Abstract
Brucella melitensis is an intracellular pathogen that establishes a replicative niche within macrophages. While the intracellular lifestyle of Brucella is poorly understood and few virulence factors have been identified, components of a quorum-sensing pathway in Brucella have recently been identified. The LuxR-type regulatory protein, VjbR, and an N-acylhomoserine lactone signaling molecule are both involved in regulating expression of the virB-encoded type IV secretion system. We have identified a second LuxR-type regulatory protein (BlxR) in Brucella. Microarray analysis of a blxR mutant suggests that BlxR regulates the expression of a number of genes, including those encoding the type IV secretion system and flagella. Confirming these results, deletion of blxR in B. melitensis reduced the transcriptional activities of promoters for the virB operon, flagellar genes, and another putative virulence factor gene, bopA. Furthermore, our data suggested that both BlxR and VjbR are positively autoregulated and cross-regulate the expression of each other. The blxR deletion strain exhibited reduced growth in macrophages, similar to that observed for a vjbR deletion strain. However, unlike the vjbR deletion, the blxR deletion did not fully attenuate virulence in mice. More strikingly, bioluminescent imaging revealed that dissemination of the blxR mutant was similar to that of wild-type B. melitensis, while the vjbR mutant was defective for systemic spread in IRF-1−/− mice, suggesting that these regulators are not functionally redundant but that they converge in a common pathway regulating bacterial processes.
Brucella species are intracellular pathogens that establish a replicative niche within macrophages (7, 22). Internalization of Brucella occurs through lipid raft-mediated macropinocytosis (28, 58). After uptake into the phagosome, Brucella-containing vesicles move by altered intracellular trafficking through membrane-bound compartments to fuse with endoplasmic reticulum (ER) membranes, where Brucella replicates in vesicles associated with the ER (7, 8, 20, 43). The type IV secretion system, encoded by the virB operon, is critical to intracellular survival (18, 26, 33, 41). Brucella VirB mutants are unable to persist within macrophages or HeLa cells, have lost the ability to manipulate the endosomal pathway to dock with the ER, and are avirulent in mice (1, 4, 5, 10, 41, 52).
Little is known about the mechanisms employed by Brucella for infection, maintenance of the intracellular lifestyle, and eventual exit from the host cell. One strategy employed by many bacteria for the coordinated expression of factors influencing interaction with their environment is gene regulation via quorum sensing, a cell density-dependent system for intracellular communication allowing coordinated gene regulation within a bacterial population. Quorum-sensing systems in both symbiotic and pathogenic bacteria regulate factors affecting interaction with the host, such as motility and chemotaxis (19, 27), biofilm formation (11, 12, 23, 27), and the production and secretion of virulence factors (25, 59).
Two proteins form the fundamental components of a typical quorum-sensing system: the autoinducer synthase, which synthesizes a small signal molecule, and a regulatory protein, which dimerizes and binds to DNA upon interaction with the signal molecule. The prototype of this system is the Vibrio fischeri LuxI/LuxR system. LuxI, the autoinducer synthase, produces the autoinducer molecule N-(3-oxohexanoyl)-homoserine lactone (14). This molecule diffuses through the cell wall, allowing the bacterial population to regulate genes coordinately in response to population density. The regulatory protein, LuxR, contains an N-terminal acyl-homoserine lactone autoinducer binding domain (24) and a C-terminal helix-turn-helix DNA binding and dimerization domain (9).
There is evidence that the type IV secretion system and the flagellum in Brucella melitensis are regulated by a quorum-sensing pathway. Although there are no luxI homologs in the B. melitensis genome, an autoinducer molecule has been identified. The autoinducer N-dodecanoyl-homoserine-lactone (C12-HSL) was purified from solvent-extracted supernatant of Brucella grown in RPMI (54). Addition of synthetic C12-HSL suppressed expression of the virB operon (54), suggesting that the quorum-sensing system of Brucella may act to repress, rather than to induce, target virulence genes in response to autoinducer accumulation. A homolog of the LuxR transcriptional regulator, VjbR, encoded at locus BMEII1116, was found to be required for expression of the virB and flagellar genes (13, 32). The vjbR deletion strain was also defective for growth in macrophages and displayed attenuated virulence in mice (13). Complementation of this strain with an autoinducer binding domain mutant vjbR allele resulted in expression of virB that was not suppressed by addition of exogenous C12-HSL (56), suggesting that this autoinducer interacts with VjbR to mediate suppression of virB.
We have identified a second LuxR homolog, termed BlxR (Brucella luxR-like regulator). Like VjbR, BlxR was required for transcription of the type IV secretion system and flagellar genes. Transcriptional activity of the vjbR and blxR promoters was reduced in both the vjbR and blxR deletion strains, indicating that expression of VjbR and BlxR are autoregulated, as well as cross-regulated. The blxR deletion mutant exhibited reduced growth in macrophages similar to that of the vjbR deletion mutant. However, deletion of blxR did not fully attenuate virulence in an IRF-1−/− mouse model of infection, unlike the vjbR deletion. Lastly, bioluminescent imaging revealed that the spread of the blxR mutant was similar to that of wild-type B. melitensis, while the vjbR mutant was defective for in vivo dissemination.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
All strains and plasmids used in this study are listed in Table S1 in the supplemental material. Brucella strains were maintained on brucella agar (Becton Dickinson, Sparks, MD) and grown in brucella broth (BB), tryptic soy broth (TSB), or minimal yeast extract medium (44) at 37°C. E. coli strains were grown in Luria-Bertani broth at 37°C. The following antibiotics were used as appropriate: kanamycin (50 μg/ml for Brucella; 20 μg/ml for E. coli), ampicillin (200 μg/ml), chloramphenicol (40 μg/ml), and zeocin (250 μg/ml for Brucella; 50 μg/ml for E. coli).
Generation of vjbR and blxR deletion mutants.
The vectors used for allelic replacement were generated in two steps as previously described (46). First, the gene of interest plus ∼1 kb of flanking DNA was amplified by PCR. All primer sequences are listed in Table S2 in the supplemental material. XbaI and KpnI sites added to the primers were used to clone each fragment into the KpnI-XbaI sites of pZErO-1. The resulting plasmids were amplified by inverse PCR to remove a fragment internal to the gene to be mutated (∼300 bp for blxR; ∼500 bp for vjbR) and to introduce BamHI sites. The kanamycin resistance (Kmr) cassette excised from pUC4K using BamHI was inserted to create the suicide vectors pAGM18 and pAGM19.
Electrocompetent B. melitensis 16M aliquots were electroporated with pAGM18 and pAGM19 (400 Ω; 25 μF; 1.8 kV). Transformants were selected on BB agar supplemented with kanamycin, and resistant colonies were screened for sensitivity to zeocin. Replacement of target genes was verified by PCR using sets of primers internal to the Kmr cassette and flanking the deleted genes (see Table S2 in the supplemental material). Southern hybridization was performed to confirm the replacement of vjbR and blxR using a DNA fragment internal to vjbR (250 bp) and blxR (273 bp) amplified from B. melitensis 16M DNA, as well as a 600-bp DNA fragment internal to the Kmr cassette, by North2South Detection (Pierce) as described previously (47).
