Abstract
Genomic analysis suggested the existence of a CO-sensing bacterial transcriptional regulator that couples an N-terminal PAS fold domain to a C-terminal DNA-binding LytTR domain. UV/visible-light spectral analyses of heterologously expressed, purified full-length proteins indicated that they contained a hexacoordinated b-type heme moiety that avidly binds CO and NO. Studies of protein variants strongly suggested that the PAS domain residues His74 and Met104 serve as the heme Fe(II) axial ligands, with displacement of Met104 upon binding of the gaseous effectors. Two RcoM (regulator of CO metabolism) homologs were shown to function in vivo as CO sensors capable of regulating an aerobic CO oxidation (cox) regulon. The genetic linkage of rcoM with both aerobic (cox) and anaerobic (coo) CO oxidation systems suggests that in different organisms RcoM proteins may control either regulon type.
Heme-containing sensor proteins exist in all kingdoms of life and regulate cellular function in response to heme iron redox poise [Fe(III)/Fe(II)] or the binding of gaseous ligands (25). In general, these sensor proteins exhibit one of the following six heme-coordinating structures: (i) the heme-nitric oxide binding form (termed H-NOX [3]), (ii) the globin fold (11), (iii) the SCHIC domain (38), (iv) the CRP/FNR family (cyclic AMP receptor protein/fumarate-nitrate reduction) CooA (bacterial CO oxidation transcriptional activator) homologues (1, 44), (v) the GAF domain (46, 47), or (vi) the extensive PAS fold family (16, 17).
These domains can exhibit remarkable effector and cofactor flexibility. In particular, the ∼7,000 identified PAS fold domains (InterPro IPR013767 [http://www.ebi.ac.uk/interpro/]) appear to maintain similar structures despite low sequence conservation over their ∼120-residue length, and they bind a variety of effectors and cofactors (23). Moreover, even the heme-containing PAS fold subclass displays only modest sequence conservation with an array of heme coordination characteristics (17). For example, the bacterial O2 sensor proteins FixL (oxygen sensor kinase) and phosphodiesterase A1 from Acetobacter xylinum (PDEA1Ax) maintain a pentacoordinate b-type heme with a single axial His ligand to the Fe(II) iron (5, 18), while the bacterial Escherichia coli direct oxygen sensor phosphodiesterase (DOSEc) and the mammalian CO sensor neuronal PAS domain protein 2 (NPAS2) incorporate a hexacoordinate b-type heme with His/Met and His/His Fe(II) axial ligands, respectively (6, 7, 21, 50). Also, a class of PAS domain response proteins bearing c-type heme has been described (34).
In addition to variations in heme coordination, these response regulators exist either as one-component proteins, where the sensor domain is covalently linked to the response domain (13, 51), or as two-component systems, where the sensor and response domains exist on two different proteins (13, 15). Most heme-containing PAS, GAF, H-NOX, SCHIC, and globin sensors belong to the latter class. In contrast, the known CO sensors are single-component proteins: the prokaryotic CooA proteins fuse an N-terminal CRP/FNR-like sensor module with a C-terminal helix-turn-helix DNA-binding domain (1, 44), and the mammalian NPAS2 proteins join an N-terminal basic helix-loop-helix DNA-binding module with a C-terminal PAS fold sensor domain (7).
An appreciation of the biological significance of CO, and the need to regulate metabolism accordingly, is increasing. Recent studies have demonstrated that CO serves as a central metabolic intermediate in anaerobic metabolism (42), as an enzyme metallocenter ligand (8, 30), and as a physiologically significant signal in higher organisms (53). A primordial role in the origin of life has also been postulated (26). CO is present at a low concentration (<1 ppm) in the bulk environment, yet numerous prokaryotes express distinct CO-regulated systems that catalyze catabolically productive aerobic and anaerobic CO oxidation (28, 42, 44). Aerobic organisms employ a high-affinity, oxygen-tolerant, molybdo-flavoprotein protein complex to effect CO oxidation (37). This system is encoded by cox genes and, in the best-studied example, from Oligotropha carboxidovorans, has been shown to be transcriptionally regulated (45). The putative regulators CoxC and CoxH, however, remain uncharacterized (12). Anaerobic utilization of CO depends on an oxygen-sensitive Ni-containing protein complex that couples CO oxidation to CO2 with the production of H2. This system is encoded by coo genes, which have been shown to be transcriptionally regulated by CooA (1, 44).
Here we describe a new prokaryotic regulator of CO metabolism (RcoM) that in different organisms appears to regulate coo or cox gene expression. Sequence similarity among putative representatives from several bacterial sources along with spectral analyses of an RcoM protein from Burkholderia xenovorans LB400 (4) indicates that these regulators join an N-terminal heme-bearing PAS fold module with a C-terminal LytTR-class DNA-binding module (39). Using a cox-linked reporter system borne in E. coli, functional analyses of two proteins from B. xenovorans demonstrate a CO-dependent response under aerobic growth conditions. Thus, RcoM-type regulators represent the third analyzed example of a one-component CO-sensing transcription factor: while it incorporates a PAS fold heme domain like the mammalian NPAS2 CO sensor, its particular heme ligation characteristics, DNA-binding modules, and domain organizations differ from those of the mammalian sensor, and both the heme- and DNA-binding domains of RcoM and CooA are dissimilar.
MATERIALS AND METHODS
Cloning and plasmid constructions.
Putative rcoM ORFs were identified by a PSI-BLAST search (threshold = 0.005, expect = 100, word size = 2, matrix = BLOSUM45, gap existence and extension values of 16 and 1, respectively) of the nonredundant GenBank database (posting dates, 24 April 2007 and 15 January 2008; http://www.ncbi.nlm.nih.gov/) with residues 1 to 140 of the RcoM proteins of Rhodospirillum rubrum (GenBank identifier [gi]:83594844), Pelobacter carbinolicus DSM 2380 (gi:77917674), Alkalilimnicola ehrlichei MLHE-1 (gi:114320715), and Geobacter uraniumreducens Rf4 (gi:148262698). We obtained chromosomal DNA samples of representative organisms and amplified regions using Pfu Turbo polymerase (Stratagene, La Jolla, CA) and primers that hybridized to the target region (purchased from the University of Wisconsin Biotechnology Center oligonucleotide synthesis facility or Integrated DNA Technologies (Coralville, IA). These primers also incorporated extensions that permitted cloning and controlled expression: the upstream primers generally appended 5′-GAATTCAGGAGGAATTTAAA-3′ to the rcoM ATG codon, thereby introducing an EcoRI restriction site (underlined) and a ribosome-binding site (35), while the downstream primer typically added a TAA stop codon and a HindIII restriction site such that the cloned region ended with 5′-TAAGCTT-3′ (HindIII site underlined). In two instances a SacI site replaced the 5′ EcoRI site, and in one case a SalI site replaced the downstream HindIII site, as dictated by the presence of EcoRI or HindIII sites in the amplified region. All constructs were cloned into appropriately digested pEXT20 (Apr, pBR322 ori [10]) to permit IPTG (isopropyl-β-d-thiogalactopyranoside)-dependent transcription of the cloned rcoM. The cloned rcoM1 and rcoM2 regions from B. xenovorans LB400 were subsequently amplified using a primer that inserted six histidine codons immediately upstream of the endogenous stop codon, generating a terminal sequence of 5′-CAT-CAC-CAT-CAC-CAT-CAC-TGA-TAA-GCTT-3′. This primer also introduced four translationally silent alterations in the final six codons of the cloned rcoM1. Variant proteins were generated by mutagenesis of rcoM1 according to the QuikChange protocol (Stratagene). Sequences of all constructs were verified using BigDye v. 3.1 reaction chemistry (Applied Biosystems, Foster City, CA) with subsequent analysis by the University of Wisconsin Biotechnology Center DNA sequencing facility.
