Abstract
The actinomycete Corynebacterium glutamicum grows as rod-shaped cells by zonal peptidoglycan synthesis at the cell poles. In this bacterium, experimental depletion of the polar DivIVA protein (DivIVACg) resulted in the inhibition of polar growth; consequently, these cells exhibited a coccoid morphology. This result demonstrated that DivIVA is required for cell elongation and the acquisition of a rod shape. DivIVA from Streptomyces or Mycobacterium localized to the cell poles of DivIVACg-depleted C. glutamicum and restored polar peptidoglycan synthesis, in contrast to DivIVA proteins from Bacillus subtilis or Streptococcus pneumoniae, which localized at the septum of C. glutamicum. This confirmed that DivIVAs from actinomycetes are involved in polarized cell growth. DivIVACg localized at the septum after cell wall synthesis had started and the nucleoids had already segregated, suggesting that in C. glutamicum DivIVA is not involved in cell division or chromosome segregation.
In rod-shaped bacteria such as Escherichia coli or Bacillus subtilis, cell division gives rise to hemispherical cell poles, which after their formation, largely remain inert (13). Bacterial cell elongation involves extension of the lateral cell wall, which in most rod-shaped bacteria occurs by intercalation of new peptidoglycan (PG) into the wall along most of its length. This process requires the assembly of MreB, an actin homologue, into a helical cytoskeleton that is distributed throughout the cell (7). In E. coli, the depletion of MreB results in the formation of coccoid cells (24). These observations suggest that the MreB-based cytoskeleton is involved in organizing or localizing enzymes that participate in cell wall assembly, such as penicillin-binding proteins, presumably through a linkage to the cell wall synthesis machinery via the proteins MreC, MreD, and RodA (6, 14, 24, 26).
A different and mreB-independent mode of cell elongation and rod shape acquisition is found among the corynebacteria. Similar to other members of the phylum Actinobacteria (gram-positive bacteria with a high G+C content in their DNA) that do not undergo sporulation (30), corynebacterial genomes are devoid of mreB homologues (8, 22, 30, 34, 43). Moreover, staining of the rod-shaped bacterium Corynebacterium glutamicum with fluorescently labeled vancomycin (vancomycin-Bodipy FL [Van-FL]) showed that PG is primarily assembled at the cell poles rather than along the lateral wall (11). In the simplest model describing the polarization of PG assembly in C. glutamicum, components of the cell division machinery are sufficient to recruit the enzymes for cell wall elongation to the new cell pole. However, the filamentous hyphal cells of another member of the Actinobacteria, Streptomyces coelicolor, grow by tip extension and incorporate new PG at the hyphal apex (18). This highly polarized mode of growth does not require cell division (31) and does not occur at the cell poles created by division. Instead, new hyphal tips are generated de novo either by branching or by the emergence of the germ tube, implying that the bacterium possesses a mechanism to establish apical cell wall assembly independent of the division process.
Observations in C. glutamicum, S. coelicolor, and, recently, in Mycobacterium smegmatis suggest that the coiled-coil protein DivIVA influences apical growth and cell shape determination (19, 33, 35). In these organisms, DivIVA has a distinct polar localization. The divIVA genes of S. coelicolor and M. smegmatis (divIVASc and divIVAMs, respectively) were found to be essential (19, 33), and this was also evidenced in C. glutamicum by the inability to obtain viable cells when the gene was deleted (19, 33, 35). Furthermore, in all three organisms, divIVA overexpression results in bloated asymmetric cells (19, 33, 35), whereas partial depletion of DivIVA in S. coelicolor affects tip extension and branching (19). In Mycobacterium tuberculosis, which also exhibits Van-FL staining patterns consistent with zonal PG biosynthesis at the poles (10), DivIVA is phosphorylated by the serine/threonine kinases PknA and PknB, and overexpression of divIVA or overexpression or partial depletion of these kinases was shown to alter the control of cell shape (23).
In this study, we addressed the basis for rod shape acquisition of C. glutamicum. The results showed that depletion of DivIVA leads to a coccoid morphology, and thus they provide evidence for the first time that DivIVA contributes to a mechanism by which cell growth and rod-shaped morphology are maintained via cell wall elongation at the poles. This function could also be fulfilled by heterologous DivIVAs from other Actinobacteria but not by those from B. subtilis and S. pneumoniae from Firmicutes, suggesting that DivIVA functions differently in the two main phylogenetic lines of gram-positive bacteria.
MATERIALS AND METHODS
Bacterial strains and general methods for nucleic acids.
Bacterial strains and plasmids are described in Table 1 (see Table S1 in the supplemental material for a description of the primers). E. coli cells were grown in Luria broth or Luria agar (20) complex medium at 37°C with aeration. C. glutamicum cells were grown in TSB (trypticase soy broth; Oxoid) or complex medium consisting of TSB containing 2% agar at 30°C. When required, the following supplements were added to the culture medium: kanamycin (50 μg/ml for E. coli; 12.5 μg/ml for corynebacteria), apramycin (50 μg/ml for E. coli; 12.5 μg/ml for corynebacteria), chloramphenicol (20 μg/ml for E. coli), ampicillin (100 μg/ml for E. coli), and tetracycline and streptomycin (12.5 μg/ml for E. coli).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype or descriptiona | Source and/or reference |
|---|---|---|
| E. coli strains | ||
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG; used for general cloning | Invitrogen |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3); used for in vivo gene expression | Novagen |
| S17-1 | pro recA; mobilizing donor strain that has an RP4 derivative integrated into the chromosome | 39 |
| XL1-Blue MRF′ | Host strain used for propagating pBT and pTRG recombinants | Stratagene |
| BacterioMatch II | Reporter strain derived from Stratagene's XL1-Blue MR, harbors lacIq and the reporter genes HIS3 and aadA on the F′ episome | Stratagene |
| C. glutamicum strains | ||
| ATCC 13869 | Wild type | ATCC |
| R31 | ATCC 13869 derivative; used as recipient in conjugation assays and as control strain | 38 |
| LACD | R31 derivative obtained by integration of plasmid pOJD; Aprr | This work |
| LACID | LACD derivative containing the bifunctional plasmid pALacI; Aprr Kanr | This work; Fig. 1B |
| R33 | R31 derivative containing divIVACg-egfp2 under the control of Pdiv (Pdiv-divIVACg-egfp2) by integration of plasmid pK18-CGE; Kanr | This work; Fig. 5B |
| GNTD | R31 derivative containing a complete copy of divIVACg under the control of PgntP by integration of plasmid pOJPD; Aprr | This work |
| GNTCG, GNTBS, GNTSC, GNTSP, GNTMT | GNTD derivatives containing the respective divIVA genes under the control of Pdiv (Pdiv-divIVA) by integration of plasmids pK18-CG, -BS, -SC, -SP, and -MT respectively; Aprr Kanr | This work; Fig. 2A |
| GNTD3 | GNTD derivative containing the plasmid pK18-3 integrated in the chromosome; Aprr Kanr | This work; Fig. 2A |