Bioluminescent vjbR and blxR deletion mutants were generated by allelic replacement of the vjbR and blxR genes in the constitutively bioluminescent B. melitensis strain GR023 (47) as described above.
RNA extraction and labeling.
Bacteria were grown in TSB until mid- to late log phase. Before RNA isolation, phenol-ethanol (1:19) was added, and the cells were pelleted by centrifugation and stored at −80°C. Total RNA was extracted from cell pellets using the MasterPure RNA purification kit (Epicenter), and DNA was removed using Turbo DNA-free (Ambion). cDNA was synthesized from 7 μg of total RNA in the presence of CyDye-3-dCTP (Amersham Biosciences). The labeled cDNA was purified using the ChipShot cDNA System (Promega). For each sample, RNA was extracted from two independently grown cultures, and cDNA was generated three times for each RNA sample.
Microarray hybridization.
For the minimicroarray, 289 potential open reading frames (ORFs) of the B. melitensis 16M genome were selected based on their hypothesized involvement in virulence. They included genes for proteins involved in type IV secretion, flagella, outer membrane biogenesis, transcriptional regulation, denitrification, iron uptake, and peptide and small-molecule transport. Oligonucleotides (70-mers) representing these 289 genes were purchased from Qiagen, resuspended in 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and spotted onto Ultra Gaps-coated slides (Corning) using the MicroGrid Microarrayer (Apogent Discoveries). The slides were desiccated in a vacuum oven with Drierite for 48 h and UV cross-linked using a Stratalinker at 600 mJ. The printing of each ORF spot was verified by hybridizing it with Cy3-labeled 9-mer (Qiagen). The slides were stored under vacuum at room temperature with Drierite pellets until further use.
16M genomic DNA (gDNA) was labeled by incorporation of CyDye-5-dCTP (Amersham Biosciences) using High Prime DNA Labeling (Roche) according to the manufacturer's instructions. Unincorporated nucleotides were removed using Micron YM-30 columns (Millipore).
Cy3-labeled cDNA was mixed with 0.75 μg Cy5-labeled gDNA, and hybridization buffer (Pronto Plus Kit; Promega) was added to a final volume of 35 μl. The entire sample was loaded onto the printed array and covered with a HybriSlip coverslip (Molecular Probes). The arrays were placed in specialized hybridization chambers (Telechem Inc., Sunnyvale, CA) and incubated at 42°C overnight. The slides were washed using the Pronto Plus Hybridization System (Promega) and dried by centrifugation at 1,000 rpm for 5 min.
Microarray scanning and data analysis.
Slides were scanned using the GenePix4000B scanner (Axon Instruments) with independent excitation of the fluorophores Cy3 and Cy5. Background intensities for each spot were calculated using GenPixPro 3.0 software (Axon Instruments) via the segmentation method. The signal intensity for each spot was calculated as the difference between the average signal intensity and the average local background intensity. Data were normalized to gDNA levels by calculating the ratios of cDNA to gDNA for each spot. The data were then log2 transformed, and the signal ratio for each spot was divided by the mean signal ratio of the entire array to validate comparisons between different arrays. Data normalization and statistical analysis were performed using The Institute for Genomic Research (TIGR) MeV (51).
Generation of a promoterless luxCDABE plasmid.
A PCR fragment containing the promoter of the B. melitensis ery (erythritol) operon (upstream of BMEII0430) was generated from B. melitensis gDNA using the eryP primer pair. Plasmid pBBR1-MCS4 (31) and the PCR fragment were digested with NsiI and BamHI and ligated together, deleting the lacZ promoter from pBBR1-MCS4 to generate plasmid pBBR-Pery. To delete the ery promoter, pBBR-Pery was digested with NsiI and PstI, resulting in identical overhangs, and religated to produce pBBR-ΔPery. The luxCDABE operon was digested from pXen-13 (Xenogen) using BamHI and SacI and cloned into pBBR-ΔPery to generate pEP3, consisting of a promoterless lux operon downstream of KpnI, ApaI, XhoI, and BamHI restriction sites.
Construction of lux transcriptional fusion plasmids.
Regions consisting of approximately 300 bp of sequence upstream of the selected ORFs were amplified by PCR using primers (Table S2) that added a KpnI site to the 5′ end and an XhoI site to the 3′ end. The PCR products were cloned into pCR2.1 (Invitrogen). After being sequenced, the promoter regions were subcloned into pEP3 using KpnI and XhoI.
Bioluminescent transcriptional assays.
Transcriptional lux fusion reporter plasmids were electroporated into B. melitensis strains 16M, 16MΔvjbR, and 16MΔblxR. Single colonies from transformants were inoculated into BB and grown for 72 h. Strains were resuspended to an optical density (OD) or A600 of 0.1 in BB and incubated at 37°C. At selected time points, 100 μl of each culture was transferred to a black 96-well plate for quantification of bioluminescence using a biophotonic imaging system (Xenogen). The values for each well, in photons/s/sr/cm2, were normalized for background luminescence by subtracting the value for a control well. The OD of the culture was determined, and the bioluminescence value was divided by the OD to normalize for the culture cell density.
To obtain a baseline for the transcriptional activities of the promoters, the bioluminescence of strain 16M harboring the different reporter plasmids was measured every 4 h for 28 h during growth in BB, with an additional sample taken at 48 h. The optimal time for measurement of promoter activity was found to be 20 h.
Growth in liquid culture.
Cultures were started from isolated colonies and grown for 2 days in BB at 37°C and then subcultured to an OD (A600) of 0.1 in BB and grown at 37°C. The OD (A600) was taken at 8, 16, 24, 32, and 48 h.
Macrophage infection.
The murine macrophage-like cell line RAW264.7 was maintained in RPMI 1640 supplemented with 10% fetal bovine serum at 37°C with 5% CO2. Prior to infection, macrophages were seeded at 5 × 105 cells/well in fresh RPMI 1640 supplemented with 10% fetal bovine serum in six-well plates and incubated for 16 h at 37°C with 5% CO2. Infection assays were performed at a multiplicity of infection of 100, as previously described (6). The cells were incubated for 90 min at 37°C with 5% CO2, and extracellular bacteria were removed by washing the cells three times with phosphate-buffered saline (PBS), followed by 30 min of incubation in RPMI containing 30 μg/ml gentamicin. Infected cells were maintained in medium containing 2 μg/ml of gentamicin. At specified times, the wells were washed three times with PBS, the macrophages were lysed using 0.1% Triton X-100, and viable intracellular bacterial counts were determined by dilution plating. All dilutions were plated in duplicate. For complementation, the vjbR and blxR ORFs plus 300 bp of upstream sequence were amplified using primers that added a KpnI site to the 5′ end and an XhoI site to the 3′ end. PCR products were cloned into pCR2.1 (Invitrogen). After being sequenced, vjbR was subcloned into pBBR1-MCS4 (31) and blxR was subcloned into pBBR1-MCS1 (31) using KpnI and XhoI. For the macrophage infection assay, B. melitensis strains harboring plasmids were dilution plated with and without antibiotic selective for plasmid maintenance to confirm that the CFU were representative of bacteria containing the complementing plasmid.