Because the B. xenovorans rcoM and coxM regions are divergently transcribed (see below), construction of a reporter system required a separate amplification and cloning of an ∼1,200-bp region that encompassed rcoM1 and the first three codons of the linked coxM1. The lacZ-Kmr cassette from pKOK6 (29) was cloned into a SalI site introduced into coxM1 during amplification. Then, taking advantage of BamHI sites in the vector multiple cloning region and within rcoM1, an ∼5,500-bp fragment was moved into the low-copy-number vector pEXT21 (Sp-Smr, IncW ori [10]) such that expression of the promoterless lacZ depended on coxM1 transcription, while the vector Ptac promoter was oriented in the same direction as the kan gene and thus in the direction opposite that of the lacZ reporter. This construct, which is compatible with the pEXT20-based rcoM expression plasmids, was introduced into E. coli strain BW29860 (54) to create the reporter strain designated UQ4972.
Assays of RcoM- and CO-dependent lacZ expression in vivo.
Cultures bearing the desired rcoM expression plasmid in the UQ4972 reporter system host were assayed for activity using MOPS [3-(N-morpholino)propanesulfonic acid]-buffered medium in which a nonfermentable carbon source (40 mM sodium acetate) replaced glucose, which was used previously (27). Growth in this medium was strictly O2 dependent (data not shown). Log-phase (optical density at 550 nm [OD550] ∼ 1) cultures in this medium were used to inoculate 120-ml serum vials containing 3 ml of medium at an OD550 of 0.05; these cultures were sealed, and 2 ml of CO was injected, as required. Cultures were incubated at 30°C with agitation (100 rpm, 2.5-cm throw), and cell pellets were collected and frozen when cultures reached an OD550 of ∼1.1. The assay yielded similar results for samples obtained between OD550s of 0.9 and 1.7, and all strains grew similarly, regardless of CO addition, and were not O2 limited (data not shown). β-Galactosidase activities were determined according to a standard protocol (43).
Protein accumulation, purification, and CO photolysis.
Expression constructs were introduced into the host strain E. coli VJS6737 (48) and cultivated at 28 to 30°C in rich medium supplemented with ferric citrate and IPTG (27). Cell pellets were resuspended in lysis buffer (50 mM Tris·HCl [pH 8.0], 500 mM NaCl) and then lysed by passage through a French press. Extract supernatants (∼200 ml) were applied to 25-ml His-Bind column (Novagen, EMD Biosciences, Madison, WI), which was washed with 200 ml of lysis buffer and then 150 ml of lysis buffer containing 50 mM imidazole. Protein was eluted using lysis buffer containing 300 mM imidazole and immediately applied to a 25-ml column containing Bio-Gel HTP hydroxyapatite resin (Bio-Rad, Hercules, CA) equilibrated in lysis buffer. This column was sequentially washed with 200 ml of lysis buffer and then ∼50 ml of lysis buffer amended with 40 mM potassium phosphate (pH 8.0), and the protein was then eluted with ∼50 ml of lysis buffer containing 100 mM potassium phosphate (pH 8.0). Protein was precipitated by the addition of (NH4)2SO4 to 55% and centrifugation. After all liquid was removed, the protein pellet was dissolved in ∼1 ml of buffer A (25 mM MOPS [pH 7.4], 500 mM KCl), applied to a buffer-equilibrated Sephadex G-25 column (GE Healthcare, Piscataway, NJ), and eluted. The desalted protein was distributed into aliquots, frozen, and stored at −80°C. Sample protein concentration was determined with a bicinchoninic acid assay (Pierce, Rockford, IL) with bovine serum albumin as the standard. Protein purities were >90% by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis. Determinations of heme content relied on formation of the pyridine hemochrome (52).
In the rich medium used for expression, RcoM proteins were typically isolated in predominantly the CO-bound form, which under aerobic conditions was stable under low illumination. The bound CO was gradually photolyzed by exposing samples to incandescent illumination at an incident intensity of 140 W/m2. Samples were air cooled and maintained at ∼25°C during illumination.
UV/Vis spectral analyses.
Room-temperature UV/visible-light (UV/Vis) absorption spectra of samples diluted in buffer A in 1-cm-path, 1.8-ml quartz cuvettes were obtained using a Shimadzu UV-2401PC spectrophotometer operated at a 75-nm/min scan speed, 1-nm slit width, and 0.2-nm data recording interval. The instrument wavelength accuracy was verified weekly. Samples (1 ml) in sealed cuvettes were reduced by flushing the headspace with Ar for 2 min and then injecting a freshly prepared anaerobic solution of sodium dithionite to 0.5 to 1 mM. CO-bound samples were prepared by flushing the cuvette headspace with CO for 2 min, followed by dithionite addition. The dithionite-free CO-bound spectrum (see Fig. 3B) was obtained by adding the minimally sufficient level of dithionite (0.2 mM final concentration) to a CO-saturated sample to obtain the CO-bound species and then desalting the 1-ml sample over a 10-ml anoxic Sephadex G-25 column. The spectrum of the desalted sample was normalized to the A280 of the Fe(III) sample and was verified to be unaffected by subsequent additions of CO and dithionite (data not shown). NO-bound RcoM was generated by the addition of excess NO gas to an anoxic sample in the presence of 1 mM ascorbate.
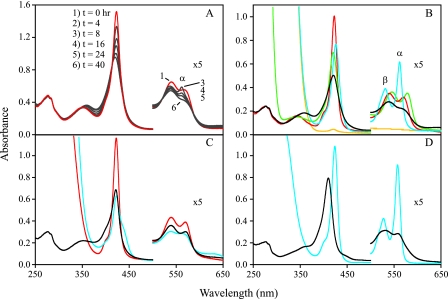
FIG. 3.