| GNTCGE, GNTBSE, GNTSCE, GNTSPE, GNTMTE | GNTD derivatives containing the respective divIV-egfp2 genes under the control of the Pdiv promoter (Pdiv-divIVA-egfp2) by integration of plasmids pK18-CGE, -BSE, -SCE, -SPE, -MTE, respectively; Aprr Kanr | This work; Fig. 2B |
| GNTDE | GNTD derivative containing egfp2 under the control of Pdiv promoter (Pdiv-egfp2) by integration of plasmid pK18-E; Aprr Kanr | This work; Fig. 2B |
| AG1 | R31 derivative containing the bifunctional mobilizable plasmid pEAG1; Kanr Catr | This work; Fig. 3 |
| AG3 | R31 derivative containing the bifunctional mobilizable plasmid pEAG3; Kanr Catr | This work; Fig. 3 |
| BS, SC, SP, MT | R31 derivatives containing the bifunctional mobilizable plasmids pEBS, pESC, pESP, and pEMT respectively; Kanr Catr | This work; Fig. 3 |
| BSE, SCE, SPE, MTE | R31 derivatives containing the bifunctional mobilizable plasmids pEBSE, pESCE, pESPE, and pEMTE respectively; Kanr Catr | This work; Fig. 3 |
| Plasmids | ||
| pET28a | E. coli expression vector; kan | Novagen |
| pETDIV | pET28a derivative carrying the His6-tagged divIVACg gene | This work |
| pGEM-TEasy | E. coli vector; bla lacI orif1 | Promega |
| pECM2 | Mobilizable plasmid capable of replicating in E. coli and C. glutamicum; kan cat | 21 |
| pEAG1 | pECM2 derivative carrying Pdiv-divIVACg | 35 |
| pEAG2 | pEAG1 derivative carrying Pdiv-divIVACg-egfp2 | 35 |
| pEAG3 | pEAG2 derivative including an extra NdeI site between divIVACg-egfp2 and Pdiv | This work |
| pEAGE | pEAG3 derivative carrying Pdiv-egfp2 and used to construct pK18-3 | This work |
| pEGFP | pECM2 derivative carrying egfp2 under the control of the Pkan and flanked by transcriptional terminators T1 and T2 | 28 |
| pEGNC | pEGFP derivative carrying the promoterless egfp2 gene | 28 |
| pEPDG | pEGFP derivative carrying PgntP-divIVACg-egfp2 | 28 |
| pOJ260 | Mobilizable plasmid carrying an E. coli origin of replication and the apr resistance determinant | 4 |
| pOJD | pOJ260 derivative carrying the 5′ end of C. glutamicum divIVACg under the control of the promoter Plac (Plac-ΔdivIVACg) | This work |
| pOJPD | pOJ260 derivative carrying the 5′ end of C. glutamicum divIVACg under the control of the promoter of the C. glutamicum gntP gene (PgntP-ΔdivIVACg) | This work |
| pAlacI | Mobilizable plasmid capable of replicating in E. coli and C. glutamicum; lacIqkan | 45 |
| pK18-3 | Mobilizable plasmid carrying an E. coli origin of replication, kan and three C. glutamicum ORFs of unknown function located downstream from ftsZ | 1 |
| pEBSE, pESCE, pESPE, pEMTE | pEAG3 derivatives carrying the respective Pdiv-divIVA-egfp2 gene fusion | This work |
| pEBS, pESC, pESP, pEMT | pECM2 derivatives carrying the respective Pdiv-divIVA gene fusion | This work |
| pK18-CGE, -BSE, -SCE, -SPE, -MTE | pK18-3 derivatives carrying the respective Pdiv-divIVA-egfp2 gene fusion | This work |
| pK18-E | pK18-3 derivative carrying Pdiv-egfp2 | This work |
| pK18-CG, -BS, -SC, -SP, -MT | pK18-3 derivatives carrying the respective Pdiv-divIVA gene fusion | This work |
| pBT | Two-hybrid system bait plasmid containing the cat gene, p15A origin of replication and λ cI ORF | Stratagene |
| pBTDCg,, pBTDBs, pBTDSc,, pBTDSp, pBTDMt | pBT derivatives carrying the respective divIVA genes | This work |
| pTRG | Two-hybrid system target plasmid containing the tet gene, ColE1 origin of replication, and RNA polymerase α subunit ORF | Stratagene |
| pTRGDCg, pTRGDBs, pTRGDSc, pTRGDSp, pTRGDMt | pTRG derivatives carrying the respective divIVA genes | This work |
Aprr, Kanr, and Catr indicate resistance to apramycin, kanamycin, and chloramphenicol, respectively; bla, kan, apr, cat, and tet are the ampicillin, kanamycin, apramycin, chloramphenicol, and tetracycline resistance genes, respectively. ORF, open reading frame.
Chromosomal DNA and plasmids were isolated, purified, digested, and labeled using conventional kits and protocols (28). Plasmids were transferred to the host by transformation (E. coli) or conjugation (C. glutamicum) following classical protocols (20, 29). Genetic constructions of C. glutamicum transconjugant strains were confirmed by Southern blot hybridization, using probes obtained by PCR amplification and labeled with digoxigenin according to the manufacturer's instructions (Boehringer Mannheim).
The correct design of plasmids expressing the different alleles used in this work required the characterization of the transcriptional control of divIVA. RNA was isolated from exponentially growing C. glutamicum in TSB medium using the RNeasy commercial kit (Qiagen). Rapid amplification of cDNA ends (RACE)-PCR experiments were carried out according to the protocol provided with a 5′/3′ RACE Kit, 2nd Generation (Roche). To identify promoters located upstream from divIVA, 2 μg of total RNA was used as a template to generate single-strand cDNA using primer race1 (see Table S1 in the supplemental material). A homopolymeric A tail was added to the 3′ end of the purified cDNA preparation using terminal transferase, and the dA-tailed cDNA was used in two further PCR amplification steps. In the first one, the primers dT/race2 (see Table S1 in the supplemental material) were used; the amplified DNA product of this reaction then served as the substrate in the second PCR amplification, using the primers dT/race3 (see Table S1 in the supplemental material). The amplified fragments were cloned into pGEM-TEasy vector (Table 1) based on a T-A cloning technique and used to transform E. coli TOP10. Ten plasmids isolated from different clones were sequenced; all of them started with the same G residue, located 80 nucleotides upstream from the ATG start codon of C. glutamicum divIVA (divIVACg).
Computer analyses were carried out with DNASTAR (DNAstar, Inc., London, United Kingdom), database similarity searches were done at the BLAST and FASTA public servers (NCBI, Bethesda, MD), and multiple alignments of sequences were carried out using Clustal W (EBI, Hinxton Hall, United Kingdom). Plasmid constructions (Table 1) were confirmed to be correct by sequencing using the dideoxy nucleotide chain termination method of Sanger (37).
Construction of recombinant plasmids.
The divIVACg gene was PCR amplified using the primers div1/2 (see Table S1 in the supplemental material). The adequate PCR product was cloned into plasmid pET28a by NdeI-EcoRI digestion, giving rise to the vector pETDIV (Table 1). pETDIV allowed the expression and purification of the C. glutamicum DivIVA (DivIVACg) protein with an N-terminal His tag, which was used for the production of polyclonal anti-DivIVACg antibodies (see below).
Plasmids pOJD and pOJPD were designed to disrupt the chromosomal copy of divIVACg and to place a second copy of divIVACg under the control of the regulated promoters Plac or PgntP. To construct pOJD, a 498-bp 5′ region of divIVACg was obtained by NdeI-NaeI digestion of the plasmid pETDIV. This DNA fragment was blunted by Klenow treatment and then ligated into the EcoRV-digested pOJ260 plasmid, yielding the vector pOJD (Table 1). To construct pOJPD, a 770-bp DNA fragment was obtained by EcoRV-NaeI digestion of plasmid pEPDG (Table 1). The product of this digestion carried the PgntP promoter fused to the 498-bp divIVACg 5′ region. This DNA fragment was subcloned into EcoRV-digested pOJ260, yielding the plasmid pOJPD (Table 1).
