnaK, as well as the CovR response regulator (19), were down-regulated in the Δrgg strain. Supporting the reliability of the slot blot Northern analysis used, the decreased mRNA levels of covR in Δrgg bacteria have been previously shown by reverse transcription-PCR (12).
FIG. 3.
Rgg is involved in transcriptional repression of an ahpCF operon. (A) Slot blot Northern analysis of the mRNA levels of putative and known (oxidative) stress-related ORFs in e-stat wt and Δrgg bacteria. The ratio of mRNA levels for any give gene refers to the mean ratio ± SD of the hybridization signal intensities in three Δrgg slots divided by the averaged hybridization signal intensity from the corresponding three wt slots. (Inset) Western analysis of Dpr expression in relation to growth phase. (B) Genomic organization of the ahpCF operon according to the contig AE006628 of the GAS M1 genome (20). The bar below ahpC specifies the DNA fragment that was used to insertionally inactivate the whole ahpCF operon. (C) Northern analysis of the ahpC and ahpF transcripts. ahpC and ahpF are transcribed in the same bicistronic mRNA, with an approximate size of 2.6 kb. The membrane was probed in sequence, first with ahpC-, then with ahpF-, and finally with 16S rRNA-specific DNA probes. (D) GFP reporter assays. Promoterless gfp was inserted into the GAS genome under the control of the ahpCF operon or dpr promoters. The cellular levels of GFP were quantified from bacteria grown to e-stat. The values represent means ± SD of the GFP-specific fluorescence counts from four independent bacterial cultures. Student's t test: ***, P < 0.005.
Since Dpr, also known as MrgA (4), is the only protein known to protect streptococci against mM level H2O2 stress (4, 34, 35), Dpr expression was further compared in wt and Rgg-deficient GAS by Western blotting. By using an anti-S. suis Dpr rabbit polyclonal antiserum (34) that is cross-reactive with GAS Dpr (amino acid identity, 64% for AAN47198 [S. suis] aligned with NP_269604 [GAS]), no differences in Dpr levels could be detected in any of the growth phases studied (Fig. 3A, inset). In addition, the transcriptional activity of the dpr promoter was not affected by the Rgg deficiency, as evidenced by studies using a genome-integrated GFP reporter (Fig. 3D). Therefore, the increased H2O2 resistance of Rgg-deficient GAS appears to be mediated by a Dpr/MrgA-independent factor(s).
One ORF with an increased amount of mRNA in the Δrgg strain was identified (Fig. 3A). Examination of the genomic sequence of serotype M1 GAS (20) revealed that this ORF, ahpC, was located only 21 nucleotides upstream of another ORF, ahpF. There were no typical −10/−35 promoter sequence motifs in the intergenic region, but a characteristic Shine-Dalgarno motif (AGGAG) was found 6 nucleotides upstream of the annotated start codon of ahpF. These data indicated that ahpC and ahpF might form a bicistronic operon. Indeed, by Northern hybridization, both ORFs were localized to a similarly migrating mRNA (∼2.6 kb) (Fig. 3C). The size estimate is in good agreement with the theoretical size (∼2.2 kb) of an ahpCF bicistronic mRNA (20).
In an independent expression analysis, a promoterless gfp was genomically integrated as a single copy under the promoter of the ahpCF operon in wt and Δrgg bacteria, and the cell-associated GFP was quantified at e-stat. As shown in Fig. 3D, the promoter of ahpCF operon was more active in the Δrgg background. The data are in good agreement with the Northern hybridization data (Fig. 3A and C) and indicate that the lack of Rgg increases the transcription of the ahpCF operon in GAS.
Up-regulated expression of AhpCF mediates the increased H2O2 decomposition in Δrgg bacteria.
The gene products of ahpC and ahpF share 89% and 75% sequence identity with AhpC (BAA25695) and AhpF (BAA25696) of Streptococcus mutans, respectively (32). These proteins form a cysteine-based bicomponent H2O2-decomposing complex, as shown by in vitro studies with recombinant proteins (32). Recently, a promising but still insignificant role was reported for AhpC in H2O2 decomposition of GAS under mM levels of H2O2 (4). We extended this in vivo study further to the physiological function of AhpCF in serotype M49 GAS, and especially its up-regulated state as an explanation for the increased H2O2 decomposition in stat Rgg-deficient GAS (Fig. 2).
