Abstract
The genome of Pseudomonas putida KT2440 encodes only five recognizable proteins belonging to the phosphoenolpyruvate (PEP)-carbohydrate phosphotransferase system (PTS). Two of these PTS constituents (FruA and FruB) form a complete system for fructose intake. The other three products, encoded by ptsP (EINtr), ptsO (NPr), and ptsN (EIIANtr), comprise a branch of the system unrelated to sugar traffic but thought to have an influence on coordination of N and C metabolism. We used a genetic approach to clarify the course of high-energy phosphate through this reduced set of PTS proteins. To this end, we monitored the phosphorylation state in vivo of the EIIANtr enzyme in various genetic backgrounds and growth conditions. Our results show that the source of phosphate available to the system is PEP and that the primary flow of phosphate through the N/C-sensing PTS proceeds from PEP to EINtr to NPr to EIIANtr. We also found that in the presence of fructose, unlike in the presence of succinate, EIIANtr can be phosphorylated in a ptsP strain but not in a ptsP fruB double mutant. This result revealed that the fructose transport system has the ability to cross talk in vivo with the N-related PTS branch. The data reported here thus document an unexpected connection in vivo between the sugar-dependent and sugar-independent PTSs.
The activity of most, if not all, bacterial promoters is subject to various layers of regulatory checks that merge specific signals (for instance, the presence of certain nutrients) with gross environmental inputs, so that expression of specific genes is coordinated with the overall physiological economy of each cell and the population as a whole (6). A number of physiological sensors are known to orchestrate the functioning of diverse cellular mechanisms in response to competing nutritional signals and physicochemical conditions (23). One widespread physiological sensor is the phosphoenolpyruvate (PEP)-carbohydrate phosphotransferase transport system (PTS) (3, 13). This system mediates the phosphorylation and concurrent uptake of a large number of carbohydrates in both gram-positive and gram-negative bacteria through a mechanism that allows sequential utilization of the sugars and interaction between their transport and other cellular functions (28, 34, 37). The fact that the PTS is a key, general component of cell function is reflected by its inclusion in proposals for a minimal bacterial cell or for a minimal genome (15).
Any canonical PTS involves a set of three major phosphotransfer catalytic activities (the so-called EI, HPr, and EII enzymes), which mediate the flow of a high-energy phosphate from PEP all the way to the sugar to be transported. However, sugar traffic-related PTSs often coexist with a protein set (PtsP/EINtr, PtsO/NPr, and PtsN/EIIANtr) which, while having the core phosphotransfer domains, lacks the EIIBC modules that make the connection to sugar intake. The functions of such a branch of a PTS, which is alien to carbohydrate transport, and the signals that bring about the phosphorylation of the corresponding proteins in vivo are uncertain. Yet they seem to be related to sensing the metabolic balance between N and C (3, 8, 27, 32, 39), and therefore this branch is designated PTSNtr.
The only known mechanistic hint of the function of PTSNtr is that PtsN checks the biosynthesis of branched-chain amino acids (20) by interacting directly with the K+ transporter TrkA (19) in Escherichia coli. However, this does not explain the great diversity of phenotypes resulting from knockout of ptsP, ptsO, or ptsN in both E. coli and non-E. coli microorganisms. For instance, in the soil bacterium Pseudomonas putida, the principal m-xylene-responsive σ54-dependent Pu promoter of the pWW0 plasmid for m-xylene catabolism is insensitive to repression by glucose in ptsN mutants (5, 7, 10, 11). In contrast, in the ptsO counterpart the promoter is permanently down-regulated, although a ptsP mutation does not affect Pu behavior (9). We have argued previously that phosphorylation of PtsN mediates the inhibition of Pu in the presence of glucose (7). However, the source of the high-energy phosphate is unclear in this case, as the system was still repressed by the sugar in a ptsP (EINtr−) strain (9). Moreover, P∼EIIANtr accumulates during growth, even in the presence of glucose in the medium (27), while the maximum Pu activity occurs at stationary phase. As a result, the mechanisms that link such PTS proteins to Pu activity remain ambiguous. In a separate set of experiments, a ptsN mutant of P. putida was observed to overaccumulate polyhydroxyalkanoates, which are typical products of carbon overflow (39). In contrast, ptsP or ptsO cells did not accumulate this polymer to any significant extent (39). This indicates either that the P-free form of EIIANtr represses polyhydroxyalkanoate formation or that the phosphorylated species of PtsO stimulates the process.
While these phenotypes are consistent with the participation of the PTSNtr branch in the physiological channeling of carbon to various destinations (8, 10, 11), the specific protein forms that account for the effects are not obvious. With this background, we set out to clarify the route of transfer of high-energy phosphate through the EINtr, NPr, and EIIANtr proteins of P. putida. To this end, we used a recently developed procedure for visualizing the relative proportions of phosphorylated and nonphosphorylated EIIANtr in intact cells (26, 27). As shown below, by using various P. putida genetic backgrounds and growth conditions, we found that the phosphorylation state of EIIANtr was a suitable descriptor of the functioning of the whole ∼P transfer chain. On this basis, the flow of phosphate between the phosphotransferases involved could be determined. Unexpectedly, we found that the N-related branch of the PTS cross talks in vivo with what has hitherto been believed to be a fructose-specific system that coexists with PTSNtr in P. putida.