To easily clone any promoterless gene under the control of the PdivCg promoter and fuse it to the egfp2 gene (encoding an enhanced green fluorescent protein) at the same time, a new NdeI site overlapping the start codon of divIVACg was generated in plasmid pEAG2 (35). This was done by PCR mutagenesis using the specific primers listed in Table S1 in the supplemental material, yielding the plasmid pEAG3. The divIVA genes, without their stop codons, from B. subtilis, S. coelicolor, S. pneumoniae, and M. tuberculosis were then PCR amplified from their corresponding chromosomal DNAs using the primers listed in Table S1 in the supplemental material. The PCR products were cloned into the NdeI-digested pEAG3 plasmid, yielding the bifunctional mobilizable vectors pEBSE (for B. subtilis divIVA [divIVABs]), pESCE (for S. coelicolor divIVASc), pESPE (for S. pneumoniae divIVA [divIVASp]), and pEMTE (for M. tuberculosis divIVA [divIVAMt]) (Table 1).
To overexpress the above-mentioned divIVA genes (hereafter divIVANN) without fusion to egfp2, these genes, including their stop codons and the promoter Pdiv sequence, were PCR amplified from plasmids pEBSE, pESCE, pESPE, and pEMTE using the primers listed in Table S1 in the supplemental material. The corresponding PCR products were then subcloned by EcoRV digestion into the pECM2 plasmid (Table 1), yielding the vectors pEBS, pESC, pESP, and pEMT.
To introduce a copy of the DNA fragments comprising Pdiv-divIVANN-egfp2 into the C. glutamicum chromosome, the plasmids pEAG3 and pEBSE, pESCE, pESPE, and pEMTE were digested with EcoRV-SmaI; the obtained DNA fragments were then subcloned into the EcoRV-digested pK18-3 plasmid, giving rise to the vectors pK18-CGE, -BSE, -SCE, -SPE, and -MTE (Table 1). Similarly, plasmids pEBS, pESC, pESP, and pEMT were digested with EcoRV, and the fragments containing only the Pdiv-divIVANN genes, i.e., not fused to the egfp2 gene, were subcloned into EcoRV-digested pK18-3 plasmid, yielding the vectors pK18-BS, -SC, -SP, and -MT (Table 1). In contrast, plasmid pK18-CG was constructed by direct cloning of a PCR-amplified fragment containing Pdiv-divIVACg into the EcoRV-digested vector pK18-3 (Table 1).
As a negative control for the gene complementation experiments, the plasmid pK18-E was constructed. To do so, plasmid pEAG3 was digested with NdeI and religated, giving rise to pEAGE (Pdiv-egfp2) (Table 1). A fragment containing Pdiv-egfp2 was obtained by EcoRV-SmaI digestion of pEAGE and then subcloned into the EcoRV-digested pK18-3 plasmid, yielding the vector pK18-E (Table 1).
Two-hybrid system assays.
To construct the plasmids for the two-hybrid assays, the divIVA genes from C. glutamicum, B. subtilis, S. coelicolor, S. pneumoniae, and M. tuberculosis were PCR amplified by using specific primers (see Table S1 in the supplemental material) and cloned separately into pBT (bait) and pTRG (target/prey) vectors (BacterioMatch II Two-Hybrid System Vector Kit; Stratagene) (Table 1), yielding a series of pBTD and pTRGD plasmids carrying the respective divIVA genes (Table 1). Bait and prey plasmids were cotransformed into the E. coli BacterioMatch II reporter strain (Table 1) and analyzed as previously described (44).
Protein isolation and manipulation.
C. glutamicum cells were disrupted using FastPROTEIN Blue Lysing Matrix (Qbiogene Inc., Carlsbad, CA) and the BIO101 Thermo Savant FastPrep FP120 (Qbiogene Inc.) as described previously (27). The amount of protein present in cell extracts was quantified by a Bradford assay (5). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (25), stained with Coomassie blue or electroblotted onto polyvinylidene difluoride membranes (Millipore), and immunostained with a 1:10,000 dilution of rabbit polyclonal antibodies raised against green fluorescent protein (GFP) or His tags (Santa Cruz Biotechnology) and with guinea pig polyclonal antibodies raised against purified His-tagged DivIVACg (see below). Anti-rabbit or anti-guinea pig immunoglobulin G-alkaline phosphatase (Santa Cruz Biotechnology) was used as the secondary antibody at a 1:10,000 dilution.
To monitor the cellular content of DivIVACg and localize the protein by immunofluorescence microscopy, antiserum was raised against the divIVACg gene product as follows: E. coli BL21(DE3) cells (Table 1) were transformed with plasmid pETDIV (Table 1) and induced with isopropyl-β-d-thiogalactopyranoside, and the His-tagged DivIVACg protein was purified by one-step Ni2+ affinity chromatography (His-Bind; Novagen). This His-tagged protein was used to immunize male guinea pigs (66-day immunization protocol) for the production of polyclonal anti-DivIVACg antibodies. The resulting polyclonal antibodies specifically recognized a protein of ∼38 kDa, the expected size of DivIVACg.
Microscopy.
C. glutamicum cells containing the constructs carrying GFP or stained with fluorescent dyes (see later) were observed with a Nikon E400 fluorescence microscope. Photographs were taken with a DN100 Nikon digital camera and assembled using Corel Draw.
Van-FL (green fluorescence) or vancomycin-Bodipy 650/665 ([Van-650/665] red fluorescence; Molecular Probes) staining was done by adding equal proportions of unlabeled vancomycin and Van-FL or Van-650/665 to growing cultures at a final concentration of 1 μg/ml (11). The culture was then incubated for 5 min to allow adsorption of the antibiotic, after which the cells were viewed directly by fluorescence microscopy.
DAPI (4′,6-diamino-2-phenylindole) staining and immunofluorescence microscopy were carried out as described previously (12), except that C. glutamicum cells were permeabilized for 4 h at 30°C with lysozyme (10 μg/ml). The permeabilized cells were then incubated with a 1:1,000 dilution of anti-DivIVACg antiserum for 1 h and then for another hour with a 1:10,000 dilution of anti-guinea pig fluorescein-conjugated secondary antibody (Santa Cruz Biotechnology).
For transmission electron microscopy, cell pellets were rinsed three times in water, resuspended in 2.5% glutaraldehyde in phosphate buffer (pH 7), and fixed for 1 h. The cells were then rinsed three times in phosphate buffer and centrifuged at 1,600 × g; the cell pellet was mixed with 1% agarose and processed as described previously (46). The samples were infiltrated with epon-araldite, blocked, and polymerized at 55°C (blocks). From the blocks, ultrathin sections (70 nm thick) were obtained using a Leica Ultracut UCT ultramicrotome and placed on 200-mesh bare copper grids. Sections were stained with 0.5% aqueous lead citrate and then with 4% aqueous uranyl acetate and examined in a LEO EM910 transmission electron microscope operated at 80 kV. Images were taken at various magnifications using a Gatan 792 BioScan digital camera (1024 by 1024 pixels).
RESULTS
Partial depletion of DivIVA causes loss of the rod shape.
In C. glutamicum, divIVA seems to be an essential gene, as in a previous study no null mutants were obtained (35). We therefore investigated the function of the divIVA gene product by conditional gene expression using two promoters able to regulate the expression of essential genes in C. glutamicum: Plac (36) and PgntP (28). Accordingly, C. glutamicum was transformed with the suicide plasmids pOJD (Plac-ΔdivIVACg) or pOJPD (PgntP-ΔdivIVACg) (Table 1). These plasmids are unable to replicate autonomously in C. glutamicum and carry truncated copies of divIVACg under the control of regulated promoters. When introduced into C. glutamicum, the plasmids integrate by homologous recombination into the chromosomal divIVA locus, leaving a disrupted copy of the gene under its natural promoter and a full-length version under the control of the promoter Plac or PgntP. Southern blot analysis of DNAs obtained from the transconjugant strains C. glutamicum LACD and GNTD (Table 1) showed the pattern expected for Campbell-type integration of plasmids pOJD and pOJPD, respectively, at the chromosomal divIVA locus (see Fig. S1 in the supplemental material).