Mouse virulence assay.
The virulence of B. melitensis 16M, 16MΔvjbR, and 16MΔblxR was evaluated using a murine model of infection (29). Groups of 6- to 9-week old female and male IRF-1−/− mice were infected intraperitoneally with 1 × 107 bacteria in 0.2 ml PBS (n = 7 for 16M; n = 8 for 16MΔvjbR and 16MΔblxR). The mice were monitored for mortality for 2 weeks.
In vivo bioluminescent imaging.
Mice were infected intraperitoneally with 1 × 107 bacteria as described above using the bioluminescent strains GR023, ARL01, and ARL02. The mice were anesthetized with isoflurane and imaged using a charge-coupled device camera every 2 days. The images were recorded and analyzed using LivingImage software (Xenogen).
Microarray data accession number.
The microarray data have been deposited in the NCBI Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) under GEO series accession number GSE10735.
RESULTS
BlxR (BMEI1758) is a LuxR homolog.
A gene with homology to members of the luxR family of transcriptional regulators was identified at locus BMEI1758 by BLAST search (http://www.ncbi.nlm.nih.gov/BLAST) and designated blxR for Brucella LuxR-like regulator (Fig. 1). The predicted amino acid sequence of BlxR was 31% identical and 45% similar to that of LuxR from V. fischeri and had the highest homology to CepR of Burkholderia cepacia (43% identity; 58% similarity). BlxR also shares 25% identity and 45% similarity with VjbR. BlxR contains the two domains present in all LuxR family members: an autoinducer binding domain and a DNA binding domain (Fig. 1) (9). Several residues shown to be important for autoinducer or DNA binding in V. fischeri LuxR are also conserved (15, 30, 55).
FIG. 1.
Amino acid sequence homology of BlxR to quorum-sensing regulators. Shown is a comparison of the BlxR sequence to those of V. fischeri LuxR and B. melitensis VjbR. The autoinducer binding domain is shown in boldface, and the DNA binding domain is underlined. The asterisks indicate residues that are highly conserved in LuxR homologs. Residues shown to be important for autoinducer binding or DNA binding in V. fischeri LuxR (15, 30, 55) are overscored. The conserved aspartic acid shown to be important for autoinducer binding in B. melitensis VjbR (56) is indicated with a box.
To study the effect of BlxR on gene expression, bacterial growth, and pathogenicity, deletion mutants were constructed in which either the blxR or vjbR gene in B. melitensis 16M was replaced with a kanamycin resistance cassette. The deletion was confirmed by PCR and Southern hybridization (data not shown).
BlxR affects multiple genes, as detected by microarray.
Since LuxR homologs in other bacteria are involved in regulating genes involved in a number of bacterial processes, including virulence, the effect of blxR deletion on the expression of B. melitensis potential virulence genes was determined using a microarray. A minimicroarray containing spotted oligonucleotides corresponding to 289 B. melitensis genes was used to identify candidate genes whose transcription is influenced by BlxR. This microarray contains genes encoding the type IV secretion system, flagella, transcriptional regulators, transporters, and proteins involved in outer membrane biogenesis, iron acquisition, and denitrification. RNA was prepared from both B. melitensis 16M and the blxR deletion strain, 16MΔblxR, grown to late log phase in TSB. Microarray analysis identified 36 genes with altered transcript levels in 16MΔblxR compared to 16M, using Welch's approximation with a critical P value of 0.01 (Table 1). The ORFs included in the microarray are listed in Tables S3 (chromosome I) and S4 (chromosome II) in the supplemental material, along with the corresponding P values. Interestingly, five flagellar genes and eight genes from the virB type IV secretion system operon were identified in this screen. Transcription from both the fliF and the virB promoters was downregulated in a vjbR mutant (13).
TABLE 1.
Genes identified with significant changes in transcript levels between 16M and the BlxR mutant
| Locus | Name | P valuea | Fold changeb |
|---|---|---|---|
| Type IV secretion system | |||
| BMEII0025 | Attachment-mediating protein VirB1 | 0.0013 | 0.16 |
| BMEII0026 | Attachment-mediating protein VirB2 | 0.0085 | 0.63 |
| BMEII0027 | Channel protein VirB3 | 8.57E−07 | 0.17 |
| BMEII0029 | Attachment-mediating protein VirB5 | 0.0020 | 0.12 |
| BMEII0030 | Channel protein VirB6 | 0.0001 | 0.16 |
| BMEII0031 | Channel protein VirB7 | 0.0001 | 0.23 |
| BMEII0032 | Channel protein VirB8 | 0.0026 | 0.14 |
| BMEII0033 | Channel protein VirB9 | 0.0066 | 0.17 |
| Flagella | |||
| BMEII0154 | Chemotaxis protein MotB | 0.0007 | 0.51 |
| BMEII0156 | Chemotaxis protein MotD | 0.0024 | 0.42 |
| BMEII0170 | Flagellar protein FlgJ | 0.0029 | 1.79 |
| BMEII1109 | Chemotaxis protein MotA | 0.0017 | 2.04 |
| BMEII1114 | Flagellar biosynthetic protein FlhB | 0.0077 | 0.30 |
| Transcriptional regulators | |||
| BMEI0387 | Transcriptional regulatory protein, IclR family | 0.0049 | 0.13 |
| BMEI1573 | Transcriptional regulatory protein, LysR family | 0.0019 | 0.46 |
| BMEI1631 | Transcriptional regulatory protein, TetR family | 0.0102 | 0.19 |
| BMEI1913 | Transcriptional regulatory protein, LysR family | 0.0087 | 0.30 |
| BMEII0104 | Transcriptional regulatory protein, AraC family | 0.0005 | 0.56 |
| BMEII0204 | Transcriptional regulatory protein, GntR family | 4.34E−06 | 0.12 |
| BMEII0545 | Transcriptional regulatory protein, RpiR family | 0.0064 | 0.32 |
| BMEII0814 | Transcriptional regulatory protein, AraC family | 0.0039 | 0.44 |
| BMEII0878 | Transcriptional regulatory protein, GntR family | 0.0001 | 0.44 |
| Cell envelope biogenesis | |||
| BMEI0026 | Oxoacyl acyl carrier protein reductase | 0.0012 | 2.08 |
| BMEI1553 | Bacteroid development protein BacA | 0.0102 | 0.23 |
| BMEII0685 | Glucosamine fuctose 6-phosphate aminotransferase | 0.0057 | 0.19 |
| BMEII0832 | UDP glucose 4-epimerase | 0.0011 | 0.30 |
| BMEII0847 | Glycosyl transferase | 0.0008 | 0.21 |
| BMEII0848 | GDP mannose 4,6-dehydratase | 0.0012 | 0.48 |
| BMEII0849 | GDP 4 dehydro d rhamnosereductase | 0.0035 | 0.38 |
| Signal transduction | |||
| BMEI0374 | Sensory transduction histidine kinase | 0.0069 | 0.41 |
| BMEII1015 | Two-component system sensor | 0.0009 | 0.25 |
| BMEI2034 | HprK/P bifunctional kinase/phosphatase | 0.0102 | 0.53 |
| Other | |||
| BMEI0500 | Soluble lytic murein transglycosylase | 0.0042 | 0.53 |
| BMEI0781 | DNA-directed RNA polymerase alpha chain | 0.0006 | 0.39 |
| BMEI0912 | Hypothetical protein | 0.0001 | 0.15 |
| BMEI1458 | Homoserine kinase | 0.0014 | 0.38 |
| BMEI1869 | Efflux protein, LysE family, putative | 0.0059 | 0.23 |
| BMEI2029 | Adenosylhomocysteinase | 0.0054 | 0.13 |
| BMEII0096 | Oxygen-independent coproporphyrinogen III oxidase iron | 0.0030 | 0.38 |
| BMEII0409 | Osmotically inducible protein C | 0.0019 | 25.00 |
| BMEII0564 | Proline dehydrogenase delta1pyrroline 5 carboxylate dehydrogenase | 0.0013 | 0.11 |
P ≤ 0.01 determined by Welch's approximation.
