Abstract
The ferric uptake regulatory protein, Fur, functions as a global regulatory protein of gene transcription in the mucosal pathogen Neisseria gonorrhoeae. We have shown previously that several N. gonorrhoeae Fur-repressed genes are expressed in vivo during mucosal gonococcal infection in men, which suggests that this organism infects in an iron-limited environment and that Fur is expressed under these conditions. In this study we have demonstrated expression of the gonococcal fur gene in vitro, in human cervical epithelial cells, and in specimens from female subjects with uncomplicated gonococcal infection. In vitro studies confirmed that the expression of the gonococcal fur gene was repressed during growth under iron-replete growth conditions but that a basal level of the protein was maintained. Using GFP transcriptional fusions constructed from specific Fur binding sequences within the fur promoter/operator region, we determined that this operator region was functional during N. gonorrhoeae infection of cervical epithelial cells. Furthermore, reverse transcription-PCR analysis, as well as microarray analysis, using a custom Neisseria Fur and iron regulon microarray revealed that several Fur- and iron-regulated genes were expressed during N. gonorrhoeae infection of cervical epithelial cells. Microarray analysis of specimens obtained from female subjects with uncomplicated gonococcal infection corroborated our in vitro findings and point toward a key role of gonococcal Fur- and iron-regulated genes in gonococcal disease.
Iron homeostasis is tightly regulated in almost all bacteria because of the toxicity that results from the formation of iron-catalyzed reactive oxygen species even though iron is essential for a number of physiological functions (53). The iron-responsive transcriptional regulator Fur (ferric uptake regulator) controls the transcription of many iron-regulated genes in several microorganisms (10, 11, 23, 24, 47, 48). Homologs of Fur are widespread in gram-negative bacteria and are also found in some gram-positive organisms (6, 9, 37). The role of the Fur protein as a repressor has been well documented where Fur forms a dimer with ferrous iron and binds to a consensus sequence (Fur-box) that overlaps the promoters of iron-regulated genes, resulting in the inhibition of transcription. The regulation of gene transcription by Fur and iron has been defined by the construction of Fur mutants in a variety of organisms, including Bacillus subtilis (2), Campylobacter jejuni (29), Escherichia coli (38), Vibrio cholerae (40), Neisseria meningitidis (10, 47), and Helicobacter pylori (17). Fur also functions to control the expression of genes required for pathogenesis (33, 37, 44, 52) in addition to controlling the transcription of iron transport genes. The Fur protein plays a role in the acid shock response (26), detoxification of oxygen radicals (14, 28), production of toxins and other virulence factors (37, 51), quorum sensing (8), and the regulation of metabolic pathways (42, 48) in a number of organisms. Furthermore, recent studies indicate that Fur can function as an activator of gene transcription through both direct and indirect mechanisms (7, 12-14, 26, 27, 30). We recently determined that pathogenic Neisseria spp. possess small RNA molecules and have identified one small RNA (NrrF) that is repressed by Fur and functions as a posttranscriptional repressor (39).
Using DNA microarray analysis, we have previously demonstrated that nearly 10% of the entire genome of N. meningitidis strain MC58 is regulated during growth under iron-replete or iron-depleted conditions. Approximately 50% of these iron-regulated genes, including iron-repressed and iron-activated genes, had the potential to be regulated by Fur. This was determined by identifying predicted Fur binding sites in the promoter regions of these iron-regulated genes and was confirmed in vitro by mobility shift assays for a subset of genes (24). In addition, Delany et al. (10) recently reported the derepression of a number of genes regulated by iron in a Fur deletion mutant of N. meningitidis. We have determined by in silico analysis that the closely related pathogen, N. gonorrhoeae, possesses a large number of diverse genes with putative Fur binding sequences within the promoter/operator regions. Binding of Fur with varying degrees of affinity to the promoter/operator region of gonococcal fur, tbpB, lbpB, fbpA, hmbR, fetA, tonB, opa, sodB, hemO, recN, and fumC genes has also been confirmed by mobility shift assays (46).
In addition to these in vitro studies, we have recently determined that several N. gonorrhoeae Fur-repressed genes are expressed in vivo during mucosal gonococcal infection in men (1), suggesting that this organism may infect in an iron-limited environment. However, gonococcal disease is distinctly different in women than men. Men who present with gonococcal urethritis often experience inflammatory symptoms accompanied by a purulent discharge marked by the infiltration of polymorphonuclear leukocytes. In contrast, women infected with gonococci may not report specific symptoms that suggest an inflammatory condition. N. gonorrhoeae infects the lower female genital tract and may invade ectocervical and endocervical epithelium without producing a significant inflammatory response but ascending, nevertheless, to the upper tract, where it may cause serious sequelae, such as endometritis and salpingitis (16).
To begin to define the expression profile of N. gonorrhoeae Fur- and iron-regulated genes during N. gonorrhoeae infection in women, we first confirmed the expression of the gonococcal Fur protein in vitro in the present study. We then examined the expression of gonococcal Fur- and iron-regulated genes during N. gonorrhoeae infection of immortalized epithelial cell lines, developed from normal human vagina, endocervix, and ectocervix. Using green fluorescent protein (GFP) transcriptional fusions constructed from specific Fur binding sequences within the fur promoter/operator region, we determined that this operator region was functional during N. gonorrhoeae infection of cervical epithelial cells. Furthermore, we also demonstrated that N. gonorrhoeae Fur- and iron-regulated genes are expressed during N. gonorrhoeae infection of human cervical epithelial cell lines and in specimens obtained from women with uncomplicated gonorrhea.
MATERIALS AND METHODS
Growth experiments in iron-replete and iron-depleted conditions.
Wild-type N. gonorrhoeae strain F62 GFP transcriptional fusion strains (F62-tonBGFP, F62-furGFP and F62-rmpGFP [described below]) were grown in chemically defined medium (CDM) aerobically for 3 h at 37°C (41) supplemented with 4.2% NaHCO3 plus 25 μM Desferal (Ciba-Geigy). Cultures were then centrifuged (5,000 × g, 4°C, 10 min), the resulting pellets were washed once with CDM, and the suspensions were recentrifuged. These bacteria served as the inocula for fresh CDM-12.5 μM Desferal cultures or CDM-100 μM ferric nitrate cultures using a starting absorbance of 660 nm (A660) of ∼0.06. Bacterial growth was monitored by measuring the absorbance every hour for 5 h; samples were removed and washed prior to reverse transcription-PCR (RT-PCR) and fluorescence-activated cell sorting (FACS) analysis (see below). E. coli cultures used for cloning and transformation were grown in Luria-Bertani (LB) agar and broth. Media were supplemented with chloramphenicol (2 μg/ml for Neisseria and 25 μg/ml for E. coli) when required.
RT-PCR analysis.
Total RNA was isolated from broth-grown N. gonorrhoeae F62 by using the Qiagen RNeasy kit (Valencia, CA). Samples were treated with DNase I enzyme prior to an RT-PCR, performed according to the manufacturer's instructions (Promega, Madison, WI). To 150 ng of total RNA was added 25 μl of 2× reaction mix, 100 ng of each primer (Table 1), 1 μl of RT-Taq mix, and diethyl pyrocarbonate-treated water to a final volume of 50 μl. cDNA synthesis was performed at 50°C for 30 min, which was followed by denaturation at 94°C for 2 min. PCR amplifications were carried out with the following parameters for each cycle: denaturation at 94°C for 45 s, annealing at 56°C for 30 s, and elongation at 72°C for 30 s. Five, fifteen, twenty-five, and thirty-five cycles were used to determine whether endpoint amplification reactions were substrate limiting as cycle numbers increased in the PCR program. To ensure the absence of DNA contamination, PCRs were performed without the addition of reverse transcriptase.
TABLE 1.
Primers used in this study
| Primer pair sequence (5′-3′)a | Amplified gonococcal sequence |
|---|---|
| F, CGGGATCCACAGGCGAGGATAGGGTTTGT | 121-bp fur promoter fragment |
| R, CCAAGCTTGACAGGGTAAAATACCGCTTA | |
| F, CGGGATCCGAATTCCTATCCGATTTGCCG | 100-bp rmp promoter fragment |
| R, CCAAGCTTTTATTCCCTCATTAGATTTGTA | |
| F, CGGGATCCCACGGTTTGCCTATGAATGG | 91-bp tonB promoter fragment |
| R, CCGGATCCGGGCATAATAATAGCAACAATTCC | |
| F, GAAACCATTTCCCTGTCTGC | 347-bp rmp internal fragment |
| R, GTTACGATGCTGCGGATTTT | |
| F, GAAATGTTATCGAATTTGTCG | 498-bp tonB internal fragment |
| R, GTAAAGGATTGCCTTTGCT | |
| F, CGCGTTTGAAGATTTTGGAT | 348-bp fur internal fragment |
| R, TGCACACGCCGTACATATAAA | |
| F, GGATAAATTGCCCGAAGGTT | 347-bp tbpA internal fragment |
| R, CACGGAGGGTGAAGTGTTTT | |
| F, TATCCGATACGCACTGCTTG | 349-bp fbpA internal fragment |
| R, CAGTGCCACCCAGTCTTTTT | |
| F, GCGGTAGTCGATGTAATAGG | 407-bp sodB internal fragment |
| R, CAAACCTACATCACCAACCT |
F, forward primer; R, reverse primer.
Quantification of N. gonorrhoeae Fur.