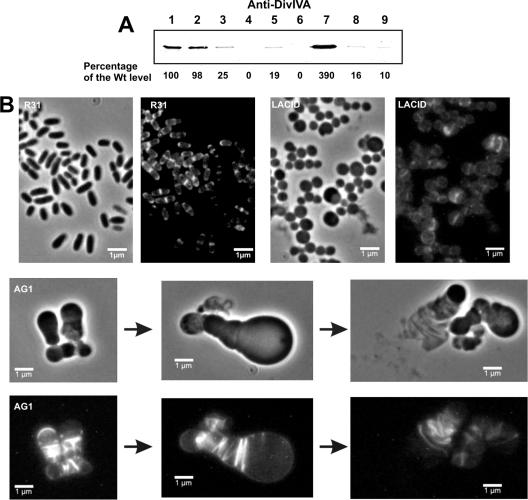
Quantitative Western blot analysis of strains LACD and GNTD revealed that the level of DivIVA was reduced to 19 and 25% of wild-type levels, respectively (Fig. 1A). DivIVA depletion was more evident when the E. coli lacIq gene present on plasmid pALacI (Table 1) was transferred to strain LACD to repress the lac promoter (yielding strain LACID) (Table 1) or when strain GNTD was grown in the presence of 4% sucrose (4%Suc), which was previously shown to strongly repress PgntP (28). For strains LACID and GNTD (the latter grown on 4%Suc), DivIVA levels were estimated to be 1.0 and 1.6%, respectively, of those normally present in the wild-type strain (Fig. 1A, lanes 4, 6, 8, and 9).
FIG. 1.
Effect of DivIVA depletion on cell shape and polar growth of Corynebacterium glutamicum. (A) Quantitative Western blot of DivIVA from different exponentially grown C. glutamicum strains. Lane 1, strain R31; lane 2, strain R31 plus 4%Suc; lane 3, strain GNTD (PgntP-divIVACg); lanes 4 and 8, strain GNTD grown in 4%Suc; lane 5, strain LACD (Plac-divIVACg); lanes 6 and 9, strain LACID (Plac-divIVACg lacIq); lane 7, strain AG1 (multicopy divIVACg). Ten micrograms of cell extract proteins was used in lanes 8 and 9; otherwise, 1 μg was loaded per lane. (B) Phase-contrast and Van-FL staining of exponentially growing cells. The typical morphology of C. glutamicum R31 was converted to a coccoid morphology (no polar growth) in strain LACID, which is depleted of DivIVA. In C. glutamicum LACID the zonal PG synthesis is absent whereas strain AG1 (multicopy divIVACg) grows apically and forms many apparently incomplete septa. Wt, wild type.
The only band detected in Western blot analyses using anti-DivIVA antibodies was the corresponding 38-kDa full-length protein. Therefore, it can be assumed that the genetic methods used to disrupt the chromosomal copy of divIVACg did not result in partial DivIVA proteins that may have interfered with the analysis. A polar effect was not expected because of the monocistronic character of the transcript (35). These data confirmed that the observed effects described below were due only to the partial depletion of DivIVA.
The C. glutamicum LACD strain, expressing divIVA from Plac, had a coccoid phenotype and thus was morphologically clearly different from the rod-shaped cells of the parent strain C. glutamicum R31. A similar phenotype was obtained when the level of DivIVA was lowered even further in the LACID strain (Fig. 1B), although the strain's growth rate was lower than that of strain LACD (see Fig. S2A in the supplemental material). The same coccoid phenotype was obtained when strain GNTD was grown in TSB alone or in TSB plus 4%Suc to repress the expression of PgntP (data not shown). The coccoid shape assumed by the cells after DivIVA depletion suggested that cell elongation was perturbed. To test this idea, cells from strains R31 and LACID were stained with Van-FL. As reported previously (11), Van-FL staining confirmed the incorporation of new PG at the cell poles and septum of C. glutamicum R31 (Fig. 1B). In sharp contrast, in strain LACID, which has only 1% of the normal level of DivIVA and exhibits a coccoid phenotype, PG synthesis was detected mainly as bands in the middle of the cells (Fig. 1B). When divIVACg was introduced in trans through the integrating plasmid pK18-CG (Table 1) into the DivIVA-depleted strain GNTD, normal divIVA expression and cell shape were restored (see below).
Function of heterologous DivIVA proteins in C. glutamicum.
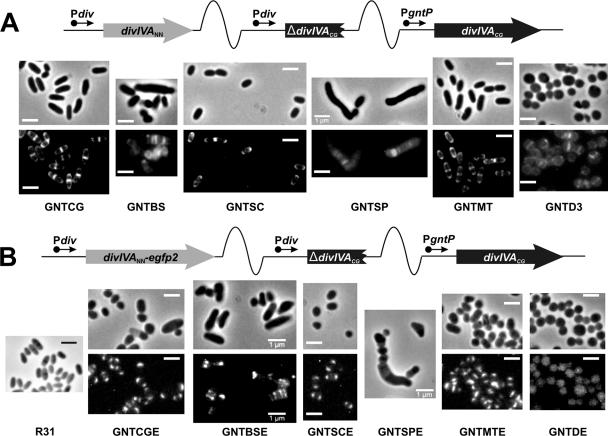
We analyzed whether DivIVA proteins from other species could restore polarized growth and a rod shape to C. glutamicum GNTD (Table 1), in which DivIVACg is depleted. These experiments could not be carried out on strain LACID due to the resistance markers present on pALacI and the plasmids used to deliver heterologous divIVA genes. The divIVA genes from two representatives of each of the two main phylogenetic lineages of gram-positive bacteria were tested (Fig. 2): divIVASc and divIVAMt from Actinobacteria and divIVABs and divIVASp from the Firmicutes. The exogenous genes were cloned behind the divIVACg promoter (Pdiv), and the construct was inserted as a single copy by homologous recombination into the region upstream from divIVA in the C. glutamicum GNTD chromosome (Fig. 2A), as previously described for other genes (1). Similar constructs carrying translational fusions between divIVA genes and egfp2 were also produced and integrated into strain GNTD (Fig. 2B). Southern blotting and PCR analysis of the transconjugants indicated that integration of the suicide plasmids took place at the expected site (see Fig. S3 in the supplemental material).
FIG. 2.