Purified RcoMBx-1 has a high affinity for CO and contains a hexacoordinate heme cofactor with His74/Met104 Fe(II) heme ligands. (A) Purified RcoMBx-1 protein (partially CO bound; red line), subjected to photolysis in aerobic buffer at ∼25°C (see Materials and Methods), gradually released bound CO and developed a metastable Fe(II) α peak (562 nm), particularly in the 4-h (not shown) and 8-h samples. (B) Spectra of photolyzed WT protein (0.24 mg protein/ml) show typical hexacoordinate features in the Fe(III) form (black line), the Fe(II) form (in the presence of dithionite; blue line), the Fe(II)-CO form (after desalting to remove dithionite; red line), and in the Fe(II)-NO form (in the presence of ascorbate; green line). The Fe(II)-CO spectrum of the H74Y variant (at 1.6 mg protein/ml, in the presence of dithionite; yellow line) indicates a heme content of <1% relative to the WT protein. (C) The spectra of Fe(III) (black line), Fe(II) (blue line), and Fe(II)-CO (red line) forms of the photolyzed M104L RcoMBx-1 protein (at 0.30 mg protein/ml) are most notable for the altered Fe(II) spectrum, with a diminished Soret band intensity (425-nm peak and prominent 439-nm shoulder) and a broad absorbance between 530 and 580 nm instead of the discrete αβ peaks of the WT protein. (D) The M104K RcoMBx-1 protein (at 0.30 mg protein/ml) exhibited hexacoordinate Fe(III) (black line) and Fe(II) (blue line) spectra, although peak positions are blue-shifted relative to the WT spectra.
RESULTS
Genomic analyses suggest that RcoM is a new single-component regulator of CO metabolism.
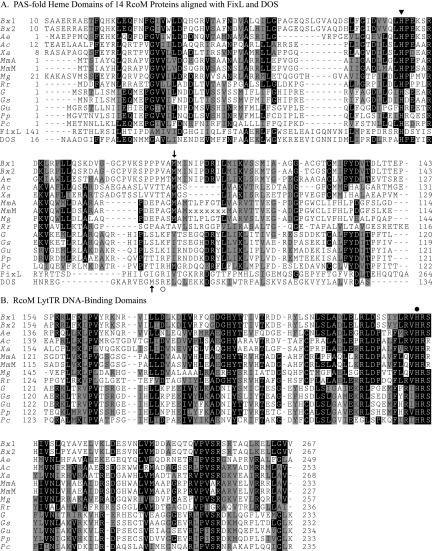
The RcoM sensor type was initially suggested by analysis of bacterial genomic sequences that showed linkage of genes encoding LytTR-type DNA-binding proteins to cox- and coo-encoded CO oxidation functions. We further probed the National Center for Biotechnology Information database using the N-terminal portion of several potential RcoM proteins (thereby avoiding the plethora of LytTR-class proteins; see Materials and Methods) and identified 14 candidates that were similar in primary sequence and domain organization: the ∼140-residue N-terminal portion aligned with the conserved PAS fold (16, 17, 23), and the C-terminal ∼100-residue portion invariably bore a highly conserved LytTR DNA-binding domain (39). Figure 1 presents an alignment of the candidate proteins, and their gi numbers are given in the legend.
FIG. 1.
Modular domain structure and alignments of RcoM proteins. RcoM proteins consist of an N-terminal ∼140-residue heme-binding PAS fold joined to a C-terminal ∼100-residue DNA-binding LytTR domain. (A) Alignment of 14 RcoM PAS fold domains and the PAS fold regions of the Bradyrhizobium japonicum FixL [FixL; gi:27351017] and E. coli DOS (DOS; gi:50513458) sensor proteins. Bx1 and Bx2, B. xenovorans LB400 (gi:91783672 and gi:91780098); Ae, A. ehrlichei MLHE-1 (gi:114320715); Ac, Azorhizobium caulinodans ORS571 (gi:158425899); Xa, Xanthobacter autotrophicus Py2 (gi:154246483); MmA, Magnetospirillum magneticum AMB-1 (gi:83310685); MmM, Magnetospirillum magnetotacticum MS-1 (gi:23014593 and 46201255; protein fragments encoded on two DNA contigs); Mg, Magnetospirillum gryphiswaldense MSR-1 (gi:144897948); Rr, R. rubrum UR2 (gi:83594844); G, Geobacter sp. strain FRC-32 (gi:110600736); Gs, Geobacter sulfurreducens PCA (gi:39997197); Gu, G. uraniumreducens Rf4 (gi:148262698); Pp, Pelobacter propionicus DSM 2379 (gi:118579919); and Pc, P. carbinolicus DSM 2380 (gi:77917674). The conserved His residues in the RcoM proteins (▾, His74 in Bx1) are aligned with the proximal heme ligands of the FixL (His200) and DOS (His77) proteins. The semiconserved Met residue that likely serves as the distal RcoM Fe(II) heme ligand is also indicated (↓, Met104 in Bx1). The Met95 distal ligand of the hexacoordinate Fe(II) heme DOS protein is indicated by an upward arrow, and a circle indicates the Arg residue which stabilizes bound O2 in FixL and DOS proteins (see Discussion). (B) Alignment of the LytTR DNA-binding domains of RcoM proteins. The conserved His residue of this domain (His218 in the Bx1 sequence) is indicated (•). Sequences were aligned using the CLUSTALW2 service (version 2.0.1, http://www.ebi.ac.uk/Tools/clustalw2/ [33]), with subsequent manual adjustment of the FixL and DOS sequences to bring the proximal His residue into alignment with the conserved RcoM His residue, as suggested by an anonymous reviewer of the manuscript. The alignment was shaded using Boxshade (version 3.21, http://www.ch.embnet.org/software/BOX_form.html; shading alignment fraction = 0.5).
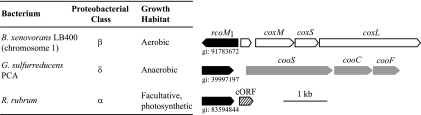
Of the candidate RcoM proteins, representatives were encoded in four of the five proteobacterial classes (α, β, γ, and δ), and most were readily grouped into one of three genomic contexts: six were adjacent to an uncharacterized but conserved ∼300-nucleotide ORF (cORF), four were adjacent to coo genes predicted to encode anaerobic CO oxidation systems, and three were linked to cox genes predicted to encode aerobic CO oxidation systems. Representative rcoM genomic contexts are illustrated in Fig. 2. The physiological significance of the juxtaposition of rcoM and the cORF is unclear, for the function of the cORF-encoded ∼100-residue acidic protein is unknown (although it appears to be regulated by CooA in some organisms [data not shown]), but otherwise the genomic contexts are appropriate: rcoM is adjacent to cox genes in organisms that grow aerobically and adjacent to coo genes in anaerobes (Fig. 2). Moreover, the genomes of anaerobes with adjacent rcoM and cooS genes (P. carbinolicus and Geobacter spp.) do not appear to encode CooA, and therefore cooS expression, if regulated, is likely controlled by RcoM. Similarly, both organisms with juxtaposed rcoM and coxM genes (B. xenovorans and A. ehrlichei) do not appear to contain coxC or coxH, making RcoM a reasonable regulator of cox expression in these systems.
