Abstract
Archaea, like Eukarya and Bacteria, are able to N glycosylate select protein targets. However, in contrast to relatively advanced understanding of the eukaryal N glycosylation process and the information being amassed on the bacterial process, little is known of this posttranslational modification in Archaea. Toward remedying this situation, the present report continues ongoing efforts to identify components involved in the N glycosylation of the Haloferax volcanii S-layer glycoprotein. By combining gene deletion together with mass spectrometry, AglE, originally identified as a homologue of murine Dpm1, was shown to play a role in the addition of the 190-Da sugar subunit of the novel pentasaccharide decorating the S-layer glycoprotein. Topological analysis of an AglE-based chimeric reporter assigns AglE as an integral membrane protein, with its N terminus and putative active site facing the cytoplasm. These finding, therefore, contribute to the developing picture of the N glycosylation pathway in Archaea.
Despite the fact that numerous examples of N-glycosylated proteins have been reported in Archaea (9, 22, 27, 28), including the surface (S)-layer glycoprotein of the haloarchaeon Halobacterium salinarum, the first noneukaryal glycoprotein to be described in detail (17, 23), little is known of the enzymatic steps involved in the archaeal version of this posttranslational modification. Recently, however, the first systematic examinations of the archaeal N glycosylation pathway have been undertaken.
After having chemically characterized the N-linked glycan moieties found on both the flagellins and S-layer glycoprotein of Methanococcus voltae (26), Jarrell and coworkers subsequently identified two gene products, i.e., AglA and AglB, involved in N glycosylation of these target proteins (7). At the same time, Abu-Qarn and Eichler (1) used bioinformatics to identify genes in the haloarchaeon Haloferax volcanii homologous to eukaryal and bacterial sequences encoding proteins known to participate in N glycosylation. By next addressing the glycosylation status of a reporter glycoprotein, i.e., the S-layer glycoprotein, in a series of H. volcanii strains deleted of genes putatively participating in N glycosylation, the involvement of two gene products in protein glycosylation, i.e., AglD and AglB, originally termed Alg5-A and Stt3, was demonstrated (2). AglD is involved in loading the final 162-Da sugar residue onto the pentasaccharide decorating at least two of the seven predicted sequons (Asn-X-Ser/Thr motifs, where X is any residue but Pro) of the H. volcanii S-layer glycoprotein. AglB, the sole component of the eukaryal oligosaccharide transferase complex thus far detected in any archaeal genome (1), is required for sugar-based modification of the same S-layer glycoprotein Asn residues. Deletion of either gene, moreover, affected S-layer structure and led to reduced cell growth as growth medium salt content increased, pointing to a role for this posttranslational modification in the ability of H. volcanii to thrive in hypersaline conditions (2).
In addition to containing genes encoding two Alg5 homologues, one of which corresponds to AglD (2), H. volcanii also encodes at least nine other putative glycosyltransferases. These include four homologues of eukaryal dolichol-phosphate mannosyl transferase 1 (Dpm1), responsible for loading activated mannose subunits onto dolichol carriers in the endoplasmic reticulum membrane (6), termed Dpm1-A through Dpm1-D in H. volcanii (1). In addition, H. volcanii also encodes a fifth Dpm-1 homologue, Hdpm-1, previously detected in Haloarcula marismortui (5) but not in eukaryal dpm-1 sequence-based homology searches of H. volcanii (1). While the genes encoding these putative H. volcanii glycosyltransferases are transcribed (1), suggesting that all encode expressed proteins, their substrate specificities and, indeed, their participation in N glycosylation, remains to be proven. Toward this end, we have generated glycosyltransferase-deleted H. volcanii strains to assess the roles of the absent proteins in the N glycosylation of the S-layer glycoprotein.
In the research reported here, we have focused on the role played by the product of the dpm1-B gene, given that dpm1-B is located only eight open reading frames upstream from aglB (http://archaea.ucsc.edu), previously shown to participate in H. volcanii N glycosylation (2). Moreover, both dpm1-B and aglB present similar transcription patterns (1). The results show that although it is not essential for H. volcanii viability, Dpm1-B is indeed involved in glycosylation of the reporter S-layer glycoprotein, participating in adding the fourth subunit of the pentasaccharide decorating at least two Asn residues of the protein. Thus, adopting the terminology of Chaban et al. (7), we rename dpm1-B as aglE. Finally, topology studies involving a AglE-based chimera point to AglE being a membrane-spanning protein with an inwardly oriented N terminus, suggesting the active site of AglE to lie on the cytoplasmic side of the plasma membrane.
MATERIALS AND METHODS
Growth conditions.
H. volcanii WR536 (H53) was obtained from Moshe Mevarech (Tel Aviv University) and grown in complete medium containing 3.4 M NaCl, 0.15 M MgSO4·7H2O, 1 mM MnCl2, 4 mM KCl, 3 mM CaCl2, 0.3% (wt/vol) yeast extract, 0.5% (wt/vol) tryptone, and 50 mM Tris-HCl (pH 7.2) at 40°C (3, 24). In Casamino Acids medium, yeast extract and tryptone were replaced by Casamino Acids (Difco, Detroit MI) at a final concentration of 0.5% (wt/vol). Escherichia coli was grown in Luria-Bertani medium.
Gene deletion.