Actinomycete DivIVAs restore polar growth and a rod shape to DivIVACg-depleted C. glutamicum cells. (A) Schematic representation of the integration of plasmids pK18-CG, -BS, -SC, -SP, and -MT carrying Pdiv-divIVA from C. glutamicum (strain GNTCG), B. subtilis (strain GNTBS), S. coelicolor (strain GNTSC), S. pneumoniae (strain GNTSP), or M. tuberculosis (strain GNTMT) as a single copy into the chromosome of C. glutamicum GNTD. Integration of the empty vector pK18-3 in the chromosome of C. glutamicum GNTD was used as a negative control (strain GNTD3). All strains were grown in 4%Suc, and fluorescence images of Van-FL staining (lower panels) and phase-contrast images of the cells (upper panels) are shown. Reversion of the coccoid morphology to yield rod-shaped cells and polar/septal rather than septal Van-FL staining was observed with Actinobacteria DivIVAs but not with Firmicutes DivIVAs. Size bar, 1 μm. (B) Schematic representation of the integration of plasmids pK18-CGE, -BSE, -SCE, -SPE, and -MTE carrying Pdiv-divIVA-egfp2 from C. glutamicum (strain GNTCGE), B. subtilis (strain GNTBSE), S. coelicolor (strain GNTSCE), S. pneumoniae (strain GNTSPE), or M. tuberculosis (strain GNTMTE) as a single copy into the chromosome of C. glutamicum GNTD. Integration of the plasmid pK18-E in the chromosome of C. glutamicum GNTD was used as a negative control (strain GNTDE). All strains were grown in 4%Suc, and fluorescence images of EGFP2 (lower panels) and phase-contrast images of the cells (upper panels) are shown. Reversion of the coccoid morphology to yield rod-shaped cells and polar localization of the fused proteins was observed with Actinobacteria DivIVAs but not with Firmicutes DivIVAs. The morphology of the parent strain R31 is provided for comparison. Size bar, 1 μm.
Strain C. glutamicum GNTD had a coccoid morphology that reverted to the typical corynebacterial shape when the gene divIVACg, divIVASc, or divIVAMt was inserted into the chromosome, resulting in strains GNTCG, GNTSC, and GNTMT, respectively (Fig. 2A; Table 1). Similar results were obtained with divIVACg, divIVASc, and divIVAMt fused to egfp2 in strains GNTCGE, GNTSCE, and GNTMTE, respectively (Fig. 2B). As negative controls, C. glutamicum GNTD was transformed with the empty vector pK18-3 or pK18-E (the latter carrying the promoter-less egfp2 gene under Pdiv) (Table 1), leading to strains GNTD3 and GNTDE, respectively (Table 1), in which the coccoid phenotype was maintained (Fig. 2A and B). Van-FL staining of polar cell wall synthesis, which was not detected in the depletion strains C. glutamicum GNTD (not shown) or GNTD3, became visible again after integration of divIVACg, divIVASc, or divIVAMt (Fig. 2A). Furthermore, distinctive localization of the EGFP2-tagged gene products of either divIVASc or divIVAMt to the cell poles and septation sites was similar to that of DivIVACg-EGFP2 (Fig. 2B).
In contrast, when divIVABs and divIVASp were introduced in the same way into strain GNTD to generate GNTBS and GNTSP, respectively (Table 1), polar Van-FL staining was not observed in either case (Fig. 2A). Moreover, the presence of these genes appeared to be toxic, such that the growth rates of these strains were reduced compared to growth of the parent strain GNTD (see Fig. S2B in the supplemental material). Although cells from GNTBS and GNTSP were elongated, this did not seem to be a result of polar growth but rather of aberrant septation and failure of the daughter cells to separate. None of the EGFP2-fused DivIVA proteins from B. subtilis or S. pneumoniae showed a polar localization in C. glutamicum. In the case of strain GNTBSE (divIVABs-egfp2), the fusion protein was mainly targeted to cell division sites, leading to modestly filamentous cells and the appearance of several septa (Fig. 2B). Strain GNTSPE (divIVASp-egfp2) showed very little fluorescence, but the morphology of this strain and the shape of the cells, together with Van-FL staining of GNTSP (Fig. 2A), suggested that divIVASp expression inhibits a late stage of cell division or cell separation in C. glutamicum. All EGFP2-fused proteins were detected in cell extracts using anti-GFP antibodies (see Fig. S3B in the supplemental material). The only exception was DivIVASp-EGFP2, which appeared to be unstable, and there was no detectable fluorescence in the transconjugant strain GNTSPE.
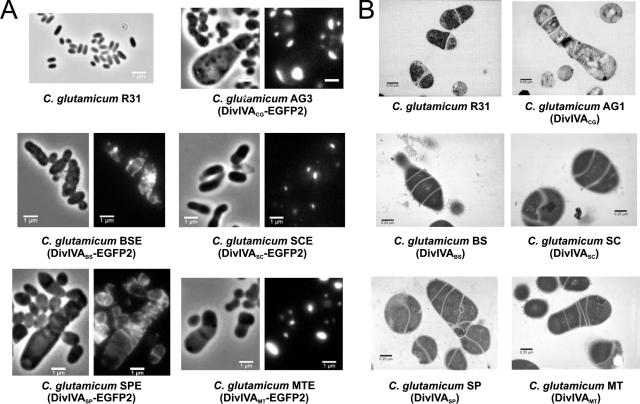
Homologous overexpression of divIVACg was previously shown to cause pronounced effects on cell morphology, with large and asymmetric swollen cells in which DivIVA-EGFP2 accumulated, often asymmetrically, at the cell poles (35). This provided a second approach to test the activities of heterologous divIVA genes in C. glutamicum, i.e., whether their expression had the same effect on cell morphology as overexpression of the native gene. PCR-amplified heterologous divIVA genes were placed under the control of the divIVACg promoter (Pdiv) by exchanging them with divIVACg in the multicopy plasmid pEAG3 (Table 1) and fused at their 3′ end to egfp2 (Pdiv-divIVACg-egfp2). These genes were also subcloned with Pdiv but without egfp2 (Pdiv-divIVACg) in the same vector, which has a copy number of 30 to 40 per cell (36). The resulting sets of plasmids (the set pEBSE, pESCE, pESPE, and pEMTE and the set pEBS, pESC, pESP, and pEMT, with and without fusion to egfp2, respectively) (Table 1) allowed multicopy expression of the different divIVA genes. The results showed that the effects on morphology were similar with or without fusion to egfp2. Overexpression of divIVACg led to large and bulky cells, often with shapes reminiscent of “ice cream cones” (Fig. 1B and 3, strains AG1 and AG3). In these cells, active assembly of PG was detected mainly at one pole and/or in multiple apparent cross-walls inside the cell (Fig. 1B). In the case of divIVAMt and divIVASc (strains GNTMT and GNTSC, respectively) (Table 1), the morphologies of the cells were similar to that resulting from divIVACg overexpression, and the EGFP2 fusion proteins (strains GNTMTE and GNTSCE) accumulated as a large cluster at one cell pole (Fig. 3A). Overexpression of divIVABs or divIVASp also led to aberrant morphologies (strains GNTBS and GNTSP) but, in contrast to the other proteins, DivIVABs or DivIVASp fused to EGFP2 localized at mid-cell or in multiple septa and never clearly accumulated at the cell poles (Fig. 3A, C. glutamicum BSE and C. glutamicum SPE).
FIG. 3.
Effect on C. glutamicum of the overproduction of DivIVA proteins from different gram-positive bacteria. (A) Fluorescence images of EGFP2 (right panels) and phase-contrast images (left panels) of the different C. glutamicum strains overexpressing divIVA fused to egfp2 from C. glutamicum (strain AG3), B. subtilis (strain BSE), S. coelicolor (strain SCE), S. pneumoniae (strain SPE), or M. tuberculosis (strain MTE). Note that polar and septal localization of DivIVA was achieved only with DivIVA-EGFP2 from actinomycetes. (B) Transmission electron micrographs of exponentially growing cells of different C. glutamicum strains overexpressing divIVA from C. glutamicum (strain AG1), B. subtilis (strain BS), S. coelicolor (strain SC), S. pneumoniae (strain SP), or M. tuberculosis (strain MT). C. glutamicum R31 was included as a control.