FIG. 2.
Taxonomic occurrence and genomic arrangement of representative rcoM ORFs. Of 14 identified rcoM genes, three are genetically linked to regions that encode the CO oxidation (cox) functions of aerobic metabolism (rcoM of A. ehrlichei; B. xenovorans rcoM1 and rcoM2 occur on different chromosomes and are linked to different coxM genes), four are adjacent to CO oxidation functions (coo encoded) of anaerobic organisms (the rcoM genes of Geobacter sp. strain FRC-32, G. sulfurreducens, G. uraniumreducens, and P. carbinolinicus), while six are immediately adjacent to a cORF that encodes an ∼100-residue acidic protein of unknown function (the rcoM genes of A. caulinodans, X. autotrophicus, M. magneticum, M. magnetotacticum, M. gryphiswaldense, and R. rubrum). The genomic organization is presented for representatives of each group, along with taxonomic and growth habitat characteristics of the relevant organism.
The RcoM proteins from B. xenovorans are hexacoordinate hemoproteins that bind CO and NO but not O2.
We cloned seven rcoM genes, including two from the multireplicon genome of B. xenovorans, and attempted their IPTG-dependent expression in E. coli (see Materials and Methods). Both the B. xenovorans rcoM1- and rcoM2-encoded proteins accumulated well, yielding dark red cell pellets, and UV/Vis spectral analysis of extracts confirmed the overproduction of hemoproteins. Under the same expression conditions, RcoM proteins from Xanthobacter autotrophicus Py2 and A. ehrlichei MLHE-1 accumulated ∼5% of the hemoprotein level of expressed B. xenovorans RcoM-1. We did not detect (≤1%) hemoprotein expression in cultures bearing rcoM genes cloned from R. rubrum, P. carbinolicus DSM 2380, or G. uraniumreducens Rf4, which could reflect poor protein accumulation or the possibility that the expressed proteins do not contain heme. The B. xenovorans RcoM-1 and RcoM-2 expression systems were subsequently modified to incorporate a C-terminal six-His tag, permitting facile protein purification, and these proteins are designated RcoMBx-1 and RcoMBx-2, respectively.
The expressed RcoMBx-1 protein contained predominantly CO-bound heme, and the bound CO persisted in the isolated His-tagged protein despite aerobic extract preparation and purification steps. The heme Fe(II)-CO species was stable for days under aerobic conditions at room temperature when maintained in the dark, but CO could be dissociated by incandescent illumination, with conversion of the heme to the Fe(III) state (Fig. 3A). As indicated by the appearance of an α peak (562 nm [Fig. 3A]), a small pool of Fe(II) heme was evident during this aerobic photolysis, suggesting that the protein does not readily bind O2. [The correspondence of this α peak to that of the Fe(II) heme was consistent with its peak position (562 nm; see below) and its immediate elimination upon ferricyanide oxidation or addition of CO, an Fe(II)-specific ligand (data not shown).] We subsequently attempted to effect oxygen binding to the Fe(II) heme after removal of reductant using a Sephadex G-25 desalting column; this preparation oxidized to the Fe(III) form over a period of 2 to 3 h at room temperature, again indicating the absence of stable O2 binding (data not shown).
UV/Vis absorption spectra of the photolyzed protein (Fig. 3B) indicated hexacoordinate heme iron ligation in all forms. The Fe(III) species showed α, β, Soret, and δ bands at 574 (shoulder), 539, 420 (ɛ = 110 mM−1 cm−1), and 360 nm, respectively, the Fe(II) spectrum exhibited intense peaks at 562 nm (ɛ = 27 mM−1 cm−1), 531 nm (ɛ = 17 mM−1 cm−1), and 426 nm (ɛ = 168 mM−1 cm−1), and the Fe(II)-CO spectrum (after dithionite removal) revealed peaks at 570, 540, 423 (ɛ = 230 mM−1 cm−1), and 344 nm, respectively. The Fe(II) protein also readily bound NO, generating a typical hexacoordinate spectrum with stable absorbance bands at 578, 545, and 421 nm (Fig. 3B). The reduced pyridine hemochromogen displayed an α peak at 557 nm, diagnostic for a b-type heme. A more extensive spectroscopic analysis of the purified B. xenovorans RcoM-2 protein, which although encoded on a different chromosome than RcoMBx-1 is likewise adjacent to a cox system (Fig. 2) and is 93% similar to RcoMBx-1 at the protein level (Fig. 1), supports the conclusion that the RcoMBx-1 and -2 proteins maintain hexacoordinate heme ligation in all forms with displacement of an endogenous axial heme ligand upon the binding of CO or NO (K. A. Marvin, R. L. Kerby, H. Youn, G. P. Roberts and J. N. Burstyn, unpublished data). Thus, the genetic and spectral data are consistent with a CO-sensing function for RcoM proteins, though a biochemical ability to sense NO cannot be discounted at present.
Identification of endogenous heme ligands.
A key to understanding heme protein function is the identification of the endogenous axial ligand(s) to the heme iron. All characterized PAS fold hemoproteins utilize a histidine proximal heme ligand, and the comparison of RcoM sequences (Fig. 1) indicated two invariant residues, His74 in the PAS fold and His218 in the LytTR domain (numbers refer to the RcoMBx proteins), as possible ligand candidates. The purified RcoMBx-1 H218A variant accumulated well and was spectrally identical to wild-type (WT) protein (data not shown), discounting this residue as a heme ligand, but the purified H74Y, H74K, and H74M variants contained <1% as much heme as WT protein (Fig. 3B and data not shown). We propose that His74 is the proximal axial heme ligand in RcoMBx-1, based on (i) the low heme content of the His74 variants, (ii) the absence of spectral changes in the H218A variant and evidence that its conservation reflects a role in DNA binding by LytTR-domain proteins (36), and (iii) the alignment of His74 with the conserved proximal His axial ligands in the structurally characterized FixL (19, 20) and DOSEc (31, 40) PAS fold domains (Fig. 1A).
