Abstract
Gene pairs specific for a toxin and its antitoxin are called toxin-antitoxin modules and are found on the chromosomes of many bacteria. The most studied of these modules is Escherichia coli mazEF, in which mazF encodes a stable toxin, MazF, and mazE encodes a labile antitoxin, MazE, which prevents the lethal effect of MazF. In a previous report from this laboratory, it was shown that mazEF-mediated cell death is a population phenomenon requiring a quorum-sensing peptide called the extracellular death factor (EDF). EDF is the linear pentapeptide NNWNN (32). Here, we further confirm that EDF is a signal molecule in a mixed population. In addition, we characterize some physiological conditions and genes required for EDF production and response. Furthermore, stress response and the gene specifying MazEF, the Zwf (glucose-6-phosphate dehydrogenase) gene, and the protease ClpXP are critical in EDF production. Significant strain differences in EDF production and response explain variations in the induction of mazEF-mediated cell death.
Over the last few years, a great deal of attention has been focused on toxin-antitoxin modules that are found on the Escherichia coli chromosome and on the chromosomes of many other bacteria, including pathogens (12, 13, 23, 38, 41). Each of these modules consists of a pair of genes, of which generally the downstream gene encodes a stable toxin and the upstream gene encodes a labile antitoxin. In E. coli, seven toxin-antitoxin systems have been described (11, 14, 15, 20, 46). Among these, one of the most studied is the chromosomal toxin-antitoxin system mazEF, which was the first to be described as regulatable and responsible for bacterial programmed cell death (3, 15). E. coli mazF is specific for the stable toxin MazF, and mazE is specific for the labile antitoxin MazE. In vivo, MazE is degraded by the ATP-dependent ClpAP serine protease (but not by the proteases ClpXP or Lon) (3). MazF is a sequence-specific endoribonuclease that preferentially cleaves single-stranded mRNAs at ACA sequences (54, 55). MazE counteracts the action of MazF. Since MazE is a labile protein, preventing MazF-mediated action requires the continuous production of MazE. Thus, any stressful condition that prevents the expression of the chromosomally borne mazEF module will lead to the reduction of MazE in the cell, permitting toxin MazF to act freely. Such conditions include (i) short-term inhibition of transcription and/or translation by antibiotics such as rifampin, chloramphenicol, and spectinomycin (44); (ii) the overproduction of ppGpp, which inhibits mazEF transcription (3, 10); and (iii) DNA damage caused by thymine starvation (45) as well as by DNA-damaging agents, such as mitomycin C or nalidixic acid (15, 25, 32). These antibiotics and stressful conditions that are well known to cause bacterial cell death (1, 9) have been found to act through the mazEF module (25, 32, 44, 45). Clearly, a system that causes any given cell to die is not advantageous to that particular cell. On the other hand, the death of an individual cell may be advantageous for the bacterial population as a whole. It was therefore suggested that mazEF-mediated cell death is a population phenomenon in which bacteria communicate with each other (11, 15).
Bacteria communicate with one another via quorum-sensing signal molecules that are also called “autoinducers” (19; reviewed in references 4, 5, 6, 17, 27, 30, and 47). Quorum sensing provides a mechanism for bacteria to monitor one another's presence and to modulate gene expression in response to population density. In the simplest scenario, the accumulation of a threshold autoinducer concentration, which is correlated with increasing population density, initiates a signal transduction cascade that culminates in a population-wide alteration in gene expression. The most studied have been four main kinds of quorum-sensing signal molecules (autoinducers) that are specific for various processes. (i) Acylated homoserine lactones (AHLs) are typically synthesized by a LuxI-type enzyme (4, 27, 47) in gram-negative bacteria. When the AHL concentration reaches the threshold level, AHLs are bound by LuxR-type protein molecules. By binding to appropriate promoters, these LuxR-AHL complexes affect the transcription of quorum-sensing regulated target genes. One of the best studied AHL systems is the lux phenotype of Vibrio spp. (reviewed in references 17, 18, and 19). (ii) In addition to a typical AHL, designated AI-1, a second autoinducer, designated AI-2, which is a furanosyl borate diester, has more recently been discovered (8) to be involved in the bioluminescent quorum sensing of the marine bacterium Vibrio harveyi; it is involved in interspecies communication (4, 16, 47, 53). (iii) 2-Heptyl-3-hydroxy-4-quinolone is produced by the opportunistic pathogen Pseudomonas aeruginosa (33, 42). (iv) Finally, short modified peptides processed from precursors are the autoinducers in gram-positive bacteria. They are involved in many systems, including the development of competence in Bacillus subtilis (35, 48, 49, 50) and the virulence response in Staphylococcus aureus (28, 29, 34, 36, 39, 40). Signal transduction occurs by a phosphorylation cascade that activates a DNA binding protein that controls the transcription of target genes. These autoinducers of gram-positive bacteria are highly specific because each oligopeptide sensor selects for a given peptide signal (reviewed in references 30, 34, and 52).