To test the essential nature of H. volcanii dpm1-B, the gene was deleted as previously described (1). The primers used to amplify regions of ∼500 bp in length flanking the dpm1-B coding sequence are as follows: forward upstream flanking region primer (forup), gggctcgagGACTTGTTGAGTGCTACGAG; reverse upstream flanking region primer (revup), cccaagcttGTAGGTGATAGTATCCCAAG; forward downstream flanking region primer (fordwn), gggggatccCCTCTCTTTCTGACATTCTTG; and reverse downstream flanking region primer (revdwn), ccctctagaGTCCTCCAGAGGCCGCAAAC. In each primer sequence, the genomic DNA sequences are in capital letters, and the introduced restriction sites, including three guanine or cytosine residues, are in lowercase letters. XhoI and HindIII sites were introduced in the forup and fordwn sequences, respectively, while BamHI and XbaI sites were introduced in the fordwn and revdwn sequences, respectively.
Mass spectrometry (MS).
For in-gel tryptic digestion of the H. volcanii S-layer glycoprotein from cells of the WR536 background strain, or the same strain depleted of dpm1-B (or aglE, see below), samples were run on 10% precast gels (Invitrogen, Paisley, United Kingdom) and stained with Novex colloidal blue stain (Invitrogen). The bands of interest were excised, destained in 400 μl of 50% (vol/vol) acetonitrile in 0.1 M ammonium bicarbonate (pH 8.4), and dried by using a SpeedVac drying apparatus. The gel slices were rehydrated in 20 μl of trypsin working solution (Promega sequencing grade modified trypsin, prepared according to the manufacturer's instructions) and incubated at 37°C overnight. The supernatant was removed, and digestion was terminated by the addition of 50 μl of 0.1% (vol/vol) trifluoroacetic acid (10 min, 37°C). The supernatant was removed, and the peptides were further extracted with 200 μl of 60% (vol/vol) acetonitrile-0.1% (vol/vol) trifluoroacetic acid (15 min, 37°C). The supernatant was again removed and pooled with the previous supernatant. Both extraction steps were then repeated, and the supernatants were pooled. The volume of the combined supernatants was subsequently concentrated by using a SpeedVac drying apparatus.
For offline liquid chromatography matrix-assisted laser desorption ionization-time of flight (MALDI-TOF)/TOF-MS analysis, protease-generated peptides were separated by using the Ultimate 3000 LC system (Dionex, Sunnyvale CA), fitted with a Pepmap analytical C-18 nanocapillary (75-μm internal diameter by 15-cm length; Dionex) analytical column. An aliquot of the digest was loaded onto the column and eluted by using solvent A (0.1% [vol/vol] trifluoroacetic acid in 2% [vol/vol] acetonitrile) and solvent B (0.1% [vol/vol] trifluoroacetic acid in 90% [vol/vol] acetonitrile) in the following gradient: 0 to 60% solvent B (0 to 36 min), 60 to 90% solvent B (36 to 37 min), 90% solvent B (37 to 40 min), and 100% solvent A (40 to 41 min) at a flow rate of 0.300 nl/min. The eluant was mixed directly with the α-cyano-hydroxy cinnamic acid matrix and spotted onto a metal MALDI target plate. MALDI-TOF/TOF-MS was performed by using an Applied Biosystems 4800 mass spectrometer in positive reflectron mode, set for delayed extraction. Tandem MS (MS/MS) was performed with the collision-induced dissociation (CID) setting turned on. Sequazyme peptide mass standards served as external calibrants.
Construction of a CBD-Dpm1-B chimera.
The dpm1-B gene was amplified from H. volcanii strain WR536 genomic DNA using primers (forward primer, gggcatATGACTTCGACTCTACCGTTTG; reverse primer, cccggtaccTCATTGTTCCTCCGATCTTAGG) designed to introduce NdeI and KpnI restriction sites (lowercase letters) on each side of the dpm1-B coding region (capital letters) and ligated into the pGEM-T Easy vector (Promega). The dpm1-B gene was then excised upon digestion with NdeI and KpnI and inserted into the pWL-CBD vector (13), also digested with the same restriction enzymes, resulting in DNA encoding the Clostridium thermocellum cellulosome cellulose-binding domain (CBD) fused to the 5′ end of the Dpm1-B-encoding gene.
Subcellular fractionation and urea treatment of H. volcanii cells.
H. volcanii cells (1 ml) were broken by sonication (2 s on and 1 s off for 90 s, 35% output; Misonix XL2020 ultrasonicator) and centrifuged at 196,000 × g for 13 min at 4°C in an ultracentrifuge (Sorvall M120). While 200 μl of the supernatant was precipitated in 15% (wt/vol) trichloroacetic acid (TCA), the pelleted membrane fraction was resuspended in 200 μl of distilled water and precipitated in 15% (wt/vol) TCA. For urea treatment, designed to distinguish between peripheral and integral membrane proteins, the pelleted membrane fraction was resuspended in 300 μl of 6 M urea in 2 M NaCl-50 mM Tris-HCl (pH 7.2), followed by incubation at room temperature for 15 min. Subsequently, the mixture was overlain onto 700 μl of a 40% (wt/vol) sucrose cushion in 2 M NaCl-50 mM Tris-HCl (pH 7.2) and centrifuged for 75 min at 196,000 × g and 4°C. The membrane-containing pellet was resuspended in 200 μl of distilled water, and proteins were precipitated in 15% (wt/vol) TCA. All TCA-precipitated samples were washed with ice-cold acetone, resuspended in sample buffer, and resolved by sodium dodecyl sulfate-12% (wt/vol) polyacrylamide gel electrophoresis (SDS-12% PAGE).