Fold change in mutant 16 MΔblxR/16M.
BlxR is involved in transcription of virB and flagellar genes.
To further investigate the effect of the blxR deletion on regulation of the type IV secretion system and flagella, lux transcriptional fusions to the virB and flagellar promoters flhB and flgJ were constructed by cloning the promoter regions of selected genes upstream of a promoterless lux operon in the Brucella shuttle vector pBBR1-MCS4 (31). A lux transcriptional fusion to the initiation factor 1 (infA) promoter region (BMEI1671) was constructed as a control, since this gene was not predicted to be under quorum-sensing control and a green fluorescent protein fusion to the IF-1 promoter was constitutively fluorescent (16). A transcriptional fusion was also constructed to the upstream region of Brucella outer protein (bopA) at locus BMEII0188, which was not included in the microarray. BopA is a hypothetical protein with homology to secreted R-factor proteins involved in host specificity in plant pathogens. BMEI0188 is located within genomic island 5 (45), a region that is not present in Brucella ovis, a species that is nonpathogenic to humans.
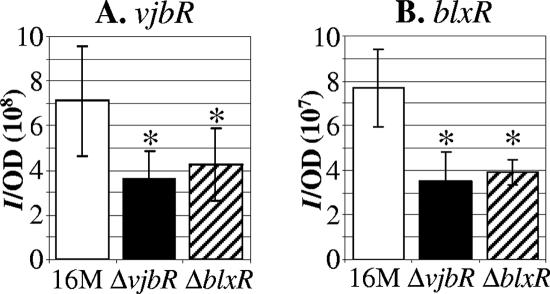
Plasmids harboring the transcriptional fusions were electroporated into wild-type B. melitensis 16M, 16MΔvjbR, and 16MΔblxR. Cultures grown for 72 h were used to inoculate fresh BB, and after 20 h of incubation at 37°C, samples were removed for measurement of bioluminescence and OD. Bioluminescence values were divided by the OD to normalize for slight differences in culture growth. As expected, transcription from the infA promoter was not reduced in the regulatory-gene mutant strains compared to wild-type B. melitensis 16M (Fig. 2A). Transcription from the virB promoter was significantly reduced in 16MΔblxR (Fig. 2B) and was also reduced in the vjbR mutant, consistent with previously published results (13). Transcription of the flagellar genes flhB and flgJ was also significantly reduced in 16MΔblxR (Fig. 2C and D), confirming the microarray result, and in 16MΔvjbR, consistent with the effect of a vjbR deletion on other flagellar genes (13). Additionally, transcription of bopA was reduced in both 16MΔvjbR and 16MΔblxR relative to expression in the wild type (Fig. 2E), suggesting that transcription of bopA is regulated through the quorum-sensing pathway.
FIG. 2.
Transcriptional activities of potential target genes. Shown are transcriptional activities of the following promoters in wild-type B. melitensis 16M and the regulatory-gene deletion strains 16MΔvjbR and 16MΔblxR: PinfA (BMEI1671) (A), PvirB (BMEI0025) (B), PflhB (BMEII1114) (C), PflgJ (BMEI1692) (D), and PbopA (BMEII0188) (E). Units of intensity (I) are photons·s−1·sr−1·cm−2. The asterisks indicate transcription levels that are significantly lower than transcription in strain 16M, as determined by paired t tests (P < 0.05). The error bars represent standard deviations for three independent experiments.
BlxR and VjbR are autoregulatory and cross-regulatory.
Several LuxR homologs regulate their own transcription (36, 39, 49), and in bacteria that contain more than one LuxR homolog, LuxR proteins can regulate the transcription of each other (35, 37, 42, 57). To determine whether autoregulation and cross talk also occur with VjbR and BlxR, we constructed lux transcriptional fusions to regions upstream of the vjbR and blxR genes. The transcriptional activity of the regulatory gene promoters was monitored in B. melitensis 16M, 16MΔvjbR, and 16MΔblxR (Fig. 3). Transcription of blxR was down-regulated in 16MΔblxR, indicating that BlxR is an activator of its own transcription, and transcription of vjbR was down-regulated in 16MΔvjbR, suggesting that VjbR is also a positive regulator of its own transcription. Likewise, blxR was downregulated in 16MΔvjbR and vjbR was downregulated in 16MΔblxR, suggesting that both transcriptional regulators cross-activate transcription of each other, although the reduction in expression was less than twofold for the vjbR promoter and about twofold for the blxR promoter. This result confirms the connection between the regulatory networks of the two quorum-sensing regulators. However, analysis of the genes directly regulated by each protein may be complicated by the fact that deletion of either the vjbR or blxR gene reduces transcription of the other gene by about half.
FIG. 3.
Autoregulation and cross talk between VjbR and BlxR. Shown are transcriptional activities of the regulatory-gene promoter-lux fusions in wild-type 16M and the regulatory-gene deletion strains. (A) vjbR activity. (B) blxR activity. The asterisks indicate transcription levels significantly lower than transcription in strain 16M, as determined by paired t tests (P < 0.05). The error bars represent standard deviations of four independent experiments for PblxR and six independent experiments for PvjbR.
Deletion of blxR reduced intracellular growth in macrophages.