In a previous report from this laboratory, it was shown that mazEF-mediated cell death is a population phenomenon requiring a quorum-sensing factor that we call the extracellular death factor (EDF) (32). We also characterized the chemical nature of EDF to be the linear pentapeptide NNWNN. Each of the five amino acids in EDF is important for its mazEF-mediated killing activity, and the terminal asparagines are the most crucial (32). The quorum-sensing process involved in mazEF-mediated cell death and the quorum-sensing peptide EDF are particularly interesting not only because no other peptide has apparently been reported to be involved in quorum sensing in E. coli but also because EDF appears to be a distinct type of molecule related to the quorum-sensing peptides of gram-positive bacteria. Here, we further confirm that EDF is a signal molecule in a mixed population. Furthermore, our experiments reveal that mazEF is required for both EDF production and response. In addition, stress response and genes encoding Zwf (glucose-6-phosphate dehydrogenase) and the protease ClpXP are involved in EDF production.
MATERIALS AND METHODS
Bacterial strains and plasmids.
We used the following sets of E. coli strains: (i) MC4100relA1 (7) and MC4100relA+ (10) and their ΔmazEF::kan derivatives (10) and E. coli strain K38 (43) and its ΔmazEF derivative (24); (ii) MC4100relA1ΔclpP, MC4100relA1ΔclpA, MC4100relA1ΔclpX, and MC4100relA1Δlon (3); (iii) W3110 and MG1655 (22) and their ΔmazEF::kan derivatives, which we constructed by P1 transduction from strain MC4100relA1ΔmazEF::kan; and (iv) MC4100relA+Δzwf, MC4100 relA+ΔclpP, and MC4100relA+ΔygeO, constructed by us using PCR deletion (32), pBAD33 carrying a chloramphenicol resistance gene (21), pQE30 (Qiagen) carrying an ampicillin resistance gene, pQE-mazF carrying mazF under the control of the lac operator and also lacZ, pKK223-mazEF carrying mazEF under the tac promoter (24), and pQEzwf and and pQEygeO, which we constructed as follows. zwf and ygeO genes were PCR amplified from strain W3110 and cloned using EcoRI and HindIII sites into the plasmid pQE32lacIq (kindly provided by the laboratory of Orna Amster-Choder) bearing an ampicillin resistance gene, downstream of the T5 promoter.
Materials and media.
Bacterial cultures were grown in liquid M9 minimal medium with 1% glucose and a mixture of amino acids (10 μg/ml each) (37) and plated on rich LB agar plates as described previously (23). Isopropyl-β-d-thiogalactopyranoside (IPTG), nalidixic acid, mitomycin C, trimethoprim, rifampin, chloramphenicol, spectinomycin, serine hydroxamate, and Trizma base were obtained from Sigma (St. Louis, MO). Ampicillin was obtained from Biochemie GmbH (Kundl, Austria). Chemically synthesized EDF peptide (98% purity) was synthesized for us by GenScript Corporation (Piscataway, NJ).
Production of supernatants from dense cultures.
A culture of an E. coli strain that served as an EDF donor was grown in M9 medium with shaking (160 rpm) at 37°C for 12 h. The cells were diluted 1:100 in M9 medium and grown with shaking (160 rpm) at 37°C to mid-logarithmic phase (optical density at 600 nm [OD600] of 0.6; 2.5 × 108 cells/ml). Cells were then centrifuged at 14,000 rpm for 5 min. The supernatant was removed and filtered through a 0.22-μm filter; the filtrates were stored at 4°C.
Induction of mazEF-mediated cell death.
E. coli cells were grown as described in the legend to each figure. When the cultures reached a density of 3 × 108 cells/ml, samples were treated and stressful conditions were induced as described in the figure legends. Samples were centrifuged at 14,000 rpm for 5 min and washed in preheated saline. The number of CFU was detected by plating the washed samples on prewarmed LB plates that were then incubated at 37°C overnight. The percentage of surviving CFU is represented by the ratio of “treated cells” to “untreated cells.”
Quantification of EDF activity.
The supernatant of MC4100relA+ (dense culture of 2.5 ×108 cells/ml), serving as a donor, was titrated for EDF activity at different dilutions in Tris buffer (pH 7.0). A diluted culture (2.5 ×104 cells/ml) of MC4100relA+ served as a recipient. A dilution factor of 25 (which is found in the linear range of the curve) permits 70% loss of viability. Therefore, 1 unit of EDF corresponds to a dilution factor of 25.
RESULTS
EDF is a signal molecule.
We wished to examine whether EDF is a signal molecule in a population context. To this aim, we constructed a new experimental system based on the observation that a brief induction with chloramphenicol causes mazEF-mediated cell death (31). In the new system, we mixed two cultures of E. coli strain MC4100relA+: a dense culture of MC4100relA+Camr, to serve as the EDF “donor,” and a diluted culture of MC4100relA+Cams, to serve as the EDF “recipient.” As shown in Fig. 1A, in the presence of EDF produced by the donor subpopulation, chloramphenicol triggered mazEF-mediated cell death in the Cams recipient subpopulation. On the other hand, mazEF-mediated cell death did not occur when the donor subpopulation was diluted (Fig. 1B). Our results suggest that the EDF produced by the dense, Camr subpopulation could act as a signal molecule for the mazEF-mediated cell death of cells in the diluted, Cams subpopulation.
FIG. 1.