Samples of broth grown N. gonorrhoeae (iron-replete and iron-depleted) were collected at different time intervals (2, 3, 4, and 5 h), resuspended in 200 μl of RLT buffer, followed by incubation at 37°C for 1 h. Two micrograms of total protein from each sample was examined by sodium dodecyl sulfate (SDS)-12% polyacrylamide gel electrophoresis (PAGE) (34). Proteins were transferred onto nitrocellulose membrane (Bio-Rad, Hercules, CA) in 30 mM 3-(cyclohexylamino)-1-propanesulfonic acid (CAPS) buffer (pH 11.0; Sigma-Aldrich, St. Louis, MO) at constant current of 0.2 A for 1 h. Western blots were performed as described previously (46) with slight modifications. Membranes were blocked overnight at room temperature in Tris-buffered saline containing 0.05% Tween 20 (TTBS) with 5% albumin, followed by incubation for 2 h with a 1:2,000 dilution of anti-Neisseria Fur serum (46) in TTBS containing 0.5% albumin. After a 2 h of incubation with 1:10,000 peroxidase-conjugated anti-rabbit immunoglobulin (Sigma-Aldrich) in TTBS containing 0.5% albumin, the membranes were washed with TTBS. Chemiluminescence was detected by using an ECL detection system (Amersham Pharmacia, Piscataway, NJ). Autoradiography films were exposed and developed. Developed films were scanned, and the intensity of Fur protein bands was quantified by using Adobe Photoshop quantification software. The concentration of gonococcal Fur was determined by linear regression analysis using a standard curve of different concentrations of gonococcal Fur protein, 0.2 to 6.4 μg, purified as described previously (46) and electrophoresed simultaneously.
DNA manipulations.
Restriction endonucleases, T4 DNA ligase, alkaline phosphatase, DNA polymerase (Klenow fragment), and DNA-modifying enzymes were purchased from Invitrogen (Carlsbad, CA) and Promega (Madison, WI) and used according to the manufacturers' instructions. Restriction endonuclease fragments were separated by electrophoresis on 1% agarose gels in Tris-acetate-EDTA (TAE) buffer. Plasmid DNA was isolated by using a Qiagen miniprep kit according to the manufacturer's instructions.
DNase I footprinting.
A DNase I protection (foot printing) technique was used to characterize the operator sequence within the fur promoter region bound by gonococcal Fur (46). Mn2+ was used as a cofactor, and the DNase I foot printing experiments were carried out as previously described (18, 19) with slight modifications. The fur and tonB promoter/operator DNA fragments were PCR amplified from N. gonorrhoeae strain F62 by colony PCR with specific primers (Table 1) and cloned into an appropriate restriction site of the plasmid pBCSK+ (Stratagene, La Jolla, CA) to generate the recombinant plasmids pBCSKfur and pBCSKtonB. DNase I footprinting analysis of the rmp promoter/operator region was not performed because in previous studies we did not observe binding of the Fur protein to the rmp promoter/operator region (46). DNA probes were labeled by 5′-end labeling M13 forward and M13 reverse primers with [γ-32P]ATP (3,000 Ci/mmol; Perkin-Elmer, Boston, MA) using 1 μl of T4 polynucleotide kinase enzyme (New England Biolabs, Inc., Beverly, MA) at 37°C for 40 min. Regions that encompassed the promoter region and the putative Fur box in the fur gene and in the tonB gene were amplified by PCR from the recombinant pBCSK plasmid, using the M13 forward (5′ labeled) and reverse primers. Approximately 40,000 cpm of labeled probe was used in each reaction for DNA footprinting experiments. Reactions were performed in 100 μl of footprinting buffer (20 mM Tris-HCl [pH 8], 40 mM KCl, 5 mM MgCl2, 6 mM CaCl2, 0.125 mM MnCl2, 1 mM dithiothreitol, 10% glycerol), containing 1 μg of sonicated salmon sperm DNA and 5 μg of acetylated bovine serum albumin for 30 min at room temperature. After the addition of DNase I enzyme (0.1 ng), digestion was carried out for 2 min at 37°C. The reaction was stopped by the addition of 100 μl of stop buffer (0.1 M EDTA [pH 8.0], 0.6 M sodium acetate, and 20 μg of sonicated salmon sperm DNA/ml). After denaturation at 95°C for 2 min, samples were electrophoresed on a 6% urea-polyacrylamide gel at 2,000 V and then dried and autoradiographed.
DNA sequencing.
Putative Fur binding sites were sequenced by manual sequencing via the dideoxy chain termination method using Sequenase version 2.0 (U.S. Biochemicals) and M13 forward and reverse primers according to the manufacturer's instructions.
Construction of GFP fusions in N. gonorrhoeae.
Transcriptional fusions in N. gonorrhoeae strain F62 were made by using GFP from Aequorea victoria. To construct the F62-furGFP fusion, we PCR amplified the promoter/operator region spanning the Fur box, the predicted −10 and −35 regions of the promoter region of the fur gene, and cloned this region upstream of the promoterless gfp gene in pLES99 (gift from Virginia Clark, University of Rochester, School of Medicine and Dentistry). These plasmid constructs with the cloned inserts were transformed initially into E. coli strain XL1-Blue. Sequencing and PCR analysis confirmed the transformation and orientation of the inserts (data not shown). They were then introduced into N. gonorrhoeae F62 by transformation (36) to yield the final constructs. Similarly, a portion of the partial promoter/operator region of the tonB gene, a positive control for an iron-regulated gene, and the rmp gene, the latter a negative control gene (not regulated in response to iron), were also amplified to construct the GFP fusion strains (N. gonorrhoeae strains F62-tonBGFP and F62-rmpGFP, respectively). Both the fur and the tonB promoter regions that had been used to construct the N. gonorrhoeae GFP fusion strains represented the protected region determined by DNA footprinting analysis (see Fig. 2). Sequencing, PCR, and fluorescence microscopy confirmed the correct insertion of the fusions into the gonococcal chromosome (data not shown).
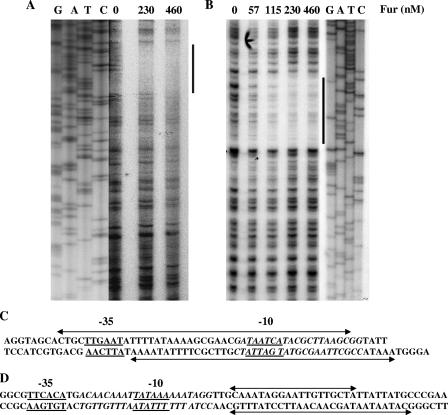
FIG. 2.
DNase I footprinting analysis of N. gonorrhoeae Fur protein with gonococcal fur and tonB promoter/operator regions. (A) DNase I footprint analysis of the fur promoter operator region. Radiolabeled DNA was incubated with increasing concentrations of Fur (0, 230, and 460 nM) prior to digestion with DNase I (indicated in the top panel). DNA standards (GATC) are shown on the left side of the panel. Fur-protected nucleotide bases are indicated by a thick line. (B) DNase I footprint analysis of the tonB promoter operator region. Radiolabeled DNA was incubated with increasing concentrations of Fur (0, 57, 115, 230, and 460 nM) prior to digestion with DNase I enzyme. The Fur-protected tonB promoter/operator region is indicated by a thick line. The Fur concentration and probes used are indicated at the top of each panel, and the DNA standards (GATC) are shown on the right side of the panel. (C) Schematic representation of a portion of the predicted fur promoter/operator region. The Fur-protected region is marked by a double-arrow line on both template and nontemplate strands. The predicted −10 and −35 promoter elements are underlined; the predicted Fur box is italicized. (D) Schematic representation of part of the predicted tonB promoter/operator region. The Fur-protected region is marked by a line with a double arrow on both template and nontemplate strands. The predicted −10 and −35 promoter elements are underlined; the predicted Fur box is italicized.
Epithelial cell cultures and adherence and invasion assays.
Vaginal (Vk2/E6E7), endocervical (End1/E6E7), and ectocervical (Ect1/E6E7) cell lines (21) were maintained in keratinocyte serum-free medium (Gibco-BRL/Life Technologies, Gaithersburg, MD) supplemented with 50 μg of bovine pituitary extract per ml, 0.1 ng of epidermal growth factor per ml, and CaCl2 to a final concentration of 0.4 mM. When necessary, penicillin and streptomycin were used at concentrations of 100 U/ml and 100 μg/ml, respectively. Cell lines (third passage) were seeded onto six-well tissue culture plates at a concentration of 2.5 × 105 epithelial cells/well and grown to confluence (106 cells/well) in antibiotic-free keratinocyte serum-free medium. Overnight cultures of N. gonorrhoeae F62, F62-tonBGFP, F62-furGFP, and F62-rmpGFP were diluted to ca. 5 × 107 CFU/ml, and 2 ml of the suspensions was added to epithelial monolayers to give a multiplicity of infection (MOI) of 100:1 (100 gonococci per epithelial cell). Cocultures were maintained at 37°C in a 5% CO2 incubator. Total bacteria associated with epithelial cells and intracellular bacteria were sampled at 1, 2, 4, and 8 h postinfection, and viable bacteria were measured as CFU by spread plating various dilutions of each sample on GCB agar plates, followed by incubation overnight at 37°C in 5% CO2 as reported previously (20). To quantify the number of intracellular bacteria at each time point (1, 2, 4, and 8 h), gonococcus-infected epithelial cells were treated with gentamicin (20 μg/ml) for an additional 1 h and then washed three times with Dulbecco phosphate-buffered saline (D-PBS) (without Ca2+ and Mg2+) (Gibco-BRL/Life Technologies), and viable intracellular bacteria were measured as described above. To quantify total bacteria associated with N. gonorrhoeae-infected epithelial cells, samples were handled similarly but without the addition of gentamicin. For FACS analysis, samples were collected as described above, and 1% paraformaldehyde was used for fixation. For RT-PCR and microarray analysis (see below), epithelial cells were infected with N. gonorrhoeae strain F62, samples that represented either total cell-associated bacteria or intracellular bacteria were collected at the time points discussed above from multiple plates, and total RNA was isolated by using a Qiagen RNeasy kit. Isolated total RNA was treated with DNase I (Invitrogen) according to the manufacturer's instructions, and the quality was assessed by agarose gel electrophoresis. In addition, PCR lacking reverse transcriptase was performed with each sample using the internal rmp gene primers to ensure that no DNA contamination was present. A 150-ng sample of isolated RNA was used for RT-PCR analysis, as described above, with the primers listed in Table 1 to amplify internal fragment of genes (rmp, fur, tonB, tbpA, fbpA, and sodB) for 25 cycles. The amplicon densities increased as greater numbers of cycles were used, ensuring that PCR amplification was not limited by substrate when a 5-, 15-, 25-, or 35-cycle PCR program was used (data not shown).