The PAS fold alignment (Fig. 1A) did not indicate a conserved distal heme ligand trans to RcoMBx-1 His74, but the PAS fold loop between the Fα and Gβ structural elements is known to be highly flexible (23). We therefore analyzed conserved RcoM residues as well possible ligands (RcoMBx Met104 and Met105) in the vicinity of the characterized distal heme pockets (19, 20, 31, 40) of FixL and DOSEc (Met95), as indicated in the alignment (Fig. 1A). The RcoMBx-1 M105L variant exhibited a typical hexacoordinate Fe(II) spectrum, with intense αβ peaks, and proteins altered at other conserved RcoM residues (Lys81, Lys160, His185, Tyr186, and His221) did not display altered Fe(II) spectra (data not shown), which rules them out as likely Fe(II) heme ligands. In contrast, the M104L variant bound CO with higher affinity than the WT protein and required a twofold-prolonged photolysis treatment to effect removal of most of the bound CO. This suggested the loss of a competitive endogenous ligand. Moreover, the Fe(II) spectrum of the M104L variant displayed a notably diminished and broadened Soret band and a broad absorbance in the 530- to 580-nm range (Fig. 3C), without the distinctive αβ peaks of the WT protein (Fig. 3B). These spectral features are consistent with a single axial heme Fe(II) ligand in the M104L variant, based upon their similarities to spectra of pentacoordinate PAS fold hemoproteins, including FixL (18) and PDEA1Ax (5), and the DOSEc protein with substitutions (Ala, Ile, or Leu) of the distal Met ligand (21, 24). It is worth noting that the CO-bound spectra of WT and M104L RcoM proteins were nearly identical, suggesting that Met104 is the protein ligand displaced by CO.
The conclusion that the pentacoordinate Fe(II) spectrum of the RcoMBx-1 M104L variant resulted from direct perturbation of a heme ligand, rather than an indirect effect of an altered protein conformation, was supported by analysis of a variant protein in which an amino acid (Lys) capable of heme ligation replaced the native Met residue. The RcoMBx-1 M104K protein was predominantly isolated in the CO-free Fe(III) state (Soret band at 410 nm [Fig. 3D]) that upon reduction exhibited sharp peaks at 558, 528, and 424 nm (Fig. 3D), consistent with a hexacoordinate heme ligation that is different from that of the WT protein. The RcoMBx-1 M104K protein bound CO slowly, transforming to the Fe(II)-CO species in several hours under the same conditions in which both the M104L and the WT proteins bind CO within the few minutes it takes to prepare and scan the sample (data not shown). The altered peak positions and slowed CO-binding properties indicate an altered heme ligation in the Fe(II) form of the M104K variant, supporting the hypothesis that the Lys residue has replaced the native Met104 distal ligand. In general accord with the alignment and as indicated by the analysis of variant proteins, we propose that the RcoMBx-1 heme Fe(II) axial ligands are the His74/Met104 pair.
In vivo evidence for CO-dependent activation of RcoM.
We next assessed the functional activities of RcoMBx-1 and -2 proteins using an E. coli host bearing two compatible plasmids that express rcoM under IPTG-dependent regulation and encode a reporter system that fused lacZ to B. xenovorans coxM1 (adjacent but divergently transcribed from rcoM1 [Fig. 2; also see Materials and Methods]). Figure 4 presents results for aerobically grown cultures treated with CO and left untreated. Under these growth conditions, β-galactosidase activity is both RcoM and CO dependent (samples A to C), with more activity for RcoMBx-1 (sample B) than RcoMBx-2 (sample C). The H74K RcoMBx-1 variant was inactive (sample D), as was a construct that expressed H74A (data not shown). However, the H74Y variant was clearly different, for its high activity was RcoM expression dependent (samples E and F) but not CO regulated (sample F). Although the mechanism of the H74Y constitutive activity remains uncertain, alterations of RcoMBx-1 His74 clearly affect both heme ligation (Fig. 3B) and CO-sensing function (Fig. 4), consistent with the role of this residue as the proximal ligand.
FIG. 4.
In vivo analysis of RcoMBx CO-sensing transcriptional activity. E. coli strains bearing compatible plasmids that express rcoM under IPTG-dependent regulation and have coxM1 of B. xenovorans LB400 fused to a lacZ reporter were grown aerobically with and without CO. Cells were assessed for RcoM- and CO-dependent lacZ expression. Activities are presented in Miller units (MU), and error bars indicate 95% confidence intervals.
The M104L distal ligand alteration eliminated CO-dependent activity in vivo (Fig. 4, sample G) without significantly affecting heme incorporation (Fig. 3C). It remains to be determined whether this inactivity results from a deleterious effect of the introduced amino acid or the lack of the native Met. Finally, the H218A variant was also inactive (sample H). This His residue is conserved in LytTR domain proteins and is essential for DNA-binding function (36). In summary, these results indicate that under conditions of aerobic growth (using acetate as a nonfermentable carbon source [see Materials and Methods]), both WT RcoMBx proteins can respond in vivo to exogenous CO and therefore act as aerobic sensors of CO.
DISCUSSION
PAS fold proteins with different heme ligation schemes and effector responses.
The PAS fold is noted for its structural and ligand-binding flexibility (16, 23), and comparisons of RcoM and other members of the PAS fold hemoprotein class indicate different heme axial ligands [in the Fe(II) state: His in FixL and PDEA1Ax, His/His in NPAS2, His/Met in DOSEc and RcoMBx] and heme environments, particularly in the distal region. The diversity of PAS fold heme ligands appears to be recapitulated among the RcoM proteins, as a residue corresponding to the Met104 distal ligand of RcoMBx-1 is conserved in only about half of the proteins identified here (Fig. 1A). Either the proteins lacking this Met residue employ different distal ligands, a distinct possibility given the flexibility of the region, or they maintain a pentacoordinate heme, similar to FixL and PDEA1Ax proteins. Of course, it is also possible that certain RcoM proteins do not contain heme, though the absolute preservation of the proximal His residue suggests otherwise.
We note that RcoMBx-1 formed a stable hexacoordinate Fe(II)-NO species (Fig. 3B), and it is possible that the protein is also activated by NO. NO would not be stable under the aerobic conditions of the in vivo assay reported here and would seem to be physiologically irrelevant for organisms growing under aerobic conditions, yet there is a recent claim of NO oxidation by the aerobic CO oxidation system of mycobacteria (41). This might indicate a physiological rationale for NO-dependent regulation of cox genes and deserves further analysis. Likewise, analysis of the ligand binding and response properties of an RcoM from an anaerobic organism is of interest insofar as NO should be more stable under anoxic conditions.
In contrast to its high affinity for CO and NO, the low affinity of RcoMBx-1 for O2 may in part result from the distal heme environment, as indicated by the PAS fold sequence alignment (Fig. 1A). A highly conserved Arg residue present in the distal pocket of O2-binding PDEA1Ax (not shown), DOSEc (Arg97), and FixL (Arg220) is not evident in this region of RcoMBx-1 and likewise is not conserved in most RcoM proteins (Fig. 1A). The distal-pocket Arg coordinates with bound O2 in DOSEc and FixL proteins (19, 40), and alterations of this residue often reduce O2 affinity (2, 9, 49). Thus, these RcoM PAS fold environments may discriminate against the binding of O2. The alignment does indicate conservation of a basic residue at the analogous position in the RcoM proteins from Pelobacter and Geobacter spp., but here the physiological relevance of O2 binding is unclear, since these anaerobic organisms would not typically encounter high O2 concentrations.