To disrupt the expression of the full ahpCF operon in GAS, a suicide plasmid was targeted to ahpC in the wt and Δrgg strains. The mRNA for ahpC, and also ahpF, was absent in the resulting strains (referred to hereafter as ΔahpCF and ΔrggΔahpCF) (Northern data not shown), which verified the simultaneous inactivation of ahpF by a polar effect. The involvement of AhpCF in H2O2 decomposition by GAS was studied kinetically in stat bacteria (Fig. 4) that were suspended in PBS. Strikingly, initial control experiments led to the finding that ΔahpCF and Δrgg ΔahpCF bacteria secreted H2O2, although the secretion of H2O2 by parental wt and Δrgg strains was negligible (Fig. 4A). The increased H2O2 secretion by the ΔahpCF and ΔrggΔahpCF strains correlated with a decreased capacity to decompose exogenously administrated H2O2 (Fig. 4B). This indicates that the bacteria had lost their endogenous capacity to decompose H2O2. Importantly, the low basal levels of H2O2 decomposition in the ΔahpCF and ΔrggΔahpCF strains were identical, although the Δrgg strain decomposed H2O2 faster than the wt (Fig. 4B). It appears, therefore, that the increased capacity of stat Δrgg bacteria to decompose H2O2 was mediated by the up-regulated expression of AhpCF. More importantly, AhpCF appears to provide an important means for catalase-negative GAS to decompose H2O2.
AhpCF is an organic hydroperoxide resistance factor.
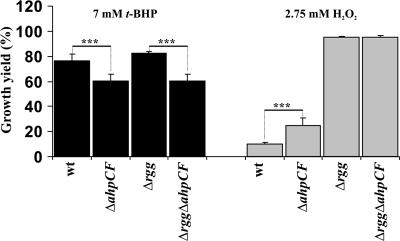
It is known that, in addition to H2O2, AhpCF of S. mutans decomposes organic hydroperoxides, as shown by in vitro studies with recombinant proteins (32). The sensitivity of GAS to organic hydroperoxide stress was assayed by measuring the ODs of O/N cultures grown in the absence or presence of t-BHP. Although the Rgg-deficient GAS grew to slightly higher ODs, there were no statistically significant differences in the densities of O/N cultures of wt and Rgg-deficient GAS in the presence of 7 mM t-BHP or any of the other tested concentrations (data not shown). However, when the ahpCF operon was inactivated, significant density decreases were detected in both wt and Δrgg backgrounds (Fig. 5). Importantly, these decreases were of the same magnitude in both backgrounds. The data provide evidence in the M49 serotype background that AhpCF is an organic hydroperoxide resistance factor in GAS.
FIG. 5.
Effects of AhpCF deficiency on H2O2 and organic hydroperoxide resistance. Bacteria from e-stat cultures were inoculated into fresh THY containing different concentrations of t-BHP or H2O2. The OD600s of standing cultures were measured after an O/N (20-h) incubation at 37°C. The results from five independent cultures are expressed as mean growth yield percentages ± SD (OD600s in the presence of t-BHP or H2O2 divided by the averaged OD600s without t-BHP or H2O2). Student's t test: ***, P < 0.005.
The role of AhpCF in H2O2 resistance.
To study the role of AhpCF in the H2O2 resistance of Rgg-deficient GAS, the ODs of O/N cultures grown in the absence or presence of H2O2 were measured. Despite the fact that stat Δrgg bacteria decomposed more H2O2 due to up-regulated AhpCF expression (Fig. 4B), AhpCF deficiency did not affect the densities of O/N Δrgg cultures under 2.75 mM H2O2 or any of the other tested concentrations (Fig. 5 and data not shown). Moreover, no significant changes due to the AhpCF deficiency were identified in the enhanced capacity of Δrgg bacteria to establish a replicative state in the presence of 750 μM H2O2 or to survive under 4 mM H2O2 stress in PBS (data not shown). It therefore appears that the increased H2O2 resistance of Δrgg bacteria under mM level, as well as μM level, H2O2 stress was unrelated to the up-regulation of AhpCF-mediated H2O2 decomposition. Given the fact that a clear phenotype for the AhpCF deficiency was observed under organic-peroxide stress (Fig. 5), an efficient AhpCF-independent H2O2 resistance factor(s) appears to be up-regulated in the Rgg-deficient GAS.
To study the role of AhpCF in the H2O2 resistance of wt GAS, the ODs of O/N cultures grown in the absence or presence of H2O2 were measured. Strikingly, AhpCF deficiency increased the densities of O/N cultures under 2.75 mM H2O2 (Fig. 5). Similar results were obtained when bacterial survival was determined under 4 mM H2O2 stress in PBS (data not shown). In this setting, the H2O2 resistance of stat ΔahpCF bacteria was comparable even to that of stat Δrgg bacteria. It is noteworthy here that the absence of AhpCF-mediated H2O2 decomposition did not increase the H2O2 resistance of Rgg-deficient GAS in any of the studied settings (Fig. 5 and data not shown). Moreover, AhpCF deficiency in B. subtilis is known to increase H2O2 resistance, similar to the AhpCF deficiency in the wt GAS background (Fig. 5), due to induction of oxidative-stress resistance pathways during prestress bacterial manipulations (7). The data point to a possibility that Rgg-mediated repression of H2O2 resistance in GAS could be derepressed by cytosolic H2O2 or by some other sources of cellular stress, which in turn are under the control of AhpCF (see “Discussion” and data below) (Fig. 6).