EDF is a signal molecule that can trigger mazEF-mediated cell death. We used strains MC4100relA+/pBAD (Camr), and MC4100relA+/pQE30 (Cams). Each strain was grown separately in M9 minimal medium containing the relevant antibiotic. When the cultures reached mid-logarithmic phase (OD600 of 0.4), they were washed and resuspended in M9 minimal medium. (A) A mixture of two strains was prepared in M9 medium such that the final concentrations were 108 cells/ml of a “donor” Camr culture (carrying Camr/pBAD) and 104 cells/ml of “recipient” Cams Ampr culture (WT or ΔmazEF carrying Ampr pQE30). (B) Both “donor” Camr culture (carrying Camr/pBAD) and the “recipient” Cams Ampr culture (carrying Ampr pQE30) were diluted in M9 medium; a mixture of the two was prepared in which the final concentration of each strain was 104 cells/ml. At various times, samples were removed and preincubated without shaking at 37°C for 10 min, after which chloramphenicol (45 μg/ml) was added to induce cell death. A culture to which no chloramphenicol was added served as a control. The cultures were washed and resuspended in preheated (37°C) saline. CFU were determined by plating on LB medium plates with either chloramphenicol or ampicillin that were then incubated at 37°C for 12 h. Here, we present only the Ampr Cams subculture survivors, which we determined by comparing the number of chloramphenicol-induced Ampr colonies versus the number of uninduced control colonies on LB medium plates with ampicillin. In this figure and in all the following figures, the results are the averages from three independent experiments that were carried out in triplicate.
mazEF affects both EDF production and response.
Stressful conditions have been previously shown to prevent the expression of chromosomally borne mazE, leading to the reduction in MazE and thereby permitting MazF to exert its toxic effect (24, 44, 45). We call the effect of such stressful conditions “activation of mazEF.” Here, we wished to compare the EDF activities of the supernatants of dense cultures that had been subjected to various stressful conditions. To this end, we developed an assay to quantify the EDF activity of the supernatant. This assay compares the dilution factor of the supernatant that permits 70% loss of viability of MC4100relA+ (wild type [WT]) that serves as a recipient (diluted culture). We triggered the mazEF module by subjecting bacterial cultures to various specific stressful conditions by subjecting the donor strain MC4100relA+ (dense culture) to them and then determined the resulting levels of EDF production by measuring the EDF activity of its supernatant. As shown, the level of EDF is significantly increased by all applied stressful conditions, although not always at the same level (Fig. 2A). Inducing mazEF by subjecting the cultures to a high temperature or to the chemical inhibitors of transcription and/or translation by rifampin or chloramphenicol led to an intermediate level (2.5 to 4.0 units/ml) of EDF production. Inducing mazEF by subjecting the cultures to agents damaging DNA either directly or indirectly, such as trimethoprim, nalidixic acid, or mitomycin C, led to a high level (6.6 to 8.0 units/ml) of EDF production (Fig. 2A). Overproducing MazF from a plasmid led to the highest level (11.0 units/ml) of EDF production (Fig. 2A). Thus, the activation of mazEF causes a significant increase in EDF production. Additional evidence for the role of mazEF in EDF production is our finding that the supernatant of logarithmic ΔmazEF cells lacked EDF activity (Fig. 2B). However, under various stressful conditions, about 10% of EDF activity was still detected in the supernatant of the ΔmazEF strain, compared to that of the WT (Fig. 2B). These results imply that EDF production is affected by the mazEF module but is not completely dependent on it.
FIG. 2.
Effect of various stressful conditions on EDF production. (A) E. coli MC4100relA+ WT and (B) MC4100relA+ΔmazEF were grown in M9 medium (containing 0.5% glucose) with shaking (160 rpm) at 37°C for 12 h. Cells were then diluted 1:100 in M9 medium and were grown with shaking (160 rpm) at 37°C to mid-logarithmic phase (OD600 of 0.6). The cells were incubated without shaking for 10 min and then one of the following stressful conditions was applied: (i) incubation at 37°C with chloramphenicol (45 μg/ml) for 20 min (Cam); rifampin (20 μg/ml) (Rif), nalidixic acid (1,000 μg/ml) (Nal), or mitomycin C (0.25 μg/ml) (Mit) for 10 min; or trimethoprim (2 μg/ml) (Tm) for 1 h; (ii) incubation for 10 min at 50°C; or (iii) overexpression of MazF. MC4100relA/pQEmazF was induced with IPTG (1 mM) at 37°C for 30 min. Supernatants were obtained as described in Materials and Methods. The supernatants from cultures that were induced by antibiotics were dialyzed in Tris buffer (1 mM) at 24°C for 8 h, followed by incubation at 4°C for 12 h. The EDF activities of the supernatants were quantified as described in Materials and Methods. The supernatants of an untreated culture (NT) served as a control.
Concerning the response to EDF, it was previously shown that it is completely dependent on mazEF at EDF concentrations of 1 to 200 μg/ml (32).
Effects of various E. coli proteases on EDF response and production.