FACS analysis.
Samples representing total and intracellular bacteria obtained from human epithelial cells infected with the three N. gonorrhoeae transcriptional fusion strains were centrifuged, washed twice with D-PBS, and fixed in filtered 1% paraformaldehyde for FACS analysis. GFP fluorescence was measured by gating on the epithelial cells, and 70,000 to 80,000 events were read by FACScan (Becton Dickinson, Franklin Lakes, NJ) using CellQuest software (version 3.3). The results were expressed as the geometric mean of GFP fluorescence of the N. gonorrhoeae F62-furGFP strain or the N. gonorrhoeae F62-tonBGFP strain divided by the geometric mean of the N. gonorrhoeae F62-rmpGFP strain.
Custom microarray protocols. (i) Design of Fur and iron regulon microarray.
A custom DNA microarray, composed of 50-mers, was designed representing 124 gonococcal genes. These included iron- and Fur-regulated genes and Neisseria genes that are known or are hypothesized to play a role in the pathogenesis and virulence of N. gonorrhoeae. A number of the genes included in this microarray had also been identified by us in a previous study that defined the complete N. meningitidis iron regulon by whole-genome microarray analysis (24). Oligonucleotides that corresponded to a subset of these genes (homologous to N. gonorrhoeae genomic sequences) were included on the microarray. Of the 124 genes represented on the microarray used in the current study, 43 were known to have a function, 35 were predicted to have a function, and the remaining 46 were hypothetical genes. We also included in this microarray oligonucleotides that corresponded to five constitutively expressed human genes as an appropriate control for potential contamination of human RNA.
(ii) Oligonucleotide synthesis and microarray slide preparation.
Oligonucleotides were synthesized on an ABI 3900 DNA synthesizer (Applied Biosystems, Foster City, CA) on a 200-nmol scale using standard phorphoramidite chemistry and 5′-DMT-mdC (TEG-FMOC) columns (Biosearch Technologies, Novato, CA). After synthesis, oligonucleotides were cleaved from the support and deprotected by using concentrated NH4OH at 65°C for 5 h. Oligonucleotides were purified on Varian TOP columns (Varian Instruments, Walnut Creek, CA) according to the manufacturer's protocol and then quantitated by using an Oligreen ssDNA quantitation kit (Molecular Probes, Eugene, OR). Oligonucleotides (50 μM in 150 mM sodium phosphate [pH 8.5]) were spotted in duplicate onto Codelink slides (Amersham Biosciences; Piscataway, NJ) at 25°C (55% relative humidity) from 384-well chilled spotting plates (15°C) using a QArraymini spotter (Genetix, Boston, MA) using Stealth SMP4 pins (Telechem, Sunnyvale, CA). After spotting, the microarrays were incubated for 24 to 48 h in a sealed container at ∼75% relative humidity. The microarray slides were then washed with 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS for 30 min at 50°C, rinsed twice with water, and dried by centrifugation at 900 × g for 10 min.
Labeling, hybridization, washing, and scanning of cDNA.
Total RNA samples isolated from uninfected or N. gonorrhoeae infected epithelial cells were labeled using the microbial RNA aminoallyl labeling for microarrays protocol from The Institute for Genomic Research (http://pfgrc.tigr.org/protocols/protocols.shtml). After RT, aminoallyl-modified cDNA was labeled with Cy3- or Cy5-NHS esters (Amersham Biosciences, Piscataway, NJ). Cy3- and Cy5-labeled cDNAs were combined, dried, and redissolved in 15 μl of hybridization cocktail (5× SSC, 0.1% SDS, 0.1 mg of salmon sperm DNA/ml). These hybridization mixtures were incubated at 95°C for 5 min and at 45°C for 5 min, cooled to RT, pipetted onto microarray slides, and covered with coverslips (9 by 22 mm). Microarrays were hybridized for 16 to 20 h at 42°C in a humidified chamber. After hybridization, the arrays were washed first in 2× SSC-0.1% SDS at 55°C for 10 min, then 0.1× SSC-0.1% SDS at room temperature for 10 min, then twice with 1× SSC at room temperature for 5 min, and finally with water at room temperature for 10 min. The microarrays were centrifuged at 900 × g for 10 min and scanned with a ScanArray Express apparatus (Perkin-Elmer).
Analysis of microarray data.
Data obtained at each time point for in vitro studies were represented as the average of three independent experiments. Net signal intensity was calculated by subtracting the local mean background intensity of each spot from the mean signal intensity of each spot. These net values from three independent experiments were normalized to eliminate labeling artifacts. Spots were not included in the study if the spot intensity was not at least one standard deviation (SD) above the background intensity. The data were expressed as the log2 of the normalized value of a spot divided by the normalized value of the spot representing the rmp gene, and the resulting ratio was termed the expression ratio of the genes. The rmp gene was used as a basis to compare the expression of N. gonorrhoeae Fur and iron regulated genes because: (i) in vitro broth-grown N. gonorrhoeae showed equivalent rmp gene expression under both iron-depleted and iron-replete conditions (1); (ii) endocervical cell-associated N. gonorrhoeae displayed equivalent levels of rmp transcripts throughout an 8-h growth period (see Results); and (iii) the antibody levels to TbpA in men who were infected for the first time with N. gonorrhoeae were correlated with tbpA gene expression when rmp gene expression was normalized, suggesting that rmp gene expression was constitutive (1). The rmp gene has also been used as a constitutive marker for N. gonorrhoeae during infection of human peripheral blood mononuclear cells (45). Furthermore, the Rmp protein is a conserved integral gonococcal outer membrane protein that possesses conserved antigenic properties and is expressed in clinical specimens (31, 32, 43).
Microarray protocols for clinical samples.
Specimens were collected from women with uncomplicated gonorrhea treated at the public health clinics at the Boston University Medical Center, Boston, MA, and the Baltimore City Health Department and the Johns Hopkins Medical Institution, Baltimore, MD. Female subjects 18 years and older, who provided signed consent, were enrolled. Cervical swab specimens were obtained from female subjects diagnosed with gonococcal infection, as evidenced by a Gram's stain of cervical secretions that showed gram-negative intracellular diplococci. The results were confirmed subsequently by the growth of N. gonorrhoeae on Thayer-Martin media or by a positive hybridization reaction using a transcription-mediated amplification assay (Gen-Probe, San Diego, CA). A separate swab for the present study was placed in 1 ml of TRIzol reagent for total RNA isolation.
Total RNA was isolated from TRIzol-preserved cervical swab specimens as previously published (1). To further purify RNA, samples were passed through an RNeasy Qiagen column prior to DNA microarray analysis. On-column DNase I digestion was also performed for each sample according to the manufacturer's instructions (Qiagen). PCRs were also performed without the addition of reverse transcriptase using internal rmp gene primers to ensure that DNA contamination was not present. RNA was labeled, hybridized, washed, and scanned for microarray analysis as discussed above for N. gonorrhoeae-infected human epithelial cells. For samples obtained from female subjects a 250-ng sample of total RNA was used for microarray analysis. Because we used a custom-array that represented a small number of genes and only two sets were spotted, we were able to use a fraction of the total amount of RNA usually required for microarray analysis. In pilot studies, we determined that 100 ng of RNA isolated from broth grown organisms was adequate for use with this custom-designed microarray (data not shown). The data were analyzed as discussed above and are expressed as the log2 of the normalized value of a spot divided by the normalized value of the spot that represented the rmp gene from the same clinical sample.
RESULTS
Expression of the gonococcal fur gene and Fur protein under iron-replete and iron-depleted conditions.
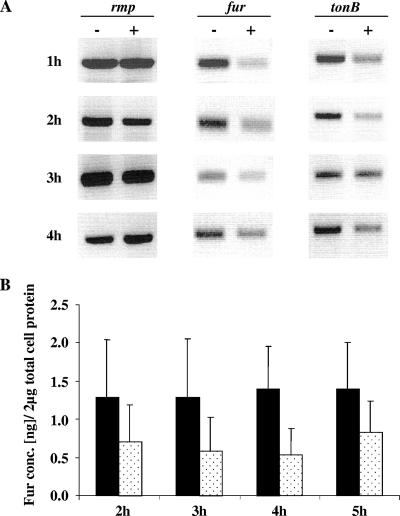
We used semiquantitative RT-PCR analysis first to examine the influence of iron on the transcription of the N. gonorrhoeae fur gene in vitro. Here we compared the transcription of the fur gene to the rmp gene because previously we showed that the rmp gene is not regulated by iron (1). We found that densities of rmp amplicons, generated by a 25-cycle PCR, were equal when broth-grown N. gonorrhoeae strain F62 was grown either in iron-depleted or iron-replete conditions (Fig. 1A). We observed increased transcript levels for the fur gene during N. gonorrhoeae growth in iron-depleted conditions compared to that observed during growth under iron-replete conditions (Fig. 1A). As expected, we also observed increased transcript levels of the iron-regulated tonB gene (15, 24, 46) during N. gonorrhoeae growth in iron-depleted conditions (Fig. 1A). Immunoblot analysis was performed next to estimate the levels of N. gonorrhoeae Fur protein produced under iron-depleted and iron-replete growth conditions (Fig. 1B). The Fur protein levels measured by immunoblot analysis were consistent with the fur transcript levels measured by RT-PCR analysis. The growth of N. gonorrhoeae under iron-depleted conditions resulted in higher levels of the Fur protein than those observed during N. gonorrhoeae growth in iron-replete conditions (Fig. 1B). However, we observed that gonococcal Fur protein was still produced when N. gonorrhoeae was grown under iron-replete conditions and throughout the 5-h period of growth, indicating that a basal level of the Fur protein was expressed even under iron-replete conditions (Fig. 1B).