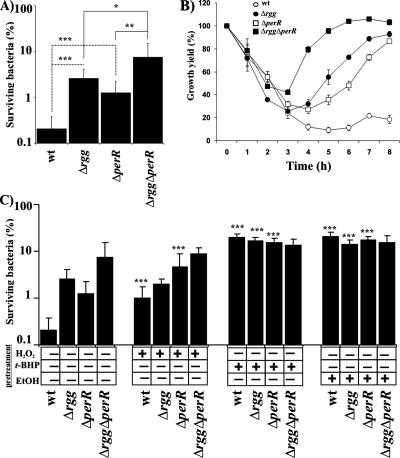
FIG. 6.
The roles of Rgg and PerR in stress-induced adaptation to H2O2 stress. (A) H2O2 sensitivity of GAS in PBS. Bacteria were harvested from e-stat and, after being washed, were exposed in PBS to 4 mM H2O2 for 2 h at 37°C. The numbers of viable bacteria were determined by plating dilution series onto THY agar. The values are mean survival percentages ± SD of six independent determinations from two independent cultures (CFU in the presence of H2O2 divided by the averaged CFU in the absence of H2O2). Student's t test: *, P < 0.05; **, P < 0.01; ***, P < 0.005. (B) Effect of PerR deficiency on the ability of GAS to establish a replicative state in the presence of H2O2. Bacteria from e-stat cultures were inoculated into fresh THY containing 750 μM H2O2. The bacteria were incubated at 37°C as standing cultures, and the OD600s were measured at 1-hour intervals. The results from five independent cultures are expressed as mean growth yield percentages ± SD (OD600s in the presence of H2O2 divided by the averaged OD600s without H2O2). Part of the data is also shown in Fig. 1B. (C) H2O2 sensitivity of stress-adapted GAS in PBS. Bacteria were grown to exp and, after the addition of H2O2, t-BHP, or ethanol (pretreatment) at a sublethal concentration, the bacteria were grown to e-stat. The bacteria were harvested and, after being washed, were exposed in PBS to 4 mM H2O2 for 2 h at 37°C. The numbers of viable bacteria were determined by plating dilution series onto THY agar. The values are mean survival percentages ± SD of six independent determinations from two independent cultures (CFU in the presence of H2O2 divided by the averaged CFU in the absence of H2O2). Significant increase in H2O2 resistance, due to the stress adaptation, is indicated by asterisks (Student's t test; ***, P < 0.005).
Rgg and PerR are divergently involved in repression of H2O2 resistance.
M1 and M6 GAS strains lacking a homolog of the B. subtilis peroxide resistance repressor PerR tolerate H2O2 better than the parental strains (26, 36). A similar phenotype was found in the current study for Rgg deficiency in the M49 strain (Fig. 1). Moreover, it has been shown that PerR-mediated repression of H2O2 resistance can be derepressed in M1 and M6 serotype strains of GAS by vigorous aeration of the cultures or by exposure of the bacteria to a sublethal concentration of H2O2 prior to lethal H2O2 stress (26, 36). To study the possible role of Rgg in (oxidative) stress-induced adaptation to H2O2 stress, wt and Δrgg M49 serotype strains that also lacked PerR were constructed. It is noteworthy that the mRNA levels of perR and one of its regulatory target dpr genes were unaffected by the lack of Rgg in e-stat (Fig. 3), which was also evidenced for Dpr by Western blotting in exp, e-stat, and stat (Fig. 3), nor did the lack of PerR affect the mRNA levels of rgg in e-stat (data not shown). This indicates that the Rgg and PerR regulons are not cross-regulated.
As reported previously for the M1 and M6 serotypes of GAS (26, 36), inactivation of perR in M49 serotype GAS increased bacterial survival under mM level H2O2 stress (Fig. 6A). Compared to the H2O2 resistance of Δrgg bacteria, PerR deficiency had a weaker effect. When both of the genes were inactivated, the double-mutant strain tolerated H2O2 approximately 35-fold better than the wt, and also better than the strains with individual perR or rgg inactivations. Therefore, Rgg and PerR appear to independently repress H2O2 resistance in GAS. The data were also supported in an independent experimental setting, where the ability of GAS strains to establish a replicative state in THY was measured in the presence of 750 μM H2O2 (Fig. 6B).
Rgg is involved in repression of stress-inducible H2O2 resistance.