Immunoblotting.
Proteins were electrotransferred from SDS-PAGE gels to nitrocellulose membranes (0.45-μm pore size; Schleicher & Schuell, Dassel, Germany) and incubated with primary and secondary antibodies at the following dilutions: polyclonal rabbit anti-CBD antibodies (obtained from Ed Bayer, Weizmann Institute of Science or Yuval Shoham, Technion-Israel Institute of Technology) were used at a 1:10,000 dilution, while polyclonal rabbit anti-S-layer glycoprotein (10) and anti-SRP54 (25) antibodies were diluted 1:1,000. Horseradish peroxidase-conjugated goat anti-rabbit antibodies (Bio-Rad), serving as secondary antibodies, were used at a 1:2,000 dilution. Detection was achieved by using the ECL Western blotting detection reagent (Amersham Biosciences, United Kingdom).
GenBank accession number.
The sequence of H. volcanii dpm1-B (listed as aglE, see below) has been deposited into the EMBL/GenBank/DDBJ databases and assigned accession number AM888352.
RESULTS
Dpm1-B is a member of a conserved protein family abundant in all three domains of life.
In an earlier study (1), dpm1-B was identified in a homology-based search of a partially completed H. volcanii genome sequence as one of the homologues of murine dpm1 encoded by the haloarchaeon. However, the dpm1-B sequence does not correspond to any of the open reading frames delineated in the recently annotated H. volcanii genome sequence (http://archaea.ucsc.edu). Upon closer analysis, dpm1-B was shown to be present on the reverse strand of the chromosomal DNA (nucleotides 1387487 to 1388401), overlapping with the latter portion of HVO_1523 (nucleotides 1386375 to 1387784) and covering most of the nonannotated region between HVO_1523 and HVO_1524 (nucleotides 1388454 to 1389884). Thus, dpm1-B is located eight genes upstream from the oligosaccharide transferase-encoding aglB gene (2), designated HVO_1530. Based on the previously presented transcription of dpm1-B (1) and the functional data presented below, it can be concluded that dpm1-B (aglE, see below) corresponds to a verified protein-coding gene, calling for revision of the current annotation of the H. volcanii genome.
The dpm1-B gene (or aglE [see below]) encodes a predicted 33.8-kDa protein and includes a glycosyltransferase 2 domain (PF00535), spanning residues 9 to 173, much like murine Dpm1, which contains the same domain, spanning residues 28 to 199 (Fig. 1). Outside these regions, the two protein sequences show little homology. This, together with documented difficulty in predicting protein function from amino acid sequence alone (8) and the broad range of functions attributed to proteins comprising the Pfam PF00535 family (pfam.sanger.ac.uk/family?acc = PF00535), suggests that H. volcanii Dpm1-B and its eukaryal homologues may not carry out the same functions. Nonetheless, sequence analysis reveals that H. volcanii Dpm1-B belongs to a universal protein family, whose members are found in Archaea, Eukarya, and Bacteria.
FIG. 1.
H. volcanii Dpm1-B (AglE) is a homologue of eukaryal Dpm1. The deduced amino acid sequence of H. volcanii Dpm1-B (AglE) was aligned with the sequence of the Mus musculus Dpm1 protein (NP_034202.1). Identical amino acids are in black boxes, while similar amino acids are shaded gray.
Dpm1-B is involved in adding the 190-Da sugar residue to the N-linked pentasaccharide decorating the S-layer glycoprotein.
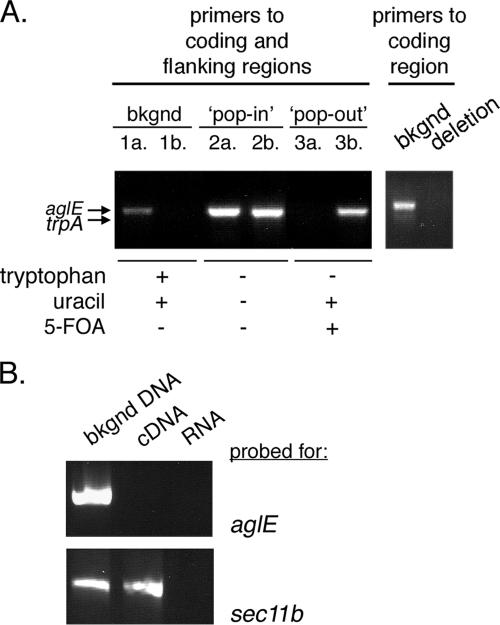
To assess the function of H. volcanii dpm1-B, the gene was deleted from the genome as described previously (3). To delete dpm1-B, 500-bp stretches flanking either side of the gene were cloned and inserted on the corresponding sides of the trpA gene, encoding tryptophan synthetase, in plasmid pTA131. The resulting vector was then used to transform H. volcanii strain WR536 cells, with integration of the construct into the chromosome being achieved via homologous recombination. To verify each step of the deletion procedure, PCR was performed using genomic DNA as a template. To follow the presence of dpm1-B (or aglE, see below), a forward primer directed at the dpm1-B 5′-flanking region and a reverse primer directed at a region within the dpm1-B gene sequence were used (Fig. 2A, left panel, lanes 1a, 2a, and 3a). The same forward primer was used together with a reverse primer directed at a region within the trpA gene sequence to confirm the presence or absence of trpA (Fig. 2A, lanes 1b, 2b, and 3b). While the WR536 background strain contains only the dpm1-B sequence (Fig. 2A, lanes 1a and 1b), the trpA gene was integrated into the chromosome of H. volcanii during the so-called “pop-in” stage (Fig. 2A, lanes 2a and 2b). Successfully transformed cells were next streaked onto Casamino plates containing uracil and 5-fluoroorotic acid but no tryptophan, conditions that select for cells in which the dpm1-B gene and plasmid pTA131 are no longer found within the genome. Cell growth could be detected in such conditions, with PCR confirming that the dpm1-B gene had indeed been replaced by the trpA sequence (Fig. 2A, lanes 3a and 3b). The absence of the dpm1-B gene in the deletion strain was further suggested by the failure to obtain a PCR product using primers directed against the coding region of this sequence (Fig. 2A, right panel). Thus, these results point to the loss of the dpm1-B gene in the deletion strain.