FIG. 1.
In vitro gene and protein expression of gonococcal Fur in iron-replete and iron-depleted growth conditions. (A) Gonococcal gene expression examined by RT-PCR analysis of total RNA isolated from samples collected from iron-depleted (−) and iron-replete (+) conditions, monitored at different time points of growth (indicated to the left). The internal fragments of the iron-regulated fur and tonB genes were amplified and compared to the rmp gene, a gene not regulated by the presence or absence of iron. The amount of RNA utilized for analysis was 150 ng. The amplified cDNA fragments isolated by RT-PCR were run on a 1% agarose gel in 1× TAE buffer with 0.5 μg of ethidium bromide/ml and then visualized under UV light. (B) Gonococcal Fur protein quantification from N. gonorrhoeae F62 grown in iron-depleted (black box) and iron-replete (dotted box) conditions. A total of 2 μg of total cell protein from different growth time points (2 to 5 h) was loaded onto an SDS-PAGE gel and detected by immunoblot analysis using polyclonal anti-Fur antiserum (1:2,000). Fur bands were quantified ± the SD using Adobe Photoshop quantification software (version 6; Adobe Systems Incorporated, San Jose, CA), and the protein concentration was determined by using regression analysis with Fur protein standards of known concentration.
Mapping of the Fur binding sequence in the gonococcal fur gene.
Previous electrophoretic mobility shift assays have demonstrated binding of N. gonorrhoeae and N. meningitidis Fur homologs to their cognate promoter/operator region(s) (3, 46, 49). We extended these findings using DNase I footprinting studies with the gonococcal Fur protein and the N. gonorrhoeae fur promoter region to localize the gonococcal Fur binding sequence. In the absence of added gonococcal Fur protein, digestion of the end-labeled template strand produced a relatively uniform distribution of the DNA fragment ladder (Fig. 2A). A protected zone of 46 nucleotides (nt) was apparent with the addition of 230 nM or 460 nM Fur protein (Fig. 2A). This zone of protection spanned the predicted N. gonorrhoeae fur promoter region from +5 to −40. A 43-nt protection zone was also observed on the nontemplate strand that overlapped the in silico identified Fur box (22) (data not shown). The protected regions of the template and nontemplate strands overlapped the predicted 21-bp Fur box sequence, and this protected sequence was homologous to the predicted Neisseria Fur-binding consensus sequence (Fig. 2C). Similarly, 19- and 28-nt protected regions spanned the template and nontemplate strands, respectively, of the tonB promoter/operator region by the Fur protein (Fig. 2B). The protected nucleotides are represented in Fig. 2D in context with the predicted −10 and −35 regions that encompass the putative “Fur box”.
Construction and characterization of transcriptional fusions with the gonococcal fur operator region.
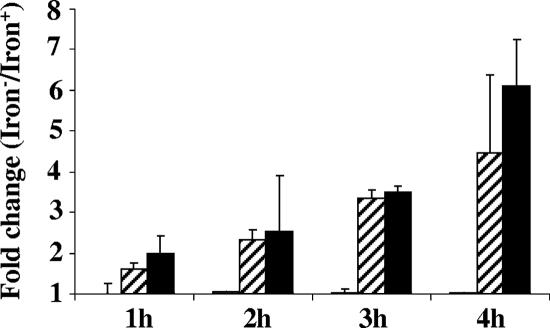
To confirm that the mapped N. gonorrhoeae fur operator sequence was functional, we next constructed transcriptional fusions in N. gonorrhoeae strain F62 using GFP fused to the N. gonorrhoeae fur operator sequences. N. gonorrhoeae GFP transcriptional fusion strains were also constructed with the operator regions of the N. gonorrhoeae tonB and rmp genes, and these served as positive and negative controls for operator regions regulated by the Fur protein, respectively. N. gonorrhoeae transcriptional fusion strains were grown under iron-depleted and iron-replete conditions, the samples were removed hourly, and GFP expression was measured by FACS analysis. Differential GFP expression for each of the transcriptional fusion was expressed as a fold change; a ratio of GFP fluorescence measured under iron-depleted conditions divided by fluorescence measured under iron-replete conditions. We observed that N. gonorrhoeae strain F62-furGFP exhibited a (1.6 ± 0.15)-fold increase in fluorescence in the first hour of growth, and this increased to 4.5 ± 1.87 by the fourth hour of growth (Fig. 3). Similarly, we observed that N. gonorrhoeae strain F62-tonBGFP exhibited a (2.0 ± 0.34)-fold increase in the first hour of growth increasing to 6.1 ± 1.1 by the fourth hour of growth. As expected, the level of fluorescence measured for N. gonorrhoeae strain F62-rmpGFP was similar in samples grown under iron-replete and iron-depleted conditions.
FIG. 3.
Characterization of gonococcal transcriptional fusions. Expression of N. gonorrhoeae rmp, fur, and tonB GFP transcriptional fusions during in vitro growth. N. gonorrhoeae strains that expressed the transcriptional fusions F62-rmpGFP, F62-furGFP, and F62-tonBGFP were grown in iron-replete and iron-depleted conditions; samples were taken at every hour (so indicated), and fluorescence was measured by FACS analysis. The fluorescence mean was measured by using CellQuest software (version 3.3) and is expressed as the ratio of rmp, fur, and tonB genes (iron depleted and iron replete). The bars indicate the fold change ratios: white bars for the rmp gene (not visible since this was 1.0), hatched bars for the fur gene, and black bars for the tonB gene Data represent mean ratios ± the SD of three independently performed experiments.
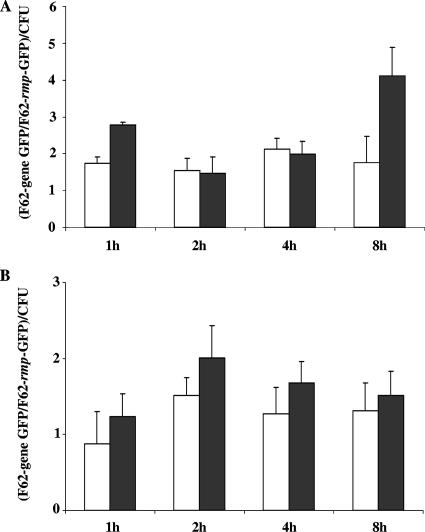
We next examined GFP expression in N. gonorrhoeae transcriptional fusion strains during infection of human cervical epithelial cells. For these studies we utilized immortalized human endocervical epithelial cells that were previously developed as a model system to examine N. gonorrhoeae interactions with epithelial cells (20). The results obtained from a standard antibiotic protection assay confirmed that the N. gonorrhoeae GFP fusion strains exhibited similar levels of total and intracellular bacteria as the N. gonorrhoeae wild-type strain F62 at each of the time points sampled (data not shown). After infection of endocervical epithelial cells with N. gonorrhoeae strain F62-furGFP or strain F62-tonBGFP, we observed an increase in GFP fluorescence in the total and intracellular bacteria, as measured throughout the infection period (Fig. 4). Similar results were obtained when N. gonorrhoeae strain F62-furGFP or strain F62-tonBGFP were used to infect ectocervical or vaginal epithelial cells (data not shown). Collectively, these results demonstrated that the mapped Fur binding site within the N. gonorrhoeae fur operator sequence was functional and responsive during in vitro iron growth conditions and during N. gonorrhoeae infection of cervical epithelial cells.
FIG. 4.
Fold change of gonococcal tonB and fur promoter region expression during N. gonorrhoeae infection of endocervical epithelial cells. N. gonorrhoeae GFP expressing strains (F62-rmpGFP, F62-furGFP, and F62-tonBGFP) were incubated with epithelial cells at an MOI of 100:1 (100 gonococci to 1 epithelial cell), and the cultures were examined at 1-, 2-, 4-, and 8-h intervals after infection for fold change in fur and tonB gene promoter expression compared to rmp gene promoter expression per each CFU. (A) Fold change determined for total bacteria (adherent plus intracellular); (B) fold change determined for intracellular bacteria only. Gene promoter expression was measured by using FACS analysis (CellQuest software, version 3.3). The fold change is expressed as the mean fold change ± the SD of three independently performed experiments (each performed in duplicate): the fur fluorescence fold change is represented by open bars, and the tonB fluorescence fold change is represented by black bars.
Expression of gonococcal genes encompassing the Fur and iron regulon during infection of endocervical epithelial cells.