Transmission electron microscopy images indicated that overexpression of the divIVA genes perturbed cell division (Fig. 3B); multiple septa were visible, but they were incomplete and were not associated with cell separation. These results were confirmed by Van-FL staining (data not shown). A negative effect on cell growth was also observed (Fig. S2C, GNTBS and GNTSP). Antibodies against GFP were used to measure protein levels in the strains carrying the DivIVANN-EGFP2 fusion. The levels were similar in all cases, except for the DivIVASp-EGFP2 hybrid, which was most likely less stable than the other fusion proteins (see above).
Interactions between heterologous DivIVA proteins.
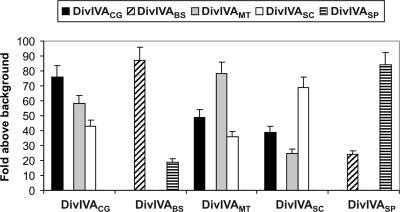
DivIVABs has been reported to interact with itself, presumably through coiled-coil regions, to form an oligomeric structure (32, 33, 42). We therefore measured the capacity for self-interaction of DivIVA using a bacterial two-hybrid system. All five tested DivIVAs self-interacted when present on both bait and prey plasmids (pBT, pTRG, and its corresponding plasmids series) (Table 1) and allowed E. coli reporter cells to grow in minimal selective medium. The results (Fig. 4) are consistent with the idea that DivIVA proteins exist as oligomers in their hosts. DivIVACg was found to interact only with actinomycete DivIVAs (S. coelicolor and M. tuberculosis) and not with B. subtilis or S. pneumoniae DivIVAs (Fig. 4), while DivIVABs and DivIVASp interacted with each other but not with any of the actinomycete DivIVA proteins (Fig. 4).
FIG. 4.
Interactions between different DivIVA proteins from gram-positive bacteria analyzed in a bacterial two-hybrid system. All combinations between bait (abscises) and prey (legends) are shown. The basal growth detected in negative controls (E. coli BacterioMatch II transformed with the empty plasmids) was considered as the background used to represent the data. The values are the means of four independent experiments; standard deviations are indicated on the bar top. Note that the DivIVA proteins from actinomycetes interact with each other, but they do not interact with DivIVAs from B. subtilis or S. pneumoniae.
Late localization of DivIVA at the septum.
Immunolocalization studies demonstrated the frequent presence of DivIVA at the cell septum. In 20% of the 500 cells obtained from an exponentially growing culture of C. glutamicum, DivIVA was localized to both cell poles and to the septum; in 77%, DivIVA was detected at both poles while in only 3% of the cells the protein was found at one pole (Fig. 5A). This distribution pattern was in agreement with the localization patterns obtained when DivIVACg was fused to the reporter protein EGFP2 (DivIVACg-EGFP2) (Fig. 5B), indicating that the hybrid protein behaved like DivIVACg.
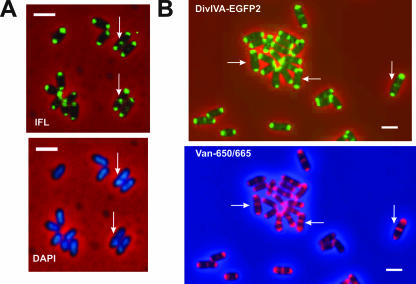
FIG. 5.
DivIVA arrives at mid-cell at a late stage of cell division in C. glutamicum. (A) Immunofluorescence (IFL) microscopy using anti-DivIVA antiserum and DAPI staining of nucleoids in C. glutamicum R31. Arrows show that DivIVA localizes at the septum in cells with visibly segregated nucleoids. Size bar, 1 μm. (B) Fluorescence microscopy (EGFP2) and Van-650/665 staining of C. glutamicum R33, carrying an extra chromosomal copy of divIVACg-egfp2 under the control of Pdiv (Pdiv-divIVACg-egfp2). Arrows indicate the cells in which the biosynthesis of new PG (visualized by Van-650/660 staining) has preceded septal localization of DivIVA-EGFP2 (EGFP2 fluorescence). Size bar, 1 μm.
DivIVA recruitment to the division site at mid-cell was examined in relation to nucleoid segregation and septal PG synthesis. Visualization of PG assembly using Van-650/665 (red fluorescence) staining showed that approximately 35% of the cells stained at mid-cell lacked detectable DivIVA-EGFP2 signal at that site (Fig. 5B, horizontal arrows). In contrast, all cells with DivIVA-EGFP2 at the putative division site were stained with Van-650/665 at mid-cell (Fig. 5B, vertical arrows). Furthermore, in DAPI-stained cells containing DivIVA at mid-cell (as detected by immunofluorescence), there was always a gap between two distinct nucleoids (Fig. 5A, vertical arrows). These observations suggested that DivIVA arrives at the septum after the beginning of PG synthesis during cell division and at a stage in which the nucleoids have already visibly segregated.
DISCUSSION
Four years ago, two independent reports presented evidence supporting a role for the divIVA gene in the cell elongation of two different actinomycetes (19, 35). Polar localization of the DivIVA protein in Streptomyces and Corynebacterium and the striking effects on cell shape following overexpression of divIVA suggested that DivIVA functioned in polar cell wall synthesis. These data were confirmed very recently in another actinomycete, M. smegmatis (33). In that study, the authors also provided convincing evidence for the oligomerization of DivIVAMs in large structures. Nevertheless, none of these three previous reports answered a fundamental question: is DivIVA essential for cell elongation in actinomycetes?
This is to our knowledge the first study showing the essential role of DivIVA in the elongation of C. glutamicum cells. C. glutamicum strains were successfully depleted of DivIVA by following two different strategies that yielded bacterial cells with similar coccoid morphologies and a lack of polar PG synthesis. These results led to the conclusion that normal levels of DivIVA are required for a rod-shaped morphology and for polarized growth by zonal PG synthesis. A similar conversion from rods to spheres is seen in E. coli mutants that have lost their cell-elongation systems, e.g., by depletion of MreB, MreC, or MreD or by mutation of rodA or pbpA (2, 24). Thus, it appears that Corynebacterium, like many other rod-shaped bacteria, has two different systems for cell-wall growth, one involving cell division and septation that can also sustain the growth of spherical cells and another for cell-wall elongation (40, 44).
Since we previously showed that divIVACg was essential in C. glutamicum and that the gene could not be deleted (35), it was surprising that the depletion of DivIVACg did not prevent growth or strongly limited viability, despite a marked loss of apical cell wall elongation. This finding suggests that the residual level of DivIVACg detected in the depletion experiments may have been sufficient to fulfill another, as-yet-unknown, essential function in C. glutamicum, even though participation of DivIVACg in cell wall elongation had been abolished. DivIVA arrives at the septum after cell division-related PG synthesis has already started and at a stage in which the nucleoids are already visibly segregated (Fig. 5). This late arrival of DivIVA at the division site seems to rule out a direct involvement of the protein in cell division and chromosome segregation. In addition, C. glutamicum cells partially depleted of DivIVA do not form chains of cells attached at the poles, as seen for streptococci (17), making it improbable that the protein is necessary for a final stage of cell division or cell-pole maturation.
The activities and polar localization of DivIVA homologues from the actinobacteria S. coelicolor and M. tuberculosis were similar to what was observed in C. glutamicum. However, the proteins from B. subtilis and S. pneumoniae accumulated at septation sites and interfered with growth and cell division. This revealed a functional distinction between the DivIVA orthologues of Actinobacteria and Firmicutes, in which only those of the former may be involved in zonal cell wall assembly at the cell poles. This conclusion is consistent with the fact that neither B. subtilis nor S. pneumoniae grows in the polarized fashion typical of many Actinobacteria (11, 18). Interestingly, overexpression of DivIVASc in C. glutamicum did not lead to branching, as it does in S. coelicolor (19). Thus, other components needed for triggering true branching in S. coelicolor are perhaps absent from Corynebacterium.