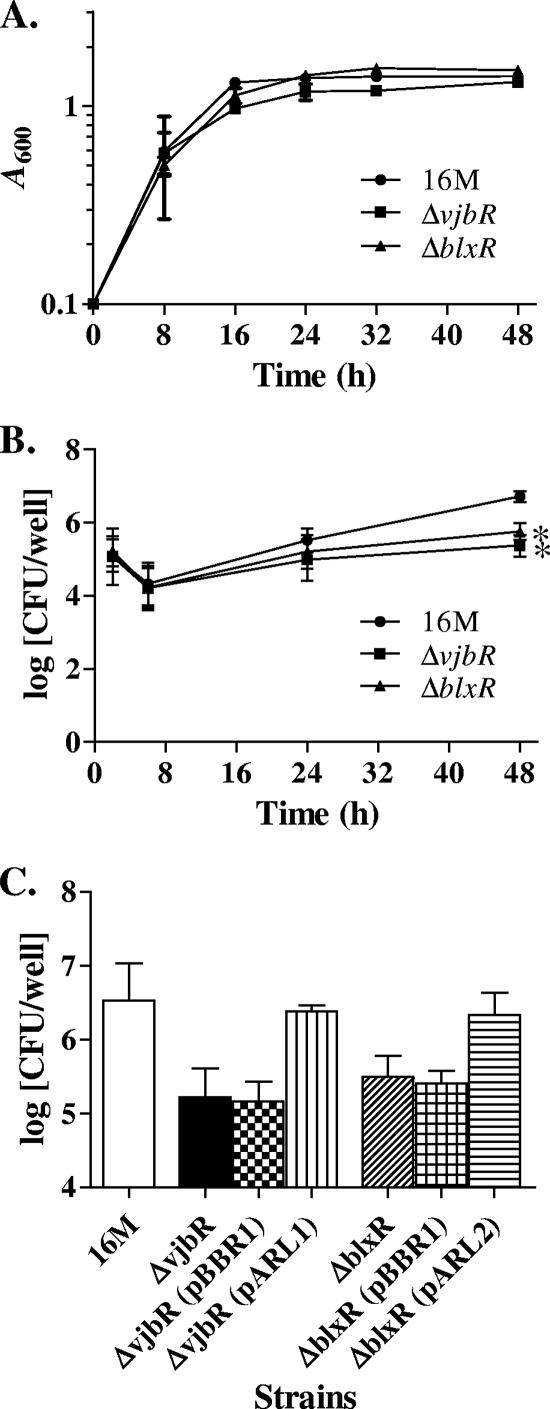
During growth in BB, the overall growth of the mutants was similar to that of wild-type B. melitensis 16M (Fig. 4A), although 16MΔvjbR appeared to transition into stationary phase earlier and reached stationary phase at a slightly lower OD (A600) than wild-type B. melitensis. The OD of 16MΔvjbR at 16 and 24 h was significantly different from that of 16M (P < 0.05), while the OD of 16MΔblxR was not significantly different from that of 16M at either time, suggesting that VjbR expression affects growth in late-log phase while BlxR expression does not.
FIG. 4.
Growth of B. melitensis 16M vjbR and blxR mutants. (A) Growth in BB. The error bars represent the standard deviations for three independent trials. (B) Growth in RAW264.7 cells. (C) Complementation of blxR and vjbR mutants. Bacterial counts were determined after 48 h of infection of RAW264.7 cells. The error bars represent the standard deviations for four independent trials.
The abilities of 16MΔvjbR and 16MΔblxR to infect and persist within host cells in vitro were evaluated using the murine macrophage cell line RAW 264.7 (Fig. 4B). Both 16MΔvjbR and 16MΔblxR entered macrophages at levels similar to that of the wild type, indicating no defect in adherence or entry. In all three strains, an initial drop in intracellular bacteria occurred, followed by a steady increase in intracellular bacteria from 6 to 48 h postinfection. Intracellular counts of strains 16MΔvjbR and 16MΔblxR increased at a slightly lower rate than that of the wild type. By 48 h, the number of intracellular bacteria was 1.34 log units lower for the vjbR mutant than for the wild type (P < 0.01), a difference consistent with previously published data for this time point (13, 56), and 0.96 log units lower for the blxR mutant (P < 0.02). The difference in intracellular growth between the two deletion strains was not statistically significant, suggesting that loss of either LuxR homolog has a similar effect on intracellular survival.
Growth in macrophages was restored to wild-type levels by complementation with the replicating plasmid pBBR1 containing the deleted gene plus 300 bp of upstream sequence but was not affected by the presence of the vector plasmid alone (Fig. 4C).
Deletion of blxR increased the mean number of days to mortality but did not affect bacterial spread in mice.
To determine whether BlxR is required for virulence in vivo, IRF-1−/− mice were infected with 1 × 107 bacteria via intraperitoneal injection. IRF-1−/− mice are highly susceptible to Brucella infection and succumb to infection with B. melitensis 16M in 7 to 10 days (29). All mice in the control group infected with B. melitensis 16M died between 7 and 9 days postinfection, whereas mice infected with 16MΔvjbR did not show any symptoms of disease and did not succumb to infection (Fig. 5). Mice infected with 16MΔblxR died between 8 and 11 days postinfection. The death of 16MΔblxR-infected mice was slightly delayed compared to 16M-infected mice (7.4 versus 10 mean days to death), indicating that BlxR may be required for full virulence, although loss of BlxR did not fully attenuate B. melitensis virulence in IRF-1−/− mice.
FIG. 5.
Virulence in IRF-1−/− mice. Mice were infected with B. melitensis strain 16M and isogenic vjbR and blxR mutants (1 × 107 CFU/mouse intraperitoneally; n = 7 for 16M; n = 8 for 16MΔvjbR and 16MΔblxR). The mice were monitored daily.
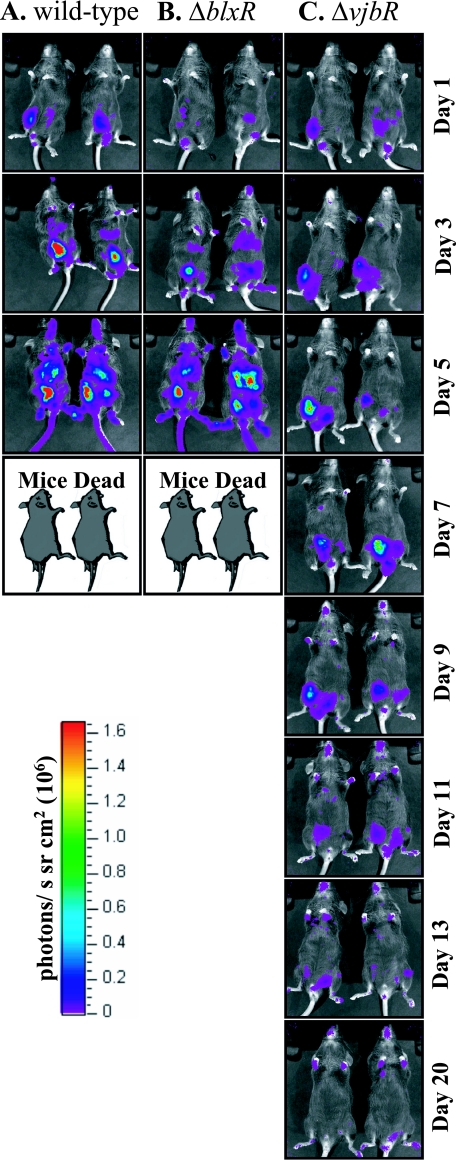
Real-time in vivo imaging was used to examine the dissemination of the mutants within the host. The vjbR and blxR genes were deleted from the constitutively bioluminescent B. melitensis strain GR023 (47) by gene replacement. The resultant B. melitensis strains ARL01, ARL02, and GR023 were used to infect IRF-1−/− mice as described above. The mice were imaged every 2 days using an in vivo imaging system (Xenogen). Dissemination of the wild-type bioluminescent strain was consistent with previously published reports for this strain in IRF-1−/− mice (47, 48), and dissemination of the blxR mutant (ARL02) was similar to that of the wild type (GR023) (Fig. 6A and B). By day 3, bioluminescence was visible in the upper abdominal cavity, indicating a bacterial load in the liver, and in the paws and submandibular area. In contrast, the vjbR mutant (ARL01) did not disseminate from the injection site during the first week of infection (Fig. 6C). Although bacterial bioluminescence was visible at the injection site from day 1 and in the paws and submandibular area from the second week of infection, bioluminescence was not detected in the upper abdominal cavity.