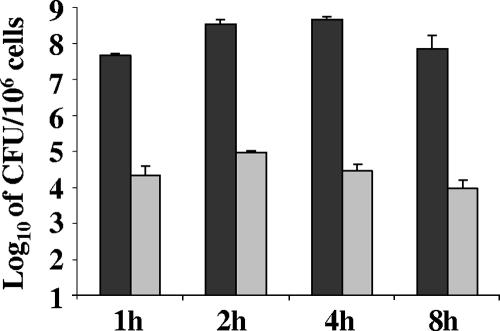
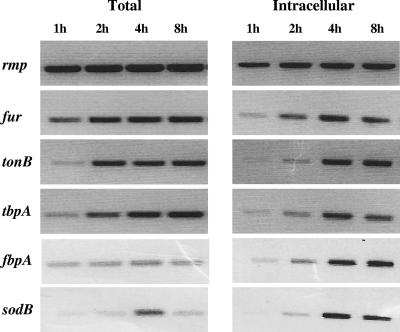
We next examined the expression of a subset of genes that encompass the gonococcal Fur and iron regulon during N. gonorrhoeae strain F62 infection of human endocervical epithelial cells by RT-PCR analysis. Total viable counts confirmed that N. gonorrhoeae strain F62 was viable in samples of total and intracellular bacteria at each time point examined (Fig. 5). Based on the initial inoculum used for infection, the percentages of total and intracellular viable bacteria for each time point were 5, 35, 46, and 7% and 0.002, 0.0097, 0.003, and 0.001%. RT-PCR analysis confirmed that equivalent amounts of rmp transcripts were produced throughout the 8-h incubation period of N. gonorrhoeae F62 with endocervical cells (Fig. 6). This was observed in both total and intracellular bacteria. However, the fur transcript was increased over the course of the 8-h incubation period of N. gonorrhoeae F62 with endocervical cells (Fig. 6). This was most pronounced in intracellular bacteria samples. Several additional genes encompassing the gonococcal Fur regulon (tonB, tbpA, fbpA, and sodB) were increased over the course of the 8-h incubation period of N. gonorrhoeae F62 with endocervical cells in both total and intracellular bacteria samples (Fig. 6).
FIG. 5.
Total and intracellular bacteria after infection of endocervical epithelial cells with N. gonorrhoeae strain F62 at an MOI of 100:1 (100 gonococci to 1 epithelial cell). The total numbers of viable cells (adherent plus intracellular gonococci) at 1, 2, 4 and 8 h are shown, indicated as CFU/106 endocervical cells (black bars) and intracellular bacteria in epithelial cells at different time points assessed by a gentamicin assay and also expressed as CFU/106 endocervical cells (gray bars). The data are expressed as the mean ± the SD, results representative of three independently performed experiments.
FIG. 6.
Gene expression of known iron-repressed and iron-activated genes during N. gonorrhoeae infection of endocervical cells. Gonococcal gene expression was examined by RT-PCR analysis with equal amounts of total RNA isolated from total and intracellular bacteria collected at different time points of infection (indicated above each lane). The internal fragments of the iron-regulated fur, tonB, tbpA, fbpA, and sodB genes were amplified in addition to the rmp gene, a gene not regulated by the presence or absence of iron. The amplified cDNA fragments isolated by RT-PCR were resolved on a 1% agarose gel in 1× TAE buffer with 0.5 μg of ethidium bromide/ml and then visualized under UV light.
To examine the expression of Fur- and iron-regulated genes during N. gonorrhoeae infection of human epithelial cells on a more global level, we next examined bacterial samples from infected cells by microarray analysis. For this analysis, we designed a custom DNA microarray that included genes encompassing the Neisseria Fur and iron regulons as determined in part by previous results obtained using a whole N. meningitidis genome microarray and N. meningitidis grown in vitro under iron-depleted and iron-replete conditions (24). The complete list of these genes included in the microarray slide is provided in Table S1 in the supplemental material.
To validate the Neisseria Fur and iron regulon microarray, we used total RNA from N. gonorrhoeae strain F62 grown under iron-replete and iron-depleted conditions. Our analysis confirmed that transcriptions of known N. gonorrhoeae iron-repressed genes (e.g., tbpAB, fbpA, and tonB) were increased in samples obtained from bacteria grown under iron-depleted conditions compared to bacteria grown under iron-replete conditions (column 2 in Tables 2 to 5). Similarly, known iron-activated genes (NG0904 [NMB1436] and NG0906 [NMB1438]) (23) exhibited increased transcription levels in samples obtained from bacteria grown under iron-depleted conditions compared to bacteria grown under iron-replete conditions. These expression profiles are reported in Tables 2 to 5 (column 2). Similar results were also observed with N. gonorrhoeae strain FA1090 (data not shown) that was grown under identical conditions.
TABLE 2.
Expression of representative gonococcal genes with a known function by microarray analysis during infection of endocervical epithelial cells by N. gonorrhoeae strain F62
| Gene (ORF)a | IRb | Gene product | Log2 ratioc at:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 h
|
2 h
|
4 h
|
8 h
|
|||||||
| TOT | INT | TOT | INT | TOT | INT | TOT | INT | |||
| tonB (NG1379) | + | Transport and binding proteins | 3.73 | 4.16 | 3.99 | 4.47 | 2.87 | 3.12 | 3.69 | 3.75 |
| fbpB (NG0216) | + | Iron-uptake permease inner membrane protein, MSD protein in ABC transport system | 3.57 | 3.47 | 3.53 | 3.46 | 2.90 | 2.53 | 3.45 | 3.21 |
| exbD (NG1377) | + | Transport protein | 3.18 | 3.32 | 3.43 | 3.50 | 2.81 | 2.56 | 3.34 | 2.86 |
| tbpA (NG1495) | + | Transferrin binding protein A | 2.15 | 2.46 | 2.41 | 3.05 | 1.71 | 2.03 | 2.28 | 2.47 |
| fbpA (NG0217) | + | Periplasmic iron-binding protein, ABC solute binding protein | 1.95 | 1.73 | 2.39 | 1.71 | 1.16 | 0.9 | 1.67 | 1.47 |
| fumC (NG1029) | + | Fumarate hydratase | 0.97 | 1.23 | 0.95 | 1.44 | 0.68 | 0.83 | 0.53 | 0.66 |
| fetA (NG2093) | + | Iron-regulated outer membrane protein | 0.44 | 0.03 | 1.16 | 0.04 | 1.50 | 0.07 | 0.82 | 0.05 |
| fur (NG1779) | + | Ferric uptake regulatory protein | 0.32 | 0.24 | 1.00 | 0.27 | 0.75 | 0.3 | 0.05 | 0.5 |
The numbers in parentheses denote the ORF numbers assigned by the gonococcal genome sequence project (http://www.genome.ou.edu/gono.html).
IR, iron response: +, iron-repressed gene.
The indicated values are the ratios expressed as the log2 of the intensity of the mean of the spots that represented the indicated gene divided by the mean of the spots that represented the rmp gene of the total associated (adhered plus intracellular) (TOT) and intracellular-only (INT) gonococci after infection of endocervical epithelial cells at 1, 2, 4, and 8 h.
TABLE 5.
Expression of hypothetical gonococcal genes by microarray analysis during uncomplicated gonorrhea in women
| ORFa | IRb | Hypothetical gene product | Median log2 ratio (mean ± SEM)c |
|---|---|---|---|
| NG0021 | + | Iron receptor; TonB dependent ferric siderophore receptor; similar to fhuE from E. coli | 2.33 (2.21 ± 0.46) |
| NG1536 | + | Conserved hypothetical protein | 1.93 (1.40 ± 0.52) |
| NG2049 | + | Neisseria-specific, integral membrane protein | 0.9 (0.80 ± 0.31) |
| NG1189 | - | hsp33; heat shock protein Hsp33 chaperonin | 2.33 (2.19 ± 0.59) |
| NG1529 | + | ftsA; cell division protein | 0.33 (0.35 ± 0.18) |
| NG1823 | - | rplO; 50S ribosomal protein L15 | 0.05 (0.13 ± 0.17) |
| NG1976 | +/- | Neisseria-specific protein | 0.70 (0.45 ± 0.18) |
| NG0869 | - | dedA; probable MSD protein in ABC transport | 0.07 (0.17 ± 0.35) |
| NG0044 | - | accC; acetyl coenzyme A carboxylase | 0.45 (0.17 ± 0.13) |
| NG1376 | + | Conserved hypothetical protein, possible ABC solute binding protein | 0.05 (0.06 ± 0.12) |
| NG0959 | + | Conserved hypothetical protein | 0.53 (0.30 ± 0.3) |
| NG0108 | + | Conserved hypothetical protein (oxidoreductase) | 0.09 (0.12 ± 0.17) |
The numbers denote the ORF numbers assigned by the gonococcal genome sequence project (http://www.genome.ou.edu/gono.html).
IR, genes regulated by iron: +, iron-repressed genes; -, iron-activated genes.
Data were obtained from clinical samples. The indicated values are the ratios expressed as log2 of the intensity of the mean of the spots that represented the indicated gene divided by the mean of spots that represented the rmp gene from the same clinical sample.
In samples obtained from N. gonorrhoeae infected endocervical epithelial cells we observed the expression of several Fur- and iron-regulated genes (Table 2). Microarray analysis revealed that the Fur and iron regulated tonB gene was highly expressed in total and intracellular bacteria throughout the entire 8-h incubation period (Table 2). While gene expression varied between the total and intracellular bacteria during infection of the endocervical epithelial cells, we did not observe significant differences in expression levels for the majority of genes in these samples (Table 2). We also detected expression of the fur gene itself in samples obtained from N. gonorrhoeae infected endocervical epithelial cells with some variability in expression over time. Interestingly, we did not detect expression of known iron-activated genes in N. gonorrhoeae infected endocervical epithelial cells (data not shown).