FIG. 2.
Deletion of the dpm1-B (aglE) gene does not affect cell viability. (A) Left panel: PCR amplification was performed by using a forward primer directed at the 5′ dpm1-B flanking region and a reverse primer directed at a sequence within the dpm1-B coding region (lanes 1a, 2a, and 3a) or a sequence within the trpA sequence (lanes 1b, 2b, and 3b), together with genomic DNA from cells of the WR536 background strain (bkgnd, lanes 1a and 1b), from plasmid-incorporating cells (pop-in, lanes 2a and 2b) or from cells that had replaced the dpm1-B gene (pop-out, lanes 3a and 3b), as a template. For the left panel, PCR amplification was performed with primers directed against the dpm1-B coding region together with genomic DNA from cells of the WR536 background strain (bkgnd) or the dpm1-B-deleted strain (deletion). (B) RT-PCR was performed with primers directed at dpm1-B (upper panel) or sec11b (lower panel) and genomic DNA from WR536 background cells or cDNA or RNA from cells with dpm1-B deleted as a template.
To address deletion of dpm1-B (or aglE, see below) at the RNA level, a reverse transcriptase PCR (RT-PCR) was performed with primers directed to the dpm1-B coding region (1) and cDNA, derived from the WR536 background or deletion strains, as a template. Although no dpm1-B band could be detected using cDNA from the dpm1-B deletion strain (upper panel, middle lane), cDNA from the same cells could serve as a template for the amplification of the unrelated sec11b gene (11) (lower panel, middle lane), using the appropriate primers, confirming the ability of the RT to generate cDNA (Fig. 2B). Finally, control experiments confirmed the ability to amplify both dpm1-B and sec11b using genomic DNA from the WR536 background strain as a template and appropriate primers (left lanes), whereas no PCR products were attained in similar reactions using RNA isolated from either strain as a template (right lanes). Thus, it can be concluded that dpm1-B is not essential for H. volcanii cell survival.
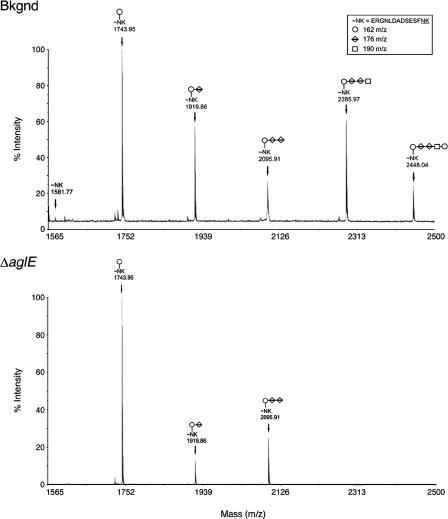
To determine whether Dpm1-B plays a role in H. volcanii N glycosylation, cells with the encoding gene deleted were examined by MS to assess the glycosylation pattern of the S-layer glycoprotein. Recent work (2) has shown the N-linked glycan decorating at least two sequons of the H. volcanii S-layer glycoprotein to be a pentasaccharide comprising two 162-Da residues, two 176-Da residues, and a single 190-Da moiety, with the two 176-Da moieties most likely corresponding to hexuronic acids and the 190-Da entity likely being a dimethylated hexose or a methyl ester of a hexuronic acid. In agreement with these earlier studies, MS analysis of the Asn13-containing, S-layer glycoprotein-derived tryptic peptide isolated from cells of the WR536 background strain revealed the presence of penta-, tetra-, tri-, di-, and monosaccharide-modified peptide peaks (Fig. 3, upper panel). In contrast, examination of the N-linked glycosylation pattern of the same S-layer glycoprotein-derived peptide isolated from cells with dpm1-B (or aglE [see below]) deleted revealed the presence of only peaks corresponding to the mono-, di-, and trisaccharide-modified peptides (Fig. 3, lower panel). Peaks corresponding to tetra- and pentasaccharide-modified peptides were completely absent. As such, it appears that Dpm1-B is involved in the addition of the 190-Da saccharide found at position 4 of the pentasaccharide. Hence, given its demonstrated role in N glycosylation, Dpm1B was renamed AglE (for archaeal glycosylation protein E), in accordance with the nomenclature proposed by Chaban et al. (7).