Growth of wt GAS under sublethal (sub-growth-inhibitory) doses of H2O2 (250 μM), ethanol (4%), and t-BHP (500 μM) prior to high-level H2O2 stress increased bacterial survival (5-, 65-, and 65-fold, respectively [Fig. 6C]). The H2O2 resistance of Δrgg bacteria could not be increased by H2O2, but significant six- and fivefold inductions were achieved with t-BHP and ethanol, respectively. The H2O2 resistance of ΔperR bacteria could be increased approximately 5-fold by H2O2 and approximately 20-fold by t-BHP and ethanol. Strikingly, the H2O2 resistance of Δrgg ΔperR bacteria could not be modulated by any of the toxic compounds. The data indicate that both Rgg and PerR are involved in repression of stress-inducible H2O2 resistance factors in GAS. Under the assay conditions in serotype M49 GAS, Rgg appears to repress the expression of H2O2-inducible H2O2 resistance factors, and PerR, together with Rgg, represses the expression of t-BHP- and ethanol-inducible H2O2 resistance factors.
The roles of Rgg and AhpCF in virulence.
Having established that Rgg is involved in adaptive H2O2 resistance and that one of its regulatory targets, AhpCF, is important for GAS to decompose H2O2, as well as to protect GAS against organic-peroxide stress, we analyzed the roles of these proteins in the virulence of GAS. The studies were also highlighted by the fact that several known virulence factors, like M protein, C5a peptidase, and streptolysins S and O, were known to be transcriptionally up-regulated in the Rgg-deficient GAS (12). However, definite in vivo proof of the increased virulence of Rgg-deficient GAS was still lacking.
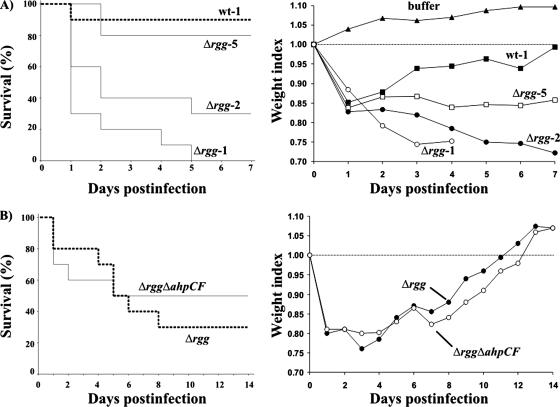
Mice were intraperitoneally infected with different GAS strains grown to exp. At a 7.3 × 108-CFU/mouse dose of wt GAS, 90% of the mice were alive at the end of the experiment on day 7 (Fig. 7A). However, when a similar dose of Rgg-deficient GAS (6.7 × 108 CFU/mouse) was used, all of the mice were dead on day 5. When the same Δrgg suspension was diluted twofold, 70% of the mice were dead on day 7. By a fivefold dilution, the virulence of Δrgg bacteria could be lowered statistically to the same level (80% survival) as with undiluted wt suspension. The high virulence of Δrgg bacteria in comparison with the wt was also reflected in the weight indexes of the surviving mice, a parameter that measured the general condition of the infected animals (Fig. 7A).
FIG. 7.
Roles of Rgg and AhpCF in virulence. (A) Rgg-deficient GAS is more virulent than wt GAS. Ten mice divided into two cages were challenged intraperitoneally with wt (7.3 × 108 CFU; wt-1), Δrgg (6.7 × 108 CFU; Δrgg-1), twofold-diluted Δrgg-1 (Δrgg-2), or fivefold-diluted Δrgg-1 (Δrgg-5) bacteria. The survival of the mice was monitored daily for up to 7 days. The survival data are presented as a Kaplan-Meier plot. (B) Up-regulation of AhpCF expression in Δrgg does not account for the increased virulence of Δrgg. Ten mice divided into two cages were challenged intraperitoneally with Δrgg (1.5 × 108 CFU) or Δrgg ΔahpCF (1.6 × 108 CFU) bacteria. The survival of the mice was monitored daily up to 14 days. The survival data are presented as a Kaplan-Meier plot. All mice were weighed daily to get a readout of the general condition of the infected animals as weight indexes.
To study whether the up-regulation of AhpCF expression contributes to the increased virulence of Rgg-deficient GAS, mice were infected with doses of 1.5 × 108 CFU/mouse of Δrgg bacteria and 1.6 × 108 CFU/mouse of Δrgg ΔahpCF bacteria, and the course of infection was followed for a period of 2 weeks. Despite the slight differences in morbidity, there were no statistical differences in the survival of the animals between the groups (Fig. 7B). This was also evidenced by the weight indexes of the surviving mice. It is therefore conclusive that, although the absence of Rgg converts GAS into a hypervirulent pathogen, this hypervirulence is not mediated by the up-regulated AhpCF expression. However, the Rgg repressor target AhpCF may still be an important tissue- or serotype-specific virulence factor in GAS. It was recently reported that AhpC is needed for virulence of the serotype M6 GAS strain in a mouse subcutaneous-infection model (4).