We also observed increased expression levels for a number of hypothetical genes represented on the microarray in total and intracellular bacteria after N. gonorrhoeae infection of epithelial cells (Table 3). Expression of the NG0021 gene (most highly expressed gene of this group) increased over time in total and intracellular bacteria after infection (Table 3). Increased expression was also observed for the NG2049 gene in both total and intracellular bacteria after N. gonorrhoeae infection of epithelial cells (Table 3). We have determined previously that the N. meningitidis NG2049 homolog (NMB0034) is expressed at high levels under iron-depleted conditions (20). In addition, we observed the expression of several iron-activated genes, including NG1189, NG1823, NG0869, and NG044, in total and intracellular bacteria after N. gonorrhoeae infection of epithelial cells. Collectively, these results indicate that both iron-repressed and iron-activated Fur-regulated genes are expressed during N. gonorrhoeae infection of endocervical cells.
TABLE 3.
Expression of hypothetical gonococcal genes by microarray analysis during infection of endocervical epithelial cells by N. gonorrhoeae strain F62
| ORFa | IRb | Hypothetical gene product | Log2 ratioc at:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 h
|
2 h
|
4 h
|
8 h
|
|||||||
| TOT | INT | TOT | INT | TOT | INT | TOT | INT | |||
| NG0021 | + | Iron receptor; TonB dependent ferric siderophore receptor; similar to fhuE from E. coli | 4.39 | 4.72 | 4.43 | 4.69 | 3.47 | 3.71 | 4.15 | 4.29 |
| NG1536 | + | Conserved hypothetical protein | 4.31 | 4.31 | 4.60 | 4.20 | 3.53 | 3.04 | 4.01 | 3.70 |
| NG2049 | + | Neisseria-specific, integral membrane protein | 3.81 | 3.97 | 4.06 | 3.68 | 3.40 | 2.77 | 3.46 | 3.25 |
| NG1189 | - | hsp33; heat shock protein Hsp33 chaperonin | 2.67 | 3.26 | 2.77 | 3.52 | 2.05 | 2.64 | 2.80 | 2.84 |
| NG1529 | + | ftsA; cell division protein | 2.42 | 2.35 | 2.31 | 2.59 | 1.75 | 1.65 | 2.20 | 2.04 |
| NG1823 | - | rplO; 50S ribosomal protein L15 | 1.83 | 1.64 | 1.37 | 1.27 | 1.59 | 1.02 | 1.09 | 0.84 |
| NG1976 | +/- | Neisseria-specific protein | 1.78 | 1.91 | 1.65 | 2.11 | 1.15 | 1.30 | 1.66 | 1.74 |
| NG0869 | - | dedA; probable MSD protein in ABC transport | 1.55 | 1.61 | 1.05 | 1.17 | 0.73 | 0.45 | 0.54 | 0.38 |
| NG0044 | - | accC; acetyl-CoA carboxylase | 1.73 | 1.85 | 2.25 | 2.12 | 1.52 | 1.27 | 1.27 | 1.25 |
| NG1376 | + | conserved hypothetical protein, possible ABC solute binding protein | 1.73 | 1.82 | 1.69 | 2.01 | 0.97 | 0.87 | 1.58 | 1.43 |
| NG0959 | + | Conserved hypothetical protein | 1.39 | 1.00 | 1.64 | 0.96 | 1.58 | 0.43 | 0.85 | 0.53 |
| NG0108 | + | Conserved hypothetical protein (oxidoreductase) | 1.19 | 1.73 | 1.33 | 1.56 | 0.47 | 0.67 | 0.16 | 0.43 |
The numbers denote the ORF numbers assigned by the gonococcal genome sequence project (http://www.genome.ou.edu/gono.html).
IR, iron response: +, iron-repressed genes; -, iron-activated genes.
The indicated values are the ratios expressed as log2 of the intensity of the mean of the spots that represented the indicated gene divided by the mean of spots that represented the rmp gene of the total associated (adhered plus intracellular) (TOT) and intracellular only (INT) gonococci after infection of endocervical epithelial cells at 1, 2, 4, and 8 h.
Expression of gonococcal genes in specimens obtained from women with mucosal gonococcal infection.
Next, using microarray analysis, we examined the expression of gonococcal Fur- and iron-regulated genes in total RNA samples isolated from cervical specimens obtained from six female subjects with uncomplicated gonorrhea. Because the Rmp protein is a biochemically, structurally, and antigenically conserved outer membrane protein expressed by N. gonorrhoeae (31, 32), we reasoned that it would serve as an appropriate control gene identically represented in the different strains of N. gonorrhoeae present in cervical specimens obtained for this analysis. Furthermore, the rmp gene also appears to be constitutively expressed in vivo (1). As observed in N. gonorrhoeae samples obtained from infected endocervical epithelial cells, we detected the expression of a number of genes that are Fur and iron regulated in samples obtained from all six specimens obtained from women with uncomplicated gonorrhea (Table 4). The expression levels of these Fur- and iron-regulated genes were similar to that observed during N. gonorrhoeae infection of epithelial cells (Table 4). For example, we detected expression of the tonB, tbpA, fbpA, fbpB, and exbD genes in all specimens obtained from women with uncomplicated gonorrhea.
TABLE 4.
Expression of representative gonococcal genes with a known function by microarray analysis during uncomplicated gonorrhea in women
| Gene (ORF)a | IRb | Gene product | Median log2 ratio (mean ± SEM)c |
|---|---|---|---|
| tonB (NG1379) | + | Transport and binding proteins | 3.59 (2.86 ± 0.43) |
| fbpB (NG0216) | + | Iron-uptake permease inner membrane protein, MSD protein in ABC transport system | 0.78 (0.48 ± 0.23) |
| exbD (NG1377) | + | Transport protein | 2.09 (2.03 ± 0.46) |
| tbpA (NG1495) | + | Transferrin binding protein A | 2.55 (2.02 ± 0.56) |
| fbpA (NG0217) | + | Periplasmic iron-binding protein, ABC solute binding protein | 0.25 (0.2 ± 0.17) |
| fumC (NG1029) | + | Fumarate hydratase | 0.83 (0.29 ± 0.28) |
| fetA (NG2093) | + | Iron-regulated outer membrane protein | 0.09 (0.13 ± 0.32) |
| fur (NG1779) | + | Ferric uptake regulatory protein | 0.2 (0.23 ± 0.35) |
The numbers in parentheses denote the ORF numbers assigned by the gonococcal genome sequence project (http://www.genome.ou.edu/gono.html).
IR, iron response: +, iron-repressed genes.
Data were obtained from clinical samples. The indicated values are the ratios expressed as log2 of the intensity of the mean of the spots that represented the indicated gene divided by the mean of spots that represented the rmp gene from the same clinical sample.
We also demonstrated that a number of N. gonorrhoeae Fur- and iron-regulated hypothetical genes were expressed in specimens obtained from women with uncomplicated gonorrhea (Table 5). The expressions of iron repressed (NG0021 and NG1536) and iron activated (NG1189) in specimens obtained from these women were similar to that observed in N. gonorrhoeae samples obtained from infected endocervical epithelial cells. Taken as a whole, our microarray results indicate that N. gonorrhoeae Fur- and iron-regulated genes are expressed during N. gonorrhoeae infection of human endocervical epithelial cells and in mucosal samples obtained from female subjects with uncomplicated gonorrhea.
DISCUSSION
In this study we demonstrated expression of the N. gonorrhoeae fur gene in vitro, in cervical epithelial cells, and in specimens obtained from female subjects with uncomplicated gonococcal infection. In vitro studies confirmed that the expression of the gonococcal fur gene was repressed during growth under iron-replete growth conditions but that a basal level of the protein was maintained. We also demonstrated that the N. gonorrhoeae fur gene was expressed during N. gonorrhoeae infection of endocervical epithelial cells in samples obtained from total and intracellular bacteria throughout the incubation period. The endocervical epithelial cell line utilized in our study was developed from normal human endocervical tissue. Endocervical tissue consists of a single layer of tall columnar epithelial cells with rare cuboidal reserve cells (21). Under healthy conditions in vivo, endocervical cells differentiate in a sterile environment. Therefore, this corresponding cell line may be representative of a normal human host environment for N. gonorrhoeae because the cells maintain the normal karyotype and stable phenotype of the tissue of origin.
Using this endocervical cell line, we also demonstrated that N. gonorrhoeae Fur- and iron-regulated genes are expressed during N. gonorrhoeae infection of endocervical epithelial cells. RT-PCR analysis revealed that the expressions of the tbpA, fbpA, and sodB genes were increased over the course of the 8-h incubation period. Furthermore, these results were validated using a custom Neisseria Fur and iron regulon microarray. Microarray analysis also identified additional N. gonorrhoeae Fur- and iron-repressed hypothetical genes, which were expressed during N. gonorrhoeae infection of cervical epithelial cells. We observed that the expression of these genes was fairly constant throughout the 8-h incubation period. In addition, while we observed some differences in the levels of expression of these genes in total bacteria compared to intracellular bacteria, in general there were no significant differences in these gene profiles.