FIG. 3.
MALDI-TOF analysis of an Asn-13-containing, H. volcanii S-layer glycoprotein-derived glycopeptide. The MALDI-TOF spectra of the Asn-13-containing tryptic peptides derived from the S-layer glycoprotein of the WR536 background (upper panel) and dpm1-B (aglE)-deleted strains (lower panel) are shown. The components of the glycopeptide-associated sugar residues are shown in the inset box, while the glycan moieties decorating the peptide peaks are marked on the MALDI-TOF spectra, accordingly.
Having demonstrated that cells deleted of aglE lack the final 190- and 162-Da sugar subunits of the S-layer glycoprotein N-linked glycan moiety, the consequences of this altered N glycosylation profile were considered. In contrast to what was observed in the case of cells lacking AglB or AglD (1, 2), cells lacking AglE showed no shift in the apparent molecular weight of the S-layer glycoprotein, nor were any differences in S-layer glycoprotein glycostaining apparent. Similarly, neither release of the S-layer glycoprotein to the medium nor proteolytic cleavage of the S-layer of aglE-deleted cells, both indicative of altered S-layer structure, differed from what was observed with cells of the WR536 background strain. Moreover, cells lacking AglE grew at the same rate as did cells of the WR536 background strain in standard laboratory growth conditions, in the presence of high or low concentrations of NaCl (4.5 or 1.75 M NaCl, respectively), at low or high temperatures (30 and 50°C, respectively), or in the presence of low concentrations of magnesium (16 mM MgCl2). In summary, the absence of AglE did not seem to alter H. volcanii cell growth or stability of the S-layer.
Topology of an H. volcanii AglE-based chimera.
Like its other mammalian counterparts, murine dolichol-phosphate mannosyltransferase exists as a complex, comprising the catalytically active Dpm1 subunit together with the regulatory Dpm2 and stabilizing Dpm3 subunits (21). BLAST-based searches of the H. volcanii genome, however, only detected homologues to the mammalian Dpm1 subunit. Thus, given the apparent absence of the membrane association-mediating Dpm3 subunit (4), the subcellular localization of H. volcanii AglE is unclear.
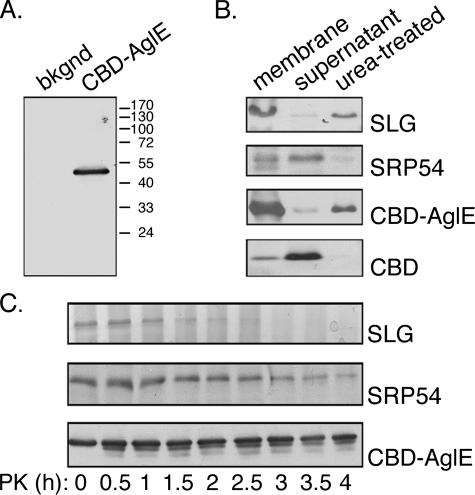
To experimentally consider both the subcellular localization and the topology of AglE, a chimera was constructed between the C. thermocellum cellulosome CBD and AglE. Accordingly, H. volcanii cells were transformed to express DNA encoding the CBD moiety fused to the predicted N terminus of AglE. Expression of the CBD-AglE chimera was confirmed by immunoblot analysis of crude cell extracts using antibodies directed against the CBD moiety. This revealed the presence of a 52-kDa band, corresponding to the combined apparent molecular masses of the 33.8-kDa AglE and the 18.4-kDa CBD moieties (Fig. 4A). No such band could be detected in the untransformed strain WR536 background cells.
FIG. 4.
Expression and topology of the CBD-AglE construct. (A) The protein contents of cells of the WR536 background strain or the same cells expressing CBD-AglE were precipitated, separated by SDS-PAGE, and immunoblotted with antibodies directed against the cellulose-binding domain. The positions of molecular mass markers are shown on the right. (B) H. volcanii cells expressing CBD-AglE were broken by sonication, membrane and supernatant fractions were obtained by ultracentrifugation, and the different fractions were immunoblotted with antibodies to the S-layer glycoprotein (SLG), SRP54 or CBD. Membrane fractions treated with 6 M urea and ultracentrifuged through a 40% sucrose cushion are shown in the right lanes of each panel. The same treatments were performed with H. volcanii cells expressing the soluble CBD moiety alone (bottom panel). (C) H. volcanii cells (1 ml) expressing CBD-AglE were subjected to proteolysis with 1 mg of proteinase K/ml at 55°C. Aliquots (100 μl) were removed every 30 min, TCA (15%) precipitated, separated by SDS-12% PAGE, and either Coomassie-stained (S-layer glycoprotein, upper panel) or immunoblotted with antibodies to SRP54 (middle panel) or CBD (lower panel) at the concentrations given in Materials and Methods. Antibody binding was detected by enhanced chemiluminescence.
To determine the subcellular localization of CBD-AglE, H. volcanii cells expressing the chimera were subjected to cell fractionation, and the resulting soluble and membrane fractions were analyzed by immunoblotting (Fig. 4B). The predominant distribution of the S-layer glycoprotein, a marker of the plasma membrane, and SRP54, a marker of the cytoplasm, to the membrane and soluble fractions, respectively, confirmed the efficacy of the subcellular fractionation. When the same subcellular fractions were probed for the presence of CBD-AglE, the chimera was almost exclusively found in the membrane fraction. In contrast, and as expected, the CBD moiety, when expressed alone in transformed H. volcanii cells, was exclusively localized to the soluble fraction.