DISCUSSION
PerR is a prototype of a transcriptional repressor that regulates the expression of peroxide defense genes in gram-positive bacteria. It was described in B. subtilis, where it modulates the expression of catalase, the alkyl hydroperoxide reductase AhpCF, and the Dps-like peroxide resistance protein Dpr/MrgA (8). Two independent studies (26, 36) of different serotype strains of GAS (M1 and M6) have shown that PerR deficiency increases H2O2 resistance. PerR is also known to regulate Dpr/MrgA expression (4). In striking contrast to B. subtilis, the PerR deficiency does not appear to influence the expression of AhpCF in GAS (4). In the current study, the PerR-deficient M49 serotype strain of GAS was more resistant to H2O2, again reinforcing the importance of PerR in the oxidative-stress resistance of GAS. More importantly, the M49 serotype strain lacking the transcriptional regulator Rgg (10) was more resistant to H2O2 than the parental strain and also decomposed H2O2 more efficiently. Furthermore, Rgg deficiency increased the H2O2 resistance of PerR-deficient GAS and did not alter the expression of the PerR-regulatory target Dpr/MrgA. The data indicate that GAS has a PerR-independent and Rgg-regulated peroxide resistance regulon.
In an attempt to identify genes of the novel peroxide resistance Rgg regulon, the current study utilized Northern analysis of GAS ORFs having similarity with known bacterial oxidative-stress resistance genes. By focusing on bacteria grown to e-stat, a significant up-regulation of a bicistronic ahpCF operon was identified in the Rgg-deficient GAS, as also evidenced by genome-integrated GFP reporters. The gene products of ahpC and ahpF share 89% and 75% amino acid sequence identity with AhpC and AhpF of S. mutans, respectively (32). In vitro, these proteins form a cysteine-based bicomponent H2O2-decomposing complex, alkyl hydroperoxide reductase (32), which was originally identified in Salmonella enterica serovar Typhimurium (24). AhpC is a peroxide-reducing component and, in addition to organic peroxides, is capable of decomposing H2O2 (17, 31). In support of these in vitro studies and one earlier in vivo study in a serotype M6 GAS background (4), AhpCF-deficient GAS decomposed H2O2 less efficiently than the parental strain. More importantly, the same basal level of H2O2 decomposition was detected in the Rgg-deficient background when the ahpCF operon was inactivated. It therefore appears that the increased H2O2 decomposition in the Rgg-deficient GAS was exclusively mediated by the up-regulated expression of the ahpCF operon. Finally, it was found that ahpCF deficiency, whether in the wt or the Rgg-deficient background, converted GAS more sensitive to organic hydroperoxide (t-BHP) stress. It is therefore conclusive that the ahpCF operon belongs to the novel peroxide resistance Rgg regulon and that the corresponding gene products provide means for catalase-negative GAS to decompose H2O2, as well as to survive under organic-peroxide stress.
PerR of B. subtilis senses H2O2 by Fe(II)-catalyzed histidine oxidation (28). This irreversible protein modification weakens the PerR-promoter interaction, and the expression of the PerR regulon is activated (28). The alkyl hydroperoxide reductase AhpCF, in addition to catalase, fine tunes this response by controlling the cytosolic H2O2 concentration (7). It was therefore of interest to see that the ahpCF-deficient GAS survived under H2O2 stress better than the parental strain. More importantly, this effect was not detected in the Rgg-deficient background. It was reasoned that this is a direct reflection of a regulatory scheme in which the Rgg-mediated repression of H2O2 resistance can be derepressed by H2O2. Indeed, when the wt and Rgg-deficient GAS were grown in the presence of a sublethal concentration of H2O2, the H2O2 resistance of the Rgg-deficient GAS did not change but the resistance of the wt GAS increased significantly. More importantly, the same level of increase was detected in the PerR-deficient GAS, but again, not in the PerR and Rgg double-deficient background. The data add proof of an H2O2-mediated induction of the peroxide resistance Rgg regulon. However, the data also create a conflict with previous knowledge about the PerR-mediated repression of H2O2 resistance. It has been reported that in serotype M6 PerR-deficient GAS, the H2O2 hyperresistance cannot be increased by growth in the presence of a sublethal concentration of H2O2, although the resistance of the parental strain increases significantly (26). This discrepancy remains to be studied and could be explained by reasons such as the following. (i) Different serotype backgrounds. It is known that genome scale variability is extensive between and even within different serotype strains of GAS (3). (ii) Experimental setups and growth conditions. It is known that the H2O2 sensitivity of B. subtilis PerR is dependent on its metal ion composition, e.g., it is weakened significantly by manganese (21, 23). Even slight differences in growth conditions and other experimental steps might create variability in GAS metal ion homeostasis, especially when different serotype strains are studied (3). (iii) Variations in the PerR and Rgg sequences and their expression levels. Natural variants of B. subtilis PerR that are as sensitive to H2O2 as the wt iron-cofactored PerR have been isolated, although they are manganese cofactored (8). Whatever the reason, the current study identified conditions under which the adaptive and H2O2-inducible H2O2 resistance of GAS can be studied independently of PerR.