FIG. 6.
Dissemination in IRF-1−/− mice. Mice were infected with 1 × 107 CFU/mouse bioluminescent B. melitensis strain GR023 or an isogenic deletion mutant, ARL01 (GR023ΔvjbR) or ARL02 (GR023ΔblxR), and imaged every 2 days beginning on day 1 (immediately postinfection). The numbers indicate days postinfection. Mice infected with GR023 and GR023ΔblxR died 7 days postinfection.
Spleens taken from two 16MΔvjbR-infected animals on day 16 contained 2,500 and <100 CFU/spleen, confirming that bacterial numbers were low. The difference in pathogenicity of the two mutant strains suggests that there are differences between the regulatory pathways of the VjbR and BlxR proteins that play an important role in vivo.
DISCUSSION
The identification of a second quorum-sensing regulatory protein suggests a greater level of complexity to the Brucella quorum-sensing network than has been previously proposed. While BlxR and VjbR are both involved in regulation of the VirB type IV secretion system and flagella, microarray analysis identified other systems that may be involved in the quorum-sensing network. They include nine transcriptional-regulatory proteins and seven proteins predicted to be involved in cell envelope biogenesis. Although the targets of the transcriptional-regulatory proteins are currently unidentified, the lysR family transcriptional regulator encoded at locus BMEI1573 is known to be involved in virulence in mice (21). BMEI1573 is located near genes for purine catabolism and may be involved in their regulation. Another gene known to be involved in pathogenicity was identified among the cell envelope biogenesis proteins. BacA (BMEI1553) is involved in very long-chain fatty acid modification of lipid A (17) and has been shown to play a role in the persistence of infection (34). These microarray results, combined with the very different effects of vjbR and blxR deletions on in vivo dissemination and pathogenicity, suggest that the quorum-sensing network of Brucella regulates numerous systems associated with host-pathogen interaction. However, much of the network remains to be elucidated.
Our data raise questions regarding the regulation of the type IV secretion system in broth culture versus macrophage infection. It was previously reported that the VirB8 protein was undetectable in a VjbR mutant during growth in broth, and it was suggested that the phenotype of the VjbR mutant was due to loss of the type IV secretion complex (13). Although virB transcription was dependent on both the BlxR and VjbR proteins, deletion of either regulatory-protein gene did not produce a phenotype consistent with complete loss of type IV secretion during macrophage infection. Intracellular bacterial counts of 16MΔblxR and 16MΔvjbR increased steadily between 6 and 48 h at a slightly lower rate than that of wild-type 16M. The numbers of intracellular bacteria were not significantly different at 24 h postinfection. However, by 48 h, the intracellular bacterial counts of 16MΔblxR were 0.96 log units lower than those of 16M, and the intracellular bacterial counts of 16MΔvjbR were 1.34 log units lower than those of 16M. These results are consistent with previously published reports for a vjbR mutant B. melitensis strain at 48 h postinfection (13, 56). However, previously published results for the growth of virB mutant B. melitensis are considerably different (46). The virB mutant exhibited a continuing drop in intracellular numbers after the initial decrease, reaching 4 log units lower than the wild-type 16M by 48 h postinfection.
A lower level of virB transcription in the mutants may still allow assembly of sufficient functional type IV secretion complexes for correct trafficking of the Brucella-containing vacuole in early infection. However, it is possible that the level of attenuation of virB expression observed in the regulatory-gene mutants grown in broth is not representative of quorum-sensing regulation during macrophage infection. The VirB proteins are expressed constitutively in B. melitensis during growth in rich media but are upregulated by incubation at pH 4.5 (50). The environment within the phagocytic vacuole is nutritionally poor (38, 40), and survival within the macrophage involves exposure to many stresses, including transient low pH (2, 3). Therefore, gene regulation may be much different within macrophages than during growth in rich medium.
Although the effects of the vjbR and blxR deletions on intracellular growth of B. melitensis were similar, the in vivo effects of these deletions were strikingly different. The vjbR mutant failed to disseminate early in infection and was fully attenuated in IRF-1−/− mice. In contrast, the blxR deletion only delayed the death of infected mice compared to infection with wild-type B. melitensis 16M and did not affect dissemination. Overall, these observations suggest that the two regulatory proteins are not redundant in function. Although the regulatory networks are convergent, their different contributions to pathogenesis demonstrate that certain elements of the two networks are independent.
An autoinducer binding domain mutation in VjbR rendered B. melitensis unresponsive to the C12-HSL autoinducer (56), demonstrating that VjbR responds to C12-HSL, but also suggesting that BlxR does not. The interaction of heterologous LuxR homologs has been reported in Erwinia carotovora, where ExpR1 and ExpR2 act cooperatively to repress the negative regulator RsmA in the presence of autoinducer signal (53). ExpR1 and ExpR2 bind preferentially to different autoinducer molecules, allowing the quorum-sensing system to respond to multiple signals (53). In the quorum-sensing pathway of Brucella, loss of either protein is sufficient for suppression of the type IV secretion system and flagellar genes, suggesting that the two transcriptional regulators may provide a way for genes at the intersection of the regulatory pathways to be suppressed in response to different environmental stimuli.
Supplementary Material
Acknowledgments
This work was supported in part by the following grants: NIH/NIAID RCE for Biodefense and Emerging Infectious Diseases Developmental Projects grant 1-U54-AI-057153, BARD US-3829-06, NIH 1-R01-AI-073558-01, and NIH 1-R21-AI-070229.