To ascertain whether AglE is integrally or peripherally associated with the cell membrane, membrane fractions were treated with 6 M urea to disrupt noncovalent protein associations. The urea-treated membrane fraction was then collected by centrifugation through a 40% (wt/vol) sucrose cushion. After such treatment (Fig. 4B), the S-layer glycoprotein remained associated with the membrane fraction, while that minor pool of SRP54 bound to the membrane (12, 19) was released into the soluble fraction. Like the S-layer glycoprotein, CBD-AglE was clearly detectable in the urea-treated membrane fraction, confirming AglE to be a membrane-spanning protein.
Efforts next addressed whether the N terminus of CBD-AglE is cytoplasmically or outwardly oriented. Accordingly, cells expressing CBD-AglE were subjected to proteolytic digestion with proteinase K at 55°C. Aliquots of the digested cells were removed every half hour over the course of 4 h, TCA precipitated, and separated by SDS-PAGE. While the S-layer glycoprotein was digested from the earliest time point and could be hardly detected after 3 h of digestion, SRP54, an intracellular marker, remained at a constant level throughout the 4-h digestion. Thus, under the conditions used, only proteins exposed at the cell surface were digested (Fig. 4C). Like SRP54, CBD-AglE was detected at a constant level throughout the period of proteolysis, implying the N terminus of the fusion construct to be cytoplasmically oriented. When, however, proteinase K digestion was performed in the presence of 1% (vol/vol) Triton X-100, thereby disrupting cell integrity, no CBD-AglE survived past the 30-min mark, confirming the ability of the protease to digest CBD-AglE, given the opportunity (not shown).
DISCUSSION
The dogma that protein N glycosylation is an exclusively eukaryal feature was first challenged in 1976, when the S-layer glycoprotein of Halobacterium salinarum was shown to contain N-linked glycans (23). Since then, various proteins experiencing this posttranslational modification, as well as lipid-linked oligosaccharide intermediates apparently involved in the pathway, have been described in Archaea (9, 16, 22). Recently, various groups have begun deciphering the archaeal N-glycosylation process in a variety of organisms, such as Methanococcus voltae, Pyrococcus furiosus, and H. volcanii (1, 2, 7, 14). Indeed, our earlier work on H. volcanii showed that the Stt3 homolog, AglB, is responsible for the attachment of a pentasaccharide to select Asn residues of the S-layer glycoprotein and that AglD is involved in addition of the final 162-Da hexose to this oligosaccharide (2). Now, our knowledge of the N-glycosylation pathway in H. volcanii can be expanded with the identification of an additional component, AglE. First identified as Dpm1-B (1), AglE is involved in the addition of the 190-Da sugar, likely to be a dimethylated hexose or a methyl ester of a hexuronic acid, found at position 4 of the pentasaccharide decorating at least two S-layer glycoprotein sequons.
Presently, the precise role assumed by AglE in the N-glycosylation process remains unknown. AglE shows similarity to eukaryal Dpm1, responsible for the addition of GDP-mannose to dolichol pyrophosphate to yield phosphodolichol mannose (15). The glycosyltransferase 2 domain (PF00535) of AglE is also found in enzymes responsible for transferring nucleotide-activated sugars to carriers (such as phosphodolichol) but not in glycosyltransferases that add sugar substrates to the growing lipid-linked oligosaccharide involved in N glycosylation. Therefore, AglE may be responsible for loading the 190-Da sugar subunit of the H. volcanii S-layer glycoprotein-modifying pentasaccharide onto a lipid carrier, from where it is then presumably transferred onto the 176-Da sugar species of the three-membered pentasaccharide precursor via the actions of a distinct transferase. On the other hand, it cannot yet be ruled out that AglE directly adds the 190-Da sugar subunit to the growing oligosaccharide. Indeed, genome searches have failed to identify any H. volcanii homologues of the eukaryal glycosyltransferases responsible for the transfer of phosphodolichol-linked sugar residues to the growing polysaccharide destined to decorate select Asn residues of the target protein, such as Alg3, Alg8, Alg9, Alg10, or Alg12 (6). The future development of in vitro assays will help in determining the true role of AglE in H. volcanii N glycosylation.
Unlike what was observed with cells lacking AglB or AglD (2), cells in which AglE was absent did not show any difference in cell growth in any of the tested conditions, in S-layer glycoprotein release, or in proteolytic susceptibility of the S-layer, relative to cells of the WR536 background strain. These earlier studies showed, however, that the extent of the perturbation of the S-layer glycoprotein glycan moiety, as realized upon deletion of either glycan-processing gene, did not necessarily correlate with an effect on the S-layer glycoprotein stability or H. volcanii cell behavior. In other words, the most pronounced shift in apparent molecular mass of the S-layer glycoprotein protein, as well as the biggest differences in S-layer architecture and protease susceptibility, could be observed with the aglD deletion strain, where only the final 162-Da sugar residue of the N-linked pentasaccharide is missing. In contrast, H. volcanii cells missing AglB, and thus lacking the entire N-linked pentasaccharide, showed less-pronounced differences relative to the WR536 background strain. The absence of the last two sugar subunits of the N-linked glycan entity decorating the S-layer glycoprotein in the aglE deletion strain, namely, the outermost 162-Da residue and the 190-Da residue found at position 4, yields cells phenotypically identical to those of the WR536 background strain. In contrast, deleting the final 162-Da saccharide alone drastically changes S-layer structure and behavior. As such, the relative importance of these sugar residues to the N-linked pentasaccharide is implied. One can thus speculate that the 162-Da sugar subunit plays a critical role in stabilizing the S-layer glycan structure, since its absence results in a less stable form of the S layer, possibly due to the exposure of a charged group on the oligosaccharide backbone. This, in turn, may explain why the unmasking of the 190-Da sugar residue but not its removal has such an impact on the cell. Efforts to identify the 162-, 190-. and 176-Da sugar species comprising the pentasaccharide decorating the S-layer glycoprotein are ongoing.