MATERIALS AND METHODS
Strains, plasmid, and growth conditions.
All Pseudomonas strains used in this work were derived from strain P. putida MAD2. This is a rifampin-resistant variant of reference strain P. putida KT2440, which has inserted in its chromosome a Pu-lacZ transcriptional fusion along with the constitutive xylRΔA+ allele of the xylR regulator of the pWW0 plasmid assembled in a mini-Tn5 vector (14). All the P. putida MAD2 mutants used in this work that lack one or more PTS genes have been described previously (9) (Table 1). In all cases, ptsN was expressed from plasmid pVLTptsNtag (27), which was conjugally transferred to the P. putida strains indicated using E. coli HB101(RK600) as the helper in a triparental mating procedure (12). Plasmid pVLTptsNHA_E is equivalent to pVLTptsNtag, except that it contains an E-tagged ptsN variant encoding an H68A change in the primary amino acid sequence (27). Unless indicated otherwise, cells were grown at 30°C either in rich LB medium or in synthetic M9 mineral medium (35) supplemented with 0.2% (wt/vol) Casamino Acids (Caa) and tetracycline at a final concentration of 5 μg/ml in the presence or absence of 0.2% (wt/vol) fructose or succinate. All experiments were performed in triplicate. For sample collection, cells were grown in 5-ml cultures until the onset of the stationary growth phase, which corresponded to an optical density at 600 nm of 0.5 to 0.6 for cells grown in M9-Caa-fructose or M9-Caa-succinate medium, of 0.6 to 0.8 for cells grown in M9-Caa medium, or of 0.7 to 0.9 for cells grown in LB medium. One-milliliter samples were collected and subjected to analysis of PtsN phosphorylation as described below. To induce transcription of ptsN in plasmid pVLTptsNtag, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the cultures to a final concentration of 1 mM.
TABLE 1.
Mutant derivatives of P. putida MAD2
| Mutant | Description | Reference |
|---|---|---|
| ptsN | Derivative of P. putida MAD2 carrying a directed chromosomal insertion of a kanamycin resistance cassette in ptsN | 7 |
| ptsN ptsO | Derivative of P. putida MAD2 carrying a directed chromosomal insertion of a xylE marker in ptsN and a kanamycin resistance cassette in ptsO | 9 |
| ptsP fruB | Derivative of P. putida MAD2 carrying a directed chromosomal insertion of a kanamycin resistance cassette in ptsP, replacing a 249-bp internal fragment, and of the xylE marker in fruB, replacing a 1.5-kb internal fragment | 9 |
| ptsP | Derivative of P. putida MAD2 carrying a directed chromosomal insertion of a kanamycin resistance cassette in ptsP, replacing a 249-bp internal fragment | 9 |
| fruB | Derivative of P. putida MAD2 carrying a directed chromosomal insertion of the xylE marker in fruB, replacing a 1.5-kb internal fragment | 9 |
Dielectric breakdown, native polyacrylamide gel electrophoresis, and Western blotting.
To detect the PtsN/EIIANtr protein and to determine the ratio of the phosphorylated and nonphosphorylated forms, protein samples were prepared and processed as described previously (26, 27). In brief, P. putida cells were grown until the beginning of the stationary phase in the medium indicated in the presence of IPTG (1 mM) in order to induce PtsN production. Bacteria were then harvested by centrifugation and resuspended in native loading dye (10% [wt/vol] glycerol, 40 mM glycine, 5 mM Tris [pH 8.9], 0.005% [wt/vol] bromophenol blue) so that a cell mass equivalent to the mass in 1 ml of culture at an optical density at 600 nm of 1 was dispersed in 200 μl of the loading buffer. Ten-microliter portions of such intact cell suspensions were directly loaded into the wells of a 10% nondenaturing gel (10% [vol/vol] acrylamide/bisacrylamide, 29:1) polymerized with 0.05% (vol/vol) N,N,N′,N′-tetramethylethylenediamine and 0.05% ammonium persulfate in 1× running buffer (200 mM glycine, 25 mM Tris; pH 8.9). Following electrophoresis at 4°C, the proteins were blotted onto a polyvinylidene difluoride membrane as described previously (18). Subsequently, the blots were probed with an anti-E tag monoclonal antibody-peroxidase conjugate and developed for luminescent detection of the PtsN-E tag fusion proteins (27).
Data analysis.
To calculate the ratio of nonphosphorylated PtsN to phosphorylated PtsN, the Western blots were developed using the VersaDoc imaging system (model 4000; Bio-Rad) with the QuantityOne software (version 4.5.1; Bio-Rad), and the bands were analyzed by using the procedures described in the supplier's manual. The trace quantity value, which was the quantity measured by the area under the intensity profile curve (intensity × millimeters), was used to calculate the ratio of PtsN phosphorylation under the various conditions.