A number of the hypothetical genes which were both iron repressed and iron activated were expressed by N. gonorrhoeae during infection of human endocervical epithelial cells and encode for conserved proteins (NG0021, NG1536, NG2049, NG1189, NG1529, NG1976, and NG1376). Two of these highly expressed genes are homologous to genes that encode membrane proteins that have a role in transport across membranes (NG0869 and NG0021). Other genes, including the hsp33 gene (NG1189), that encodes the heat shock protein Hsp33, the rplO gene (NG1823) that encodes the 50S ribosomal protein S5, and the ftsA gene (NG1529) encoding a cell division protein were also expressed during gonococcal infection in vivo. Of potential interest in this group of genes was the NG0021 gene, which has been reported previously to be expressed during N. gonorrhoeae infection of endocervical cells (50). The NG0021 (tdfF) gene was identified in a computer-generated search of N. gonorrhoeae strain FA1090 genome for unknown members of the TonB-dependent family (Tdf) and is predicted to be a TonB-dependent outer membrane ferric siderophore receptor, similar to the siderophore receptors fhuE (E. coli), fpvA (Pseudomonas aeruginosa), and pupB (P. putida). It was recently reported that TdfF was important for the intracellular survival of N. gonorrhoeae in human cervical epithelial cells (25).
Interestingly, although we did detect the expression of a few hypothetical genes that are activated by iron (i.e., NG1189, NG1823, and NG0869) during infection of endocervical epithelial cells by N. gonorrhoeae, the expression of additional iron-activated genes represented on the array was not detected. Bonnah et al. (4, 5) have shown previously that human epithelial cells infected with N. gonorrhoeae or N. meningitidis have reduced levels of transferrin receptor mRNA and cycling transferrin receptors. Using DNA microarray analysis, these investigators reported altered expression levels of a number of host genes during Neisseria infection of epithelial cells, and these researchers suggested that bacterial growth in epithelial cells represents growth in an iron-limited environment. Larson et al. (35) showed that epithelial cells must degrade ferritin in response to infection by N. meningitidis in order to access limited iron. These results support the tenet that gonococcal infection of epithelial cells occurs in an iron-limited environment and is fairly consistent with our findings.
Using DNA footprinting analysis we confirmed the binding of the gonococcal Fur protein to the operator region of the fur gene encompassing 46 nt. Prokaryotic repressors typically bind 10 bp per repressor unit per turn of DNA helix; therefore, a repressor dimer occupies two consecutive turns, leading to a protected region with a size of ∼20 bp (12, 47). Using this paradigm, the extent of the fur operator sequence (46 nt), as defined by our footprinting analysis, suggests the binding of two repressor dimers. We also determined that the 46-nt sequence within the fur operator was sufficient for iron-mediated regulation of the N. gonorrhoeae fur gene in vitro. In addition, we demonstrated that the Fur binding sequences within the fur operator sequence as determined by DNA footprint analysis mediated transcriptional regulation during N. gonorrhoeae infection of human epithelial cells.
A recent study reported on the differential gene expression pattern of in vitro-grown N. gonorrhoeae strain FA1090 in iron-replete and iron-depleted conditions using a pan-Neisseria DNA microarray (25). These investigators concluded that regulated genes in gonococci and meningococci were unique to each organism (15, 24). A contributing factor to the specificity of gene expression profiles of in vitro grown iron-depleted and iron-replete N. meningitidis (24) and N. gonorrhoeae (15) may have been differences in the treatment of growth media used in these two separate studies. For iron-depleted conditions, Ducey et al. (15) used Chelex-treated CDM for growth of N. gonorrhoeae. Our previous studies with N. meningitidis (24), as well as the studies reported here with N. gonorrhoeae F62, were performed with bacterial cultures in which Desferal was added to CDM to chelate external iron and minimize its availability for in vitro bacterial growth.
A major observation from our studies was that the gene expression profiles of specimens obtained from female subjects with uncomplicated gonococcal infection corroborated our in vitro findings. These results are in agreement with our previous studies in which we observed expression of several gonococcal iron- and Fur-regulated genes in clinical specimens obtained directly from men who had contracted uncomplicated gonococcal infection (1). However, a limitation of these previous studies was that only a small number of Fur-regulated genes were examined. The use of a Neisseria Fur and iron regulon microarray in the present study has enabled us to confirm our previous results with male specimens and extend that study further to more importantly include specimens from gonococcus-infected female subjects to demonstrate that additional Fur- and iron-regulated genes are expressed in vivo during mucosal gonococcal infection. Our previous study (1) and the results presented here are the only studies to date to examine the expression of genes encompassing the Fur regulon during in vivo infection in humans. Interestingly, while we did detect expression of the fur gene itself in specimens obtained from female subjects with uncomplicated gonococcal infection, the level of expression was fairly low. Nonetheless, it appears that the level of Fur protein must be sufficient to function in the regulation of genes encompassing the Fur regulon.
In conclusion, our studies indicate that while the Neisseria fur gene is regulated by iron and Fur under in vitro growth conditions, a basal level of the global regulator is always maintained. The ability of pathogenic Neisseria to maintain basal levels of the Fur protein may enable these bacteria to respond to changes in iron availability. Furthermore, we have established that numerous gonococcal genes that encompass the Fur and iron regulon are expressed during N. gonorrhoeae infection of human cervical epithelial cells. In addition, we have validated these studies using cervical specimens from women with uncomplicated gonococcal infection. Collectively, these results indicate that the increased expression of iron- and Fur-repressed genes during N. gonorrhoeae infection is indicative of an environment where iron is limited.
Supplementary Material
Acknowledgments
This study was supported by grants AI048611 to C.A.G. and AI038515 to P.A.R. from the NIH/NIAID.
We appreciate the gift of plasmids pLES98 and pLES99 from Virginia L. Clark at the University of Rochester. We gratefully acknowledge Mark Lenburg and Norman Gerry for their invaluable help with DNA microarray work and analysis and J. R. Mellin for critical review of the manuscript.
Footnotes
Published ahead of print on 29 February 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Agarwal, S., C. A. King, E. K. Klein, D. E. Soper, P. A. Rice, L. M. Wetzler, and C. A. Genco. 2005. The gonococcal Fur-regulated tbpA and tbpB genes are expressed during natural mucosal gonococcal infection. Infect. Immun. 734281-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baichoo, N., T. Wang, R. Ye, and J. D. Helmann. 2002. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol. Microbiol. 451613-1629. [DOI] [PubMed] [Google Scholar]
- 3.Berish, S. A., S. Subbarao, C. Y. Chen, D. L. Trees, and S. A. Morse. 1993. Identification and cloning of a fur homolog from Neisseria gonorrhoeae. Infect. Immun. 614599-4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonnah, R. A., S. W. Lee, B. L. Vasquez, C. A. Enns, and M. So. 2000. Alteration of epithelial cell transferrin-iron homeostasis by Neisseria meningitidis and Neisseria gonorrhoeae. Cell Microbiol. 2207-218. [DOI] [PubMed] [Google Scholar]
- 5.Bonnah, R. A., M. U. Muckenthaler, H. Carlson, B. Minana, C. A. Enns, M. W. Hentze, and M. So. 2004. Expression of epithelial cell iron-related genes upon infection by Neisseria meningitidis. Cell Microbiol. 6473-484. [DOI] [PubMed] [Google Scholar]
- 6.Bsat, N., and J. D. Helman. 1999. Interaction of Bacillus subtilis Fur (ferric uptake repressor) with the operator in vitro and in vivo. J. Bacteriol. 1814299-4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campoy, S., M. Jara, N. Busquets, A. M. de Rozas, I. Badiola, and J. Barbe. 2002. Intracellular cyclic AMP concentration is decreased in Salmonella typhimurium fur mutants. Microbiology 1481039-1048. [DOI] [PubMed] [Google Scholar]
- 8.Cha, J. Y., J. S. Lee, J. I. Oh, J. W. Choi, and H. S. Baik. 2008. Functional analysis of the role of Fur in the virulence of Pseudomonas syringae pv. tabaci 11528: Fur controls expression of genes involved in quorum-sensing. Biochem. Biophys. Res. Commun. 366281-287. [DOI] [PubMed] [Google Scholar]
- 9.Crosa, J. H. 1997. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol. Mol. Biol. Rev. 61319-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delany, I., R. Grifantini, E. Bartolini, R. Rappuoli, and V. Scarlato. 2006. Effect of Neisseria meningitidis fur mutations on global control of gene transcription. J. Bacteriol. 1882483-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delany, I., R. Rappuoli, and V. Scarlato. 2004. Fur functions as an activator and as a repressor of putative virulence genes in Neisseria meningitidis. Mol. Microbiol. 521081-1090. [DOI] [PubMed] [Google Scholar]
- 12.de Lorenzo, V., S. Wee, M. Herrero, and J. B. Neilands. 1987. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J. Bacteriol. 1692624-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubrac, S., and D. Touati. 2002. Fur-mediated transcriptional and posttranscriptional regulation of FeSOD expression in Escherichia coli. Microbiology 148147-156. [DOI] [PubMed] [Google Scholar]