Since DivIVABs was previously found to localize promiscuously at the cell poles of E. coli or Schizosaccharomyces pombe (16), the finding that it was not located at the cell poles of C. glutamicum was unexpected. One potential explanation for this result is that oligomerization of heterologous DivIVA with DivIVACg may be needed for targeting to the poles of C. glutamicum. Alternatively, the low levels of DivIVACg in C. glutamicum GNTD could have been sufficient to compete with DivIVABs and exclude it from the poles. It may also be the case that heterologous DivIVAs do not fold properly in C. glutamicum.
During synthesis of the bacterial cell wall, some form of internal support is required. In most rod-shaped bacteria, the Z-ring (during cell division) and the MreB helicoidal structures (during cell wall elongation) comprise a cytoskeleton that sustains PG synthesis (41). DivIVA is essential for polar growth in C. glutamicum, and the cellular localization of DivIVA in all Actinobacteria tested coincides exactly with the sites of PG synthesis during cell growth (Fig. 5B) (19, 33). Therefore, DivIVA oligomers may serve as the cytoskeletal framework for polar PG synthesis in these bacteria. In C. glutamicum, the late localization of DivIVA to the division septum suggests that this protein recognizes the nascent cell poles rather than an early component of the septal machinery (Fig. 5); this is consistent with the idea that DivIVA promotes the switch from septal to polar PG synthesis. Supporting this hypothesis, overexpression of DivIVA stimulates localized cell wall synthesis and increases the diameter of cell poles in C. glutamicum (Fig. 1B), S. coelicolor (19), and M. tuberculosis (33), implying that DivIVA is able to recruit some proteins for PG synthesis at the cell pole. We propose that when DivIVA is limiting in the C. glutamicum cell, it cannot efficiently form oligomers and thus cannot efficiently recruit the PG synthesis machinery. This forces the septal machinery to drive most of the PG synthesis, resulting in septal vancomycin staining and coccoid cells. When DivIVA is deleted completely, the septal machinery is forced to drive all PG synthesis but cannot support sufficient PG synthesis for cell viability. One possibility is that increased levels of FtsZ may help to drive increased septal PG synthesis under these conditions, as has been shown for spherical mutants of E. coli (3).
Although the above speculation suggests an interplay between FtsZ and DivIVA in C. glutamicum, DivIVA does not seems to be required for proper localization of the Z-ring in this species, in contrast to B. subtilis (9, 15). Since C. glutamicum lacks a MinCD system or any other known positive and negative regulators of bacterial cell division (i.e., ezrA, ftsA, min, noc, slmA, sulA, zipA, or zapA) (27) and does not undergo nucleoid occlusion (36), how C. glutamicum is nonetheless able to divide almost symmetrically remains unknown. Additional studies will be needed to better understand this and other aspects of cell division and cell growth in rod-shaped C. glutamicum.
Supplementary Material
Acknowledgments
M. Letek and E. Ordóñez were beneficiaries of fellowships from the Ministerio de Educación y Ciencia and the Junta de Castilla y León, respectively. This work was funded by grants from the Junta de Castilla y León (reference LE040A07), University of León (ULE 2001-08B), and Ministerio de Ciencia y Tecnología (BIO2002-03223 and BIO2005-02723).
We thank Carlos Martín (Universidad de Zaragoza, Spain), Jeff Errington (University of Newcastle, United Kingdom), David A. Hopwood (John Innes Institute, Norwich, United Kingdom), and Orietta Massidda (University of Cagliari, Italy) for the chromosomal DNAs from the wild-type strains M. tuberculosis H37Rv, B. subtilis subsp. subtilis strain 168, S. coelicolor A3(2), and S. pneumoniae R6, respectively.
Footnotes
Published ahead of print on 22 February 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Adham, S. A., A. B. Campelo, A. Ramos, and J. A. Gil. 2001. Construction of a xylanase-producing strain of Brevibacterium lactofermentum by stable integration of an engineered xysA gene from Streptomyces halstedii JM8. Appl. Environ. Microbiol. 675425-5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Begg, K. J., B. G. Spratt, and W. D. Donachie. 1986. Interaction between membrane proteins PBP3 and RodA is required for normal cell shape and division in Escherichia coli. J. Bacteriol. 1671004-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bendezu, F. O., and P. A. de Boer. 2008. Conditional lethality, division defects, membrane involution and endocytosis in mre and mrd shape mutants of Escherichia coli. J. Bacteriol. 1901792-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 11643-49. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein using the principles of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 6.Cabeen, M. T., and C. Jacobs-Wagner. 2005. Bacterial cell shape. Nat. Rev. Microbiol. 3601-610. [DOI] [PubMed] [Google Scholar]
- 7.Carballido-Lopez, R. 2006. The bacterial actin-like cytoskeleton. Microbiol. Mol. Biol. Rev. 70888-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cerdeno-Tarraga, A. M., A. Efstratiou, L. G. Dover, M. T. Holden, M. Pallen, S. D. Bentley, G. S. Besra, C. Churcher, K. D. James, Z. A. De, T. Chillingworth, A. Cronin, L. Dowd, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Moule, M. A. Quail, E. Rabbinowitsch, K. M. Rutherford, N. R. Thomson, L. Unwin, S. Whitehead, B. G. Barrell, and J. Parkhill. 2003. The complete genome sequence and analysis of Corynebacterium diphtheriae NCTC13129. Nucleic Acids Res. 316516-6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cha, J. H., and G. C. Stewart. 1997. The divIVA minicell locus of Bacillus subtilis. J. Bacteriol. 1791671-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chauhan, A., H. Lofton, E. Maloney, J. Moore, M. Fol, M. V. Madiraju, and M. Rajagopalan. 2006. Interference of Mycobacterium tuberculosis cell division by Rv2719c, a cell wall hydrolase. Mol. Microbiol. 62132-147. [DOI] [PubMed] [Google Scholar]
- 11.Daniel, R. A., and J. Errington. 2003. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113767-776. [DOI] [PubMed] [Google Scholar]
- 12.Daniel, R. A., E. J. Harry, and J. Errington. 2000. Role of penicillin-binding protein PBP 2B in assembly and functioning of the division machinery of Bacillus subtilis. Mol. Microbiol. 35299-311. [DOI] [PubMed] [Google Scholar]
- 13.de Pedro, M. A., J. C. Quintela, J. V. Holtje, and H. Schwarz. 1997. Murein segregation in Escherichia coli. J. Bacteriol. 1792823-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Divakaruni, A. V., R. R. Loo, Y. Xie, J. A. Loo, and J. W. Gober. 2005. The cell-shape protein MreC interacts with extracytoplasmic proteins including cell wall assembly complexes in Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 10218602-18607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edwards, D. H., and J. Errington. 1997. The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol. Microbiol. 24905-915. [DOI] [PubMed] [Google Scholar]