RESULTS
Rationale for determining the flow of high-energy phosphate through the PTS.
The genome of P. putida encodes as few as five proteins with PTS domains (Fig. 1) (39). Two of these proteins (FruA and FruB) are homologous to their E. coli counterparts for fructose intake, except that the P. putida FruB protein is fused to an EI domain. These circumstances limited the proteins studied to the three other proteins (PtsP, PtsO, and PtsN), which form the N-related PTS branch. The analyses described below for exposing the various phosphotransfer events between these proteins were based on (i) the availability of nonpolar mutants with mutations in the fruB, ptsP, ptsO, and ptsN genes individually and with a ptsP fruB double mutation (Table 1), (ii) the use of media with distinct compositions known to activate or not activate the fructose-specific branch of the PTS, and (iii) the expression in vivo of an epitope-tagged version of PtsN, a phosphorylation state which can be monitored by immunoblotting native cell extracts with a nondenaturing polyacrylamide gel system (26, 27). The reference strain used in this context was a P. putida ptsN mutant transformed with plasmid pVLTptsNtag, which expresses the E-tagged EIIANtr protein under the control of an inducible Ptac promoter (27). Finally, we reported previously that the onset of the stationary phase is the growth stage in which the two forms of PtsN could be best detected (27) and in which the PtsN-dependent C inhibition of Pu reaches a maximum (38). We thus planned to sample the cells at that point for every condition tested (see Materials and Methods). On this basis, we set out to examine the occurrence of the two PtsN forms in cells with diverse genetic backgrounds grown in appropriate media.
FIG. 1.
Distribution of EI, HPr, and EII domains in PTS-related proteins of P. putida. The diagrams show the types of modules and subregions distinctively found in the five proteins of the PTS of P. putida (39). The upper panel shows the domain organization of the three proteins that belong to the N-related PTS (PTSNtr). It includes PtsP (EIIANtr), with a GAF domain (137 amino acids) and an EI domain (561 amino acids), as well as the single-module proteins PtsO (NPr) having a single 83-amino-acid HPr domain and PtsN (EIIANtr) consisting of a unique 146-amino-acid EIIA moiety. The lower panel shows the domain structure of the two proteins that form the fructose-specific PTS of P. putida. This PTS includes the multiphosphoryl transfer protein (MTP) FruB having an EIIAFru domain (140 amino acids), an HPr domain (86 amino acids), and an EI domain (548 amino acids) and the bimodule fructose-specific permease FruA composed of an EIIB domain (83 amino acids) and an EIIC domain (340 amino acids). The identities of the proteins are indicated on the left.
Inference of the phosphorylation flow of the PTSNtr proteins in carbohydrate-free medium.
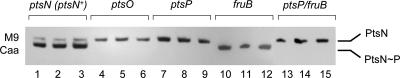
The first series of experiments for determining the course of high-energy phosphate transfer through the PTS proteins of P. putida were performed with an M9 mineral medium with 0.2% (wt/vol) Caa as the only C and N sources. By using these conditions, in principle, interference with carbohydrate catabolism and the many phosphorylated intermediates is avoided, while easily assimilable nutrients are provided. Under such growth conditions (specifically, in the absence of fructose), the PTSFru should not be active, and therefore, the phosphorylation state of the PTSNtr products may reflect only the metabolic signaling associated with this branch of the system. Figure 2 shows the distribution of the two forms of the E-tagged PtsN protein in P. putida cells grown in triplicate cultures and collected at the onset of the stationary phase (A600, 0.6 to 0.8). The results for the control conditions are shown in lanes 1 to 3, which contained extracts from the P. putida ptsN reference strain containing pVLTptsNtag, in which the loss of the chromosomal copy of the gene was compensated for by the plasmid-encoded product. This reference strain was thus equivalent, in terms of the PTS, to wild-type P. putida. In this setting, this reference strain produced the two EIIANtr forms, and the phosphorylated protein was the predominant form (Fig. 2, lanes 1 to 3).
FIG. 2.
Monitoring nonphosphorylated and phosphorylated PtsN fractions in P. putida cells at the onset of stationary phase. Cells were grown at 30°C in M9 mineral medium amended with 0.2% (wt/vol) Caa as the sole C and N source, and samples were analyzed using a native polyacrylamide gel electrophoresis system. The results of triplicate experiments are shown for each of the mutants indicated transformed with pVLTptsN-tag. The reference strain was a P. putida ptsN::Km strain complemented with pVLTptsN-tag, which was equivalent to the wild type. The position of each of the PtsN protein species is indicated on the right.