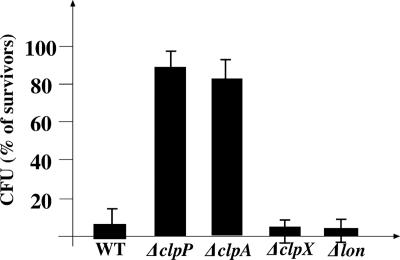
It was previously demonstrated that (i) activation of mazEF is required for the EDF response (32) and that (ii) ClpAP is responsible for MazE degradation (3); therefore, this protease permits MazF to act freely. So we were not surprised to find that neither ΔclpP nor ΔclpA recipient cultures responded to the chemically synthesized EDF (Fig. 3). On the other hand, the response to the chemically synthesized EDF (Fig. 3) was not affected in strains from which we deleted clpX or lon. These results further support the previous finding that neither ClpX nor Lon is responsible for MazE degradation in vivo (3).
FIG. 3.
Effect of various E. coli proteases on the EDF response. E. coli strains MC4100relA1 (WT), MC4100relA1ΔclpP (ΔclpP), MC4100relA1ΔclpA (ΔclpA), MC4100relA1ΔclpX (ΔclpX), and MC4100relA1Δlon (Δlon) were grown as described in Materials and Methods. When the cultures reached a density of 2.5 × 108 cells/ml, duplicate samples were removed and diluted to 3 × 104 cells/ml in prewarmed (37°C) M9 medium containing chemically synthesized EDF (0.05 μg/ml). The samples were incubated without shaking at 37°C for 10 min. Samples were further incubated without shaking at 37°C with rifampin (25 μg/ml). For the rest of the experiment, see Materials and Methods.
To study the involvement of several E. coli proteases in the production of EDF, we prepared supernatants of untreated dense cultures of E. coli strain MC4100relA1 (WT) and of its derivatives with deletions of clpP, clpA, clpX, or lon (see Materials and Methods). We observed EDF production in the supernatants from WT and Δlon donor cultures but not in the supernatants from ΔclpP, ΔclpA, or ΔclpX donor cultures (Fig. 4A), suggesting that Lon is not involved in EDF production. Since we found that the activation of mazEF, in which ClpAP is involved, is required for EDF production (Fig. 2), we were not surprised to see that deleting clpP or clpA from the dense donor cells resulted in supernatants with no EDF activity (Fig. 4A). However, since ClpX is not involved in mazEF activity (3), we were surprised to see that the supernatant from a ΔclpX donor also had no EDF activity (Fig. 4A). To test if ClpX might affect EDF production independently of the action of the mazEF module, we used donor strains that could overproduce MazF: the same E. coli strains described above, but this time harboring plasmid pQE-mazF, so that inducing them with IPTG resulted in an overproduction of MazF (see legend to Fig. 2). In the WT strain, overproducing MazF led to 10 times more EDF production than in an untreated culture (Fig. 2, compare NT with MazF). As we expected, we observed a similarly high level of EDF in the supernatant of the Δlon culture (Fig. 4B) but, again as expected, we observed no EDF activity in the supernatant of the ΔclpP or ΔclpX donor cultures (Fig. 4B). These results support our hypothesis that ClpXP is required for EDF production but is independent of mazEF activation. In a ΔclpA culture that overproduces MazF, EDF production was reduced compared to that in the WT culture (Fig. 4B). However, the ΔclpA culture still produced significant amounts of EDF when MazF was overproduced (Fig. 4B). Therefore, it seems that ClpA does not directly participate in EDF production and that its effect is indirect.
FIG. 4.
Effect of various E. coli proteases on EDF production. (A) Supernatants of dense cultures of E. coli MC4100relA1 (WT), MC4100relA1 ΔclpP (ΔclpP), MC4100relA1ΔclpA (ΔclpA), MC4100relA1ΔclpX (ΔclpX), and MC4100relA1Δlon (Δlon) grown to mid-logarithmic phase (OD600 of 0.6) were prepared (see legend to Fig. 2), and EDF activities were quantified (see Materials and Methods). (B) Strains were as in panel A, but each of them harbored pQE-mazF to allow the overproduction of MazF (+MazF). These strains were grown in M9 with ampicillin to mid-logarithmic phase (as described for panel A). Cells were washed with and resuspended in prewarmed M9 and incubated without shaking at 37°C for 10 min. To induce mazF, IPTG (1 mM) was added, and the cells were incubated without shaking at 37°C for an additional 30 min. Supernatants were obtained from these cells as described for panel A.
Zwf, but not YgeO, is necessary for cell death and EDF production.
The previously described data analysis and genetic experiments indicate that the genes zwf (encoding glucose-6-phosphate dehydrogenase) and ygeO (encoding an unknown protein) are involved in EDF production (32). It was suggested Zwf is the EDF precursor and YgeO is indirectly involved in EDF production (32). In order to support this idea, we cloned zwf and ygeO under the control of the lac operator and performed a series of complementation experiments using strains with deletions of either chromosomal zwf or ygeO. We studied loss of viability (Fig. 5A) and EDF production (Fig. 5B). As shown, overproduction of Zwf complemented these characteristics in both a Δzwf strain and a ΔygeO strain (Fig. 5). On the other hand, overproduction of YgeO which, as expected, complemented the chromosomal deletion of ygeO, did not complement a chromosomal deletion of zwf (Fig. 5). In addition, we have shown that the overproduction of neither Zwf nor YgeO was able to complement a chromosomal deletion of clpP (Fig. 5). These results indicate that Zwf and ClpP are involved primarily in EDF production, while YgeO has only a secondary role.