Footnotes
Published ahead of print on 29 February 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alvarez-Martinez, M. T., J. Machold, C. Weise, H. Schmidt-Eisenlohr, C. Baron, and B. Rouot. 2001. The Brucella suis homologue of the Agrobacterium tumefaciens chromosomal virulence operon chvE is essential for sugar utilization but not for survival in macrophages. J. Bacteriol. 1835343-5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arenas, G. N., A. S. Staskevich, A. Aballay, and L. S. Mayorga. 2000. Intracellular trafficking of Brucella abortus in J774 macrophages. Infect. Immun. 684255-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bellaire, B. H., R. M. Roop II, and J. A. Cardelli. 2005. Opsonized virulent Brucella abortus replicates within nonacidic, endoplasmic reticulum-negative, LAMP-1-positive phagosomes in human monocytes. Infect. Immun. 733702-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boschiroli, M. L., S. Ouahrani-Bettache, V. Foulongne, S. Michaux-Charachon, G. Bourg, A. Allardet-Servent, C. Cazevieille, J. P. Lavigne, J. P. Liautard, M. Ramuz, and D. O'Callaghan. 2002. Type IV secretion and Brucella virulence. Vet. Microbiol. 90341-348. [DOI] [PubMed] [Google Scholar]
- 5.Boschiroli, M. L., S. Ouahrani-Bettache, V. Foulongne, S. Michaux-Charachon, G. Bourg, A. Allardet-Servent, C. Cazevieille, J. P. Liautard, M. Ramuz, and D. O'Callaghan. 2002. The Brucella suis virB operon is induced intracellularly in macrophages. Proc. Natl. Acad. Sci. USA 991544-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canavessi, A. M., J. Harms, N. de Leon Gatti, and G. A. Splitter. 2004. The role of integrase/recombinase xerD and monofunctional biosynthesis peptidoglycan transglycosylase genes in the pathogenicity of Brucella abortus infection in vitro and in vivo. Microb. Pathog. 37241-251. [DOI] [PubMed] [Google Scholar]
- 7.Celli, J., C. de Chastellier, D. M. Franchini, J. Pizarro-Cerda, E. Moreno, and J. P. Gorvel. 2003. Brucella evades macrophage killing via VirB-dependent sustained interactions with the endoplasmic reticulum. J. Exp. Med. 198545-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Celli, J., S. P. Salcedo, and J. P. Gorvel. 2005. Brucella coopts the small GTPase Sar1 for intracellular replication. Proc. Natl. Acad. Sci. USA 1021673-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi, S. H., and E. P. Greenberg. 1992. Genetic dissection of DNA binding and luminescence gene activation by the Vibrio fischeri LuxR protein. J. Bacteriol. 1744064-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Comerci, D. J., M. J. Martinez-Lorenzo, R. Sieira, J. P. Gorvel, and R. A. Ugalde. 2001. Essential role of the VirB machinery in the maturation of the Brucella abortus-containing vacuole. Cell Microbiol. 3159-168. [DOI] [PubMed] [Google Scholar]
- 11.Croxatto, A., V. J. Chalker, J. Lauritz, J. Jass, A. Hardman, P. Williams, M. Camara, and D. L. Milton. 2002. VanT, a homologue of Vibrio harveyi LuxR, regulates serine, metalloprotease, pigment, and biofilm production in Vibrio anguillarum. J. Bacteriol. 1841617-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280295-298. [DOI] [PubMed] [Google Scholar]
- 13.Delrue, R. M., C. Deschamps, S. Leonard, C. Nijskens, I. Danese, J. M. Schaus, S. Bonnot, J. Ferooz, A. Tibor, X. De Bolle, and J. J. Letesson. 2005. A quorum-sensing regulator controls expression of both the type IV secretion system and the flagellar apparatus of Brucella melitensis. Cell Microbiol. 71151-1161. [DOI] [PubMed] [Google Scholar]
- 14.Eberhard, A., A. L. Burlingame, C. Eberhard, G. L. Kenyon, K. H. Nealson, and N. J. Oppenheimer. 1981. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry 202444-2449. [DOI] [PubMed] [Google Scholar]
- 15.Egland, K. A., and E. P. Greenberg. 2001. Quorum sensing in Vibrio fischeri: analysis of the LuxR DNA binding region by alanine-scanning mutagenesis. J. Bacteriol. 183382-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eskra, L., A. Canavessi, M. Carey, and G. Splitter. 2001. Brucella abortus genes identified following constitutive growth and macrophage infection. Infect. Immun. 697736-7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferguson, G. P., A. Datta, J. Baumgartner, R. M. Roop II, R. W. Carlson, and G. C. Walker. 2004. Similarity to peroxisomal-membrane protein family reveals that Sinorhizobium and Brucella BacA affect lipid-A fatty acids. Proc. Natl. Acad. Sci. USA 1015012-5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foulongne, V., G. Bourg, C. Cazevieille, S. Michaux-Charachon, and D. O'Callaghan. 2000. Identification of Brucella suis genes affecting intracellular survival in an in vitro human macrophage infection model by signature-tagged transposon mutagenesis. Infect. Immun. 681297-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glessner, A., R. S. Smith, B. H. Iglewski, and J. B. Robinson. 1999. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of twitching motility. J. Bacteriol. 1811623-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorvel, J. P., and E. Moreno. 2002. Brucella intracellular life: from invasion to intracellular replication. Vet. Microbiol. 90281-297. [DOI] [PubMed] [Google Scholar]
- 21.Haine, V., A. Sinon, F. Van Steen, S. Rousseau, M. Dozot, P. Lestrate, C. Lambert, J. J. Letesson, and X. De Bolle. 2005. Systematic targeted mutagenesis of Brucella melitensis 16M reveals a major role for GntR regulators in the control of virulence. Infect. Immun. 735578-5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halliburton, B. L., and R. D. Hinsdill. 1972. Recall of acquired cellular resistance in mice by antigens from killed Brucella. Infect. Immun. 542-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammer, B. K., and B. L. Bassler. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50101-104. [DOI] [PubMed] [Google Scholar]
- 24.Hanzelka, B. L., and E. P. Greenberg. 1995. Evidence that the N-terminal region of the Vibrio fischeri LuxR protein constitutes an autoinducer-binding domain. J. Bacteriol. 177815-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henke, J. M., and B. L. Bassler. 2004. Quorum sensing regulates type III secretion in Vibrio harveyi and Vibrio parahaemolyticus. J. Bacteriol. 1863794-3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong, P. C., R. M. Tsolis, and T. A. Ficht. 2000. Identification of genes required for chronic persistence of Brucella abortus in mice. Infect. Immun. 684102-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huber, B., K. Riedel, M. Hentzer, A. Heydorn, A. Gotschlich, M. Givskov, S. Molin, and L. Eberl. 2001. The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology 1472517-2528. [DOI] [PubMed] [Google Scholar]
- 28.Kim, S., M. Watarai, S. Makino, and T. Shirahata. 2002. Membrane sorting during swimming internalization of Brucella is required for phagosome trafficking decisions. Microb. Pathog. 33225-237. [DOI] [PubMed] [Google Scholar]
- 29.Ko, J., A. Gendron-Fitzpatrick, and G. A. Splitter. 2002. Susceptibility of IFN regulatory factor-1 and IFN consensus sequence binding protein-deficient mice to brucellosis. J. Immunol. 1682433-2440. [DOI] [PubMed] [Google Scholar]
- 30.Koch, B., T. Liljefors, T. Persson, J. Nielsen, S. Kjelleberg, and M. Givskov. 2005. The LuxR receptor: the sites of interaction with quorum-sensing signals and inhibitors. Microbiology 1513589-3602. [DOI] [PubMed] [Google Scholar]