We next transferred the reporter plasmid pVLTptsNtag to the ptsO, ptsP, and fruB strains and to the ptsP fruB double mutant. Figure 2 shows the effects of the various mutations compared with the reference strain. First, neither the ptsO genetic background (lanes 4 to 6) nor the ptsP genetic background (lanes 7 to 9) supported the phosphorylation of PtsN. This indicated that PtsO is the only phosphate-delivering protein for PtsN under these conditions. Since the corresponding EINtr and NPr proteins seemed to be necessary for the increase in the amount of PtsN∼P, the most logical explanation was that the primary flow of the phosphate group was from EINtr to NPr to EIIANtr. PtsP is thus essential for delivering the high-energy phosphate all the way from PEP to PtsN, surely via PtsO, as this was shown previously in vitro for the homologous E. coli proteins (29). This information is noteworthy, as it supports the hypothesis that NPr is the prime phosphate donor to EIIANtr rather than a receptor of phosphate from P∼EIIANtr. Yet these data are compatible with the observation made with purified proteins of E. coli that the transfer of phosphate between NPr and P∼EIIANtr is a reversible reaction (29).
Finally, the same test was performed with a fruB background (i.e., in the absence of the only other protein having a PTS EI domain [Fig. 1]). As shown in Fig. 2 (lanes 10 to 12), FruB does not seem to play a major role in phosphate delivery to EIIANtr, as the protein is phosphorylated in the fruB mutant. However, FruB seems to influence to some extent phosphotransfer to PtsN, as in the fruB mutant (in contrast to the reference strain [lanes 1 to 3]) not even a small fraction of nonphosphorylated PtsN could be detected. This was intriguing, as one would expect that the absence of fructose in the medium would isolate the PTSNtr proteins from any other influence. As expected, a double mutant with mutations in the genes encoding the two EI-like enzymes (ptsP fruB) had a ptsP-like phenotype (Fig. 2, lanes 13 to 15). These experiments not only allowed us to clearly define the minimal set of proteins for the phosphotransfer from PEP to PtsN and to follow the phosphotransfer, but also provided the first hint of cross talk between the PTSNtr and PTSFru branches in P. putida.
FruB phosphorylates EIIANtr in vivo.
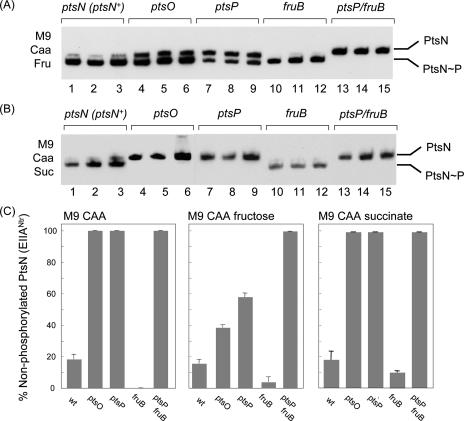
In order to explore the possible communication between the PTSNtr and PTSFru of P. putida, we replicated the experimental setup used to visualize the two PtsN (EIIANtr) species in various mutants but added 0.2% (wt/vol) fructose to the M9-Caa growth medium. In these conditions, fructose is a predominant C source, which triggers the expression and activation of the PTSFru for intake of the carbohydrate (39). The ensuing flow of ∼P through the two-domain FruA product and the multiple phosphotransfer protein FruB (Fig. 1) is predicted to result in a more dynamic situation that may favor the side transfer of phosphate to other receiver proteins. The results of these experiments are shown in Fig. 3A and C. In the reference strain, PtsN is present mainly in its phosphorylated form, and only ≤15% is the nonphosphorylated species (Fig. 3A, lanes 1 to 3). While such a ratio was comparable to that found in the culture without fructose (Fig. 2, lanes 1 to 3), the situation changed when the ratio in the mutants was examined. Specifically, there was a considerable fraction of PtsN∼P in cells lacking ptsO (Fig. 3A, lanes 4 to 6) or ptsP (lanes 7 to 9). This meant that PtsN could be phosphorylated through an alternative route alien to its partner phosphotransferases in the PTSNtr branch. The finding that the ptsP fruB double mutant did not support any phosphorylation of PtsN (Fig. 3A, lanes 13 to 15) revealed that the alternative kinase of PtsN under these growth conditions had to be FruB. On the other hand, loss of FruB made phosphorylated PtsN almost the only form of the protein (as was the case in M9-Caa medium) (Fig. 3, lanes 10 to 12).
FIG. 3.
Effect of C source addition on the nonphosphorylated and phosphorylated PtsN shares in P. putida cells. Experiments were performed in conditions identical to those described in the legend to Fig. 2, except that the M9-Caa growth medium was supplemented with either 0.2% (wt/vol) fructose (A) or 0.2% (wt/vol) succinate (B). (C) Quantification of the PtsN/PtsN∼P ratios. The bars show selected data from the experiments described above for a minimum of three separate assays. Note that the bacteria lacking both ptsP and fruB are the only bacteria that contained 100% nonphosphorylated PtsN under all the conditions tested. wt, wild type.