FIG. 5.
zwf and clpP but not ygeO are required for cell death (A) and EDF production (B). (A) E. coli strains MC4100relA+Δzwf (Δzwf), MC4100relA+ΔygeO (ΔygeO), and MC4100relA+ΔclpP (ΔclpP) with or without plasmids pQEzwf (+Zwf) or pQEygeO (+YgeO) were grown to mid-logarithmic phase (OD600 of 0.6) as described for Fig. 4. When the cultures reached a density of 2.5 × 108 cells/ml, duplicate samples were removed, and cells containing the plasmids were induced using 1 mM of IPTG and incubated without shaking at 37°C for 10 min. Samples were further incubated without shaking at 37°C with rifampin (10 μg/ml). For the rest of the experiment, see Materials and Methods. (B) Cultures of the E. coli strains described for panel A were grown in M9 with or without ampicillin to mid-logarithmic phase. In order to examine the effect of YgeO or Zwf, these proteins were induced in a manner similar to that of MazF (as described for Fig. 4B). The EDF activity of the collected supernatants was examined as described for Fig. 4B.
EDF production and response in various E. coli strains.
Having done most of our work with E. coli strains developed from the parent strain MC4100, we also wished to compare EDF production and response levels in dense donor and dilute cultures of E. coli strains K38 (32), W3110, and MG1655 (Fig. 6 and 7). We found similar EDF responses and production levels for E. coli strains MC4100relA+, K38, and W3110 (Fig. 6 and 7). However, E. coli strain MG1655 was found to be defective in both EDF response (Fig. 6) and production (Fig. 7). In this strain, mazEF-mediated cell death did not occur when we applied to its diluted culture a supernatant of a dense culture of MC4100relA+ (Fig. 6A). EDF response was observed only with the addition of 0.05 μg/ml of chemically synthesized EDF (Fig. 6C), which is five times higher than the concentration required for an EDF response in other tested E. coli strains (Fig. 6B). In addition, in contrast to the other tested strains, EDF activity in the supernatant of untreated MG1655 culture was not observed (Fig. 7A). Only the overproduction of MazF, which induced very high levels of EDF activity in other strains, permitted the detection of EDF activity in the supernatant of E. coli strain M61655 (Fig. 7B). The defect of E. coli strain M61655 in both EDF response and production explains our failure (Fig. 8) and that of others (20, 51) to show mazEF-mediated cell death in MG1655. These results (Fig. 6 and 7) were obtained with MG1655 from our strain collection. Similar results for EDF production and response were obtained with MG1655 received from the laboratories of Marlene Belfort (Albany, NY) and Orna Amster-Choder (Jerusalem, Israel) (data not shown).
FIG. 6.
Comparison of the EDF responses of various E. coli strains. E. coli strains MC4100relA+, K-38, MG1655, and W3110 were grown to mid-logarithmic phase (OD600 of 0.6) (2.5 × 108 cells/ml). Duplicate samples were removed and diluted to 3 × 104 cells/ml in preheated supernatant of a dense culture of MC4100relA+ (WT) (A), with chemically synthesized EDF in concentrations of 0.01 μg/ml (B) or 0.05 μg/ml (C). After incubation without shaking at 37°C for 10 min, rifampin (10 μg/ml) was added, and the culture was incubated without shaking at 37°C for an additional 10 min. For the rest of the experiment, see Materials and Methods.
FIG. 7.
Comparison of EDF production levels of various E. coli strains. E. coli strains MC4100relA+, K-38, MG1655, and W3110 were grown to mid-logarithmic phase (OD600 of 0.6) as described in the legend to Fig. 2. Supernatants of these untreated dense cultures (NT) were obtained (see legend to Fig. 2), and their activities were quantified (see Materials and Methods) (A). Supernatants were also obtained from cultures of the parallel strains harboring pQE-mazF in which MazF was overproduced (+MazF), which were grown in M9 medium with added ampicillin to mid-logarithmic phase, and mazF was induced (see the legend to Fig. 4) (B). Finally, supernatants were obtained and EDF activities were quantified (see Materials and Methods). After incubation without shaking at 37°C for 10 min, rifampin (10 μg/ml) was added, and the culture was incubated without shaking at 37°C for an additional 10 min. For the rest of the experiment, see the legend to Fig. 2.
FIG. 8.
Comparison of mazEF-mediated cell death in various E. coli strains. E. coli strains MC4100relA+, K38, MG1655, and W3110 were grown to mid-logarithmic phase (OD600 of 0.6) (2.5 × 108 cells/ml) as described in the legend to Fig. 2. When the cultures reached this density, duplicate samples were removed; rifampin (10 μg/ml) was added, and the culture was incubated without shaking at 37°C for an additional 10 min. For the rest of the experiment, see Materials and Methods.
Though both MG1655 and W3110 are considered to be the strains most closely related to E. coli strain K-12 (22), we found that signaling by EDF required very high concentrations of EDF only in strain MG1655 but not in strain W3110. We are currently studying the nature of this difference between strains MG1655 and W3110.