The regulation of H2O2 resistance in GAS based on environmental stimuli other than H2O2 remains poorly known. It has been reported that in serotype M1 PerR-deficient GAS, H2O2 hyperresistance cannot be increased by growth under vigorous aeration, although resistance of the parental strain increases significantly (36). This indicates that oxygen may act as one adaptation stimulus. It has also been reported that ethanol (an activator of the general stress response pathway) (40) induces higher levels of H2O2 resistance than H2O2 in a serotype M6 background (26). The current study identified t-BHP (a mimic of cellular organic peroxides) and also ethanol as extremely potent inducers of H2O2 resistance. Interestingly, the data indicate that Rgg and PerR coordinately regulate all the H2O2 resistance factors in serotype M49 GAS that are inducible by t-BHP or ethanol. The data are remarkable, given that ethanol was reported to induce an additional level of H2O2 resistance in a PerR-deficient M6 serotype strain (26). The authors suggested that an additional regulatory component independent of PerR is responsible for this phenotype (26). The current data indicate that Rgg might have been the missing regulatory component.
The current study was unable to identify the Rgg-repressive target(s) that mediates H2O2 resistance in GAS. Therefore, the identification of all the genes of the novel peroxide resistance Rgg regulon remains an important aspect of further studies. Some conclusions can already be drawn based on the available genome-wide expression profiles of wt and Rgg-deficient GAS (9, 16). HtrA (SPy2216/DegP) up-regulation, in addition to AhpC, has been witnessed at the protein level in Rgg-deficient GAS (9). This serine protease is involved in the surveillance of protein misfolding, and isogenic DegP-deficient GAS is more sensitive to thermal and oxidative stress, as well as less virulent, in a mouse infection model (25). Another candidate is ClpL (SPy0888), which has been shown to be up-regulated (at the protein level) in the Rgg-deficient GAS (9). Although there are no data available on the function of this protein in GAS, its chaperone activity and protective function under heat stress are well described in Streptococcus pneumoniae (27). Transcriptional up-regulation of a putative heat shock protein, HSP33 (SPy123) (16), in a Rgg-deficient background also merits further investigation.
From a regulatory perspective, it is interesting that H2O2-mediated induction of the Rgg-regulatory target ahpCF operon was not detected by using the genome-integrated GFP reporters (data not shown). Indeed, growth in the presence of a sublethal concentration of H2O2 did not increase the H2O2 resistance of wt GAS to the level of Rgg-deficient GAS. Therefore, the novel peroxide resistance Rgg regulon appears to contain genes that have different sensitivities to the H2O2 stimulus. The exact sensory mechanisms of Rgg for H2O2, t-BHP, and ethanol are completely unknown at the moment, and other regulatory components might be involved. However, as a cytosolic 35-kDa protein, Rgg contains an unexpectedly high number (a total of 10) of cysteine residues, some of which could serve as redox switches in H2O2 and/or t-BHP sensing (33). Finally, the finding that the H2O2-hyperresistant Rgg-deficient GAS is more virulent in a murine intraperitoneal-infection model creates a promising possibility to study the still elusive correlation between the peroxide resistance and virulence mechanisms of GAS.
Acknowledgments
The study was financially supported by the Academy of Finland and the Turku Graduate School of Biomedical Sciences (TuBS).
We gratefully acknowledge Michael Chaussee and Jeremy Brown for kindly providing us bacterial strains and plasmids. Miikka Korja is acknowledged for assistance with the animal experiment statistics. Vuokko Loimaranta is acknowledged for critical reading of the manuscript.
Footnotes
Published ahead of print on 29 February 2008.