A few conclusions can be drawn from the data shown in Fig. 3A and C, and the most salient conclusion is that when induced in a medium with fructose, both the EI and HPr domains of FruB (Fig. 1) can functionally replace the PtsP and NPr proteins of the PTSNtr branch in vivo. In addition, the levels of the PtsN∼P form in the ptsO mutant could be enhanced by inhibition of the back-flow of phosphate when NPr was missing (29). In any case, the phosphate delivered to PtsN in the ptsO mutant had to originate from FruB. These results are noteworthy, as they mean that the kinase activity of the EI and HPr domains of FruB can act on substrates not related to fructose acquisition. Although the experimental setup does not keep the natural stoichiometry between the various PTS proteins, it is quite unlikely that the considerable phosphorylation of PtsN by FruB results from residual regulatory noise, and this ought to mean that there is significant cross talk between the two PTS sets.
Phosphorylation of EIIANtr by FruB is caused by the presence of fructose in the medium.
The results presented above show that addition of fructose to the medium brings about phosphorylation of EIIANtr by FruB. However, fructose addition also changes the overall physiology of cells, as bacteria have access to an extra supply of a C source that is easily metabolizable by conversion to fructose-1-phosphate by FruA and further channeling through the Embden-Meyerhof pathway (21, 36, 38). The effect of addition of fructose is thus expression of its transport system along with vigorous metabolism of it. To ensure that the conditions that cause phosphorylation of PtsN involve fructose consumption and not simply a generic stimulation of carbon metabolism, the distribution of forms of PtsN in the collection of P. putida strains described above grown in M9-Caa medium supplemented with succinate was examined. These conditions provided extra carbon for cell metabolism at the level of the tricarboxylic acid cycle without inducing the glycolytic enzymes responsible for breakdown of sugars to pyruvate. Comparison of the resulting PtsN forms (Fig. 3B) showed that in this growth setting, the phosphorylation patterns were virtually identical to those observed with cells grown with only Caa (compare Fig. 2 and 3B) but different from those observed with cells grown with both Caa and fructose (Fig. 3A and C). Specifically, in the reference strain and the fruB mutant the bulk of PtsN was in a phosphorylated form when cells were grown with Caa and succinate, while no phosphorylation was observed with the ptsO, ptsP, or fruB ptsP mutant in the same conditions. The corollary of the comparison of the results shown in Fig. 3B with the results shown in Fig. 2 and 3A is that fruB-mediated phosphorylation of PtsN occurs in cells grown in the presence of fructose but not in cells grown with only Caa or with Caa plus succinate. On this basis, we argue that induction and functioning of the PTSFru products brought about by the presence of fructose, and not the mere stimulation of C metabolism, are required for FruB to phosphorylate PtsN.
H68 residue of PtsN is the sole phosphorylation target of PtsO and FruB.
The unexpected cross talk between PTSFru and PTSNtr reported above suggested the possibility that both PtsO and FruB could phosphorylate PtsN, but at different sites in the protein structure. We have shown previously (27) that the conserved H68 residue of PtsN is the residue with the phosphotransfer ability in vivo in the wild-type P. putida strain. However, the PtsN sequence contains four His residues in addition to six Thr sites, seven Ser sites, and one Tyr site, several of which are exposed on the surface of a three-dimensional model based on the known structure of the EIIANtr protein of E. coli (4; data not shown). Although unlikely, the possibility that such sites could be phosphorylated by alternative kinases could not be ruled out from the data presented so far. As this was an important piece of information, we set out to resolve this question by examining the mobility of an H68A variant of PtsN in which the H68 residue is replaced by Ala. This protein is locked in a dephosphorylated state at the conserved site that is the standard target of HPr-type kinases (27). The H68A variant migrated exclusively to the position of the nonphosphorylated form under all conditions tested (data not shown). These results indicated that for both FruB and PtsO H68 of PtsN was the only phosphorylation target.
DISCUSSION
Posttranslational protein modification, specifically phosphorylation, is one of the key regulatory events in bacteria, as it seems to be involved in a wide variety of metabolic pathways. Recently, phosphorylation sites on almost all glycolytic and tricarboxylic acid cycle enzymes were reported for Bacillus subtilis (22), supporting the emerging view that phosphotransfer is a fundamental regulatory process in prokaryotes. The fructose-related and nitrogen-related PTSs (PTSFru and PTSNtr) form part of the cellular phosphorelay systems available to P. putida for interconnecting different metabolic modules. In this work, we followed the course of the phosphotransfer by monitoring the in vivo phosphorylation state of PtsN in different mutants. In this way, we elucidated the main flow of high-energy phosphate from PtsP via PtsO to PtsN. In fact, our in vivo data are similar to data obtained with in vitro assays performed with isolated proteins. Powell et al. (29) showed that the single-domain EI enzyme of E. coli phosphorylated NPr, albeit at a low rate, while the housekeeping HPr protein was a good EIIANtr kinase. Interestingly, in E. coli back transfer of phosphate from EIIANtr∼P to NPr was demonstrated in vitro as well (29). If the process is conserved in P. putida, this last finding could explain the differences in PtsN phosphorylation between the ptsO and ptsP mutants in fructose-grown cells (see below); higher levels of PtsN∼P may accumulate in ptsO mutants as no back transfer of phosphate from EIIANtr∼P to NPr would be possible.