Finally, through the use of a chimeric CBD-AglE reporter, the present study predicts AglE to be a membrane-spanning protein with a cytoplasmically oriented N terminus. Although it can be argued that the 18-kDa CBD moiety fused to the N terminus of AglE inadvertently influenced the topology of CBD-AglE, bioinformatics analysis of the amino acid sequence of AglE with Signal-P (http://www.cbs.dtu.dk/services/SignalP), which is software designed to detect the existence of signal peptides, failed to predict the presence of an N-terminal signal sequence in AglE that would have been masked by the N-terminally fused CBD moiety, thereby rendering topological perturbation of AglE by the fused CBD moiety unlikely. Given that AglE is predicted to be a membrane-spanning protein with a cytoplasmically oriented N terminus, the PF00535 glycosyltransferase 2 domain (residues 9 to 173), likely comprising the active site of AglE, would face the cytoplasm. In contrast, the active site of the archaeal oligosaccharide transferase was described as being outwardly oriented, since externally added sequon-containing non-membrane-permeating peptides were glycosylated by H. salinarum cells (18). Therefore, it appears that as in Eukarya and Bacteria, the N-glycosylation process in H. volcanii, and likely other Archaea, can also be divided into two parts. The assembly of a lipid-linked oligosaccharide (16) likely transpires on the cytoplasmic side of the membrane, while the transfer of the glycan moiety to its target seemingly occurs on the external side of the membrane. It remains, however, to be determined whether transfer of the completed glycan to the target protein takes place after reorientation of the oligosaccharide to the external surface of the plasma membrane, as in the bacterium Campylobacter jejuni (20), or whether, as in higher eukaryotes (6), additional sugar residues are added to the lipid-linked glycan after its “flipping” across the membrane, prior to its transfer to target Asn residues.
Acknowledgments
J.E. is supported by the U.S. Air Force Office for Scientific Research (grant FA9550-07-10057) and the Israel Science Foundation (grant 30/07). A.D. and H.R.M. are supported by the Biotechnology and Biological Sciences Research Council and Wellcome Trust. A.D. is a Biotechnology and Biological Sciences Research Council Professorial Fellow.
Footnotes
Published ahead of print on 29 February 2008.
REFERENCES
- 1.Abu-Qarn, M., and J. Eichler. 2006. Protein N glycosylation in Archaea: defining Haloferax volcanii genes involved in S-layer glycoprotein glycosylation. Mol. Microbiol. 61511-525. [DOI] [PubMed] [Google Scholar]
- 2.Abu-Qarn, M., S. Yurist, A. Giordano, A. Trauner, H. R. Morris, P. Hitchen, O. Medalia, A. Dell, and J. Eichler. 2007. Haloferax volcanii AglB and AglD are involved in N glycosylation of the S-layer glycoprotein and proper assembly of the surface layer. J. Mol. Biol. 3741224-1236. [DOI] [PubMed] [Google Scholar]
- 3.Allers, T., H. P. Ngo, M. Mevarech, and R. G. Lloyd. 2004. Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl. Environ. Microbiol. 70943-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashida, H., Y. Maeda, and T. Kinoshita. 2006. DPM1, the catalytic subunit of dolichol-phosphate mannose synthase, is tethered to and stabilized on the endoplasmic reticulum membrane by DPM3. J. Biol. Chem. 281896-904. [DOI] [PubMed] [Google Scholar]
- 5.Baliga, N. S., R. Bonneau, M. T. Tacciotti, M. Pan, G. Glusman, E. W. Deutsch, P. Shannon, Y. Chui, R. S. Weng, R. R. Gan, P. Hung, S. V. Date, E. Marcotte, L. Hood, and W. V. Ng. 2004. Genome sequence of Haloarcula marismortui: a halophilic archaeon from the Dead Sea. Genome Res. 142221-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burda, P., and M. Aebi. 1999. The dolichol pathway of N-linked glycosylation. Biochim. Biophys. Acta 1426239-257. [DOI] [PubMed] [Google Scholar]
- 7.Chaban, B., S. Voisin, J. Kelly, S. M. Logan, and K. F. Jarrell. 2006. Identification of genes involved in the biosynthesis and attachment of Methanococcus voltae N-linked glycans: insight into N-linked glycosylation pathways in Archaea. Mol. Microbiol. 61259-268. [DOI] [PubMed] [Google Scholar]
- 8.Coutinho, P. M., E. Deleury, G. J. Davies, and B. Henrissat. 2003. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 328307-317. [DOI] [PubMed] [Google Scholar]