REFERENCES
- 1.Bartilson, M., A. Marra, J. Christine, J. S. Asundi, W. P. Schneider, and A. E. Hromockyj. 2001. Differential fluorescence induction reveals Streptococcus pneumoniae loci regulated by competence stimulatory peptide. Mol. Microbiol. 39126-135. [DOI] [PubMed] [Google Scholar]
- 2.Bates, C. S., C. Toukoki, M. N. Neely, and Z. Eichenbaum. 2005. Characterization of MtsR, a new metal regulator in group A streptococcus, involved in iron acquisition and virulence. Infect. Immun. 735743-5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beres, S. B., E. W. Richter, M. J. Nagiec, P. Sumby, S. F. Porcella, F. R. DeLeo, and J. M. Musser. 2006. Molecular genetic anatomy of inter- and intraserotype variation in the human bacterial pathogen group A Streptococcus. Proc. Natl. Acad. Sci. USA 1037059-7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenot, A., K. Y. King, and M. G. Caparon. 2005. The PerR regulon in peroxide resistance and virulence of Streptococcus pyogenes. Mol. Microbiol. 55221-234. [DOI] [PubMed] [Google Scholar]
- 5.Brenot, A., K. Y. King, B. Janowiak, O. Griffith, and M. G. Caparon. 2004. Contribution of glutathione peroxidase to the virulence of Streptococcus pyogenes. Infect. Immun. 72408-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brenot, A., B. F. Weston, and M. G. Caparon. 2007. A PerR-regulated metal transporter (PmtA) is an interface between oxidative stress and metal homeostasis in Streptococcus pyogenes. Mol. Microbiol. 631185-1196. [DOI] [PubMed] [Google Scholar]
- 7.Bsat, N., L. Chen, and J. D. Helmann. 1996. Mutation of the Bacillus subtilis alkyl hydroperoxide reductase (ahpCF) operon reveals compensatory interactions among hydrogen peroxide stress genes. J. Bacteriol. 1786579-6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bsat, N., A. Herbig, L. Casillas-Martinez, P. Setlow, and J. D. Helmann. 1998. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol. Microbiol. 29189-198. [DOI] [PubMed] [Google Scholar]
- 9.Chaussee, M. A., E. A. Callegari, and M. S. Chaussee. 2004. Rgg regulates growth phase-dependent expression of proteins associated with secondary metabolism and stress in Streptococcus pyogenes. J. Bacteriol. 1867091-7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaussee, M. S., D. Ajdic, and J. J. Ferretti. 1999. The rgg gene of Streptococcus pyogenes NZ131 positively influences extracellular SPE B production. Infect. Immun. 671715-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaussee, M. S., G. A. Somerville, L. Reitzer, and J. M. Musser. 2003. Rgg coordinates virulence factor synthesis and metabolism in Streptococcus pyogenes. J. Bacteriol. 1856016-6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaussee, M. S., G. L. Sylva, D. E. Sturdevant, L. M. Smoot, M. R. Graham, R. O. Watson, and J. M. Musser. 2002. Rgg influences the expression of multiple regulatory loci to coregulate virulence factor expression in Streptococcus pyogenes. Infect. Immun. 70762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaussee, M. S., R. O. Watson, J. C. Smoot, and J. M. Musser. 2001. Identification of Rgg-regulated exoproteins of Streptococcus pyogenes. Infect. Immun. 69822-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cleary, P. P., and A. Larkin. 1979. Hyaluronic acid capsule: strategy for oxygen resistance in group A streptococci. J. Bacteriol. 1401090-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 17333-38. [DOI] [PubMed] [Google Scholar]
- 16.Dmitriev, A. V., E. J. McDowell, K. V. Kappeler, M. A. Chaussee, L. D. Rieck, and M. S. Chaussee. 2006. The Rgg regulator of Streptococcus pyogenes influences utilization of nonglucose carbohydrates, prophage induction, and expression of the NAD-glycohydrolase virulence operon. J. Bacteriol. 1887230-7241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ellis, H. R., and L. B. Poole. 1997. Roles for the two cysteine residues of AhpC in catalysis of peroxide reduction by alkyl hydroperoxide reductase from Salmonella typhimurium. Biochemistry 3613349-13356. [DOI] [PubMed] [Google Scholar]