Finally, the mammalian NPAS2 sensor is reportedly specific for CO (7). However, the mechanism of this selectivity is not well understood, because the His residue displaced upon ligand binding remains uncertain (50), and structural data are presently unavailable. It would be unsurprising if the PAS fold structural differences influenced binding of different exogenous ligands and portend somewhat different mechanisms of signal transduction among the hemoprotein regulators. For the RcoM and NPAS2 proteins, more specific conclusions must await further biochemical analyses and structural characterizations.
RcoM and CooA: dual nonhomologous mechanisms of prokaryotic CO-dependent regulation.
Numerous and diverse prokaryotes bear coo- and cox-encoded CO oxidation functions (28, 42, 44). In contrast to extensive studies of CooA-dependent coo expression (44), cox operon regulation has not been extensively analyzed, and it is generally thought to depend on the CoxC (or the similar CoxH) protein (12, 39, 45). Here, however, the cox systems of B. xenovorans LB400 and A. ehrlichei MLHE-1 are the exception, as tBLAST genome searches using the O. carboxidovorans CoxC or CoxH as a probe yields matches to the RcoM proteins instead of probable CoxC/H proteins. Like RcoM proteins, the predicted CoxC/H proteins from O. carboxidovorans exhibit a C-terminal LytTR domain, but these larger (∼400 residues) membrane-spanning proteins remain biochemically uncharacterized. An alignment of CoxC and RcoM proteins indicates both the conserved LytTR region and some similarity between one CoxC MHYT motif, a conserved residue sequence thought to represent a sensor domain (14), and the region with the RcoM-proximal His residue (data not shown). The apparent conservation of a His residue might indicate that CoxC is likewise a hemoprotein, though if CO is its sole effector, and DNA binding its sole response, then the significance of its integral membrane conformation is curious, as CO is readily diffusible. Indeed, the ability of CO to transverse cellular boundaries is critical for its roles in eukaryotic signal transduction (53).
The RcoM proteins are fundamentally different from the CooA regulators. Instead of the PAS fold, the CooA heme domain is structurally similar to the cyclic AMP-binding region of CRP with a short deletion and appropriate residues that permit heme binding (44). The DNA-binding domains of CooA and RcoM are also dissimilar. The CooA/CRP/FNR helix-turn-helix DNA-binding domains recognize well-characterized inverted repeats (22, 32, 44), whereas the RcoM LytTR domain interaction with DNA likely involves multiple imperfect direct repeats (36, 39). Unfortunately, neither the structure of any LytTR domain (∼1200 representatives are indicated in the InterPro database [IPR007492]) nor its precise DNA interactions have been determined (13).
Functionally, compared to CooA, RcoM proteins appear to regulate a more diverse array of CO oxidation regulons, including aerobic cox expression in selected organisms (Fig. 2 and 4). The fact that the metabolically versatile aerobe B. xenovorans (4) contains two RcoM paralogues could indicate differential expression of the associated cox regulons in response to undetermined environmental cues. Similarly, the fact that R. rubrum contains both RcoM and CooA may indicate differential regulation of cORF (by RcoM [Fig. 2]) and the coo genes (by CooA [44]), and we are investigating this regulation using appropriate reporter fusions constructed in R. rubrum. Alternatively, RcoM and CooA may simply represent two nonhomologous mechanisms for prokaryotic CO-dependent transcriptional regulation, with similar functional properties. Indeed, the fact that certain anaerobes appear to harbor coo-encoded CO-oxidation functions and RcoM (Fig. 2), but not CooA, suggests that here RcoM has supplanted CooA.
Acknowledgments
This work was supported by the College of Agricultural and Life Sciences of the University of Wisconsin-Madison and by a National Institutes of Health Grant GM53228 (to G.P.R.).
Searches utilized the database and analysis functions of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/), a component of the U.S. National Library of Medicine and the National Institutes of Health. Chromosomal DNA samples were graciously provided by Gary King, Louisiana State University (A. ehrlichei MLHE-1), Deborah Himes, Michigan State University (B. xenovorans LB400), Evgenya Shelobolina, University of Wisconsin—Madison (G. uraniumreducens Rf4), Derek Lovley, University of Massachusetts (P. carbinolicus DSM 2380), Yaoping Zhang, University of Wisconsin-Madison (R. rubrum UR2), and Scott Ensign, Utah State University (X. autotrophicus Py2). Rapid DNA sequencing analyses were performed at the University of Wisconsin Biotechnology Center Sequencing facility, and we thank Jose Serate for protein purifications and Katherine Marvin and Judith Burstyn (Department of Chemistry, University of Wisconsin—Madison) for helpful discussions.
Footnotes
Published ahead of print on 7 March 2008.