- 16.Edwards, D. H., H. B. Thomaides, and J. Errington. 2000. Promiscuous targeting of Bacillus subtilis cell division protein DivIVA to division sites in Escherichia coli and fission yeast. EMBO J. 192719-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fadda, D., A. Santona, V. D'Ulisse, P. Ghelardini, M. G. Ennas, M. B. Whalen, and O. Massidda. 2007. Streptococcus pneumoniae DivIVA: localization and interactions in a MinCD free context. J. Bacteriol. 1891288-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flardh, K. 2003. Growth polarity and cell division in Streptomyces. Curr. Opin. Microbiol. 6564-571. [DOI] [PubMed] [Google Scholar]
- 19.Flardh, K. 2003. Essential role of DivIVA in polar growth and morphogenesis in Streptomyces coelicolor A3(2). Mol. Microbiol. 491523-1536. [DOI] [PubMed] [Google Scholar]
- 20.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166557-580. [DOI] [PubMed] [Google Scholar]
- 21.Jager, W., A. Schafer, A. Puhler, G. Labes, and W. Wohlleben. 1992. Expression of the Bacillus subtilis sacB gene leads to sucrose sensitivity in the gram-positive bacterium Corynebacterium glutamicum but not in Streptomyces lividans. J. Bacteriol. 1745462-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalinowski, J., B. Bathe, D. Bartels, N. Bischoff, M. Bott, A. Burkovski, N. Dusch, L. Eggeling, B. J. Eikmanns, L. Gaigalat, A. Goesmann, M. Hartmann, K. Huthmacher, R. Kramer, B. Linke, A. C. McHardy, F. Meyer, B. Mockel, W. Pfefferle, A. Puhler, D. A. Rey, C. Ruckert, O. Rupp, H. Sahm, V. F. Wendisch, I. Wiegrabe, and A. Tauch. 2003. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J. Biotechnol. 1045-25. [DOI] [PubMed] [Google Scholar]
- 23.Kang, C. M., D. W. Abbott, S. T. Park, C. C. Dascher, L. C. Cantley, and R. N. Husson. 2005. The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: substrate identification and regulation of cell shape. Genes Dev. 191692-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kruse, T., J. Bork-Jensen, and K. Gerdes. 2005. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol. Microbiol. 5578-89. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 26.Leaver, M., and J. Errington. 2005. Roles for MreC and MreD proteins in helical growth of the cylindrical cell wall in Bacillus subtilis. Mol. Microbiol. 571196-1209. [DOI] [PubMed] [Google Scholar]
- 27.Letek, M., E. Ordonez, M. Fiuza, M. P. Honrubia-Marcos, J. Vaquera, J. A. Gil, and L. M. Mateos. 2007. Characterization of the promoter region of ftsZ from Corynebacterium glutamicum and controlled overexpression of FtsZ. Int. Microbiol. 10271-282. [PubMed] [Google Scholar]
- 28.Letek, M., N. Valbuena, A. Ramos, E. Ordonez, J. A. Gil, and L. M. Mateos. 2006. Characterization and use of catabolite-repressed promoters from gluconate genes in Corynebacterium glutamicum. J. Bacteriol. 188409-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mateos, L. M., A. Schafer, J. Kalinowski, J. F. Martin, and A. Puhler. 1996. Integration of narrow-host-range vectors from Escherichia coli into the genomes of amino acid-producing corynebacteria after intergeneric conjugation. J. Bacteriol. 1785768-5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mazza, P., E. E. Noens, K. Schirner, N. Grantcharova, A. M. Mommaas, H. K. Koerten, G. Muth, K. Flardh, G. P. van Wezel, and W. Wohlleben. 2006. MreB of Streptomyces coelicolor is not essential for vegetative growth but is required for the integrity of aerial hyphae and spores. Mol. Microbiol. 60838-852. [DOI] [PubMed] [Google Scholar]
- 31.McCormick, J. R., E. P. Su, A. Driks, and R. Losick. 1994. Growth and viability of Streptomyces coelicolor mutant for the cell division gene ftsZ. Mol. Microbiol. 14243-254. [DOI] [PubMed] [Google Scholar]
- 32.Muchova, K., E. Kutejova, D. J. Scott, J. A. Brannigan, R. J. Lewis, A. J. Wilkinson, and I. Barak. 2002. Oligomerization of the Bacillus subtilis division protein DivIVA. Microbiology 148807-813. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen, L., N. Scherr, J. Gatfield, A. Walburger, J. Pieters, and C. J. Thompson. 2007. Antigen 84, an effector of pleiomorphism in Mycobacterium smegmatis. J. Bacteriol. 1897896-7910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishio, Y., Y. Nakamura, Y. Kawarabayasi, Y. Usuda, E. Kimura, S. Sugimoto, K. Matsui, A. Yamagishi, H. Kikuchi, K. Ikeo, and T. Gojobori. 2003. Comparative complete genome sequence analysis of the amino acid replacements responsible for the thermostability of Corynebacterium efficiens. Genome Res. 131572-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramos, A., M. P. Honrubia, N. Valbuena, J. Vaquera, L. M. Mateos, and J. A. Gil. 2003. Involvement of DivIVA in the morphology of the rod-shaped actinomycete Brevibacterium lactofermentum. Microbiology 1493531-3542. [DOI] [PubMed] [Google Scholar]
- 36.Ramos, A., M. Letek, A. B. Campelo, J. Vaquera, L. M. Mateos, and J. A. Gil. 2005. Altered morphology produced by ftsZ expression in Corynebacterium glutamicum ATCC 13869. Microbiology 1512563-2572. [DOI] [PubMed] [Google Scholar]
- 37.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 745463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santamaria, R. I., J. A. Gil, and J. F. Martin. 1985. High-frequency transformation of Brevibacterium lactofermentum protoplasts by plasmid DNA. J. Bacteriol. 162463-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schafer, A., J. Kalinowski, R. Simon, A. H. Seep-Feldhaus, and A. Puhler. 1990. High-frequency conjugal plasmid transfer from gram-negative Escherichia coli to various gram-positive coryneform bacteria. J. Bacteriol. 1721663-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scheffers, D. J., L. J. Jones, and J. Errington. 2004. Several distinct localization patterns for penicillin-binding proteins in Bacillus subtilis. Mol. Microbiol. 51749-764. [DOI] [PubMed] [Google Scholar]
- 41.Shih, Y. L., and L. Rothfield. 2006. The bacterial cytoskeleton. Microbiol. Mol. Biol. Rev. 70729-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stahlberg, H., E. Kutejova, K. Muchova, M. Gregorini, A. Lustig, S. A. Muller, V. Olivieri, A. Engel, A. J. Wilkinson, and I. Barak. 2004. Oligomeric structure of the Bacillus subtilis cell division protein DivIVA determined by transmission electron microscopy. Mol. Microbiol. 521281-1290. [DOI] [PubMed] [Google Scholar]
- 43.Tauch, A., O. Kaiser, T. Hain, A. Goesmann, B. Weisshaar, A. Albersmeier, T. Bekel, N. Bischoff, I. Brune, T. Chakraborty, J. Kalinowski, F. Meyer, O. Rupp, S. Schneiker, P. Viehoever, and A. Puhler. 2005. Complete genome sequence and analysis of the multiresistant nosocomial pathogen Corynebacterium jeikeium K411, a lipid-requiring bacterium of the human skin flora. J. Bacteriol. 1874671-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valbuena, N., M. Letek, E. Ordonez, J. A. Ayala, R. A. Daniel, J. A. Gil, and L. M. Mateos. 2007. Characterization of HMW-PBPs from the rod-shaped actinomycete Corynebacterium glutamicum: peptidoglycan synthesis in cells lacking actin-like cytoskeletal structures. Mol. Microbiol. 66643-657. [DOI] [PubMed] [Google Scholar]
- 45.Valbuena, N., M. Letek, A. Ramos, J. Ayala, D. Nakunst, J. Kalinowski, L. M. Mateos, and J. A. Gil. 2006. Morphological changes and proteome response of Corynebacterium glutamicum to a partial depletion of FtsI. Microbiology 1522491-2503. [DOI] [PubMed] [Google Scholar]
- 46.Wood, J. I., and K. L. Klomparens. 1993. Characterization of agarose as an encapsulation medium for particulate specimens for transmission electron microscopy. Microsc. Res. Tech. 25267-275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.