Apart from clarifying the flow of P transfer between proteins of PTSNtr, the unanticipated result of this work was the finding that in cells exposed to fructose (but not in cells exposed to succinate), FruB is able to phosphorylate PtsN. This has to be due not only to the induction of the fruBKA operon of this bacterium by the product of the divergent fruR (cra) gene upon fructose addition (39) but also to the inherent ability of the multidomain FruB protein (Fig. 1) to deliver phosphate to PTS proteins not related to fructose. Specifically, the results shown in Fig. 3A and C allowed us to deduce that FruB can deliver phosphate also to NPr, as PtsN acquires ∼P in different proportions in ptsP and ptsO mutants. The considerable contribution of FruB to the phosphorylation of PtsN and the fact that this phosphorylation occurs in media with fructose but not in media with succinate (Fig. 3) suggested that the effect was not residual regulatory noise but reflected significant cross-regulation between the two branches of the PTS. Figure 4 shows a summary of such cross talk. The hypothesis that NPr and EIIANtr can be phosphorylated by either PtsP or FruB is consistent with the finding that the chromosomal location of ptsP is not adjacent to the gene cluster to which ptsN and ptsO belong (39). This lack of genomic contiguity (2) probably reflects the functional tolerance of the PtsN-PtsO pair for either of the two possible EI enzymes available as cognate kinases in P. putida. Physiological and metabolic signals can thus enter the N-related phosphotransfer chain system through either of the two EI enzymes available in P. putida. The cross talk may not happen, however, in the opposite direction; FruB mutants cannot transport fructose through the two-domain (EIIBFru and EIICFru) FruA permease (39), meaning that PtsN cannot replace the EIIAFru domain of FruB for phosphorylation of FruA.
FIG. 4.
Summary of high-energy phosphate traffic between PTSFru and PTSNtr in P. putida. The upper panel shows the standard ∼P traffic from PEP to fructose through the various modules of the PTSFru proteins FruA and FruB. The lower panel shows the same traffic among the PTSNtr constituents, as revealed by the data shown in Fig. 2. The yellow arrows between the two panels were inferred from the data shown in Fig. 2 and 3, while the dashed arrow from EIIAFru to NPr is a prediction based on available in vitro data for E. coli proteins. In addition, the delivery of ∼P from NPr to EIIANtr is reversible in vitro but not been proven to be reversible in vivo yet. The only metabolic signal available to the FruB protein seems to be PEP, while PtsP might sense also other inputs (perhaps N and/or O2) through its GAF domain. The approximate relative sizes of the modules are indicated by the sizes of the circles.
One way or the other, the data described above indicate that cross talk between the PTSFru and PTSNtr of P. putida does occur in vivo, at least under some growth conditions. But which signals are the signals that bring about the transfer of high-energy phosphate from FruB to PtsN? FruB is a multiphosphoryl transfer protein consisting of a classic EI domain fused to an equally standard HPr module and one EIIAFru module. Apart from the induction of the fruBKA operon of this bacterium by glycolytic intermediates via the divergently encoded FruR (cra) gene when fructose is added (33, 39), PEP is the only metabolic signal that could be readily available to such an multiphosphoryl transfer protein. It is possible that the carbon flux through glycolysis, as reflected in the PEP/pyruvate ratio, is eventually translated as one input to the phosphorylation state of PtsN via FruB. This possibility is not without precedent, as phosphorylation of the E. coli EIIAGlc protein in vivo is determined by the intracellular PEP-pyruvate fraction in the absence of PTS carbon sources (17). Therefore, only under glycolytic conditions may there be enough FruB protein to carry out the side phosphorylation of PtsN. Unfortunately, the actual regulation of the fruB operon and the many effects of Cra in P. putida have not been studied so far, and therefore this regulatory issue still needs substantial clarification.
On the other hand, the ptsP gene encodes a typical EI moiety (Fig. 1) fused to a distinctive GAF domain. Such an extra module, which occurs at the N terminus of PtsP (30, 31), belongs to one of the largest families of small-molecule binding units present in nature which seem to generally act as receptors of cyclic GMP (cGMP) (16) and, in some cases, 2-oxoglutarate (24). This is intriguing, as no bona fide guanidyl cyclases are encoded in the chromosome of P. putida, and thus cGMP can hardly be an intracellular signal molecule; it may well be that not all GAF domains bind cGMP. Yet the similarity of such a domain to sequences of the N-terminal, regulatory module of NifA (a transcriptional sensor of oxygen and nitrogen availability [24, 25]) suggests that there is a different class of physiological signals that could be channeled into phosphorylation of EIIANtr via PtsP. The ptsN and ptsO genes are in fact located in the same operon as rpoN encoding the σ54 factor involved in regulation of many nitrogen-dependent genes. PtsN is a key player of carbon repression of m-xylene catabolism mediated by the pWW0 plasmid of P. putida mt-2 (1, 7). Furthermore, in E. coli, PtsN is involved in regulation of biosynthesis of branched-chain amino acids (20), and it regulates the K+ transporter TrkA by direct interaction in E. coli (19). In both cases the dephosphorylated form of PtsN is the regulatory active species, a fact that is consistent with our observations (27). Our next analyses will therefore involve attempts to identify possible targets of PtsN that might be subject to regulation through protein-protein interactions.