REFERENCES
- 1.Aono, S. 2003. Biochemical and biophysical properties of the CO-sensing transcriptional activator CooA. Acc. Chem. Res. 36825-831. [DOI] [PubMed] [Google Scholar]
- 2.Balland, V., L. Bouzhir-Sima, L. Kiger, M. C. Marden, M. H. Vos, U. Liebl, and T. A. Mattioli. 2005. Role of arginine 220 in the oxygen sensor FixL from Bradyrhizobium japonicum. J. Biol. Chem. 28015279-15288. [DOI] [PubMed] [Google Scholar]
- 3.Cary, S. P. L., J. A. Winger, E. R. Derbyshire, and M. A. Marletta. 2006. Nitric oxide signaling: no longer simply on or off. Trends Biochem. Sci. 31231-239. [DOI] [PubMed] [Google Scholar]
- 4.Chain, P. S. G., V. J. Denef, K. T. Konstantinidis, L. M. Vergez, L. Agulló, V. L. Reyes, L. Hauser, M. Córdova, L. Gómez, M. González, M. Land, V. Lao, F. Larimer, J. J. LiPuma, E. Mahenthiralingam, S. A. Malfatti, C. J. Marx, J. J. Parnell, A. Ramette, P. Richardson, M. Seeger, D. Smith, T. Spilker, W. J. Sul, T. V. Tsoi, L. E. Ulrich, I. B. Zhulin, and J. M. Tiedje. 2006. Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. Proc. Natl. Acad. Sci. USA 10315280-15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, A. L., J. R. Tuckerman, G. Gonzalez, R. Mayer, H. Weinhouse, G. Volman, D. Amikam, M. Benziman, and M.-A. Gilles-Gonzalez. 2001. Phosphodiesterase A1, a regulator of cellulose synthesis in Acetobacter xylinum, is a heme-based sensor. Biochemistry 403420-3426. [DOI] [PubMed] [Google Scholar]
- 6.Delgado-Nixon, V. M., G. Gonzalez, and M.-A. Gilles-Gonzalez. 2000. Dos, a heme-binding PAS protein from Escherichia coli, is a direct oxygen sensor. Biochemistry 392685-2691. [DOI] [PubMed] [Google Scholar]
- 7.Dioum, E. M., J. Rutter, J. R. Tuckerman, G. Gonzalez, M.-A. Gilles-Gonzalez, and S. L. McKnight. 2002. NPAS2: a gas-responsive transcription factor. Science 2982385-2387. [DOI] [PubMed] [Google Scholar]
- 8.Drennan, C. L., J. Heo, M. D. Sintchak, E. Schreiter, and P. W. Ludden. 2001. Life on carbon monoxide: X-ray structure of Rhodospirillum rubrum Ni-Fe-S carbon monoxide dehydrogenase. Proc. Natl. Acad. Sci. USA 9811973-11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunham, C. M., E. M. Dioum, J. R. Tuckerman, G. Gonzalez, W. G. Scott, and M.-A. Gilles-Gonzalez. 2003. A distal arginine in oxygen-sensing heme-PAS domains is essential to ligand binding, signal transduction, and structure. Biochemistry 427701-7708. [DOI] [PubMed] [Google Scholar]
- 10.Dykxhoorn, D. M., R. St. Pierre, and T. Linn. 1996. A set of compatible tac promoter expression vectors. Gene 177133-136. [DOI] [PubMed] [Google Scholar]
- 11.Freitas, T. A. K., J. A. Saito, S. Hou, and M. Alam. 2005. Globin-coupled sensors, protoglobins, and the last universal common ancestor. J. Inorg. Biochem. 9923-33. [DOI] [PubMed] [Google Scholar]
- 12.Fuhrmann, S., M. Ferner, T. Jeffke, A. Henne, G. Gottschalk, and O. Meyer. 2003. Complete nucleotide sequence of the circular megaplasmid pHCG3 of Oligotropha carboxidovorans: function in the chemolithoautotrophic utilization of CO, H2 and CO2. Gene 32267-75. [DOI] [PubMed] [Google Scholar]
- 13.Galperin, M. Y. 2006. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J. Bacteriol. 1884169-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galperin, M. Y., T. A. Gaidenko, A. Y. Mulkidjanian, M. Nakano, and C. W. Price. 2001. MHYT, a new integral membrane sensor domain. FEMS Microbiol. Lett. 20517-23. [DOI] [PubMed] [Google Scholar]
- 15.Gao, R., T. R. Mack, and A. M. Stock. 2007. Bacterial response regulators: versatile regulatory strategies from common domains. Trends Biochem. Sci. 32225-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilles-Gonzalez, M.-A., and G. Gonzalez. 2004. Signal transduction by heme-containing PAS-domain proteins. J. Appl. Physiol. 96774-783. [DOI] [PubMed] [Google Scholar]
- 17.Gilles-Gonzalez, M.-A., and G. Gonzalez. 2005. Heme-based sensors: defining characteristics, recent developments, and regulatory hypotheses. J. Inorg. Biochem. 991-22. [DOI] [PubMed] [Google Scholar]
- 18.Gilles-Gonzalez, M. A., G. Gonzalez, M. F. Perutz, L. Kiger, M. C. Marden, and C. Poyart. 1994. Heme-based sensors, exemplified by the kinase FixL, are a new class of heme protein with distinctive ligand binding and autoxidation. Biochemistry 338067-8073. [DOI] [PubMed] [Google Scholar]
- 19.Gong, W., B. Hao, and M. K. Chan. 2000. New mechanistic insights from structural studies of the oxygen-sensing domain of Bradyrhizobium japonicum FixL. Biochemistry 393955-3962. [DOI] [PubMed] [Google Scholar]
- 20.Gong, W., B. Hao, S. S. Mansy, G. Gonzalez, M. A. Gilles-Gonzalez, and M. K. Chan. 1998. Structure of a biological oxygen sensor: a new mechanism for heme-driven signal transduction. Proc. Natl. Acad. Sci. USA 9515177-15182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez, G., E. M. Dioum, C. M. Bertolucci, T. Tomita, M. Ikeda-Saito, M. R. Cheesman, N. J. Watmough, and M.-A. Gilles-Gonzalez. 2002. Nature of the displaceable heme-axial residue in the EcDos protein, a heme-based sensor from Escherichia coli. Biochemistry 418414-8421. [DOI] [PubMed] [Google Scholar]
- 22.He, Y., D. Shelver, R. L. Kerby, and G. P. Roberts. 1996. Characterization of a CO-responsive transcriptional activator from Rhodospirillum rubrum. J. Biol. Chem. 271120-123. [DOI] [PubMed] [Google Scholar]
- 23.Hefti, M. H., K.-J. Françoijs, S. C. de Vries, R. Dixon, and J. Vervoort. 2004. The PAS fold. A redefinition of the PAS domain based upon structural prediction. Eur. J. Biochem. 2711198-1208. [DOI] [PubMed] [Google Scholar]
- 24.Hirata, S., T. Matsui, Y. Sasakura, S. Sugiyama, T. Yoshimura, I. Sagami, and T. Shimizu. 2003. Characterization of Met95 mutants of a heme-regulated phosphodiesterase from Escherichia coli. Optical absorption, magnetic circular dichroism, circular dichroism, and redox potentials. Eur. J. Biochem. 2704771-4779. [DOI] [PubMed] [Google Scholar]
- 25.Hou, S., M. F. Reynolds, F. T. Horrigan, S. H. Heinemann, and T. Hoshi. 2006. Reversible binding of heme to proteins in cellular signal transduction. Acc. Chem. Res. 39918-924. [DOI] [PubMed] [Google Scholar]
- 26.Huber, C., and G. Wächtershäuser. 2006. α-Hydroxy and α-amino acids under possible Hadean, volcanic origin-of-life conditions. Science 314630-632. [DOI] [PubMed] [Google Scholar]