- 18.Eriksson, A., and M. Norgren. 2003. Cleavage of antigen-bound immunoglobulin G by SpeB contributes to streptococcal persistence in opsonizing blood. Infect. Immun. 71211-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Federle, M. J., K. S. McIver, and J. R. Scott. 1999. A response regulator that represses transcription of several virulence operons in the group A streptococcus. J. Bacteriol. 1813649-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 984658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuangthong, M., A. F. Herbig, N. Bsat, and J. D. Helmann. 2002. Regulation of the Bacillus subtilis fur and perR genes by PerR: not all members of the PerR regulon are peroxide inducible. J. Bacteriol. 1843276-3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibson, C. M., T. C. Mallett, A. Claiborne, and M. G. Caparon. 2000. Contribution of NADH oxidase to aerobic metabolism of Streptococcus pyogenes. J. Bacteriol. 182448-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helmann, J. D., M. F. Wu, A. Gaballa, P. A. Kobel, M. M. Morshedi, P. Fawcett, and C. Paddon. 2003. The global transcriptional response of Bacillus subtilis to peroxide stress is coordinated by three transcription factors. J. Bacteriol. 185243-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobson, F. S., R. W. Morgan, M. F. Christman, and B. N. Ames. 1989. An alkyl hydroperoxide reductase from Salmonella typhimurium involved in the defense of DNA against oxidative damage. Purification and properties. J. Biol. Chem. 2641488-1496. [PubMed] [Google Scholar]
- 25.Jones, C. H., T. C. Bolken, K. F. Jones, G. O. Zeller, and D. E. Hruby. 2001. Conserved DegP protease in gram-positive bacteria is essential for thermal and oxidative tolerance and full virulence in Streptococcus pyogenes. Infect. Immun. 695538-5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.King, K. Y., J. A. Horenstein, and M. G. Caparon. 2000. Aerotolerance and peroxide resistance in peroxidase and PerR mutants of Streptococcus pyogenes. J. Bacteriol. 1825290-5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwon, H. Y., S. W. Kim, M. H. Choi, A. D. Ogunniyi, J. C. Paton, S. H. Park, S. N. Pyo, and D. K. Rhee. 2003. Effect of heat shock and mutations in ClpL and ClpP on virulence gene expression in Streptococcus pneumoniae. Infect. Immun. 713757-3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, J. W., and J. D. Helmann. 2006. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature 440363-367. [DOI] [PubMed] [Google Scholar]
- 29.Lyon, W. R., C. M. Gibson, and M. G. Caparon. 1998. A role for trigger factor and an rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 176263-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malke, H., R. Starke, H. E. Jacob, and W. Kohler. 1974. Bacteriocine-like activity of group-A streptococci due to the production of peroxide. J. Med. Microbiol. 7367-374. [DOI] [PubMed] [Google Scholar]
- 31.Poole, L. B., and H. R. Ellis. 1996. Flavin-dependent alkyl hydroperoxide reductase from Salmonella typhimurium. Biochemistry 3556-64. [DOI] [PubMed] [Google Scholar]
- 32.Poole, L. B., M. Higuchi, M. Shimada, M. L. Calzi, and Y. Kamio. 2000. Streptococcus mutans H2O2-forming NADH oxidase is an alkyl hydroperoxide reductase protein. Free Radic. Biol. Med. 28108-120. [DOI] [PubMed] [Google Scholar]
- 33.Poole, L. B., P. A. Karplus, and A. Claiborne. 2004. Protein sulfenic acids in redox signaling. Annu. Rev. Pharmacol. Toxicol. 44325-347. [DOI] [PubMed] [Google Scholar]
- 34.Pulliainen, A. T., S. Haataja, S. Kähkönen, and J. Finne. 2003. Molecular basis of H2O2 resistance mediated by Streptococcal Dpr. J. Biol. Chem. 2787996-8005. [DOI] [PubMed] [Google Scholar]
- 35.Pulliainen, A. T., A. Kauko, S. Haataja, A. C. Papageorgiou, and J. Finne. 2005. Dps/Dpr ferritin-like protein: insights into the mechanism of iron incorporation and evidence for a central role in cellular iron homeostasis in Streptococcus suis. Mol. Microbiol. 571086-1100. [DOI] [PubMed] [Google Scholar]
- 36.Ricci, S., R. Janulczyk, and L. Bjorck. 2002. The regulator PerR is involved in oxidative stress response and iron homeostasis and is necessary for full virulence of Streptococcus pyogenes. Infect. Immun. 704968-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saito, M., S. Ohga, M. Endoh, H. Nakayama, Y. Mizunoe, T. Hara, and S. Yoshida. 2001. H2O2-nonproducing Streptococcus pyogenes strains: survival in stationary phase and virulence in chronic granulomatous disease. Microbiology 1472469-2477. [DOI] [PubMed] [Google Scholar]
- 38.Tao, L., D. J. LeBlanc, and J. J. Ferretti. 1992. Novel streptococcal-integration shuttle vectors for gene cloning and inactivation. Gene 120105-110. [DOI] [PubMed] [Google Scholar]
- 39.Wentworth, P., Jr., L. H. Jones, A. D. Wentworth, X. Zhu, N. A. Larsen, I. A. Wilson, X. Xu, W. A. Goddard III, K. D. Janda, A. Eschenmoser, and R. A. Lerner. 2001. Antibody catalysis of the oxidation of water. Science 2931806-1811. [DOI] [PubMed] [Google Scholar]
- 40.Voelker, U., A. Voelker, B. Maul, M. Hecker, A. Dufour, and W. G. Haldenwang. 1995. Separate mechanisms activate sigma B of Bacillus subtilis in response to environmental and metabolic stresses. J. Bacteriol. 1773771-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamamoto, Y., M. Higuchi, L. B. Poole, and Y. Kamio. 2000. Role of the dpr product in oxygen tolerance in Streptococcus mutans. J. Bacteriol. 1823740-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]