Acknowledgments
This work was supported in part by grants from the EU 7th Framework Program, by the CONSOLIDER scheme of the Spanish Ministry of Education and Science, and by ERANET project PSYSMO.
Footnotes
Published ahead of print on 22 February 2008.
REFERENCES
- 1.Aranda-Olmedo, I., J. L. Ramos, and S. Marques. 2005. Integration of signals through Crc and PtsN in catabolite repression of Pseudomonas putida TOL plasmid pWW0. Appl. Environ. Microbiol. 714191-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aravind, L. 2000. Guilt by association: contextual information in genome analysis. Genome Res. 101074-1077. [DOI] [PubMed] [Google Scholar]
- 3.Barabote, R. D., and M. H. Saier, Jr. 2005. Comparative genomic analyses of the bacterial phosphotransferase system. Microbiol. Mol. Biol. Rev. 69608-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bordo, D., R. L. van Monfort, T. Pijning, K. H. Kalk, J. Reizer, M. H. Saier, Jr., and B. W. Dijkstra. 1998. The three-dimensional structure of the nitrogen regulatory protein IIANtr from Escherichia coli. J. Mol. Biol. 279245-255. [DOI] [PubMed] [Google Scholar]
- 5.Cases, I., and V. de Lorenzo. 2000. Genetic evidence of distinct physiological regulation mechanisms in the σ54 Pu promoter of Pseudomonas putida. J. Bacteriol. 182956-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cases, I., and V. de Lorenzo. 2005. Promoters in the environment: transcriptional regulation in its natural context. Nat. Rev. Microbiol. 3105-118. [DOI] [PubMed] [Google Scholar]
- 7.Cases, I., J. Perez-Martin, and V. de Lorenzo. 1999. The IIANtr (PtsN) protein of Pseudomonas putida mediates the C source inhibition of the sigma 54-dependent Pu promoter of the TOL plasmid. J. Biol. Chem. 27415562-15568. [DOI] [PubMed] [Google Scholar]
- 8.Cases, I., F. Velázquez, and V. de Lorenzo. 2007. The ancestral role of the phosphoenolpyruvate-carbohydrate phosphotransferase system (PTS) as exposed by comparative genomics. Res. Microbiol. 158666-670. [DOI] [PubMed] [Google Scholar]
- 9.Cases, I., F. Velázquez, and V. de Lorenzo. 2001. Role of ptsO in carbon-mediated inhibition of the Pu promoter belonging to the pWW0 Pseudomonas putida plasmid. J. Bacteriol. 1835128-5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.del Castillo, T., and J. L. Ramos. 2007. Simultaneous catabolite repression between glucose and toluene metabolism in Pseudomonas putida is channeled through different signaling pathways. J. Bacteriol. 1896602-6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.del Castillo, T., J. L. Ramos, J. J. Rodriguez-Herva, T. Fuhrer, U. Sauer, and E. Duque. 2007. Convergent peripheral pathways catalyze initial glucose catabolism in Pseudomonas putida: genomic and flux analysis. J. Bacteriol. 1895142-5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in Gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235386-405. [DOI] [PubMed] [Google Scholar]
- 13.Deutscher, J., C. Francke, and P. W. Postma. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70939-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernandez, S., V. de Lorenzo, and J. Perez-Martin. 1995. Activation of the transcriptional regulator XylR of Pseudomonas putida by release of repression between functional domains. Mol. Microbiol. 16205-213. [DOI] [PubMed] [Google Scholar]
- 15.Gil, R., F. J. Silva, J. Pereto, and A. Moya. 2004. Determination of the core of a minimal bacterial gene set. Microbiol. Mol. Biol. Rev. 68518-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ho, Y.-S., L. M. Burden, and J. H. Hurley. 2000. Structure of GAF domain, a ubiquitous signaling motif and new class of cyclic GMP receptor. EMBO J. 195288-5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hogema, B. M., J. C. Arents, R. Bader, K. Eijkemans, H. Yoshida, H. Takahashi, H. Aiba, and P. W. Postma. 1998. Inducer exclusion in Escherichia coli by non-PTS substrates: the role of the PEP to pyruvate ratio in determining the phosphorylation state of enzyme IIAGlc. Mol. Microbiol. 30487-498. [DOI] [PubMed] [Google Scholar]
- 18.Jurado, P., D. Ritz, J. Beckwith, V. de Lorenzo, and L. A. Fernandez. 2002. Production of functional single-chain Fv antibodies in the cytoplasm of Escherichia coli. J. Mol. Biol. 3201-10. [DOI] [PubMed] [Google Scholar]