- 9.Eichler, J., and M. W. W. Adams. 2005. Post-translational protein modification in Archaea. Microbiol. Mol. Biol. Rev. 69393-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eichler, J. 2000. Novel glycoproteins of the halophilic archaeon Haloferax volcanii. Arch. Microbiol. 173445-448. [DOI] [PubMed] [Google Scholar]
- 11.Fine, A., V. Irihimovitch, I. Dahan, Z. Konrad, and J. Eichler. 2006. Cloning, expression, and purification of functional Sec11a and Sec11b, type I signal peptidases of the archaeon Haloferax volcanii. J. Bacteriol. 1881911-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haddad, A., R. W. Rose, and M. Pohlschroder. 2005. The Haloferax volcanii FtsY homolog is critical for haloarchaeal growth but does not require the A domain. J. Bacteriol. 1874015-4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irihimovitch, V., and J. Eichler. 2003. Post-translational secretion of fusion proteins in the halophilic archaea Haloferax volcanii. J. Biol. Chem. 27812881-12887. [DOI] [PubMed] [Google Scholar]
- 14.Kohda, D., M. Yamada, M. Igura, J. Kamishikiryo, and K. Maenaka. 2007. New oligosaccharyltransferase assay method. Glycobiology 171175-1182. [DOI] [PubMed] [Google Scholar]
- 15.Kornfeld, R., and S. Kornfeld. 1985. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 54631-664. [DOI] [PubMed] [Google Scholar]
- 16.Kuntz, C., J. Sonnenbichler, I. Sonnenbichler, M. Sumper, and R. Zeitler. 1997. Isolation and characterization of dolichol-linked oligosaccharides from Haloferax volcanii. Glycobiology 7897-904. [DOI] [PubMed] [Google Scholar]
- 17.Lechner, J., and M. Sumper. 1987. The primary structure of a procaryotic glycoprotein. Cloning and sequencing of the cell surface glycoprotein gene of halobacteria. J. Biol. Chem. 2629724-9729. [PubMed] [Google Scholar]
- 18.Lechner, J., F. Wieland, and M. Sumper. 1985. Biosynthesis of sulfated saccharides N-glycosidically linked to the protein via glucose. Purification and identification of sulfated dolichyl monophosphoryl tetrasaccharides from halobacteria. J. Biol. Chem. 260860-866. [PubMed] [Google Scholar]
- 19.Lichi, T., G. Ring, and J. Eichler. 2004. Membrane binding of SRP pathway components in the halophilic archaea Haloferax volcanii. Eur. J. Biochem. 2711382-1390. [DOI] [PubMed] [Google Scholar]
- 20.Linton, D., N. Dorell, P. G. Hitchen, S. Amber, A. V. Karlyshev, H. R. Morris, A. Dell, M. A. Valvano, M. Aebi, and B. W. Wren. 2005. Functional analysis of the Campylobacter jejuni N-linked protein glycosylation pathway. Mol. Microbiol. 551695-1703. [DOI] [PubMed] [Google Scholar]
- 21.Maeda, Y., S. Tanaka, J. Hino, K. Kangawa, and T. Kinoshita. 2000. Human dolichol-phosphate-mannose synthase consists of three subunits, DPM1, DPM2 and DPM3. EMBO J. 192475-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mescher, M. F., U. Hansen, and J. L. Strominger. 1976. Formation of lipid-linked sugar compounds in Halobacterium salinarium, presumed intermediates in glycoprotein synthesis. J. Biol. Chem. 2517289-7294. [PubMed] [Google Scholar]
- 23.Mescher, M. F., and J. L. Strominger. 1976. Purification and characterization of a prokaryotic glycoprotein from the cell envelope of Halobacterium salinarium. J. Biol. Chem. 2512005-2014. [PubMed] [Google Scholar]
- 24.Mevarech, M., and R. Werczberger. 1985. Genetic transfer in Halobacterium volcanii. J. Bacteriol. 162461-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tozik, I., Q. Huang, C. Zwieb, and J. Eichler. 2002. Reconstitution of the signal recognition particle of the halophilic archaeon Haloferax volcanii. Nucleic Acids Res. 304166-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voisin, S., R. S. Houliston, J. Kelly, J. R. Brisson, D. Watson, S. L. Bardy, K. F. Jarrell, and S. M. Logan. 2005. Identification and characterization of the unique N-linked glycan common to the flagellins and S-layer glycoprotein of Methanococcus voltae. J. Biol. Chem. 28016586-16593. [DOI] [PubMed] [Google Scholar]
- 27.Zhu, B. C., R. R. Drake, H. Schweingruber, and R. A. Laine. 1995. Inhibition of glycosylation by amphomycin and sugar nucleotide analogs PP36 and PP55 indicates that Haloferax volcanii beta-glucosylates both glycoproteins and glycolipids through lipid-linked sugar intermediates: evidence for three novel glycoproteins and a novel sulfated dihexosyl-archaeol glycolipid. Arch. Biochem. Biophys. 319355-364. [DOI] [PubMed] [Google Scholar]
- 28.Zhu, B. C., and R. A. Laine. 1996. Dolichyl-phosphomannose synthase from the archae Thermoplasma acidophilum. Glycobiology 6811-816. [DOI] [PubMed] [Google Scholar]