- 31.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166175-176. [DOI] [PubMed] [Google Scholar]
- 32.Leonard, S., J. Ferooz, V. Haine, I. Danese, D. Fretin, A. Tibor, S. de Walque, X. De Bolle, and J. J. Letesson. 2007. FtcR is a new master regulator of the flagellar system of Brucella melitensis 16M with homologs in Rhizobiaceae. J. Bacteriol. 189131-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lestrate, P., R. M. Delrue, I. Danese, C. Didembourg, B. Taminiau, P. Mertens, X. De Bolle, A. Tibor, C. M. Tang, and J. J. Letesson. 2000. Identification and characterization of in vivo attenuated mutants of Brucella melitensis. Mol. Microbiol. 38543-551. [DOI] [PubMed] [Google Scholar]
- 34.LeVier, K., R. W. Phillips, V. K. Grippe, R. M. Roop II, and G. C. Walker. 2000. Similar requirements of a plant symbiont and a mammalian pathogen for prolonged intracellular survival. Science 2872492-2493. [DOI] [PubMed] [Google Scholar]
- 35.Lupp, C., and E. G. Ruby. 2004. Vibrio fischeri LuxS and AinS: comparative study of two signal synthases. J. Bacteriol. 1863873-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malott, R. J., A. Baldwin, E. Mahenthiralingam, and P. A. Sokol. 2005. Characterization of the cciIR quorum-sensing system in Burkholderia cenocepacia. Infect. Immun. 734982-4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marketon, M. M., S. A. Glenn, A. Eberhard, and J. E. Gonzalez. 2003. Quorum sensing controls exopolysaccharide production in Sinorhizobium meliloti. J. Bacteriol. 185325-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marquis, H., H. G. Bouwer, D. J. Hinrichs, and D. A. Portnoy. 1993. Intracytoplasmic growth and virulence of Listeria monocytogenes auxotrophic mutants. Infect. Immun. 613756-3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minogue, T. D., M. Wehland-von Trebra, F. Bernhard, and S. B. von Bodman. 2002. The autoregulatory role of EsaR, a quorum-sensing regulator in Pantoea stewartii ssp. stewartii: evidence for a repressor function. Mol. Microbiol. 441625-1635. [DOI] [PubMed] [Google Scholar]
- 40.Mintz, C. S., J. X. Chen, and H. A. Shuman. 1988. Isolation and characterization of auxotrophic mutants of Legionella pneumophila that fail to multiply in human monocytes. Infect. Immun. 561449-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Callaghan, D., C. Cazevieille, A. Allardet-Servent, M. L. Boschiroli, G. Bourg, V. Foulongne, P. Frutos, Y. Kulakov, and M. Ramuz. 1999. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol. Microbiol. 331210-1220. [DOI] [PubMed] [Google Scholar]
- 42.Pesci, E. C., J. P. Pearson, P. C. Seed, and B. H. Iglewski. 1997. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 1793127-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pizarro-Cerda, J., S. Meresse, R. G. Parton, G. van der Goot, A. Sola-Landa, I. Lopez-Goni, E. Moreno, and J. P. Gorvel. 1998. Brucella abortus transits through the autophagic pathway and replicates in the endoplasmic reticulum of nonprofessional phagocytes. Infect. Immun. 665711-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plommet, M. 1991. Minimal requirements for growth of Brucella suis and other Brucella species. Zentralbl. Bakteriol. 275436-450. [DOI] [PubMed] [Google Scholar]
- 45.Rajashekara, G., J. D. Glasner, D. A. Glover, and G. A. Splitter. 2004. Comparative whole-genome hybridization reveals genomic islands in Brucella species. J. Bacteriol. 1865040-5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rajashekara, G., D. A. Glover, M. Banai, D. O'Callaghan, and G. A. Splitter. 2006. Attenuated bioluminescent Brucella melitensis mutants GR019 (virB4), GR024 (galE), and GR026 (BMEI1090-BMEI1091) confer protection in mice. Infect. Immun. 742925-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rajashekara, G., D. A. Glover, M. Krepps, and G. A. Splitter. 2005. Temporal analysis of pathogenic events in virulent and avirulent Brucella melitensis infections. Cell Microbiol. 71459-1473. [DOI] [PubMed] [Google Scholar]
- 48.Rajashekara, G., M. Krepps, L. Eskra, A. Mathison, A. Montgomery, Y. Ishii, and G. Splitter. 2005. Unraveling Brucella genomics and pathogenesis in immunocompromised IRF-1−/− mice. Am. J. Reprod. Immunol. 54358-368. [DOI] [PubMed] [Google Scholar]
- 49.Reverchon, S., M. L. Bouillant, G. Salmond, and W. Nasser. 1998. Integration of the quorum-sensing system in the regulatory networks controlling virulence factor synthesis in Erwinia chrysanthemi. Mol. Microbiol. 291407-1418. [DOI] [PubMed] [Google Scholar]
- 50.Rouot, B., M. T. Alvarez-Martinez, C. Marius, P. Menanteau, L. Guilloteau, R. A. Boigegrain, R. Zumbihl, D. O'Callaghan, N. Domke, and C. Baron. 2003. Production of the type IV secretion system differs among Brucella species as revealed with VirB5- and VirB8-specific antisera. Infect. Immun. 711075-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saeed, A. I., V. Sharov, J. White, J. Li, W. Liang, N. Bhagabati, J. Braisted, M. Klapa, T. Currier, M. Thiagarajan, A. Sturn, M. Snuffin, A. Rezantsev, D. Popov, A. Ryltsov, E. Kostukovich, I. Borisovsky, Z. Liu, A. Vinsavich, V. Trush, and J. Quackenbush. 2003. TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34374-378. [DOI] [PubMed] [Google Scholar]
- 52.Sieira, R., D. J. Comerci, D. O. Sanchez, and R. A. Ugalde. 2000. A homologue of an operon required for DNA transfer in Agrobacterium is required in Brucella abortus for virulence and intracellular multiplication. J. Bacteriol. 1824849-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sjoblom, S., G. Brader, G. Koch, and E. T. Palva. 2006. Cooperation of two distinct ExpR regulators controls quorum sensing specificity and virulence in the plant pathogen Erwinia carotovora. Mol. Microbiol. 601474-1489. [DOI] [PubMed] [Google Scholar]
- 54.Taminiau, B., M. Daykin, S. Swift, M. L. Boschiroli, A. Tibor, P. Lestrate, X. De Bolle, D. O'Callaghan, P. Williams, and J. J. Letesson. 2002. Identification of a quorum-sensing signal molecule in the facultative intracellular pathogen Brucella melitensis. Infect. Immun. 703004-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trott, A. E., and A. M. Stevens. 2001. Amino acid residues in LuxR critical for its mechanism of transcriptional activation during quorum sensing in Vibrio fischeri. J. Bacteriol. 183387-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uzureau, S., M. Godefroid, C. Deschamps, J. Lemaire, X. De Bolle, and J. J. Letesson. 2007. Mutations of the quorum sensing-dependent regulator VjbR lead to drastic surface modifications in Brucella melitensis. J. Bacteriol. 1896035-6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wagner, V. E., L. L. Li, V. M. Isabella, and B. H. Iglewski. 2007. Analysis of the hierarchy of quorum-sensing regulation in Pseudomonas aeruginosa. Anal. Bioanal Chem. 387469-479. [DOI] [PubMed] [Google Scholar]
- 58.Watarai, M., S. Makino, Y. Fujii, K. Okamoto, and T. Shirahata. 2002. Modulation of Brucella-induced macropinocytosis by lipid rafts mediates intracellular replication. Cell Microbiol. 4341-355. [DOI] [PubMed] [Google Scholar]
- 59.Zhu, J., M. B. Miller, R. E. Vance, M. Dziejman, B. L. Bassler, and J. J. Mekalanos. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 993129-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.