- 19.Lee, C. R., S. H. Cho, M. J. Yoon, A. Peterkofsky, and Y. J. Seok. 2007. Escherichia coli enzyme IIANtr regulates the K+ transporter TrkA. Proc. Natl. Acad. Sci. USA 1044124-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, C. R., B. M. Koo, S. H. Cho, Y. J. Kim, M. J. Yoon, A. Peterkofsky, and Y. J. Seok. 2005. Requirement of the dephospho-form of enzyme IIA for derepression of Escherichia coli K-12 ilvBN expression. Mol. Microbiol. 58334-344. [DOI] [PubMed] [Google Scholar]
- 21.Lessie, T. G., and P. V. Phibbs, Jr. 1984. Alternative pathways of carbohydrate utilization in pseudomonads. Annu. Rev. Microbiol. 38359-388. [DOI] [PubMed] [Google Scholar]
- 22.Macek, B., I. Mijakovic, J. V. Olsen, F. Gnad, C. Kumar, P. R. Jensen, and M. Mann. 2007. The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol. Cell. Proteomics 6697-707. [DOI] [PubMed] [Google Scholar]
- 23.Martinez-Antonio, A., and J. Collado-Vides. 2003. Identifying global regulators in transcriptional regulatory networks in bacteria. Curr. Opin. Microbiol. 6482-489. [DOI] [PubMed] [Google Scholar]
- 24.Martinez-Argudo, I., R. Little, and R. Dixon. 2004. Role of the amino-terminal GAF domain of the NifA activator in controlling the response to the antiactivator protein NifL. Mol. Microbiol. 521731-1744. [DOI] [PubMed] [Google Scholar]
- 25.Martinez-Argudo, I., R. Little, N. Shearer, P. Johnson, and R. Dixon. 2004. The NifL-NifA system: a multidomain transcriptional regulatory complex that integrates environmental signals. J. Bacteriol. 186601-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pflüger, K., I. di Bartolo, F. Velázquez, and V. de Lorenzo. 2007. Non-disruptive release of Pseudomonas putida proteins by in situ electric breakdown of intact cells. J. Microbiol. Methods 71179-185. [DOI] [PubMed] [Google Scholar]
- 27.Pflüger, K., and V. de Lorenzo. 2007. Growth-dependent phosphorylation of the PtsN (EIINtr) protein of Pseudomonas putida. J. Biol. Chem. 28218206-18211. [DOI] [PubMed] [Google Scholar]
- 28.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57543-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Powell, B., D. Court, T. Inada, Y. Nakamura, V. Michotey, X. Cui, A. Reizer, M. Saier, and J. Reizer. 1995. Novel proteins of the phosphotransferase system encoded within the rpoN operon of Escherichia coli. Enzyme IIANtr affects growth on organic nitrogen and the conditional lethality of an erats mutant. J. Biol. Chem. 2704822-4839. [DOI] [PubMed] [Google Scholar]
- 30.Rabus, R., J. Reizer, I. Paulsen, and M. H. Saier, Jr. 1999. Enzyme I(Ntr) from Escherichia coli. A novel enzyme of the phosphoenolpyruvate-dependent phosphotransferase system exhibiting strict specificity for its phosphoryl acceptor, NPr. J. Biol. Chem. 27426185-26191. [DOI] [PubMed] [Google Scholar]
- 31.Reizer, J., A. Reizer, M. J. Merrick, G. Plunkett III, D. J. Rose, and M. H. Saier, Jr. 1996. Novel phosphotransferase-encoding genes revealed by analysis of the Escherichia coli genome: a chimeric gene encoding an enzyme I homologue that possesses a putative sensory transduction domain. Gene 181103-108. [DOI] [PubMed] [Google Scholar]
- 32.Reizer, J., A. Reizer, M. H. Saier, Jr., and G. R. Jacobson. 1992. A proposed link between nitrogen and carbon metabolism involving protein phosphorylation in bacteria. Protein Sci. 1722-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saier, M. H., Jr., and T. M. Ramseier. 1996. The catabolite repressor/activator (Cra) protein of enteric bacteria. J. Bacteriol. 1783411-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saier, M. H., Jr., and J. Reizer. 1994. The bacterial phosphotransferase system: new frontiers 30 years later. Mol. Microbiol. 13755-764. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 36.Sawyer, M. H., L. Baumann, S. M. Berman, J. L. Canovas, and R. H. Berman. 1977. Pathways of d-Fructose catabolism in species of Pseudomonas. Arch. Microbiol. 11249-55. [DOI] [PubMed] [Google Scholar]
- 37.Tchieu, J. H., V. Norris, J. S. Edwards, and M. H. Saier, Jr. 2001. The complete phosphotranferase system in Escherichia coli. J. Mol. Microbiol. Biotechnol. 3329-346. [PubMed] [Google Scholar]
- 38.Velázquez, F., I. di Bartolo, and V. de Lorenzo. 2004. Genetic evidence that catabolites of the Entner-Doudoroff pathway signal C source repression of the σ54 Pu promoter of Pseudomonas putida. J. Bacteriol. 1868267-8275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Velázquez, F., K. Pflüger, I. Cases, L. I. De Eugenio, and V. de Lorenzo. 2007. The phosphotransferase system (PTS) formed by PtsP, PtsO, and PtsN proteins controls production of polyhydroxyalkanoates in Pseudomonas putida. J. Bacteriol. 1894529-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]