DISCUSSION
In a previous report from this laboratory, it was shown (32) that mazEF-mediated cell death is a population phenomenon that depends on the density of the bacterial culture; death occurs in a dense culture, while not in a diluted one. In addition, mazEF-mediated cell death is dependent on a quorum-sensing factor that we named EDF. It was demonstrated that (i) EDF is required when the mazEF-mediated cell death is triggered by all studied stressful conditions, (ii) EDF is the linear pentapeptide NNWNN, and (iii) each of the five EDF amino acids is important for its mazEF-mediated killing activity (32). Here, we further showed that EDF is a signal molecule capable of conveying a signal from a dense subpopulation to a diluted subpopulation (Fig. 1).
Here, we were also interested to note that there was a positive feedback loop between the mazEF module and EDF signaling: EDF permitted the activation of MazF, and EDF action was dependent on the presence of the mazEF module in the recipient cells (32). We showed that MazF activation led to an increase in the production of EDF (Fig. 2), resulting in an increase in cell death. That EDF is an integral part of the mazEF system is supported by our results showing that both EDF response and EDF production were dependent on ClpAP (Fig. 3 and 4), the protease that is responsible for MazE degradation (3). Our finding of EDF response (reflected by mazEF-mediated cell death) as well as EDF production in a Δlon strain (Fig. 3 and 4), further supports our previous results that Lon protease does not participate in MazE degradation (3).
Our results also suggest that ClpXP is involved in EDF production: in the supernatant of a ΔclpX strain, we found no EDF activity (Fig. 4), but we did observe an EDF response in a ΔclpX strain (Fig. 3A). Since ClpXP does not participate in MazF activation (3), this result is significant. Even when we used a ΔclpX donor culture that overproduced MazF, we observed no EDF activity (Fig. 4). Based on preliminary results (32), the zwf gene product (glucose-6-phosphate-dehydrogenase), carrying the amino acid sequence NNWDN may be the precursor of EDF. A subsequent amidation step may generate the full NNWNN sequence. Amidation may occur before or after cleavage of the precursor by one of the E. coli proteases (32). The herein-described results indicate that Zwf (Fig. 5) and ClpXP protease (Fig. 3 to 5) have a primary role in EDF production. We suggest that ClpXP is the Zwf-cleaving protease involved in the generation of EDF. The biochemical steps involved in the generation of EDF as well as the involvement of ClpXP are under current investigation in our laboratory.
Our previous results (32) and the results described herein clearly show that mazEF-mediated cell death is absolutely dependent on EDF. Therefore, we emphasize that studies on E. coli mazEF-mediated cell death (and maybe that of other bacteria as well) should use a protocol in which an active EDF is produced at the required concentrations. Therefore, the density of the bacterial culture (3 × 108 to 5 × 108 cells/ml) is crucial for the success of these experiments. This was one of our experimental conditions that was not followed by Tsilibaris and colleagues; therefore, they failed to show E. coli mazEF-mediated cell death (51). In addition, our experiments revealed that in contrast to E. coli strains MC4100, K38, and W3110, strain MG1655 does not produce EDF under normal growth conditions (Fig. 7A) and only weekly responds to EDF (Fig. 6). Therefore, mazEF-mediated cell death does not occur in this strain (Fig. 8). We found that in regard to EDF, MG1655 is defective. It may be that the laboratory strain MG1655 was derived by selection against genes promoting cell death.
So far, bioluminescence, virulence factor expression, biofilm formation, sporulation, mating, and competence for DNA uptake have all been described as being regulated by quorum sensing (see reviews mentioned above). The results of the work that we describe here add to this list another important biological phenomenon: bacterial programmed cell death. In previous work, we showed that mazEF prevents the spread of phage infection (26). Previously (32) and here, we show that E. coli mazEF-directed death is mediated by the communication factor EDF, which is the pentapeptide NNWNN. These findings firmly support our view that bacterial programmed cell death is a fundamental characteristic of the multicellular behavior of bacteria (15). Programmed cell death is unproductive when undertaken by an individual bacterium, but it might be beneficial as a group strategy in which a subpopulation of cells die and release nutrients (15) and/or signaling molecules (32). Extracellular signals such as EDF may be very helpful in coordinating or regulating such a group strategy.
Acknowledgments
We thank F. R. Warshaw-Dadon (Jerusalem, Israel) for her critical reading of the manuscript. We thank Shahar Amitai for his help. We thank M. Belfort (Albany, NY) and O. Amster-Choder (Jerusalem, Israel) for kindly providing us E. coli MG1655 from their strain collection.
This research was supported by grant 938/04 from the Israel Science Foundation (ISF) administrated by the Israel Academy of Science and Humanities, by grant 2005029 from United States-Israel Binational Science Foundation, and by grant GM069509 from the National Institutes of Health.
Footnotes
Published ahead of print on 29 February 2008.
REFERENCES
- 1.Ahmad, S. I., S. H. Kirk, and A. Eisenstark. 1998. Thymine metabolism and thymineless death in prokaryotes and eukaryotes. Annu. Rev. Microbiol. 52591-625. [DOI] [PubMed] [Google Scholar]
- 2.Reference deleted.