- 14.Dubrac, S., and D. Touati. 2000. Fur positive regulation of iron superoxide dismutase in Escherichia coli: functional analysis of the sodB promoter. J. Bacteriol. 1823802-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ducey, T. F., M. B. Carson, J. Orvis, A. P. Stintzi, and D. W. Dyer. 2005. Identification of the iron-responsive genes of Neisseria gonorrhoeae by microarray analysis in defined medium. J. Bacteriol. 1874865-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edwards, J. L., and M. A. Apicella. 2004. The molecular mechanisms used by Neisseria gonorrhoeae to initiate infection differ between men and women. Clin. Microbiol. Rev. 17965-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ernst, F. D., S. Bereswill, B. Waidner, J. Stoof, U. Mader, J. G. Kusters, E. J. Kuipers, M. Kist, A. H. van Vliet, and G. Homuth. 2005. Transcriptional profiling of Helicobacter pylori Fur- and iron-regulated gene expression. Microbiology 151533-546. [DOI] [PubMed] [Google Scholar]
- 18.Escolar, L., V. de Lorenzo, and J. Perez-Martin. 1997. Metalloregulation in vitro of the aerobactin promoter of Escherichia coli by the Fur (ferric uptake regulation) protein. Mol. Microbiol. 26799-808. [DOI] [PubMed] [Google Scholar]
- 19.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 1998. Binding of the Fur (ferric uptake regulator) repressor of Escherichia coli to arrays of the GATAAT sequence. J. Mol. Biol. 283537-547. [DOI] [PubMed] [Google Scholar]
- 20.Fichorova, R. N., P. J. Desai, F. C. Gibson III, and C. A. Genco. 2001. Distinct proinflammatory host responses to Neisseria gonorrhoeae infection in immortalized human cervical and vaginal epithelial cells. Infect. Immun. 695840-5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fichorova, R. N., J. G. Rheinwald, and D. J. Anderson. 1997. Generation of papillomavirus-immortalized cell lines from normal human ectocervical, endocervical, and vaginal epithelium that maintain expression of tissue-specific differentiation proteins. Biol. Reprod. 57847-855. [DOI] [PubMed] [Google Scholar]
- 22.Genco, C. A., and P. J. Desai. 1996. Iron acquisition in the pathogenic Neisseria. Trends Microbiol. 4179-184. [DOI] [PubMed] [Google Scholar]
- 23.Grifantini, R., E. Frigimelica, I. Delany, E. Bartolini, S. Giovinazzi, S. Balloni, S. Agarwal, G. Galli, C. Genco, and G. Grandi. 2004. Characterization of a novel Neisseria meningitidis Fur and iron-regulated operon required for protection from oxidative stress: utility of DNA microarray in the assignment of the biological role of hypothetical genes. Mol. Microbiol. 54962-979. [DOI] [PubMed] [Google Scholar]
- 24.Grifantini, R., S. Sebastian, E. Frigimelica, M. Draghi, E. Bartolini, A. Muzzi, R. Rappuoli, G. Grandi, and C. A. Genco. 2003. Identification of iron-activated and -repressed Fur-dependent genes by transcriptome analysis of Neisseria meningitidis group B. Proc. Natl. Acad. Sci. USA 1009542-9547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagen, T. A., and C. N. Cornelissen. 2006. Neisseria gonorrhoeae requires expression of TonB and the putative transporter TdfF to replicate within cervical epithelial cells. Mol. Microbiol. 621144-1157. [DOI] [PubMed] [Google Scholar]
- 26.Hall, H. K., and J. W. Foster. 1996. The role of fur in the acid tolerance response of Salmonella typhimurium is physiologically and genetically separable from its role in iron acquisition. J. Bacteriol. 1785683-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hantke, K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4172-177. [DOI] [PubMed] [Google Scholar]
- 28.Hassett, D. J., M. L. Howell, U. A. Ochsner, M. L. Vasil, Z. Johnson, and G. E. Dean. 1997. An operon containing fumC and sodA encoding fumarase C and manganese superoxide dismutase is controlled by the ferric uptake regulator in Pseudomonas aeruginosa: fur mutants produce elevated alginate levels. J. Bacteriol. 1791452-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holmes, K., F. Mulholland, B. M. Pearson, C. Pin, J. McNicholl-Kennedy, J. M. Ketley, and J. M. Wells. 2005. Campylobacter jejuni gene expression in response to iron limitation and the role of Fur. Microbiology 151243-257. [DOI] [PubMed] [Google Scholar]
- 30.Horsburgh, M. J., E. Ingham, and S. J. Foster. 2001. In Staphylococcus aureus, fur is an interactive regulator with PerR, contributes to virulence, and Is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J. Bacteriol. 183468-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Judd, R. C. 1982. 125I-peptide mapping of protein III isolated from four strains of Neisseria gonorrhoeae. Infect. Immun. 37622-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Judd, R. C. 1982. Surface peptide mapping of protein I and protein III of four strains of Neisseria gonorrhoeae. Infect. Immun. 37632-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karjalainen, T. K., D. G. Evans, D. J. Evans, Jr., D. Y. Graham, and C. H. Lee. 1991. Iron represses the expression of CFA/I fimbriae of enterotoxigenic Escherichia coli. Microb. Pathog. 11317-323. [DOI] [PubMed] [Google Scholar]
- 34.Laemmli, U. K., F. Beguin, and G. Gujer-Kellenberger. 1970. A factor preventing the major head protein of bacteriophage T4 from random aggregation. J. Mol. Biol. 4769-85. [DOI] [PubMed] [Google Scholar]
- 35.Larson, J. A., H. L. Howie, and M. So. 2004. Neisseria meningitidis accelerates ferritin degradation in host epithelial cells to yield an essential iron source. Mol. Microbiol. 53807-820. [DOI] [PubMed] [Google Scholar]
- 36.Lewis, L. A., M. Gipson, K. Hartman, T. Ownbey, J. Vaughn, and D. W. Dyer. 1999. Phase variation of HpuAB and HmbR, two distinct haemoglobin receptors of Neisseria meningitidis DNM2. Mol. Microbiol. 32977-989. [DOI] [PubMed] [Google Scholar]
- 37.Litwin, C. M., and S. B. Calderwood. 1993. Role of iron in regulation of virulence genes. Clin. Microbiol. Rev. 6137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McHugh, J. P., F. Rodriguez-Quinones, H. Abdul-Tehrani, D. A. Svistunenko, R. K. Poole, C. E. Cooper, and S. C. Andrews. 2003. Global iron-dependent gene regulation in Escherichia coli: a new mechanism for iron homeostasis. J. Biol. Chem. 27829478-29486. [DOI] [PubMed] [Google Scholar]
- 39.Mellin, J. R., S. Goswami, S. Grogan, B. Tjaden, and C. A. Genco. 2007. A novel Fur- and iron-regulated small RNA, NrrF, is required for indirect fur-mediated regulation of the sdhA and sdhC genes in Neisseria meningitidis. J. Bacteriol. 1893686-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mey, A. R., E. E. Wyckoff, V. Kanukurthy, C. R. Fisher, and S. M. Payne. 2005. Iron and Fur regulation in Vibrio cholerae and the role of Fur in virulence. Infect. Immun. 738167-8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morse, S. A., and L. Bartenstein. 1980. Purine metabolism in Neisseria gonorrhoeae: the requirement for hypoxanthine. Can. J. Microbiol. 2613-20. [DOI] [PubMed] [Google Scholar]
- 42.Ochsner, U. A., and M. L. Vasil. 1996. Gene repression by the ferric uptake regulator in Pseudomonas aeruginosa: cycle selection of iron-regulated genes. Proc. Natl. Acad. Sci. USA 934409-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plummer, F. A., H. Chubb, J. N. Simonsen, M. Bosire, L. Slaney, I. Maclean, J. O. Ndinya-Achola, P. Waiyaki, and R. C. Brunham. 1993. Antibody to Rmp (outer membrane protein 3) increases susceptibility to gonococcal infection. J. Clin. Investig. 91339-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prince, R. W., C. D. Cox, and M. L. Vasil. 1993. Coordinate regulation of siderophore and exotoxin A production: molecular cloning and sequencing of the Pseudomonas aeruginosa fur gene. J. Bacteriol. 1752589-2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rarick, M., C. McPheeters, S. Bright, A. Navis, J. Skefos, P. Sebastiani, and M. Montano. 2006. Evidence for cross-regulated cytokine response in human peripheral blood mononuclear cells exposed to whole gonococcal bacteria in vitro. Microb. Pathog. 40261-270. [DOI] [PubMed] [Google Scholar]
- 46.Sebastian, S., S. Agarwal, J. R. Murphy, and C. A. Genco. 2002. The gonococcal fur regulon: identification of additional genes involved in major catabolic, recombination, and secretory pathways. J. Bacteriol. 1843965-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shaik, Y. B., S. Grogan, M. Davey, S. Sebastian, S. Goswami, B. Szmigielski, and C. A. Genco. 2007. Expression of the iron-activated nspA and secY genes in Neisseria meningitidis group B by Fur-dependent and -independent mechanisms. J. Bacteriol. 189663-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stojiljkovic, I., A. J. Baumler, and K. Hantke. 1994. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a fur titration assay. J. Mol. Biol. 236531-545. [DOI] [PubMed] [Google Scholar]
- 49.Thomas, C. E., and P. F. Sparling. 1994. Identification and cloning of a fur homologue from Neisseria meningitidis. Mol. Microbiol. 11725-737. [DOI] [PubMed] [Google Scholar]
- 50.Turner, P. C., C. E. Thomas, I. Stojiljkovic, C. Elkins, G. Kizel, D. A. Ala'Aldeen, and P. F. Sparling. 2001. Neisserial TonB-dependent outer-membrane proteins: detection, regulation and distribution of three putative candidates identified from the genome sequences. Microbiology 1471277-1290. [DOI] [PubMed] [Google Scholar]
- 51.Vasil, M. L., and U. A. Ochsner. 1999. The response of Pseudomonas aeruginosa to iron: genetics, biochemistry, and virulence. Mol. Microbiol. 34399-413. [DOI] [PubMed] [Google Scholar]
- 52.Vasil, M. L., U. A. Ochsner, Z. Johnson, J. A. Colmer, and A. N. Hamood. 1998. The fur-regulated gene encoding the alternative sigma factor PvdS is required for iron-dependent expression of the LysR-type regulator ptxR in Pseudomonas aeruginosa. J. Bacteriol. 1806784-6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weinberg, E. D. 1993. The development of awareness of iron-withholding defense. Perspect. Biol. Med. 36215-221. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.