- 27.Kerby, R. L., H. Youn, M. V. Thorsteinsson, and G. P. Roberts. 2003. Repositioning about the dimer interface of the transcription regulator CooA: a major signal transduction pathway between the effector and DNA-binding domains. J. Mol. Biol. 325809-823. [DOI] [PubMed] [Google Scholar]
- 28.King, G. M., and C. F. Weber. 2007. Distribution, diversity and ecology of aerobic CO-oxidizing bacteria. Nat. Rev. Microbiol. 5107-118. [DOI] [PubMed] [Google Scholar]
- 29.Kokotek, W., and W. Lotz. 1989. Construction of a lacZ-kanamycin-resistance cassette, useful for site-directed mutagenesis and as a promoter probe. Gene 84467-471. [DOI] [PubMed] [Google Scholar]
- 30.Korbas, M., S. Vogt, W. Meyer-Klaucke, E. Bill, E. J. Lyon, R. K. Thauer, and S. Shima. 2006. The iron-sulfur cluster-free hydrogenase (Hmd) is a metalloenzyme with a novel iron binding motif. J. Biol. Chem. 28130804-30813. [DOI] [PubMed] [Google Scholar]
- 31.Kurokawa, H., D.-S. Lee, M. Watanabe, I. Sagami, B. Mikami, C. S. Raman, and T. Shimizu. 2004. A redox-controlled molecular switch revealed by the crystal structure of a bacterial heme PAS sensor. J. Biol. Chem. 27920186-20193. [DOI] [PubMed] [Google Scholar]
- 32.Lanzilotta, W. N., D. J. Schuller, M. V. Thorsteinsson, R. L. Kerby, G. P. Roberts, and T. L. Poulos. 2000. Structure of the CO sensing transcription activator CooA. Nat. Struct. Biol. 7876-880. [DOI] [PubMed] [Google Scholar]
- 33.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 232947-2948. [DOI] [PubMed] [Google Scholar]
- 34.Londer, Y. Y., I. S. Dementieva, C. A. D'Ausilio, P. R. Pokkuluri, and M. Schiffer. 2006. Characterization of a c-type heme-containing PAS sensor domain from Geobacter sulfurreducens representing a novel family of periplasmic sensors in Geobacteraceae and other bacteria. FEMS Microbiol. Lett. 258173-181. [DOI] [PubMed] [Google Scholar]
- 35.MacFerrin, K. D., M. P. Terranova, S. L. Schreiber, and G. L. Verdine. 1990. Overproduction and dissection of proteins by the expression-cassette polymerase chain reaction. Proc. Natl. Acad. Sci. USA 871937-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McGowan, S., I. S. Lucet, J. K. Cheung, M. M. Awad, J. C. Whisstock, and J. I. Rood. 2002. The FxRxHrS motif: a conserved region essential for DNA binding of the VirR response regulator from Clostridium perfringens. J. Mol. Biol. 322997-1011. [DOI] [PubMed] [Google Scholar]
- 37.Meyer, O., L. Gremer, R. Ferner, M. Ferner, H. Dobbek, M. Gnida, W. Meyer-Klaucke, and R. Huber. 2000. The role of Se, Mo and Fe in the structure and function of carbon monoxide dehydrogenase. Biol. Chem. 381865-876. [DOI] [PubMed] [Google Scholar]
- 38.Moskvin, O. V., S. Kaplan, M.-A. Gilles-Gonzalez, and M. Gomelsky. 2007. Novel heme-based oxygen sensor with a revealing evolutionary history. J. Biol. Chem. 28228740-28748. [DOI] [PubMed] [Google Scholar]
- 39.Nikolskaya, A. N., and M. Y. Galperin. 2002. A novel type of conserved DNA-binding domain in the transcriptional regulators of the AlgR/AgrA/LytR family. Nucleic Acids Res. 302453-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park, H., C. Suquet, J. D. Satterlee, and C. H. Kang. 2004. Insights into signal transduction involving PAS domain oxygen-sensing heme proteins from the X-ray crystal structure of Escherichia coli Dos heme domain (Ec DosH). Biochemistry 432738-2746. [DOI] [PubMed] [Google Scholar]
- 41.Park, S. W., T. Song, S. Y. Kim, E. Kim, J.-I. Oh, C.-Y. Eom, and Y. M. Kim. 2007. Carbon monoxide dehydrogenase in mycobacteria possesses a nitric oxide dehydrogenase activity. Biochem. Biophys. Res. Commun. 362449-453. [DOI] [PubMed] [Google Scholar]
- 42.Ragsdale, S. W. 2004. Life with carbon monoxide. Crit. Rev. Biochem. Mol. Biol. 39165-195. [DOI] [PubMed] [Google Scholar]
- 43.Reynolds, A., and V. Lundblad. 1995. Assay for β-galactosidase in liquid cultures, p. 13-30. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, K. Struhl (ed.), Short protocols in molecular biology: a compendium of methods from current protocols in molecular biology, 3rd ed. Wiley, New York, NY.
- 44.Roberts, G. P., H. Youn, and R. L. Kerby. 2004. CO-sensing mechanisms. Microbiol. Mol. Biol. Rev. 68453-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santiago, B., U. Schübel, C. Egelseer, and O. Meyer. 1999. Sequence analysis, characterization and CO-specific transcription of the cox gene cluster on the megaplasmid pHCG3 of Oligotropha carboxidovorans. Gene 236115-124. [DOI] [PubMed] [Google Scholar]
- 46.Sardiwal, S., S. L. Kendall, F. Movahedzadeh, S. C. G. Rison, N. G. Stoker, and S. Djordjevic. 2005. A GAF domain in the hypoxia/NO-inducible Mycobacterium tuberculosis DosS protein binds haem. J. Mol. Biol. 353929-936. [DOI] [PubMed] [Google Scholar]
- 47.Sousa, E. H. S., J. R. Tuckerman, G. Gonzalez, and M.-A. Gilles-Gonzalez. 2007. DosT and DevS are oxygen-switched kinases in Mycobacterium tuberculosis. Prot. Sci. 161-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stewart, V., Y. Lu, and A. J. Darwin. 2002. Periplasmic nitrate reductase (NapABC enzyme) supports anaerobic respiration by Escherichia coli K-12. J. Bacteriol. 1841314-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanaka, A., H. Takahashi, and T. Shimizu. 2007. Critical role of the heme axial ligand, Met95, in locking catalysis of the phosphodiesterase from Escherichia coli (Ec DOS) toward cyclic diGMP. J. Biol. Chem. 28221301-21307. [DOI] [PubMed] [Google Scholar]
- 50.Uchida, T., E. Sato, A. Sato, I. Sagami, T. Shimizu, and T. Kitagawa. 2005. CO-dependent activity-controlling mechanism of heme-containing CO-sensor protein, neuronal PAS domain protein 2. J. Biol. Chem. 28021358-21368. [DOI] [PubMed] [Google Scholar]
- 51.Ulrich, L. E., E. V. Koonin, and I. B. Zhulin. 2005. One-component systems dominate signal transduction in prokaryotes. Trends Microbiol. 1352-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waterman, M. R. 1978. Spectral characterization of human hemoglobin and its derivatives. Methods Enzymol. 52456-463. [DOI] [PubMed] [Google Scholar]
- 53.Wu, L., and R. Wang. 2005. Carbon monoxide: endogenous production, physiological functions, and pharmacological applications. Pharmacol. Rev. 57585-630. [DOI] [PubMed] [Google Scholar]
- 54.Zhou, L., X.-H. Lei, B. R. Bochner, and B. L. Wanner. 2003. Phenotype microarray analysis of Escherichia coli K-12 mutants with deletions of all two-component systems. J. Bacteriol. 1854956-4972. [DOI] [PMC free article] [PubMed] [Google Scholar]