- 3.Aizenman, E., H. Engelberg-Kulka, and G. Glaser. 1996. An Escherichia coli chromosomal “addiction module” regulated by guanosine 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. USA 936059-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bassler, B. L. 2002. Small talk: cell-to-cell communication in bacteria. Cell 109421-424. [DOI] [PubMed] [Google Scholar]
- 5.Bassler, B. L., and R. Losick. 2006. Bacterially speaking. Cell 125237-246. [DOI] [PubMed] [Google Scholar]
- 6.Camilli, A., and B. L. Bassler. 2006. Bacterial small-molecule signaling pathways. Science 3111113-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casadaban, M. J., and S. N. Cohen. 1979. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc. Natl. Acad. Sci. USA 764530-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, X., S. Schauder, N. Potier, A. Van Dorsselaer, I. Pelczer, B. L. Bressler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415545-549. [DOI] [PubMed] [Google Scholar]
- 9.Davies, J., and V. Webb. 1998. Antibiotic resistance in bacteria, p. 239-273. In R. M. Kraus (ed.), Emerging infections. Academic Press, New York, NY.
- 10.Engelberg-Kulka, H., M. Reches, S. Narasimhan, R. Schoulaker-Schwarz, Y. Klemes, E. Aizenman, and G. Glaser. 1998. rexB of bacteriophage lambda is an anti-cell death gene. Proc. Natl. Acad. Sci. USA 9515481-15486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engelberg-Kulka, H., and G. Glaser. 1999. Addiction modules and programmed cell death and anti-death in bacterial cultures. Annu. Rev. Microbiol. 5343-70. [DOI] [PubMed] [Google Scholar]
- 12.Engelberg-Kulka, H., B. Sat, and R. Hazan. 2002. Bacterial programmed cell death and antibiotics. ASM News 67617-625. [Google Scholar]
- 13.Engelberg-Kulka, H., M. Reches, B. Sat, S. Amitai, and R. Hazan. 2004. Bacterial programmed cell death as a target for antibiotics. Trends Microbiol. 1266-71. [DOI] [PubMed] [Google Scholar]
- 14.Engelberg-Kulka, H., R. Hazan, and S. Amitai. 2005. mazEF: a chromosomal toxin-antitoxin module that triggers programmed cell death in bacteria. J. Cell Sci. 1184327-4332. [DOI] [PubMed] [Google Scholar]
- 15.Engelberg-Kulka, H., S. Amitai, I. Kolodkin-Gal, and R. Hazan. 2006. Programmed cell death and multicellular behavior in bacteria. PLoS Genet. 21518-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Federle, M. J., and B. L. Bassler. 2003. Interspecies communication in bacteria. J. Clin. Investig. 1121291-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuqua, C., S. C. Winans, and E. P. Greenberg. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulator. Annu. Rev. Microbiol. 50727-751. [DOI] [PubMed] [Google Scholar]
- 18.Fuqua, C., and E. P. Greenberg. 1998. Cell-to-cell communication in Escherichia coli and Salmonella typhimurium: they may be talking, but who's listening? Proc. Natl. Acad. Sci. USA 956571-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuqua, W. C., S. C. Winans, and S. C. Greenberg. 1994. Quorum sensing in bacteria: the LuxR/LuxI family of cell density responsive transcriptional regulators. J. Bacteriol. 176269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerdes, K., S. K. Christensen, and A. Lobner-Olesen. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3371-382. [DOI] [PubMed] [Google Scholar]
- 21.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 1774121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayashi, K., N. Morooka, Y. Yamamoto, K. Fujita, K. Isono, S. Choi, E. Ohtsubo, T. Baba, B. L. Wanner, H. Mori, and T. Horiuchi. 2006. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol. Syst. Biol. 22006.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayes, F. 2003. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 3011496-1499. [DOI] [PubMed] [Google Scholar]
- 24.Hazan, R., B. Sat, M. Reches, and H. Engelberg-Kulka. 2001. Postsegregational killing mediated by the P1 phage “addiction module” phd-doc requires the Escherichia coli programmed cell death system mazEF. J. Bacteriol. 1832046-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hazan, R., B. Sat, and H. Engelberg-Kulka. 2004. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J. Bacteriol. 1863663-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hazan, R., and H. Engelberg-Kulka. 2004. Escherichia coli mazEF mediated cell death as a defense mechanism that prevents spreading of phage P1. Mol. Genet. Genomics 272227-234. [DOI] [PubMed] [Google Scholar]
- 27.Henke, J. M., and B. L. Bassler. 2004. Bacterial social engagements. Trends Cell Biol. 14648-656. [DOI] [PubMed] [Google Scholar]
- 28.Ji, G., R. Beavis, and R. P. Novick. 1995. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. USA 9212055-12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ji, G., R. Beavis, and R. P. Novick. 1997. Bacterial interference caused by autoinducing peptide variants. Science 2762027-2030. [DOI] [PubMed] [Google Scholar]
- 30.Kleerebezem, M., L. E. N. Quadri, O. P. Kuipers, and W. M. de Vos. 1997. Quorum sensing by peptide pheromones and two-component signal transduction system in gram-positive bacteria. Mol. Microbiol. 24895-904. [DOI] [PubMed] [Google Scholar]
- 31.Kolodkin-Gal, I., and H. Engelberg-Kulka. 2006. Induction of Escherichia coli chromosomal mazEF by stressful conditions causes an irreversible loss of viability. J. Bacteriol. 1883420-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolodkin-Gal, I., R. Hazan, A. Gaathon, S. Carmeli, and H. Engelberg-Kulka. 2007. A linear penta-peptide is a quorum sensing factor required for mazEF-mediated cell death in Escherichia coli. Science 318652-655. [DOI] [PubMed] [Google Scholar]
- 33.Lazdunski, I., I. Ventre, and J. N. Sturgis. 2004. Regulatory circuit and communication in Gram-negative bacteria. Nat. Rev. Microbiol. 2581-592. [DOI] [PubMed] [Google Scholar]
- 34.Lyon, G. J., and R. P. Novick. 2004. Peptide signaling in Staphylococcus aureus and other Gram-positive bacteria. Peptides 251389-1403. [DOI] [PubMed] [Google Scholar]
- 35.Magnuson, R., J. Solomon, and A. D. Grossman. 1994. Biochemical and genetic characterization of a competence pheromone from B. subtilis. Cell 77207-216. [DOI] [PubMed] [Google Scholar]
- 36.Mayville, P., G. Ji, R. Beavis, H. Yang, M. Goger, R. P. Novick, and T. M. Muir. 1999. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc. Natl. Acad. Sci. USA 961218-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, J. H. 1972. Experiments in molecular genetics, p. 431. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 38.Mittenhuber, G. 1999. Occurrence of MazEF-like antitoxin/toxin systems in Bacteria. J. Mol. Microbiol. Biotechnol. 1295-302. [PubMed] [Google Scholar]
- 39.Novick, R. P., S. J. Projan, J. Kornblum, H. F. Ross, G. Ji, B. Kreiswirth, F. Vandersch, and S. Moghazeh. 1995. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol. Gen. Genet. 248446-458. [DOI] [PubMed] [Google Scholar]
- 40.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococci virulence. Mol. Microbiol. 481429-1449. [DOI] [PubMed] [Google Scholar]
- 41.Pandey, D. P., and K. Gerdes. 2005. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33966 976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pesci, E. C., J. B. Milbank, J. P. Pearson, S. McKnight, A. S. Kende, E. P. Greenberg, and B. H. Iglewski. 1999. Quinolone signaling in cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 9611229-11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Russel, M., and P. Model. 1984. Replacement of the fip gene of Escherichia coli by an inactive gene cloned on a plasmid. J. Bacteriol. 1591034-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sat, B., R. Hazan, T. Fisher, H. Khaner, G. Glaser, and H. Engelberg-Kulka. 2001. Programmed cell death in Escherichia coli: some antibiotics can trigger mazEF lethality. J. Bacteriol. 1832041-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sat, B., M. Reches, and H. Engelberg-Kulka. 2003. The Escherichia coli mazEF suicide module mediates thymineless death. J. Bacteriol. 1851803-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmidt, O., V. J. Schuenemann, N. J. Hand, T. J. Silhavy, J. Martin, A. N. Lupas, and S. Djuranovic. 2007. prlF and yhaV encode a new toxin-antitoxin system in Escherichia coli. J. Mol. Biol. 372894-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taga, M. F., and B. L. Bassler. 2003. Chemical communication among bacteria. Proc. Natl. Acad. Sci. USA 10014549-14554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tortosa, P., and D. Dubnau. 1999. Competence for transformation: a matter of taste. Curr. Opin. Microbiol. 2588-592. [DOI] [PubMed] [Google Scholar]
- 49.Tortosa, P., L. Logsdon, B. Kreiger, Y. Itoh, I. Mandic-Mulec, and D. Dubnau. 2001. Specificity of genetic polymorphism of the Bacillus competence quorum sensing system. J. Bacteriol. 183451-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tran, L. S., T. Nagai, and Y. Itoh. 2000. Divergent structure of the ComQXPA quorum-sensing components: molecular basis of strain-specific communication mechanism in Bacillus subtilis. Mol. Microbiol. 371159-1171. [DOI] [PubMed] [Google Scholar]
- 51.Tsilibaris, V., G. Maenhaut-Michel, N. Mine, and L. Van Melderen. 2007. What is the benefit to Escherichia coli of having multiple toxin-antitoxin systems in its genome? J. Bacteriol. 1896101-6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waters, M., and B. L. Bassler. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21319-346. [DOI] [PubMed] [Google Scholar]
- 53.Xavier, K. B., and B. L. Bassler. 2003. LuxS quorum sensing: more than just a numbers game. Curr. Opin. Microbiol. 6191-197. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, Y., J. Zhang, K. P. Hoeflich, M. Ikura, G. Quing, and M. Inouye. 2003. MazF cleaves cellular mRNA specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 12913-923. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, Y., J. Zhang, H. Hara, I. Kato, and M. Inouye. 2005. Insight into mRNA cleavage mechanism by MazF, an mRNA interferase. J. Biol. Chem. 2803143-3150. [DOI] [PubMed] [Google Scholar]